SYNOPSIS
Nrf2 is a transcription factor that activates transcription of a battery of cytoprotective genes by binding to the antioxidant response element (ARE). Nrf2 is repressed by the cysteine-rich Keap1 protein, which targets Nrf2 for ubiquitination and subsequent degradation by a Cul3-mediated ubiquitination complex. We find that modification of C151 of human Keap1 by mutation to a tryptophan relieves the repression by Keap1 and allows activation of the ARE by Nrf2. Keap1 C151W has a decreased affinity for Cul3, and can no longer serve to target Nrf2 for ubiquitination, though it retains its affinity for Nrf2. A series of 12 mutant Keap1 proteins, each containing a different residue at position 151, was constructed to explore the chemistry required for the effect. The series reveals that the extent to which Keap1 loses the ability to target Nrf2 for degradation, and hence the ability to repress ARE activation, correlates well with the partial molar volume of the residue. Other physico-chemical properties do not appear to contribute significantly to the effect. Based on this finding, a structural model is proposed whereby large residues at position 151 cause steric clashes that lead to alteration of the Keap1-Cul3 interaction. This model has significant implications for how electrophiles, which modify C151, disrupt the repressive function of Keap1.
Keywords: Keap1, Nrf2, partial molar volume, antioxidant response element, cysteine, tryptophan
INTRODUCTION
A promising strategy for prevention of numerous types of diseases is the induction of a battery of cytoprotective enzymes. These enzymes include NAD(P)H:quinone oxidoreductase 1 (NQO1) and glutathione S-transferase, whose induction leads to detoxification and elimination of carcinogens, antioxidant enzymes such as heme oxygenase-1, and enzymes that regulate the reducing environment of the cell including the NADPH-regenerating enzyme glucose 6-phosphate dehydrogenase. The regulatory regions for each of these inducible genes contain an antioxidant response element (ARE) [1], which is activated upon binding of the Nrf2 (nuclear factor-erythroid 2 p45 subunit-related factor 1) transcription factor protein. The key role of Nrf2 in the transcription of cytoprotective genes and in disease prevention has been illustrated in a rapidly-increasing number of studies. Some of the disease states that are believed to be mitigated by Nrf2 activation include carcinogenesis, hepatotoxicity, neurodegenerative disorders, sepsis, and pulmonary inflammatory diseases, as well as the aging process (reviewed in [2]). Nrf2 is activated by a wide range of molecules, herein termed ARE inducers. A large number of ARE inducers have been identified, and while no canonical structures have been found, most are electrophilic in nature and capable of modifying thiols, including cysteines [3]. Many ARE inducers have been discovered from plant-based sources, including commonly consumed foods [4]. For example, sulforaphane was first isolated from broccoli as a potent inducer of NQO1 in murine hepatoma cells [5], and has since been shown to be effective in animal models of cancer prevention [6–9]. Based on the large number of diseases that could be prevented or attenuated by Nrf2 activation, and the potential to activate Nrf2 through dietary changes, understanding the mechanism of its activation by ARE inducers is of great interest.
The mechanism of Nrf2 regulation and the responses of the regulatory system to ARE inducers are elaborate (reviewed in [4]). Briefly, under basal conditions, Nrf2 resides mainly in the cytoplasm and at low levels overall in the cell, primarily through the interaction of the N-terminal Neh2 domain of Nrf2 with Keap1 (Kelch-like ECH-associated protein 1). Keap1 contains a Crm1-dependent nuclear export sequence, which prevents Nrf2 nuclear localization [10–12], otherwise mediated by a nuclear localization signal in Nrf2 [13]. In addition, Keap1 serves as a bridge between Nrf2 and the Cul3-based E3-ligase ubiquitination complex [14–16]. While details of the overall stoichiometry of the Keap1-Nrf2-Cul3 complex are unknown, there are two binding sites in the Neh2 domain of Nrf2, termed the ETGE and DLG motifs, that each bind to separate Kelch-repeat domains in a Keap1 dimer [17, 18]. The binding of each motif to a Kelch domain, proposed as the ‘two-site’ model, is required for ubiquitination of seven lysines located between the motifs, as shown both by site-directed mutagenesis experiments and somatic mutations in cancer patients [18, 19]. This Nrf2 ubiquitination leads to rapid degradation of Nrf2 by the 26S proteasome and low basal levels of Nrf2 in the cell, contributing to low basal ARE activity. Several phosphorylation events have also been shown to repress Nrf2 activation of the ARE under basal conditions, including phosphorylation of Nrf2 by GSK-3β, which leads to Nrf2 cytoplasmic localization [20], and phosphorylation of Nrf2 by p38, which causes enhanced interaction with Keap1 [21].
Numerous mechanisms have been implicated in sensing electrophilic ARE inducers. These can be largely grouped into two categories, modulation of protein kinase pathways and modification of Keap1 cysteines. While many protein kinases have been shown to play a role in Nrf2 activation, only a few of the mechanisms involved have been studied in detail. For example, the phosphatidylinositol 3-kinase pathway, activated by a variety of ARE inducers [22–24], was shown to lead to inactivation of GSK-3β and to Nrf2 nuclear localization [20]. In addition, sulforaphane was shown to inhibit the MKK3/6 kinase upstream of p38, leading to decreased phosphorylation of Nrf2, and subsequent disruption of the Keap1-Nrf2 interaction [21]. However, other groups have found that activation of p38 MAPK, rather than inhibition, leads to ARE induction [25–27]. Phosphorylation of serine 40 in Nrf2 by PKC isoforms is also proposed to disrupt the Keap1-Nrf2 interaction [28], leading to Nrf2 activation [29].
Human Keap1 contains 27 cysteines, 25 of which are highly conserved among Keap1 homologs. The large number of cysteines in Keap1 and the electrophilic nature of the vast majority of the inducers is suggestive of their importance in sensing ARE inducers. Three Keap1 mutants in particular, C151S, C273S and C288S, have phenotypes that suggest they play a role in sensing inducers. It was first shown that cells overexpressing Keap1 C151S are much less responsive to the ARE inducers tert-butylhydroquinone (tBHQ) and sulforaphane as compared to wt Keap1, although in the absence of inducers cells expressing the Keap1 C151S protein have the same phenotype as those expressing wt Keap1 [30]. Conversely, it was shown that cells overexpressing Keap1 C288S or C273S have constitutive ARE activation in the absence of ARE inducers, as compared to wt Keap1, due to the fact that Nrf2 was no longer ubiquitinated or degraded [30]. Later, transgenic expression of Keap1 C273A or Keap1 C288A protein in Keap1 null mice confirmed the inability of these Keap1 molecules to target Nrf2 for ubiquitination, while mice expressing Keap1 C151S were largely unresponsive to tBHQ, emphasizing the biological importance of C151 in sensing ARE inducers [31]. Using mass spectrometry to detect modification of human Keap1 cysteines in vitro, we previously determined that, while these three cysteines are highly reactive, only Keap1 C151 was highly and consistently modified by the molecules tested, including biotinylated iodoacetamide (BIA) [32] and 1-biotinamido-4-(4′-[maleimidoethyl-cyclohexane]-carboxamido) butane (BMCC) [33], as well as the natural product ARE inducers xanthohumol from hops, isoliquiritigenin from licorice and 10-shogaol from ginger [33].
An early model suggested that modification of Keap1 cysteines by ARE inducers leads to dissociation of the Keap1-Nrf2 interaction [34, 35]. However, subsequent studies both in vitro and in vivo [16, 36, 37] indicate the Keap1-Nrf2 interaction is not disrupted upon modification of Keap1 cysteines. In the proposed ‘two-site’ model, the lower affinity DLG site alone would be disrupted, maintaining a Keap1-Nrf2 interaction but disallowing Nrf2 ubiquitination [18, 38]. Alternatively, modification of Keap1 cysteines appears to decrease the interaction of Keap1 and Cul3, leading to downregulation of Nrf2 ubiquitination. Co-purification assays showed a decreased association between Keap1 and Cul3 in transiently transfected mammalian cells [16], and a similar result was observed in vitro using proteins purified from Escherichia coli [39]. In both studies, there was significantly less disruption of the interaction between the Keap1 C151S mutant and Cul3, compared to wt Keap1, implying that modification of C151 by an ARE inducer is important for the disruption. Cysteine 151 is located in the BTB domain of Keap1, which is important for the Keap1-Cul3 interaction [15, 40]. However, it is unknown whether modification of other Keap1 cysteines in addition to C151 is required to alter the Keap1-Cul3 interaction.
To attempt to mimic a modification of C151 by an ARE inducer, in this work we substituted C151 of Keap1 with the largest natural amino acid, tryptophan. We find that modification of C151 to a tryptophan does lead to ARE activation, by altering the Keap1-Cul3 interaction and downregulating Nrf2 ubiquitination. Twelve other amino acids were substituted at position 151 to explore the physico-chemical properties required at this position to alter Keap1’s ability to catalyze Nrf2 ubiquitination. The results offer an insight into how C151 modification by an electrophile might alter Keap1-Cul3 binding and hence Nrf2 ubiquitination.
EXPERIMENTAL
Construction of Recombinant DNA Molecules
Plasmids expressing wild-type (wt) Keap1 tagged with a chitin binding domain (CBD) in pcDNA3, or hemagglutinin (HA)-tagged Cul3 protein in the pCI vector, have been previously described [16], along with the plasmids expressing HA-Nrf2 in the pCI vector and Gal4-Neh2 protein in pcDNA3 [30] and HA-ubiquitin in the pCI vector [41]. For expression of a non-tagged version of the Keap1 protein, the full-length Keap1 gene [36] was directionally cloned using PCR into the HindIII/XhoI sites of pcDNA3 (Invitrogen). Site-directed mutagenesis was conducted using standard oligo-directed mutagenesis techniques. The entire sequence of each gene in the expression plasmids was verified by dideoxy sequencing by the UIC DNA Sequencing Facility.
Cell Culture, Transfections, and Chemical Reagents
MDA-MB-231 cells were purchased from the ATCC. Cells were maintained in Eagle’s minimal essential medium in the presence of 10% fetal bovine serum. Plasmid DNA transfections were performed with Lipofectamine Plus (Invitrogen) according to the manufacturer’s instructions. Sulforaphane was purchased from MP Biomedicals, and the 1-[2-cyano-3-,12-dioxooleana-1,9(11)-dien-28-oyl]imidazole (CDDO-Im) was a generous gift from Michael Sporn (Dartmouth).
Reporter Gene Assays
The ARE TATA-Inr luciferase reporter plasmid, pARE-Luc, and the Renilla luciferase expression plasmid, pRL-TK, have been previously described [30, 42]. MDA-MB-231 cells grown on 24-well plates were transfected with 100 ng of pARE-Luc, 10 ng of pRL-TK reporter plasmid, 100 ng of the Nrf2 expression plasmid, and 50 ng of either the wt or mutant Keap1 expression plasmid. The total amount of DNA was maintained at 260 ng with pcDNA3. Both firefly and Renilla luciferase activities were measured 24 h after transfection with the dual luciferase reporter assay system (Promega). Firefly luciferase activity was normalized to Renilla luciferase activity to control for sample-to-sample variations in transfection efficiency.
Antibodies, Immunoblot Analysis and Co-immunoprecipitation Assays
Primary antibodies against tubulin, Nrf2, and Keap1, and horseradish peroxidase-coupled secondary antibodies, were purchased from Santa Cruz Biotechnology. An antibody against the HA epitope was purchased from Covance.
For detection of protein expression in total cell lysates, cells in 24 well plates were transfected with expression vectors for Nrf2 (100 ng) and either wt or mutant Keap1 (50 ng). Cells were washed with 1xPBS and lysed in MPER buffer (Thermo Scientific) supplemented with Complete Protease Inhibitor Mix (Roche) at 24 h post-transfection. For co-immunoprecipitation assays, cells in 35 mm dishes were transfected with expression vectors for HA-Cul3 (500 ng) and either wt or mutant Keap1-CBD (500 ng). Cells were washed with 1xPBS and lysed in MPER buffer supplemented with Complete Protease Inhibitor, 1 mM DTT, and 150 mM NaCl 48 h post-transfection. After centrifugation, cell lysate supernatants were incubated with chitin beads (New England Biolabs) for 4 h at 4°C. Protein complexes on the beads were washed once with lysis buffer and twice with wash buffer (20 mM Tris HCl pH 8.0, 500 mM NaCl, 0.1% Triton X100, 1 mM EDTA, 1 mM DTT and Complete Protease Inhibitor Mix). Samples were analyzed by SDS gel electrophoresis, transferred to polyvinylidene fluoride (PVDF) membranes, and immunoblotted with the appropriate antibody.
Ubiquitination of Nrf2 in cells
For detection of ubiquitinated Nrf2 in vivo, cells in 35 mm dishes were transfected with expression vectors for HA-ubiquitin (200 ng), HA-Cul3 (100 ng), Gal4-Neh2 (400 ng) and either the wt or mutant Keap1 (133 ng). After 48h, the transfected cells were exposed to 10 μM MG132 (Boston Biochem) for 3 h. Cells were washed with 1xPBS and lysed by boiling in a buffer containing 20 mM Tris-HCl, pH 7.6, 137 mM NaCl, 2% SDS and 10 mM N-ethylmaleimide. This procedure inactivates cellular ubiquitin hydrolases, preserving ubiquitin-Nrf2 conjugates. Lysates were incubated on ice to precipitate the SDS, then centrifuged, and supernatants were diluted fivefold in buffer containing 20 mM Tris-HCl, pH 7.6, 137 mM NaCl and 1% Triton X-100. After preclearing with A/G beads (Santa Cruz Biotechnology), lysates were incubated with anti-Gal4 antibodies for one hour at 4°C, followed by a 2 h incubation at 4°C with A/G beads. Beads were washed once with buffer containing 20 mM Tris-HCl, pH 7.6, 137 mM NaCl and 0.5 M LiCl, and twice in the same buffer without LiCl. Immunoprecipitated protein was analyzed by SDS gel electrophoresis, transferred to PVDF membranes, and immunoblotted with the anti-HA antibody.
RESULTS
ARE activation by potentially therapeutic ARE inducers is dependent on Keap1 C151
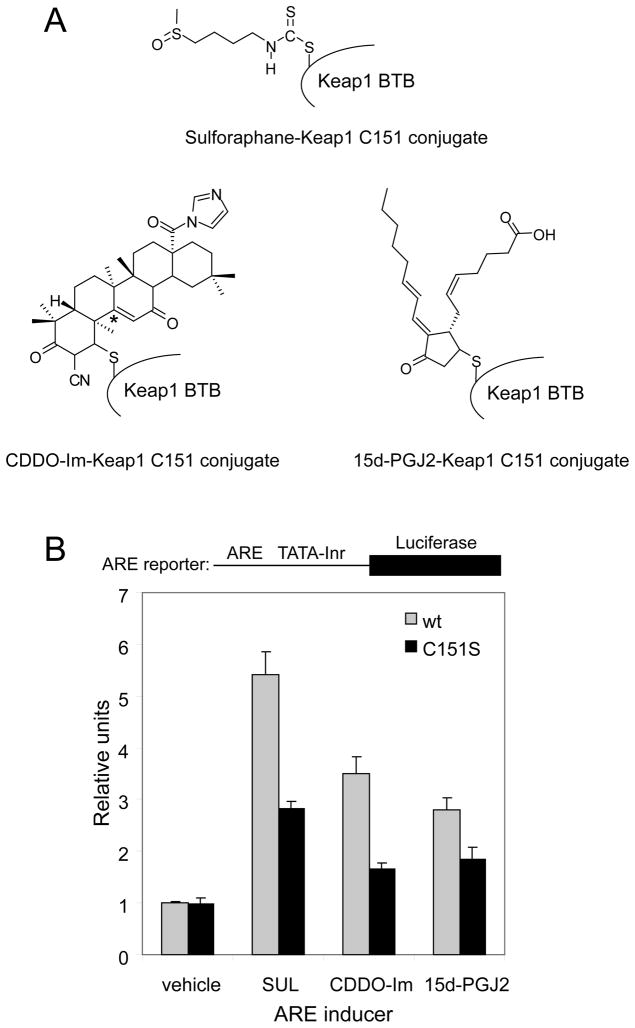
Keap1 C151 has been shown to be important for ARE activation by several ARE inducers, including sulforaphane (Figure 1A) [30]. We set out to recapitulate this result, and, to further establish the relevance of C151 in sensing other ARE inducers of potential clinical relevance, the C151 dependence of ARE activation by CDDO-Im was determined (Figure 1A). Activation of Nrf2 by CDDO-Im has shown promise in prevention or attenuation of disease in several mouse and rat models, including cigarette smoke-induced emphysema [43], acute inflammatory liver injury [44], aflatoxin-induced tumorigenesis [45], and sepsis-induced lethality [46], as well as a preclinical ex vivo study on the LPS-induced inflammatory response in sepsis [47]. We also included in the study 15d-deoxy-Δ12,14-prostagladin J2 (15d-PGJ2), a lipid oxidation product that accumulates during acute inflammation. This endogenous molecule has come to be appreciated recently as an electrophilic mediator of cellular signaling [48]. 15d-PGJ2 has been shown to induce nuclear accumulation of Nrf2 [49], and has been shown to bind to native Keap1 in vivo by using rat hepatocyte cells [50]. Further, it was demonstrated that Nrf2 mediates inflammatory processes induced by 15d-PGJ2 in a mouse model of carrageenan-induced pleurisy [50]. Previously, we observed modification of Keap1 by 15d-PGJ2 in vitro by mass spectrometry [36].
FIGURE 1. Keap1 C151 dependence of ARE activation by sulforaphane, CDDO-Im and 15d-PGJ2.
(A) Structures of the ARE inducers used in panel B, shown as conjugates to Keap1 C151. The regioselectivity of the thiol addition at the endocyclic β-position of 15d-PGJ2 within the cyclopentenone ring has been established and is reviewed elsewhere [48]. Both Michael acceptor positions within CDDO-Im have been shown to contribute to ARE activation [61]. Only one conjugation is shown for simplicity, and the other Michael acceptor site is marked with an asterisk. (B) MDA-MB-231 cells were transfected with expression vectors for Nrf2, either wild-type Keap1 or Keap1 C151S, and a plasmid containing an ARE-dependent firefly luciferase reporter gene. A plasmid containing a constitutively expressed Renilla luciferase was included to normalize for plasmid levels. The transfected cells were exposed to either DMSO (vehicle), 5 μM sulforaphane, 50 nM CDDO-Im or 25 μM 15d-PGJ2 for 16 h prior to analysis of firefly and Renilla luciferase activities in cell lysates. The y-axis is the ratio of the firefly to Renilla values, and all data are normalized to wt Keap1 with DMSO.
An ARE-dependent firefly luciferase reporter gene assay was used to determine ARE activation by sulforaphane, CDDO-Im and 15d-PGJ2 in MDA-MB-231 cells overexpressing human Nrf2 protein and either human wt Keap1 or Keap1 C151S protein. A titration of each inducer was first performed in order to determine the concentrations of each that resulted in maximal ARE activation (data not shown), and these concentrations were then used to test the response of Keap1 C151S to each molecule. As shown in Figure 1B, the ARE activation by both sulforaphane and CDDO-Im is largely attenuated when C151 is substituted with a serine. The level of attenuation is consistent with that observed previously with sulforaphane in this cell line [51]. The ARE activation by 15d-PGJ2 is also reduced for the C151S mutant compared to wt Keap1, but to a lesser extent. These results suggest that modification of C151 by sulforaphane or CDDO-Im is important for activation of the ARE in vivo, but do not address whether modification of other cysteines is also required.
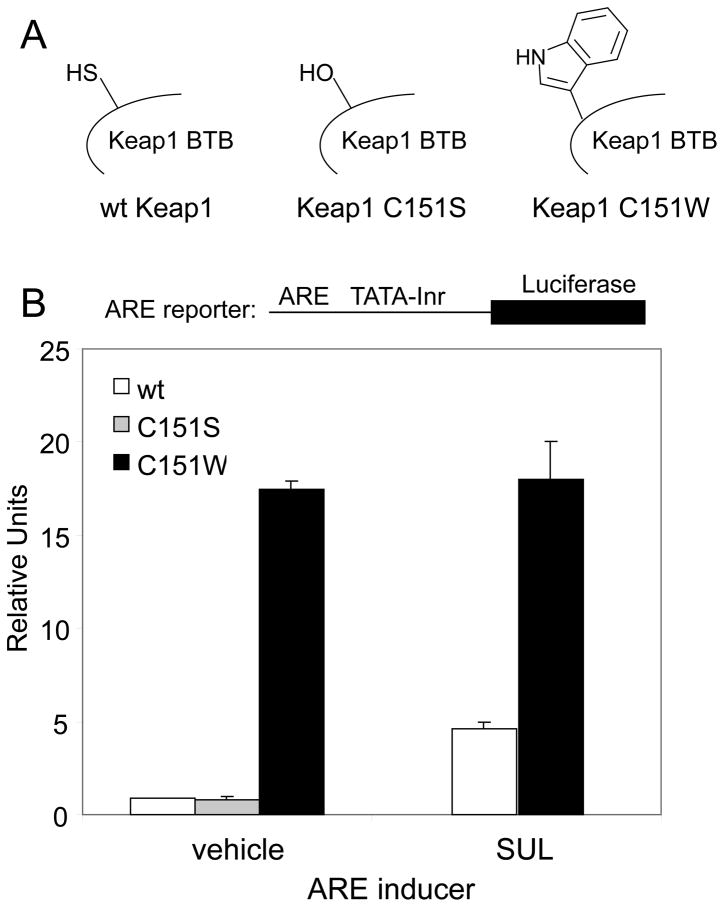
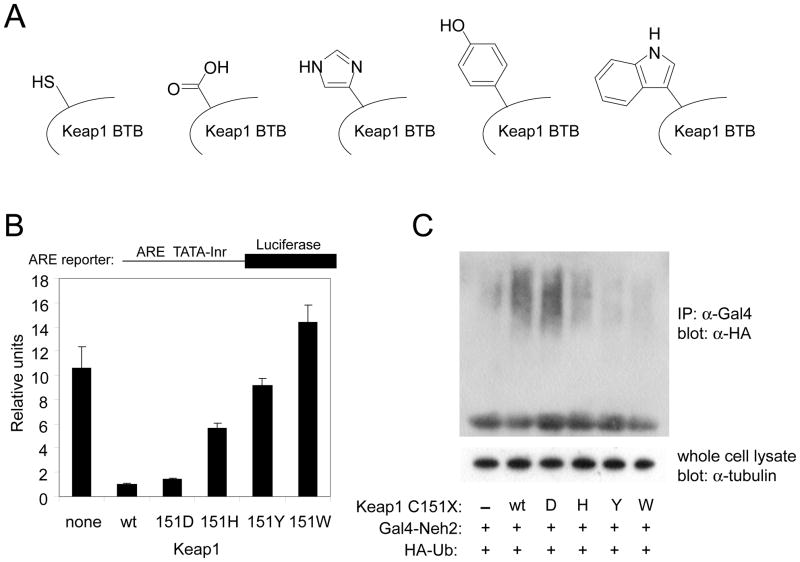
Substitution of C151 with a bulky side chain is sufficient for ARE activation
Based on the fact that various small molecule inducers depend on Keap1 C151 in order to fully activate the ARE, and the high reactivity of C151 in response to various ARE inducers [32, 33], it was of interest to see if the sole modification of C151 is sufficient to activate the ARE response. Due to the large number of cysteine residues in human Keap1, mutation of all 27 cysteines except C151 was impractical. Furthermore, mutation of C288 and C273 to serine or alanine results in constitutive ARE activation, by downregulating Nrf2 ubiquitination [30, 37]. Therefore we chose to mutate C151 to the amino acid with the largest partial molar volume (PMV), tryptophan (Figure 2A), in the hope of mimicking sterically the modification of C151 by electrophilic molecules. The ARE assay in MDA-MB-231 cells using transfected proteins was used to determine the level of ARE activation in cells expressing the Keap1 C151W mutant protein. Remarkably, the level of ARE activation by Nrf2 in the presence of Keap1 C151W was substantially higher than that obtained in the presence of wt Keap1 (Figure 2B). This result indicates that a tryptophan at position 151 does indeed mimic electrophile modification in that it causes Keap1 to lose its ability to repress Nrf2. As shown in Figure 2B, the effect of the tryptophan mutation is in stark contrast to effect of the serine mutation, which had a level of repression similar to wt Keap1. Interestingly, the level of ARE activation in cells transfected with Keap1 C151W was approximately 5 times higher than that obtained in cells transfected with wt Keap1 and treated with the potent ARE inducer sulforaphane.
FIGURE 2. Effect of C151W mutation on ability of Keap1 to repress ARE activation, and the effect of sulforaphane.
(A) Structures of Cys, Ser and Trp, shown as a side chain at position C151. (B) Transient transfection reporter gene assays were conducted essentially as described for Figure 1. Cells were exposed to DMSO or sulforaphane for 16 h. The data for wt Keap1 and Keap1 C151S are the same as in Figure 1, shown here on a different scale, to compare with the ARE activity of the Keap1 C151W protein.
The high level of ARE activation in the presence of Keap1 C151W, compared to that seen with sulforaphane, suggests that negative feedback pathways may be activated by sulforaphane that attenuate the amount of ARE activation possible. To test this, cells expressing the Keap1 C151W protein and Nrf2 were treated with sulforaphane. No attenuation of the level of ARE activation was observed (Figure 2B), indicating that negative feedback pathways are not activated by sulforaphane under these conditions.
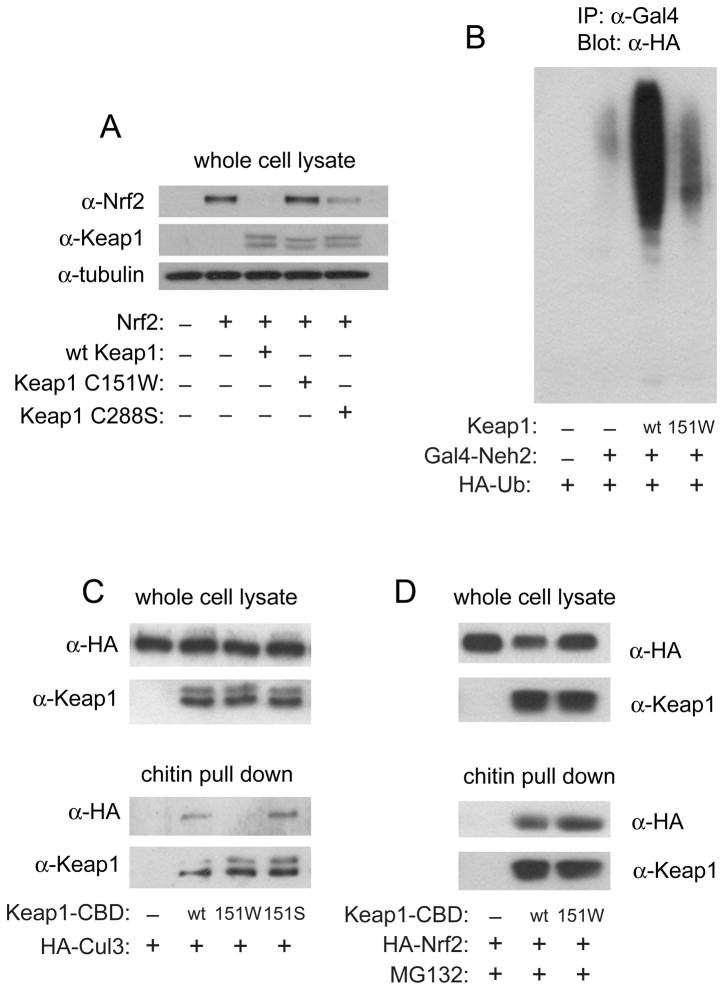
Modification of C151 to a tryptophan leads to ARE activation by increasing Nrf2 protein levels, through destabilization of the Keap1-Cul3 interaction and decreased Nrf2 ubiquitination
Previous work comparing the wt Keap1 protein to the Keap1 C151S protein showed that C151 participates in downregulating Nrf2 ubiquitination in response to sulforaphane or tBHQ in cells [30]. The use of the C151S protein in this experiment [30] meant that all cysteines in Keap1 except C151 were available for modification by the ARE inducers, raising the question of whether modification of other Keap1 cysteines was required for downregulation of Nrf2 ubiquitination. The Keap1 C151W protein allows this question to be addressed directly. The levels of Nrf2 protein and the extent of ubiquitination in the presence of Keap1 C151W were compared with those of wt Keap1 in MDA-MB-231 cells in the absence of ARE inducers (Figure 3A and 3B). Co-transfection of wt Keap1 and Nrf2 was observed to significantly decrease the level of Nrf2 protein as expected (Figure 3A). Transfection of Keap1 C151W, however, resulted in a level of Nrf2 that was similar to that observed in the absence of overexpressed Keap1. The level of Nrf2 observed in the presence of the Keap1 C151W protein was also compared to that found in the presence of Keap1 C288S. Keap1 protein containing serine or alanine substitutions for C288 have the opposite phenotype of the C151S protein, in that they are deficient in their ability to target Nrf2 for ubiquitination and to repress Nrf2-dependent gene expression in transfected cells in the absence of inducers [30, 52]. Nrf2 protein was stabilized in cells expressing Keap1 C288S, though not to the same extent as cells expressing Keap1 C151W (Figure 3A).
FIGURE 3. Effect of Keap1 C151W on Nrf2 levels and ubiquitination, and the interaction of Keap1 with Cul3 and with Nrf2.
(A) MDA-MB-231 cells were transfected with expression vectors for Nrf2 and either wild-type Keap1, Keap1 C151W or Keap1 C288S. Equivalent amounts of each cell lysate were analyzed by immunoblot with anti-Nrf2 antibody (top panel). The blot was stripped and reprobed with anti-Keap1 antibody (middle panel), then anti-tubulin antibody (bottom panel). (B) MDA-MB-231 cells were transfected with expression vectors for Keap1, HA-ubiquitin, Cul3 and Gal4-Neh2. Cells were treated with 10 μM MG132 for 3 h prior to lysis. Equivalent aliquots of each lysate were subjected to immunoprecipitation with anti-Gal4 antibodies, then analyzed by immunoblot with anti-HA antibodies to detect ubiquitinated forms of Gal4-Neh2. (C) MDA-MB-231 cells were transfected with expression vectors for HA-Cul3 and either wt, C151W or C151S Keap1-CBD. In the upper two panels, equivalent amounts of each cell lysate were analyzed by immunoblot with anti-HA antibody (top panel), followed by stripping and reprobing of the blot with anti-Keap1 antibody (bottom panel). In the bottom two panels, the same lysates were subjected to affinity purification with chitin beads. Proteins that remained bound to the beads after washing were analyzed by immunoblot with anti-HA antibody (top panel), and the blot was stripped and reprobed with anti-Keap1 antibody (bottom panel). (D) The experiment was carried out as described for panel C, except that an expression vector for HA-Nrf2 was transfected, instead of one for HA-Cul3, and cells were treated with MG132 for 4.5 hours.
Next, the cellular ubiquitination of Nrf2 in the presence of wt or C151W Keap1 was examined. The assay was conducted using the Gal4-Neh2 construct. The Neh2 domain of Nrf2 contains the seven lysine residues that are targeted for Cul3-mediated ubiquitination by Keap1 [16]. MDA-MB-231 cells were transfected with expression vectors for Gal4-Neh2, HA-ubiquitin, Cul3 and either wt Keap1 or Keap1 C151W. Gal4-immunoprecipitated proteins were immunoblotted with anti-HA antibodies to determine ubiquitinated forms (Figure 3B). Wt Keap1 strongly catalyzed the ubiquitination of Nrf2 lysines, as shown by the large increase in higher molecular weight species. Keap1 C151W was largely unable to catalyze Nrf2 ubiquitination, with levels similar to those seen in the absence of transfected Keap1.
The mechanism behind the ability of wt Keap1, but not Keap1 C151S, to downregulate Nrf2 ubiquitination in response to sulforaphane and tBHQ was shown in MDA-MB-231 cells to involve a decrease of the interaction between Keap1 and Cul3 [16]. A similar result was observed in vitro in a recent study using purified proteins from E. coli in a co-purification assay [39]. The Keap1 C151W protein offers an opportunity to determine whether modification of C151 alone leads to a decreased association of Keap1 and Cul3, or whether modification of other cysteines in addition to C151 by electrophiles is required. A chitin binding domain, fused to the C terminus of Keap1 (Keap1-CBD), was used to enable Keap1 purification, as the CBD does not alter the ability of Keap1 to associate with either Cul3 or Nrf2 or change the responsiveness of Keap1 to ARE inducers [16]. Expression vectors for Keap1-CBD and HA-Cul3 were transfected into MDA-MB-231 cells, and Keap1 was purified by use of chitin beads. Cul3 co-purified with wt Keap1, as shown in Figure 3C. The amount of Cul3 co-purified with Keap1 C151W was observed to be significantly less, indicating that modification of C151 alone is sufficient to alter the Keap1-Cul3 interaction. The extent of interaction between Keap1 C151S and Cul3 was determined as a control, and confirms that only modification to tryptophan, and not serine, decreases the interaction.
It is possible that the Keap1 C151W can no longer catalyze Nrf2 ubiquitination due to a disruption of the Keap1-Nrf2 interaction. It seemed unlikely, as we have previously shown using isothermal titration calorimetry that modification of Keap1 C151 by BIA does not alter the affinity of Keap1 for Nrf2 [36]. However, to ensure that Keap1 C151W is properly folded and functional within the cell, we examined the ability of Keap1 C151W to bind to Nrf2. The same method described above to detect changes in the Keap1-Cul3 interaction was used, except that cells were treated with MG132 before harvesting to stabilize Nrf2. Wt and C151W Keap1 were found to co-purify with Nrf2 in an identical manner, i.e. no differences were observed (Figure 3D). Therefore, modification of C151 does not dissociate the Keap1-Nrf2 complex, consistent with our previous results [36].
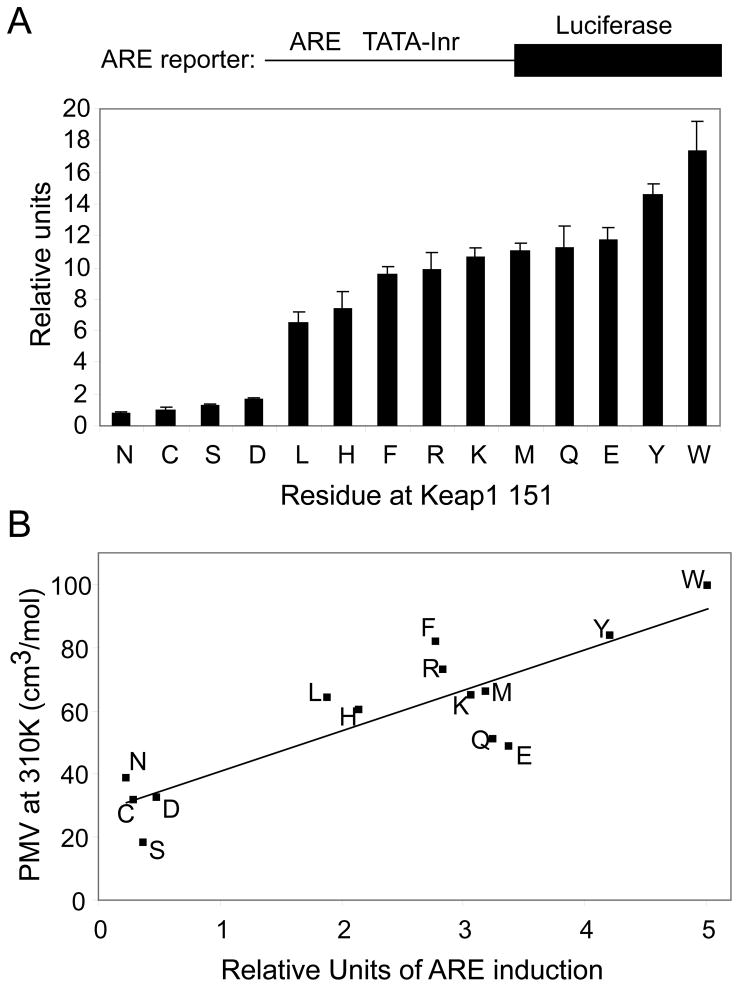
Activation of the ARE is dependent on the PMV of the residue at position 151 of the Keap1 sequence
The loss of the ability of Keap1 to repress ARE activation caused by the introduction of a tryptophan at position C151 prompted us to examine the effects of introducing residues with different physico-chemical properties at this position on ARE activation. A series of site-directed mutants were therefore created to span the various properties of naturally occurring amino acids such as charge, PMV and hydrophobicity. In addition to serine and tryptophan, eleven mutations were introduced at position C151, and the effect on ARE activation was determined (Figure 4A). All of the mutant Keap1 proteins were more effective than the Keap1 C151W protein at repression of ARE-dependent reporter gene activity. The highest repression was seen with the residues of smallest PMV, N, C, S and D, and the lowest repression with those of highest PMV, Y and W. This trend indicates that the PMV at position 151 may be a major determinant of the extent of ARE repression. To further investigate this correlation, ARE activity was plotted against the PMV of each amino acid residue [53], calculated at 310° K, the temperature at which the cells were maintained during the experiment. A good correlation was found between the ARE activity and the PMV of the residue occupying position 151 (Figure 4B). Other physico-chemical properties such as charge or hydrophobicity do not appear to contribute significantly to the extent of ARE activation. For example, the Keap1 phenylalanine mutant had a somewhat high repression of ARE activity for its PMV, indicating hydrophobicity might increase the extent of ARE repression. However, the other hydrophobic residues (leucine, tyrosine and tryptophan) showed a good correlation of ARE activity and PMV. In addition, while the Keap1 glutamate mutant showed a low level of ARE repression for its PMV, indicating the negative charge of the residue may in some way enhance the effect of its size, the ability of the aspartate mutant to repress the ARE activation correlated well with its PMV.
FIGURE 4. ARE activation by Nrf2 in the presence of Keap1 C151X.
(A) Transient transfection reporter gene assays were conducted essentially as described for Figure 1. (B) The data from panel A was plotted against the PMV of each residue at position 151 of Keap1, which was calculated at 310K as described in the text. Each data point is labeled with its corresponding residue abbreviation.
The PMV of the residue at position 151 in Keap1 determines the extent to which the Keap1 protein can catalyze Nrf2 ubiquitination
The studies above support the idea that repression of ARE activation correlates with the PMV of the residue at position 151 of Keap1, with a tryptophan showing the least repression (Figure 4). The low level of repression by Keap1 C151W observed is due to the inability of the protein to efficiently catalyze the ubiquitination of Nrf2 lysines (Figure 3B). In order to verify that the ability of the other Keap1 C151 mutants to catalyze Nrf2 ubiquitination also correlates with the PMV of the residue at position 151, a subset of the thirteen mutants was selected. In addition to C151W, Keap1 C151D, C151H and C151Y (Figure 5A) were chosen as representative of the spectrum of PMV of the side-chains. Parallel experiments were performed where one experiment determined the extent of ubiquitination of Nrf2 lysines as described for Figure 3B, and the second experiment simultaneously determined the extent of ARE activity associated with co-transfection of each Keap1 mutant along with Nrf2. As shown in Figure 5B, the ARE repression associated with these Keap1 mutants decreases steadily as the PMV of the residue increases. Correspondingly, the extent of ubiquitination of Nrf2 lysines catalyzed by Keap1 steadily decreases as the PMV of the Keap1 C151 residue increases (Figure 5C), indicating that the PMV of the residue at position 151 is a major determinant of the ability of Keap1 to catalyze Nrf2 ubiquitination.
FIGURE 5. Nrf2 ubiquitination catalyzed by Keap1 C151 mutants of increasing PMV.
(A) Structures of Cys, Asp, His, Tyr and Trp, shown as a side chain at position C151. (B) Transient transfection reporter gene assays were conducted essentially as described for Figure 1. (C) Gal4-Neh2 ubiquitination assays were performed essentially as described for Figure 3B.
DISCUSSION
We find that modification of human Keap1 C151 by mutation to a bulky residue is sufficient to downregulate Nrf2 ubiquitination and significantly activate the ARE in cells. The effect appears to be mediated by altering the interaction between Keap1 and the Cul3 protein, which targets Nrf2 for ubiquitination, as the Keap1 C151W mutation leads to a decrease in the extent of interaction between Keap1 and Cul3. Further, we find that the PMV of the residue at position 151 correlates well with the degree of ARE activation, and the loss of ability of the Keap1 protein to catalyze Nrf2 ubiquitination. No other physico-chemical properties appear to contribute significantly.
No disruption of the Keap1-Nrf2 interaction by the Keap1 C151W mutation is observed. However, it is possible that the C151W mutation alters the interaction of Keap1 with the Nrf2 DLG site, which would disrupt Nrf2 ubiquitination, as illustrated by the two-site model [18, 38]. The tryptophan mutation in the Keap1 BTB dimerization domain could alter the conformation of the Keap1 dimer, thereby decreasing the interaction of the Keap1 Kelch domain with the lower affinity DLG binding site on Nrf2. Moreover, the ability of Keap1 C151W to bind to Nrf2 indicates that the C151W mutation causes a specific conformational change in the Keap1 protein affecting its interaction with Cul3, rather than a general inactive conformation of Keap1.
The importance of modification of Keap1 C151 for Nrf2 activation agrees well with our previous mass spectrometry studies of the human proteins, in which C151 was the only cysteine consistently and highly modified by a variety of ARE inducers [32, 33]. In addition, during the preparation of this manuscript, Kobayashi et al. showed in zebrafish embryos that expression of zebrafish Keap1 with a tryptophan mutation at the cysteine position corresponding to C151 in humans leads to constitutive upregulation of an ARE-regulated gene in the absence of ARE inducers [54]. Therefore, across species, modification of this key cysteine residue alone appears to be important and sufficient for ARE activation.
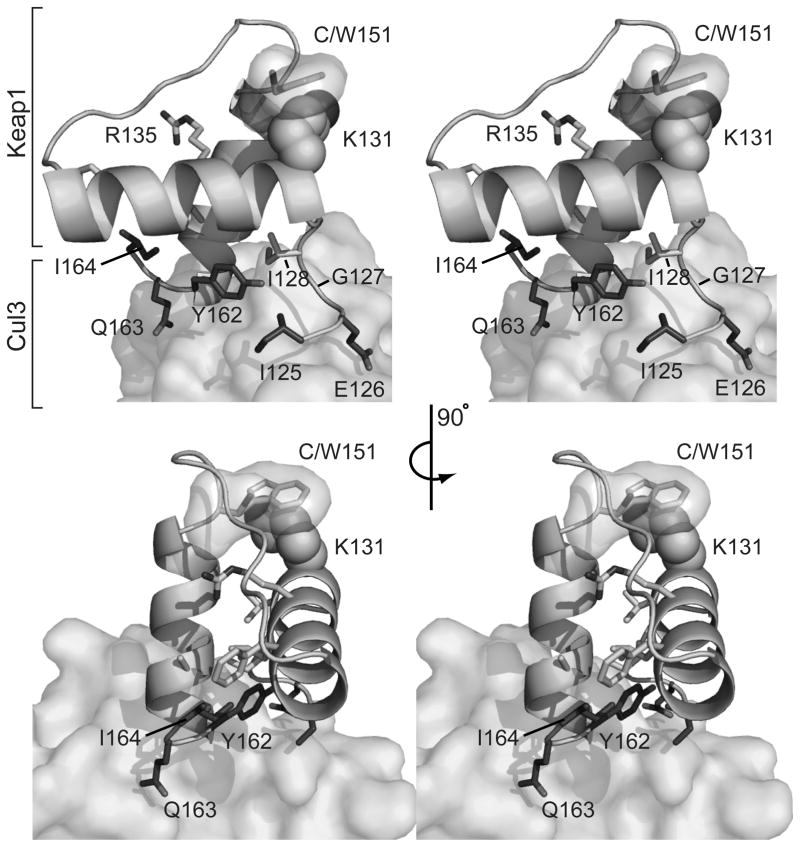
The series of mutations at position 151 of Keap1 reveals that a major contribution to the effects observed is the PMV of the residue at that position. To gain further insight as to how an increased PMV at position 151 could alter the interaction between Keap1 and Cul3, we examined a homology model of the interaction of the BTB domain of Keap1 with Cul3 (Figure 6). In earlier work, we had noted from the model that C151 is not located at the predicted Keap1-Cul3 interface [36]. Keap1 residues 125–127 and 162–164, colored in dark grey in Figure 6, are predicted to interact with Cul3 based on those required for association of MEI-26 with the Caenorhabditis elegans Cul3 protein [16, 55]. Mutant Keap1 proteins containing alanine substitutions for either 125–127 or 162–164 had similar phenotypes to the Keap1 C151W mutant in that they were unable to catalyze ubiquitination of the Neh2 domain of Nrf2 [16]. In our earlier study, we noted that C151 is close to the BTB homodimerization interface, and we predicted that modification of Keap1 C151 would perturb the homodimerization interface, thereby disrupting Nrf2 ubiquitination [36]. While a bulky modification at position 151 may alter the protein conformation at the homodimerization interface, the model in Figure 6 instead indicates that a bulky modification at that position would impose specific structural effects that would alter Keap1-Cul3 binding. In the model, substitution of a tryptophan residue at position 151 causes a significant steric clash of the tryptophan side chain with K131. The R135 residue, depending on its orientation, is also close enough to clash with the tryptophan. These residues reside on an α–helix that forms an α-helix-β-strand-α-helix motif with the α–helix containing C151. Importantly, residues K131 and R135 are conserved in sequences of Keap1 proteins from divergent species (Figure 7), suggesting their importance to Keap1 function. Interestingly, K131 is not conserved in the zebrafish Keap1 isoform (Keap1a) that has been shown to have an inactive cysteine at the position corresponding to 151 in human Keap1 [54]. However, K131 is conserved in the active Keap1b form. We hypothesize that a bulky residue at position 151 alters the interaction between these two helices via a steric clash with K131, thereby forcing the α–helix containing K131 away from position 151. Movement of this helix would then affect the positioning of residues 125–127 relative to Cul3, altering Keap1-Cul3 binding. We predict the positioning of Y162 of the other binding site, which forms a hydrophobic interaction with I128, would be altered as well.
FIGURE 6. Stereo view of a model of the putative Keap1 BTB-Cul3 interaction interface and the location of residue 151 of Keap1.
A portion of the modeled BTB domain of Keap1 is shown as a ribbon diagram, and a portion of the modeled Cul3 structure is shown in surface representation. Keap1 residue 151 in the BTB domain is shown in transparent surface representation and in stick representation, modeled both as a cysteine (sulfur atom in dark grey) and as a tryptophan (all atoms in light grey). Lysine 131 is shown in space filling representation. The Keap1 residues 125–127 and 162–164, in the putative Cul3 binding region, are shown as all atoms in dark grey.
FIGURE 7. Conservation of key residues among Keap1 proteins with a reactive C151 residue.

The sequences of six vertebrate Keap1 proteins were aligned using clustalw 3.5c. Residues are highlighted in dark gray if identical among all six proteins and in light gray if chemically conserved or identical across the species. Conservation was diagrammed using BOXSHADE 3.3.1. The GenBank accession numbers used in the alignment, respectively, are 3894323, 16924016, 1280415, 51870493, 164698434, 56118572, and 109639157. The numbering shown corresponds to human Keap1. The cysteine in zebrafish Keap1a that corresponds to C151 in human Keap1 is not reactive, due to an adjacent threonine instead of a lysine [54]. The labeled residues are those highlighted in the model in Figure 6 and are all conserved in Keap1 proteins with a reactive C151.
The effect of increasing the PMV of a residue at a key position in a signaling protein on its function has been observed in studies on rhodopsin [56], the photoreceptor molecule of the vertebrate retina and a member of the G-protein-coupled receptor superfamily [57, 58]. In these studies, the effect of changing the PMV at position 125 was explored. The original L125R mutation showed impaired signal transduction and an inability to bind 11-cis-retinal [59]. A series of 10 site-directed mutations were introduced at position 125 to study which physico-chemical properties were required for the effect [56]. As we found for position 151 of Keap1, Andres et al. found that the PMV of the residue at position 125 of rhodopsin was the major determinant of the extent of the effect, with the residues of greatest PMV showing the greatest impairment in both binding to 11-cis-retinal and downstream activation of transducin. In the case of rhodopsin, the mutations appear to cause a change in protein conformation since the thermal stability of the mutant proteins was reduced. Just as C151 is located away from the Keap1-Cul3 binding site, the crystal structure of rhodopsin [60] shows that L125 is located four α-helix turns away from the residues directly involved in chromophore binding. The data indicate that mutation of Leu125 to a bulky residue does not affect the chromophore interaction directly, but rather alters the optimal conformation of the retinal binding pocket.
The linear trend observed in Figure 4, where residues of increasing PMV at Keap1 position 151 lead to a decrease in Keap1’s ability to repress Nrf2, would suggest that electrophiles that are larger than tryptophan would induce at least as much ARE activation as the tryptophan mutant if not more upon reacting with C151. However, the extent of activation of the ARE by Nrf2 in the presence of Keap1 C151W is much higher than in the presence of wt Keap1 and the potent ARE inducers sulforaphane and CDDO-Im (Figures 1 and 2). Comparing the ARE activation by these molecules to that from other C151 mutations, the extent of activation is less than that observed for ten of the mutants, up to and including the Keap1 C151L protein (Figures 1B and 4A). Therefore, it seems unlikely that the physico-chemical properties of the mutant side chains at position 151 are more suited to cause an effect on the Keap1 protein than modification of C151 by an electrophile. A more probable explanation is that a significant population of Keap1 molecules in a cell are not modified at position 151 by ARE inducers, and remain capable of forming an active ubiquitin ligase complex. A likely reason is that the concentration of an electrophile required to modify a high percentage of Keap1 molecules at C151 is probably not attainable in the reducing cellular environment, which actively reduces electrophiles to prevent toxicity, including conjugation with glutathione. Another possibility is that access to C151 is partially obstructed in the cellular environment, perhaps due to the Keap1-Nrf2-Cul3 complex associating with other protein factors. A final possibility would be that negative feedback pathways are activated by the electrophiles, possibly by modification of other Keap1 cysteines, or alteration of phosphorylation pathways. These could limit the ARE activation attainable by an electrophilic inducer by dampening the effect of modification of C151. However, when we tested this possibility by adding sulforaphane to the experiment with Keap1 C151W, we found no difference in the response (Figure 2). Therefore, activation of pathways that downregulate the ARE response by electrophilic inducers seems unlikely.
The trend observed in Figures 4 and 5 also suggests that if any electrophile of sufficient size modifies C151, then Nrf2 ubiquitination will be disrupted, and the ARE will be activated. However, a few inducers have very recently been shown to either be independent of a requirement for C151, or to exhibit much less dependence. Wang et al. showed that ARE activation by both sodium arsenite [As(III)] and monomethylarsonous acid [MMA(III)] is independent of Keap1 C151, using the C151S mutant protein in transfected MDA-MB-231 cells [51]. The PMV of either As(III) or MMA(III) is presumably similar to that of the smaller amino acids, which do not show significant ARE activation (Figure 4A). It seems likely that modification of C151 by these agents does not lead to ARE activation due to insufficient size, and ARE activation occurs through other mechanisms. However, ARE activation by the much larger electrophile, 15d-PGJ2, was shown by Kobayashi et al. to be largely independent of C151 in zebrafish embryos transfected with mouse Keap1 C151S [54]. This result agrees with our finding that ARE activation by 15d-PGJ2 in human cells is much less dependent on Keap1 C151 than that of sulforaphane or CDDO-Im (Figure 1B). The PMV of 15d-PGJ2 is greater than that of tryptophan and sulforaphane, and modification of C151 by this molecule would be predicted to cause downregulation of Nrf2 ubiquitination and ARE activation. One possibility is that 15d-PGJ2, when conjugated to C151, adopts a conformation that does not cause a steric clash with K131 or R135. Another explanation would be that 15d-PGJ2 is simply much less reactive towards C151 than the majority of inducers whose ARE activation is C151-dependent, including sulforaphane, CDDO-Im, and tBHQ. In support of this possibility, Kobayashi et al. did not detect modification of mouse Keap1 C151 in vitro by 15d-PGJ2, while the ARE inducer diethylmaleate was found to be both C151-dependent and able to modify C151 in vitro [54]. Further studies are required to determine whether the trend of C151 dependence in vivo reflecting C151 modification in vitro is consistent for a larger number of ARE inducers. The mechanism by which the highly reactive Keap1 C151 is excluded from reacting with electrophiles in vitro such as 15d-PGJ2 is puzzling and of interest to investigate.
In conclusion, we hypothesize that modification of C151 by an electrophile of sufficient PMV alters the interaction of the modeled α-helix-β-strand-α-helix motif of Keap1 through steric clashes with K131 and possibly R135, as shown in Figure 6. These steric clashes would then affect the positioning of the Keap1 residues that interact with Cul3, translating to altered Keap1-Cul3 binding and decreased Nrf2 ubiquitination. The Keap1 C151W protein promises to be a useful tool to test effects of Keap1 C151 modification.
Acknowledgments
We thank Dr. Shih-Ching Lo of Genentech for helpful advice on the in vivo ubiquitination assay and the chitin-binding assay. We also acknowledge Dr. Michael B. Sporn of Dartmouth College for his generous gift of CDDO-Im.
FUNDING
This work was supported by the National Institutes of Health, by Program Project P01 CA48112 (to A.D.M.), by Grant Number R03 CA128095-01A1 (to A.L.E.), by Grant Number RO1 AT003389 (to M. H.) and by Grant Number P50 AT 000155 from the National Center for Complementary and Alternative Medicine and the Office of Dietary Supplements (to A.L.E.). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Center for Complementary and Alternative Medicine, the Office of Dietary Supplements, or the National Institutes of Health.
Abbreviations used in this paper
- NQO1
NAD(P)H:quinone oxidoreductase 1
- ARE
antioxidant response element
- tBHQ
tert-butylhydroquinone
- BIA
biotinylated iodoacetamide
- BMCC
1-biotinamido-4-(4′-[maleimidoethyl-cyclohexane]-carboxamido) butane
- CBD
chitin binding domain
- HA
hemagglutinin
- CDDO-Im
1-[2-cyano-3-,12-dioxooleana-1,9(11)-dien-28-oyl]imidazole
- wt
wild-type
- PVDF
polyvinylidene fluoride
- PMV
partial molar volume
- 15d-PGJ2
15d-deoxy-Δ12,14-prostagladin J2
- As(III)
sodium arsenite
- and MMA(III)
monomethylarsonous acid
Footnotes
Author contributions: A.L.E., M.H. and A.D.M. designed research; A.L.E. performed research; A.L.E., M.H., E.S. and A.D.M. analyzed data; and A.L.E., M.H., E.S. and A.D.M. wrote the paper.
References
- 1.Nioi P, McMahon M, Itoh K, Yamamoto M, Hayes JD. Identification of a novel Nrf2-regulated antioxidant response element (ARE) in the mouse NAD(P)H:quinone oxidoreductase 1 gene: reassessment of the ARE consensus sequence. Biochem J. 2003;374:337–348. doi: 10.1042/BJ20030754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kensler TW, Wakabayashi N, Biswal S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu Rev Pharmaco Toxicol. 2007;47:89–116. doi: 10.1146/annurev.pharmtox.46.120604.141046. [DOI] [PubMed] [Google Scholar]
- 3.Dinkova-Kostova AT, Massiah MA, Bozak RE, Hicks RJ, Talalay P. Potency of Michael reaction acceptors as inducers of enzymes that protect against carcinogenesis depends on their reactivity with sulfhydryl groups. Proc Natl Acad Sci USA. 2001;98:3404–3409. doi: 10.1073/pnas.051632198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eggler AL, Gay KA, Mesecar AD. Molecular mechanisms of natural products in chemoprevention: Induction of cytoprotective enzymes by Nrf2. Mol Nutr Food Res. 2008;52:S84–S94. doi: 10.1002/mnfr.200700249. [DOI] [PubMed] [Google Scholar]
- 5.Zhang Y, Talalay P, Cho CG, Posner GH. A major inducer of anticarcinogenic protective enzymes from broccoli: isolation and elucidation of structure. Proc Natl Acad Sci USA. 1992;89:2399–2403. doi: 10.1073/pnas.89.6.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chung FL, Conaway CC, Rao CV, Reddy BS. Chemoprevention of colonic aberrant crypt foci in Fischer rats by sulforaphane and phenethyl isothiocyanate. Carcinogenesis. 2000;21:2287–2291. doi: 10.1093/carcin/21.12.2287. [DOI] [PubMed] [Google Scholar]
- 7.Kuroiwa Y, Nishikawa A, Kitamura Y, Kanki K, Ishii Y, Umemura T, Hirose M. Protective effects of benzyl isothiocyanate and sulforaphane but not resveratrol against initiation of pancreatic carcinogenesis in hamsters. Cancer Lett. 2006;241:275–280. doi: 10.1016/j.canlet.2005.10.028. [DOI] [PubMed] [Google Scholar]
- 8.Fahey JW, Haristoy X, Dolan PM, Kensler TW, Scholtus I, Stephenson KK, Talalay P, Lozniewski A. Sulforaphane inhibits extracellular, intracellular, and antibiotic-resistant strains of Helicobacter pylori and prevents benzo[α]pyrene-induced stomach tumors. Proc Natl Acad Sci USA. 2002;99:7610–7615. doi: 10.1073/pnas.112203099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerhauser C, You M, Liu J, Moriarty RM, Hawthorne M, Mehta RG, Moon RC, Pezzuto JM. Cancer chemopreventive potential of sulforamate, a novel analogue of sulforaphane that induces phase 2 drug-metabolizing enzymes. Cancer Res. 1997;57:272–278. [PubMed] [Google Scholar]
- 10.Karapetian RN, Evstafieva AG, Abaeva IS, Chichkova NV, Filonov GS, Rubtsov YP, Sukhacheva EA, Melnikov SV, Schneider U, Wanker EE, Vartapetian AB. Nuclear Oncoprotein Prothymosin α Is a Partner of Keap1: Implications for Expression of Oxidative Stress-Protecting Genes. Mol Cell Biol. 2005;25:1089–1099. doi: 10.1128/MCB.25.3.1089-1099.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Velichkova M, Hasson T. Keap1 Regulates the Oxidation-Sensitive Shuttling of Nrf2 into and out of the Nucleus via a Crm1-Dependent Nuclear Export Mechanism. Mol Cell Biol. 2005;25:4501–4513. doi: 10.1128/MCB.25.11.4501-4513.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun Z, Zhang S, Chan JY, Zhang DD. Keap1 Controls Postinduction Repression of the Nrf2-Mediated Antioxidant Response by Escorting Nuclear Export of Nrf2. Mol Cell Biol. 2007;27:6334–6349. doi: 10.1128/MCB.00630-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jain AK, Bloom DA, Jaiswal AK. Nuclear Import and Export Signals in Control of Nrf2. J Biol Chem. 2005;280:29158–29168. doi: 10.1074/jbc.M502083200. [DOI] [PubMed] [Google Scholar]
- 14.Cullinan SB, Gordan JD, Jin J, Harper JW, Diehl JA. The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: oxidative stress sensing by a Cul3-Keap1 ligase. Mol Cell Biol. 2004;24:8477–8486. doi: 10.1128/MCB.24.19.8477-8486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kobayashi A, Kang MI, Okawa H, Ohtsuji M, Zenke Y, Chiba T, Igarashi K, Yamamoto M. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol Cell Biol. 2004;24:7130–7139. doi: 10.1128/MCB.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang DD, Lo SC, Cross JV, Templeton DJ, Hannink M. Keap1 Is a Redox-Regulated Substrate Adaptor Protein for a Cul3-Dependent Ubiquitin Ligase Complex. Mol Cell Biol. 2004;24:10941–10953. doi: 10.1128/MCB.24.24.10941-10953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tong KI, Katoh Y, Kusunoki H, Itoh K, Tanaka T, Yamamoto M. Keap1 Recruits Neh2 through Binding to ETGE and DLG Motifs: Characterization of the Two-Site Molecular Recognition Model. Mol Cell Biol. 2006;26:2887–2900. doi: 10.1128/MCB.26.8.2887-2900.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McMahon M, Thomas N, Itoh K, Yamamoto M, Hayes JD. Dimerization of Substrate Adaptors Can Facilitate Cullin-mediated Ubiquitylation of Proteins by a “Tethering” Mechanism: A TWO-SITE INTERACTION MODEL FOR THE Nrf2-Keap1 COMPLEX. J Biol Chem. 2006;281:24756–24768. doi: 10.1074/jbc.M601119200. [DOI] [PubMed] [Google Scholar]
- 19.Shibata T, Ohta T, Tong KI, Kokubu A, Odogawa R, Tsuta K, Asamura H, Yamamoto M, Hirohashi S. Cancer related mutations in NRF2 impair its recognition by Keap1-Cul3 E3 ligase and promote malignancy. Proc Natl Acad Sci USA. 2008;105:13568–13573. doi: 10.1073/pnas.0806268105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salazar M, Rojo AI, Velasco D, de Sagarra RM, Cuadrado A. Glycogen Synthase Kinase-3beta Inhibits the Xenobiotic and Antioxidant Cell Response by Direct Phosphorylation and Nuclear Exclusion of the Transcription Factor Nrf2. J Biol Chem. 2006;281:14841–14851. doi: 10.1074/jbc.M513737200. [DOI] [PubMed] [Google Scholar]
- 21.Keum YS, Yu S, Chang PPJ, Yuan X, Kim JH, Xu C, Han J, Agarwal A, Kong ANT. Mechanism of Action of Sulforaphane: Inhibition of p38 Mitogen-Activated Protein Kinase Isoforms Contributing to the Induction of Antioxidant Response Element-Mediated Heme Oxygenase-1 in Human Hepatoma HepG2 Cells. Cancer Res. 2006;66:8804–8813. doi: 10.1158/0008-5472.CAN-05-3513. [DOI] [PubMed] [Google Scholar]
- 22.Nakaso K, Yano H, Fukuhara Y, Takeshima T, Wada-Isoe K, Nakashima K. PI3K is a key molecule in the Nrf2-mediated regulation of antioxidative proteins by hemin in human neuroblastoma cells. FEBS Lett. 2003;546:181–184. doi: 10.1016/s0014-5793(03)00517-9. [DOI] [PubMed] [Google Scholar]
- 23.Kang KW, Lee SJ, Park JW, Kim SG. Phosphatidylinositol 3-kinase regulates nuclear translocation of NF-E2-related factor 2 through actin rearrangement in response to oxidative stress. Mol Pharmacol. 2002;62:1001–1010. doi: 10.1124/mol.62.5.1001. [DOI] [PubMed] [Google Scholar]
- 24.Li J, Johnson D, Calkins M, Wright L, Svendsen C, Johnson J. Stabilization of Nrf2 by tBHQ Confers Protection against Oxidative Stress-Induced Cell Death in Human Neural Stem Cells. Toxicol Sci. 2004;83:313–328. doi: 10.1093/toxsci/kfi027. [DOI] [PubMed] [Google Scholar]
- 25.Zipper LM, Mulcahy RT. Erk Activation Is Required for Nrf2 Nuclear Localization during Pyrrolidine Dithiocarbamate Induction of Glutamate Cysteine Ligase Modulatory Gene Expression in HepG2 Cells. Toxicol Sci. 2003;73:124–134. doi: 10.1093/toxsci/kfg083. [DOI] [PubMed] [Google Scholar]
- 26.Alam J, Wicks C, Stewart D, Gong P, Touchard C, Otterbein S, Choi AM, Burow ME, Tou J. Mechanism of heme oxygenase-1 gene activation by cadmium in MCF-7 mammary epithelial cells. Role of p38 kinase and Nrf2 transcription factor. J Biol Chem. 2000;275:27694–27702. doi: 10.1074/jbc.M004729200. [DOI] [PubMed] [Google Scholar]
- 27.Kang KW, Ryu JH, Kim SG. The Essential Role of Phosphatidylinositol 3-Kinase and of p38 Mitogen-Activated Protein Kinase Activation in the Antioxidant Response Element-Mediated rGSTA2 Induction by Decreased Glutathione in H4IIE Hepatoma Cells. Mol Pharmacol. 2000;58:1017–1025. doi: 10.1124/mol.58.5.1017. [DOI] [PubMed] [Google Scholar]
- 28.Huang HC, Nguyen T, Pickett CB. Phosphorylation of Nrf2 at Ser-40 by protein kinase C regulates antioxidant response element-mediated transcription. J Biol Chem. 2002;277:42769–42774. doi: 10.1074/jbc.M206911200. [DOI] [PubMed] [Google Scholar]
- 29.Numazawa S, Ishikawa M, Yoshida A, Tanaka S, Yoshida T. Atypical protein kinase C mediates activation of NF-E2-related factor 2 in response to oxidative stress. Am J Physiol Cell Physiol. 2003;285:C334–342. doi: 10.1152/ajpcell.00043.2003. [DOI] [PubMed] [Google Scholar]
- 30.Zhang DD, Hannink M. Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Mol Cell Biol. 2003;23:8137–8151. doi: 10.1128/MCB.23.22.8137-8151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamamoto T, Suzuki T, Kobayashi A, Wakabayashi J, Maher J, Motohashi H, Yamamoto M. Physiological significance of reactive cysteine residues of Keap1 in determining Nrf2 activity. Mol Cell Biol. 2008;28:2758–2770. doi: 10.1128/MCB.01704-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eggler AL, Luo Y, van Breemen RB, Mesecar AD. Identification of the highly reactive cysteine 151 in the chemopreventive agent-sensor Keap1 protein is method-dependent. Chem Res Toxicol. 2007;20:1878–1884. doi: 10.1021/tx700217c. [DOI] [PubMed] [Google Scholar]
- 33.Luo Y, Eggler AL, Liu D, Liu G, Mesecar AD, van Breemen RB. Sites of alkylation of human Keap1 by natural chemoprevention agents. J Am Soc Mass Spectrom. 2007;18:2226–2232. doi: 10.1016/j.jasms.2007.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Itoh K, Wakabayashi N, Katoh Y, Ishii T, Igarashi K, Engel JD, Yamamoto M. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 1999;13:76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dinkova-Kostova AT, Holtzclaw WD, Cole RN, Itoh K, Wakabayashi N, Katoh Y, Yamamoto M, Talalay P. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc Natl Acad Sci USA. 2002;99:11908–11913. doi: 10.1073/pnas.172398899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eggler AL, Liu G, Pezzuto JM, van Breemen RB, Mesecar AD. Modifying specific cysteines of the electrophile-sensing human Keap1 protein is insufficient to disrupt binding to the Nrf2 domain Neh2. Proc Natl Acad Sci USA. 2005;102:10070–10075. doi: 10.1073/pnas.0502402102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kobayashi A, Kang MI, Watai Y, Tong KI, Shibata T, Uchida K, Yamamoto M. Oxidative and Electrophilic Stresses Activate Nrf2 through Inhibition of Ubiquitination Activity of Keap1. Mol Cell Biol. 2006;26:221–229. doi: 10.1128/MCB.26.1.221-229.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tong KI, Padmanabhan B, Kobayashi A, Shang C, Hirotsu Y, Yokoyama S, Yamamoto M. Different Electrostatic Potentials Define ETGE and DLG Motifs as Hinge and Latch in Oxidative Stress Response. Mol Cell Biol. 2007;27:7511–7521. doi: 10.1128/MCB.00753-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rachakonda G, Xiong Y, Sekhar KR, Stamer SL, Liebler DC, Freeman ML. Covalent Modification at Cys151 Dissociates the Electrophile Sensor Keap1 from the Ubiquitin Ligase CUL3. Chem Res Toxicol. 2008;21:705–710. doi: 10.1021/tx700302s. [DOI] [PubMed] [Google Scholar]
- 40.Furukawa M, Xiong Y. BTB protein Keap1 targets antioxidant transcription factor Nrf2 for ubiquitination by the Cullin 3-Roc1 ligase. Mol Cell Biol. 2005;25:162–171. doi: 10.1128/MCB.25.1.162-171.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Treier M, Staszewski LM, Bohmann D. Ubiquitin-dependent c-Jun degradation in vivo is mediated by the [delta] domain. Cell. 1994;78:787–798. doi: 10.1016/s0092-8674(94)90502-9. [DOI] [PubMed] [Google Scholar]
- 42.Wasserman WW, Fahl WE. Functional antioxidant responsive elements. Proc Natl Acad Sci USA. 1997;94:5361–5366. doi: 10.1073/pnas.94.10.5361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sussan TE, Rangasamy T, Blake DJ, Malhotra D, El-Haddad H, Bedja D, Yates MS, Kombairaju P, Yamamoto M, Liby KT, Sporn MB, Gabrielson KL, Champion HC, Tuder RM, Kensler TW, Biswal S. Targeting Nrf2 with the triterpenoid CDDO- imidazolide attenuates cigarette smoke-induced emphysema and cardiac dysfunction in mice. Proc Natl Acad Sci USA. 2009;106:250–255. doi: 10.1073/pnas.0804333106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Osburn WO, Yates MS, Dolan PD, Chen S, Liby KT, Sporn MB, Taguchi K, Yamamoto M, Kensler TW. Genetic or Pharmacologic Amplification of Nrf2 Signaling Inhibits Acute Inflammatory Liver Injury in Mice. Toxicol Sci. 2008;104:218–227. doi: 10.1093/toxsci/kfn079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yates MS, Kwak MK, Egner PA, Groopman JD, Bodreddigari S, Sutter TR, Baumgartner KJ, Roebuck BD, Liby KT, Yore MM, Honda T, Gribble GW, Sporn MB, Kensler TW. Potent Protection against Aflatoxin-Induced Tumorigenesis through Induction of Nrf2-Regulated Pathways by the Triterpenoid 1-[2-Cyano-3-,12-Dioxooleana-1,9(11)-Dien-28-Oyl]Imidazole. Cancer Res. 2006;66:2488–2494. doi: 10.1158/0008-5472.CAN-05-3823. [DOI] [PubMed] [Google Scholar]
- 46.Thimmulappa RK, Scollick C, Traore K, Yates M, Trush MA, Liby KT, Sporn MB, Yamamoto M, Kensler TW, Biswal S. Nrf2-dependent protection from LPS induced inflammatory response and mortality by CDDO-Imidazolide. Biochem Biophys Res Commun. 2006;351:883–889. doi: 10.1016/j.bbrc.2006.10.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thimmulappa RK, Fuchs RJ, Malhotra D, Scollick C, Traore K, Bream JH, Trush MA, Liby KT, Sporn MB, Kensler TW, Biswal S. Preclinical Evaluation of Targeting the Nrf2 Pathway by Triterpenoids (CDDO-Im and CDDO-Me) for Protection from LPS-Induced Inflammatory Response and Reactive Oxygen Species in Human Peripheral Blood Mononuclear Cells and Neutrophils. Antioxid Redox Signal. 2007;9:1963–1970. doi: 10.1089/ars.2007.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Uchida K, Shibata T. 15-Deoxy-Delta(12,14)-prostaglandin J2: an electrophilic trigger of cellular responses. Chem Res Toxicol. 2008;21:138–144. doi: 10.1021/tx700177j. [DOI] [PubMed] [Google Scholar]
- 49.Levonen AL, Landar A, Ramachandran A, Ceaser EK, Dickinson DA, Zanoni G, Morrow JD, Darley-Usmar VM. Cellular mechanisms of redox cell signalling: role of cysteine modification in controlling antioxidant defences in response to electrophilic lipid oxidation products. Biochem J. 2004;378:373–382. doi: 10.1042/BJ20031049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Itoh K, Mochizuki M, Ishii Y, Ishii T, Shibata T, Kawamoto Y, Kelly V, Sekizawa K, Uchida K, Yamamoto M. Transcription factor Nrf2 regulates inflammation by mediating the effect of 15-deoxy-Delta(12,14)-prostaglandin j(2) Mol Cell Biol. 2004;24:36–45. doi: 10.1128/MCB.24.1.36-45.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang XJ, Sun Z, Chen W, Li Y, Villeneuve NF, Zhang DD. Activation of Nrf2 by arsenite and monomethylarsonous acid is independent of Keap1-C151: enhanced Keap1-Cul3 interaction. Toxicol Appl Pharmacol. 2008;230:383–389. doi: 10.1016/j.taap.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wakabayashi N, Dinkova-Kostova AT, Holtzclaw WD, Kang MI, Kobayashi A, Yamamoto M, Kensler TW, Talalay P. Protection against electrophile and oxidant stress by induction of the phase 2 response: fate of cysteines of the Keap1 sensor modified by inducers. Proc Natl Acad Sci USA. 2004;101:2040–2045. doi: 10.1073/pnas.0307301101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hackel M, Hinz HJ, Hedwig GR. Partial molar volumes of proteins: amino acid side-chain contributions derived from the partial molar volumes of some tripeptides over the temperature range 10–90 degrees C. Biophys Chem. 1999;82:35–50. doi: 10.1016/s0301-4622(99)00104-0. [DOI] [PubMed] [Google Scholar]
- 54.Kobayashi M, Li L, Iwamoto N, Nakajima-Takagi Y, Kaneko H, Nakayama Y, Eguchi M, Wada Y, Kumagai Y, Yamamoto M. The Antioxidant Defense System Keap1-Nrf2 Comprises a Multiple Sensing Mechanism for Responding to a Wide Range of Chemical Compounds. Mol Cell Biol. 2009;29:493–502. doi: 10.1128/MCB.01080-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu L, Wei Y, Reboul J, Vaglio P, Shin TH, Vidal M, Elledge SJ, Harper JW. BTB proteins are substrate-specific adaptors in an SCF-like modular ubiquitin ligase containing CUL-3. Nature. 2003;425:316–321. doi: 10.1038/nature01985. [DOI] [PubMed] [Google Scholar]
- 56.Andrés A, Kosoy A, Garriga P, Manyosa J. Mutations at position 125 in transmembrane helix III of rhodopsin affect the structure and signalling of the receptor. Eur J Biochem. 2001;268:5696–5704. doi: 10.1046/j.0014-2956.2001.02509.x. [DOI] [PubMed] [Google Scholar]
- 57.Ji TH, Grossmann M, Ji I. G Protein-coupled Receptors. I DIVERSITY OF RECEPTOR-LIGAND INTERACTIONS. J Biol Chem. 1998;273:17299–17302. doi: 10.1074/jbc.273.28.17299. [DOI] [PubMed] [Google Scholar]
- 58.Hargrave PA, McDowell JH. Rhodopsin and phototransduction: a model system for G protein-linked receptors. FASEB J. 1992;6:2323–2331. doi: 10.1096/fasebj.6.6.1544542. [DOI] [PubMed] [Google Scholar]
- 59.Sung CH, Davenport CM, Nathans J. Rhodopsin mutations responsible for autosomal dominant retinitis pigmentosa. Clustering of functional classes along the polypeptide chain. J Biol Chem. 1993;268:26645–26649. [PubMed] [Google Scholar]
- 60.Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Trong IL, Teller DC, Okada T, Stenkamp RE, Yamamoto M, Miyano M. Crystal Structure of Rhodopsin: A G Protein-Coupled Receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 61.Dinkova-Kostova AT, Liby KT, Stephenson KK, Holtzclaw WD, Gao X, Suh N, Williams C, Risingsong R, Honda T, Gribble GW, Sporn MB, Talalay P. Extremely potent triterpenoid inducers of the phase 2 response: Correlations of protection against oxidant and inflammatory stress. Proc Natl Acad Sci USA. 2005;102:4584–4589. doi: 10.1073/pnas.0500815102. [DOI] [PMC free article] [PubMed] [Google Scholar]