Abstract
Skeletal muscle comprises approximately 40 % of total body mass and, as such, contributes to maintenance of human health. In this review we will discuss the current state of knowledge regarding the role of molecular clocks in skeletal muscle. In addition we discuss a new function for exercise as a time setting cue for muscle and other peripheral tissues.
Keywords: circadian, skeletal muscle, exercise, peripheral tissues, zeitgeber
Introduction
Circadian rhythms are the approximate 24-hour biological cycles that function to prepare the organism for daily environmental changes. Almost all organisms ranging from single cell bacteria to plants and animals, exhibit behavioral, physiological and biochemical rhythms termed circadian rhythms.(7, 9) Underlying circadian rhythms is a molecular clock mechanism found in most, if not all, cell types including skeletal muscle. At the cellular level, the presence of a molecular clock is argued to be a necessary timekeeping mechanism to prepare the cell for daily changes in environmental conditions. The ability to synchronize the molecular clock and intracellular physiology with external day-night cycles denotes an evolutionarily conserved adaptation to environmental conditions.(7, 9)
Environmental stimuli capable of shifting the timing of molecular rhythms are critically important to understanding the synchronization of oscillators to the environment and to each other within a multicellular system. The most well understood circadian time cue (termed zeitgeber) is the photic or light time cue. While lighting conditions have been extensively studied in many species more recently, non-photic zeitgebers such as time of feeding and time of physical activity/exercise have been shown to influence molecular rhythms and behavior.(25, 31) The goal of this review is to present the latest findings demonstrating that the molecular clock exists in skeletal muscle and support for the concept that exercise plays a role as a time cue for clocks in peripheral tissues. The existing data, although limited, suggests the importance of exercise and more importantly, the timing of exercise in adding stability to the daily rhythms of the circadian system. These observations add a new function for the role of exercise in human health and suggest new concepts for the benefits of exercise in potentially postponing or preventing the development of chronic diseases.(25, 31)
The Molecular Clock in Mammals
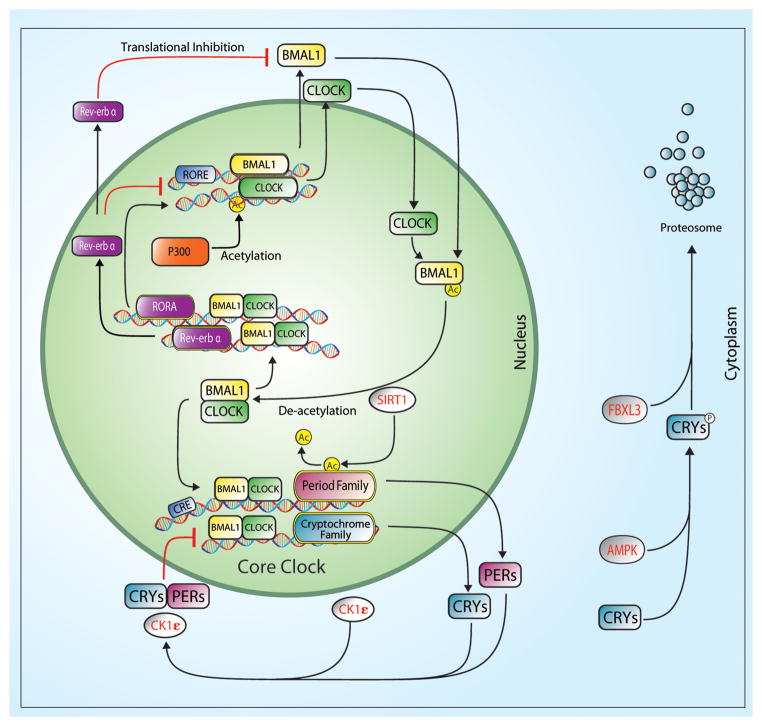
The molecular mechanism underlying circadian rhythms is a gene regulatory network composed of transcriptional-translational feedback loops referred to as the core clock.(11) The molecular clock components comprising the positive arm of the core clock are two members of the PAS-bHLH family of transcription factors, Clock (Circadian locomotor output control kaput) and Bmal1 (Brain muscle arnt-like1). BMAL1 protein is expressed in a circadian pattern in both the SCN and peripheral tissues.(4, 8) CLOCK protein levels do not oscillate in the SCN and only in some peripheral tissues.(15) However, the nuclear to cytosolic distribution of CLOCK is circadian in its pattern with highest levels of CLOCK in the nucleus occurring in the light phase in mice.(18, 28) The CLOCK:BMAL1 heterodimer activates transcription of additional core clock genes Period (Per1, Per2 and Per3) and Cryptochrome (Cry1 and Cry2) by binding to E-box (CACGTG) sequences in the regulatory region of these genes. The CRY and PER proteins constitute the negative arm of the core molecular clock by forming multimers that inhibit CLOCK:BMAL1 transcriptional activity upon translocation to the nucleus. A schematic of the molecular clock mechanism is shown in Figure 1.
Figure 1.
The core circadian clock is an autoregulatory transcriptional feedback loop comprised of a transcription factors, CLOCK and BMAL1 and their target genes; Per1, Per2, Cry1, and Cry2. The CLOCK:BMAL1 heterodimer also functions to drive transcription of Rev-erbα and Rorα which in turn either represses or activates Bmal1 transcription respectively. The CLOCK:BMAL1 heterodimer also regulates downstream targets known as clock-controlled genes (CCG). The kinases, casein kinase 1ε (CK1ε) and AMP kinase (AMPK), phosphorylate PER(CK1ε) and CRY(AMPK) proteins to promote polyubiquitination by E3 ubiquitin ligase complexes. PER and CRY proteins are then degraded by the 26S proteasome complex.
Additional components to the core molecular clock family include the orphan nuclear receptors Rora (RAR-related orphan receptor-α) and Rev-erb α/β. These gene products function to link the feedback loops by activating (Rora) or repressing (Rev-erb) Bmal1 transcription.(21, 24) Most recently, studies have added new elements, known as E3 ligases (e.g. Fbxl3), to the core molecular clock and these elements function by changing stability of the PER and CRY proteins.(33) In addition to their role in timekeeping, components of the core clock (Bmal1 and Clock) have also been shown to transcriptionally regulate the expression of genes that do not function in timekeeping and these genes are designated as clock-controlled genes (CCGs).
While the identity of all the direct clock controlled genes in a specific tissue, like skeletal muscle, have not been defined, they often encode transcription factors (e.g. MyoD1) or proteins that control rate-limiting steps in cell physiology (e.g. PBEF, the rate-limiting enzyme in the NAD+ salvage pathway).(17, 18) For more detailed reviews of the molecular clock mechanism there are several recent reviews by other groups.(16)
Central/Peripheral Clocks in Mammals and Zeitgebers
The first suggestion that the central clock (circadian pacemaker) was located in the suprachiasmatic nucleus (SCN) came when it was discovered that surgical ablation of the SCN resulted in arrhythmic behavior patterns.(27) SCN ablation in hamsters creates an actively arrhythmic animal and transplantation of a healthy SCN (from either a hamster or mouse) back into the lesioned hamsters restores activity rhythms with rhythms matching that of the donor animal (26) demonstrating the role of the SCN as a system-wide circadian synchronizer. Cell autonomy of the molecular clock in peripheral tissue was first established almost 15 years ago.(3) The development of a mouse model in which the luciferase cDNA was knocked into the Per2 coding region to generate a chimeric protein provided the powerful resource (PER2:LUC mice) to directly test the cell autonomy of the clock mechanism.(32) For these studies, tissue explants from the SCN, liver and lung from these PER2:LUC mice were placed in cell culture in the presence of luciferin and real time light emission was monitored for up to two weeks. While oscillations in luminescence were expected from the central clock (SCN slice) these findings demonstrated that the clock mechanism from peripheral tissue explants maintained normal 24hr circadian periodicity in the absence of systemic neural or humoral factors.(32) These studies also validated the use of the PER2::LUC mouse as a valuable resource to study molecular clock function in multiple tissues including skeletal muscle. (31, 35)
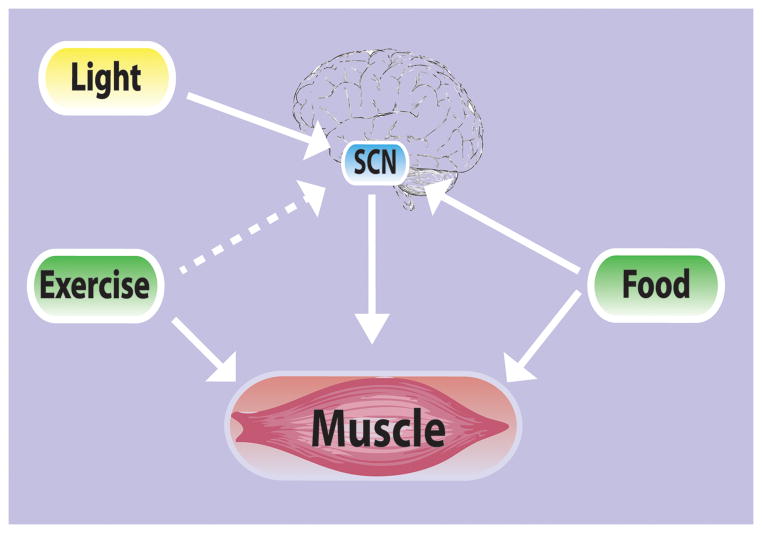
While studies have established that the molecular clock mechanism is intrinsic to each cell and that it can run in a cell autonomous manner, a critical feature is that the phase of the molecular clock can be set or reset by cues from the environment. The ability to reset the circadian clock is a critical function to be able to adapt to environmental changes. The timing or phase of the central clock is primarily entrained by cues from light.(29) Recent studies have demonstrated that the molecular clock in many peripheral tissues, including skeletal muscle can be dissociated from the rhythm of the SCN by restricting time of feeding.(4) In addition, Zambon et al.(34) reported that there is an interaction between time of day and contraction on expression of clock genes in human muscle, suggesting contractile activity might be a zeitgeber for the molecular clock mechanism in skeletal muscle. In this study, microarray analysis was used to determine the effects of resistance exercise on gene expression in the quadriceps at 6 and 18 hours after an acute bout exercise. Gene expression was also examined in the non-exercised leg at the same time points for comparison. These results indicate that feeding, and potentially contractile activity, can act as dominant zeitgebers for setting clocks in peripheral tissues. (Figure 2) They also demonstrate that circadian rhythms in peripheral tissues can be dissociated from the SCN through non-photic environmental signals.
Figure 2.
Central and peripheral clocks can respond to both photic (light) and non-photic (feeding and exercise) zeitgebers. This schematic demonstrates the interplay between the different zeitgebers and central and peripheral clocks. It also suggests that central and peripheral clocks may have differential responses depending upon the zeitgeber present.
The timing of the molecular clock can also be modulated by post-translational mechanisms using phosphorylation, acetylation and/or ubiquitination pathways. These modifications impact the stability and/or translocation of core molecular clock components with the greatest impact on period length (time for completion of 1 cycle (24h)). The most well-known regulator of clock function through changing phosphorylation status is casein kinase1ε (CK1ε). This serine/threonine kinase has been the subject of studies in both hamster and humans in which the mutations of either CK1ε or PER2 affect period length. The tau mutant hamster, has a point mutation (Arg178Cys) in the catalytic region of CK1ε, termed the tau mutation, which results in a hamster that exhibits short circadian periods.(23) In these hamsters, CK1ε is unable to phosphorylate PER proteins. With diminished levels of phosphorylation, PER translocates to the nucleus leading to a more rapid repression of the CLOCK: BMAL1 mediated transcription, effectively shortening the circadian period to 20 hours. (10) In humans, a mutation in PER2 that affects a phosphorylation site (target of CK1ε) results in the clinical condition, familial advance sleep phase syndrome (FASPS). Studies of these individuals determined that they have a shortened period length of approximately 20 hours compared to the normal 24 hours observed in most humans and is associated with early awake and early to sleep.(30)
Acetylation of histones unfolds chromatin to expose promoter and regulatory regions of genes and is associated with activation of gene expression while acetylation results in silencing of gene expression. An example for a circadian role in acetylation comes from studies of the histone acetyltransferase p300. The p300 protein associates with CLOCK in a circadian manner, implicating p300 as a component of the core clock transactivation complex. In addition, H3 histone acetylation at the promoter region of Per1 and Per2 shows robust circadian rhythms in phase with the Per1/Per2 mRNA and p300:CLOCK complex formation. p300 is inhibited by the CRY protein leading to a decrease in CLOCK: BMAL1 mediated transcription at the Per1 promoter. These data suggest that the repressor action of CRY is mediated in part by its actions on chromatin structure.(6) The most frequently studied deacetylase involved in circadian regulation is SirtT1. SirT1 deacetylates histone H3. This activity shows a strong circadian rhythm antiphase to the rhythm of histone H3 acetylation.(17) This antiphase oscillation of acetylation/deacetylation suggest an important direct or permissive mechanism for controlling clock gene expression and propose roles for acetylation/deacetylation in the initiation, duration and termination of both the activating and repressing phases of the circadian cycle.
The Molecular Clock in Skeletal Muscle
The skeletal muscle circadian transcriptome was first identified by Miller and colleagues.(14) This work was followed by a publication which identified ~215 mRNAs that were expressed in a circadian pattern in the gastrocnemius muscle of wildtype C57BL6 mice. This list included known core molecular clock components Bmal1, Per2 and Cry1.(13) Gene ontology analysis identified enrichment in multiple cellular processes including but not limited to metabolic, transcriptional and degradative processes. The tissue specificity of the circadian transcriptome in skeletal muscle was underscored by the inclusion of known muscle specific genes such as Myod1, Ucp3, Fbxo32/atrogin and Myh1(MyHC IIX). The identification of Myod1 as a gene expressed in a circadian manner was exciting as it is well-characterized as a transcription factor involved in the skeletal muscle lineage. The fact that it is expressed in a circadian manner in adult tissue and is under direct control of CLOCK:BMAL1(1) suggest that it may be critical for daily muscle maintenance. (Figure 1) To further understand the implications of molecular clock disruption in skeletal muscle McCarthy et al,(13) compared gene expression changes between skeletal muscle from wildtype C57Bl6 mice to those from Clock mutant mice. The Clock mutant mice have a mutation that results in the deletion of exon 19 of CLOCK and this is associated with longer behavioral rhythms of 27 hours.(2) We found that several core clock genes including Bmal1 and Per2, no longer oscillated in the muscle of Clock mutant mice. Oscillation was also lost in several known clock controlled genes in the skeletal muscle such as Tef, Dbp, Myod1 and Pgc1β. In addition to analysis of the circadian genes, McCarthy et al, found that approximately 35% of all the expressed genes were differentially expressed in the muscle of Clock mutant mice demonstrating the substantial impact disruption of the core clock machinery has on gene expression with implications for normal cellular function and health. With continued development of analysis tools for time course gene expression studies, the list of genes expressed in a circadian manner in skeletal muscle has grown to greater than 800. Pizarro et al (2013)(20) have established a valuable website to allow for queries into circadian expression from time course expression experiments in tissues and cell lines (http://circadb.org). These new data will open many exciting avenues of research in the study of skeletal muscle circadian physiology. However, one area that is still understudied is whether the circadian transcriptome differs among individual skeletal muscles of different developmental origin (e.g. limb vs. facial muscle) or whether they differ among muscles with markedly different fiber type, metabolic function and mechanical function.
Exercise and circadian rhythms
Wheel running or cage activity has long been utilized as a read-out of circadian behavior but the idea that physical activity/exercise and more importantly the timing of exercise could serve as an entrainment cue is relatively new. (25, 31) Studies in the late 1980’s and early 90’s were the first to show that novel wheel access at different times of day was sufficient to shift the phase of circadian activity rhythms in mice and hamsters. (5) Further studies demonstrated that exercise is a sufficient environmental cue to effect clock gene expression in the SCN (central clock) located in the hypothalamus of the brain. (12) These studies established that activity, in the form of access to a novel running wheel, during light conditions decreased peak expression of the clock genes Per1 and Per2 in the SCN. Schroeder et al took this idea further examining both timing of exercise as well as multiple bouts of exercise. They explored effects on the central clock utilizing a scheduled exercise paradigm in control and mutant mice in which the central molecular clock mechanism was weakened.(25) In attempts to better mimic the timing of exercise in the human population, mice were allowed free access to a wheel, no access to a wheel, or access limited to 6 hour time frames at the beginning or end of the dark or active phase for a minimum of 16 days. Similar to previous studies examining single bouts of exercise, they observed changes in the properties of the molecular clock in the SCN after 16 days in the control mice suggesting the phase shifts observed in previous studies were not solely an acute phase response. Moreover, PER2:LUC amplitude was damped in mice with wheel access scheduled early in the dark phase but unaffected with scheduled activity late in the dark phase or with free access to wheel running suggesting that the timing of exercise may be critical for the maintenance of molecular rhythms in the SCN. Utilizing the vasoactive intestinal polypeptide (VIP) knock-out mouse, shown to have an unstable clock mechanism, they found that scheduled exercise functioned to enhance the stability of both activity and heart rate rhythms.
The core molecular clock gene Clock has also been demonstrated to be critical for healthy skeletal muscle as Clock mutant mice exhibit ~30% reductions in normalized maximal force at both the muscle and single-fiber level. (1) In addition, myofiber architecture is disrupted and mitochondrial volume is diminished. Recent work from Pastore et al supports these results. (19) They demonstrated that CLOCK protein is critical for mitochondrial maintenance in skeletal muscle. Taking this a step further, they examined the ability of clock mutant mice to adapt to chronic exercise and found that despite the pathology as a result of the mutant Clock gene the ability of these mice to adapt to chronic exercise was not changed. Utilizing endurance training they were able to partially rescue the metabolic defects resulting from loss of functional CLOCK protein.
Most studies of exercise and shifting of circadian rhythms have relied on endurance exercise paradigms. Less is known about the potential for resistance exercise but Zambon et al., reported that one bout of 60 contractions was associated with changes in molecular clock gene expression in skeletal muscle of humans.(34) Experiments in neonatal cardiomyocytes have shown that contractile activity may modulate the molecular clock through the actions of the CLOCK protein. Histological and biochemical analysis demonstrated that CLOCK localizes to the Z-disk in neonatal cardiomyocytes and translocates to the nucleus to influence gene expression in response to contractile activity. CLOCK localization at the Z-disk puts CLOCK in an appropriate location to sense mechanical function associated with contractile activity. (22) While these studies are suggestive that resistance exercise can also modulate molecular clock function in muscle, there is still much to be determined.
With over 600 different muscles in the human body, comprising approximately 40 percent total body mass, understanding the effects of exercise on the molecular rhythms in individual skeletal muscles may provide critical insight into systemic mechanisms contributing to daily rhythms. At this stage, there is only one study that has examined more than one muscle in mice exposed to scheduled bouts of either voluntary or involuntary endurance exercise for 2 hours/day in the light phase four hours after lights on. In this study, the authors found a significant shift in clock gene expression (PER2:LUC bioluminescence) in three different skeletal muscles and the lung from exercised mice, whereas the molecular clock in the SCN remained unshifted demonstrating that scheduled exercise can alter the molecular clock in peripheral tissues. In addition, one of the muscles examined, the flexor digitorum brevis (FDB) was phase advanced more than the other two muscles (soleus and extensor digitorum longus) suggesting the potential for differential regulation of the molecular clock in individual muscles.(31) Lumicycle data in combination with data demonstrating rescue of phenotype resulting from exercise,(19, 25, 31) implicates exercise as a non-photic time cue in peripheral tissues and suggests that the molecular clock in all muscle tissues may not respond in a similar manner to non-photic cues. Since muscle is such a large contributor to systems physiology, these data have broad implications for human health and disease suggesting the power of exercise, and more specifically the interaction of exercise and muscle, as a therapeutic strategy to help stabilize/realign molecular clocks throughout the body.
Summary
Circadian rhythms and molecular clocks in skeletal muscle is a new and rapidly emerging area of research. Studies in the last 15 years have demonstrated that molecular clocks exist in skeletal muscle and more importantly, studies have demonstrated that the phase of the clocks in skeletal muscle can be reset by altering time of exercise or time of feeding independent of the central clock in the brain. In parallel studies it has been shown that skeletal muscles from mice in which the core clock genes have been disrupted are weaker and exhibit decreased mitochondrial content and function. These findings suggest a potential new role for exercise in human health through its role in providing timing information to skeletal muscle and other peripheral tissue clocks. Although difficult to extrapolate to humans, exercise studies in mice do suggest an optimal time of day for exercise to enhance the robust nature of circadian rhythms at the molecular level. Until an optimal time is determined, consistently timed daily exercise may be a viable alternative. These studies highlight the recognition that time of day matters for maintenance of proper molecular clock function and is important for issues of muscle strength and endurance.
Summary.
This review discusses the relationship between the molecular clock, skeletal muscle, and exercise.
Acknowledgments
This work was supported by the following NIH grants RC1ES018636 and R01AR55246 (KAE).
The authors would like to thank Jonathan England for his help with the figures in the preparation of this manuscript.
Footnotes
Disclosures: No conflicts of interest, financial or otherwise, are declared by the author(s).
Bibliography
- 1.Andrews JL, Zhang X, McCarthy JJ, et al. CLOCK and BMAL1 regulate MyoD and are necessary for maintenance of skeletal muscle phenotype and function. Proceedings of the National Academy of Sciences. 107(44):19090–5. doi: 10.1073/pnas.1014523107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antoch MP, Song EJ, Chang AM, et al. Functional Identification of the Mouse Circadian Clock Gene by Transgenic BAC Rescue. Cell. 1997;89(4):655–67. doi: 10.1016/s0092-8674(00)80246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balsalobre A, Damiola F, Schibler U. A Serum Shock Induces Circadian Gene Expression in Mammalian Tissue Culture Cells. Cell. 1998;93(6):929–37. doi: 10.1016/s0092-8674(00)81199-x. [DOI] [PubMed] [Google Scholar]
- 4.Damiola F, Le Minh N, Preitner N, Kornmann B, Fleury-Olela F, Schibler U. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 2000;14:2950–61. doi: 10.1101/gad.183500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Edgar D, Dement W. Regularly scheduled voluntary exercise synchronizes the mouse circadian clock. Am J Physiol. 1991;261:R928–R33. doi: 10.1152/ajpregu.1991.261.4.R928. [DOI] [PubMed] [Google Scholar]
- 6.Etchegaray JP, Lee C, Wade PA, Reppert SM. Rhythmic histone acetylation underlies transcription in the mammalian circadian clock. Nature. 2003;421(6919):177–82. doi: 10.1038/nature01314. [DOI] [PubMed] [Google Scholar]
- 7.Idda ML, Bertolucci C, Vallone D, Gothilf Y, Sánchez-Vázquez FJ, Foulkes NS. Chapter 3 - Circadian clocks: Lessons from fish. In: Andries Kalsbeek MMTR, Russell GF, editors. Progress in Brain Research. Elsevier; 2012. pp. 41–57. [DOI] [PubMed] [Google Scholar]
- 8.Lee Y, Chen R, Lee H, Lee C. Stoichiometric Relationship among Clock Proteins Determines Robustness of Circadian Rhythms. Journal of Biological Chemistry. 286(9):7033–42. doi: 10.1074/jbc.M110.207217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Loudon Andrew SI. Circadian Biology: A 2.5 Billion Year Old Clock. Current biology : CB. 2012;22(14):R570–R1. doi: 10.1016/j.cub.2012.06.023. [DOI] [PubMed] [Google Scholar]
- 10.Lowrey P, Shimomura K, Antoch M, et al. Positional syntenic cloning and functional characterization of the mammalian circadian mutation tau. Science. 2000;288(5465):483–92. doi: 10.1126/science.288.5465.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lowrey PL, Takahashi JS. MAMMALIAN CIRCADIAN BIOLOGY: Elucidating Genome-Wide Levels of Temporal Organization. Annual Review of Genomics and Human Genetics. 2004;5(1):407–41. doi: 10.1146/annurev.genom.5.061903.175925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maywood E, Mrosovsky N, Field M, Hastings M. Rapid down-regulation of mammalian period genes during behavioral resetting of the circadian clock. Proc Natl Acad Sci USA. 1999;96:15211–6. doi: 10.1073/pnas.96.26.15211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCarthy JJ, Andrews JL, McDearmon EL, et al. Identification of the circadian transcriptome in adult mouse skeletal muscle. Physiological Genomics. 2007;31(1):86–95. doi: 10.1152/physiolgenomics.00066.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller BH, McDearmon EL, Panda S, et al. Circadian and CLOCK-controlled regulation of the mouse transcriptome and cell proliferation. Proceedings of the National Academy of Sciences. 2007;104(9):3342–7. doi: 10.1073/pnas.0611724104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miyazaki M, Schroder E, Edelmann SE, et al. Age-Associated Disruption of Molecular Clock Expression in Skeletal Muscle of the Spontaneously Hypertensive Rat. PLoS ONE. 2011;6(11):e27168. doi: 10.1371/journal.pone.0027168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mohawk JA, Green CB, Takahashi JS. Central and Peripheral Circadian Clocks in Mammals. Annual Review of Neuroscience. 2012;35(1):445–62. doi: 10.1146/annurev-neuro-060909-153128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakahata Y, Kaluzova M, Grimaldi B, et al. The NAD+-Dependent Deacetylase SIRT1 Modulates CLOCK-Mediated Chromatin Remodeling and Circadian Control. Cell. 2008;134(2):329–40. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Panda S, Antoch MP, Miller BH, et al. Coordinated Transcription of Key Pathways in the Mouse by the Circadian Clock. Cell. 2002;109(3):307–20. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 19.Pastore S, Hood DA. Endurance training ameliorates the metabolic and performance characteristics of circadian Clock mutant mice. Journal of Applied Physiology. 2013;114(8):1076–84. doi: 10.1152/japplphysiol.01505.2012. [DOI] [PubMed] [Google Scholar]
- 20.Pizarro A, Hayer K, Lahens NF, Hogenesch JB. CircaDB: a database of mammalian circadian gene expression profiles. Nucleic Acids Research. 2013;41(D1):D1009–D13. doi: 10.1093/nar/gks1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Preitner N, Damiola F, Lopez-Molina L, et al. The orphan nuclear receptor REV-ERBα controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–60. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- 22.Qi L, Boateng SY. The circadian protein CLOCK localizes to the sarcomeric Z-disk and is a sensor of myofilament cross-bridge activity in cardiac myocytes. Biochemical and Biophysical Research Communications. 2006;351(4):1054–9. doi: 10.1016/j.bbrc.2006.10.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ralph MR, Menaker M. A mutation of the circadian system in golden hamsters. Science. 1988;241(4870):1225–7. doi: 10.1126/science.3413487. [DOI] [PubMed] [Google Scholar]
- 24.Sato TK, Panda S, Miraglia LJ, et al. A Functional Genomics Strategy Reveals Rora as a Component of the Mammalian Circadian Clock. Neuron. 2004;43(4):527–37. doi: 10.1016/j.neuron.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 25.Schroeder AM, Truong D, Loh DH, Jordan MC, Roos KP, Colwell CS. Voluntary scheduled exercise alters diurnal rhythms of behaviour, physiology and gene expression in wild-type and vasoactive intestinal peptide-deficient mice. The Journal of Physiology. 2012;590(23):6213–26. doi: 10.1113/jphysiol.2012.233676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sollars P, Kimble D, Pickard G. Restoration of circadian behavior by anterior hypothalamic heterografts. The Journal of Neuroscience. 1995;15(3):2109–22. doi: 10.1523/JNEUROSCI.15-03-02109.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stephan F, Zucker I. Circadian rhythms in drinking behavior and locomotor activity of rats are eliminated by hypothalamic lesions. Proc Natl Acad Sci U S A. 1972;69(6):1583–6. doi: 10.1073/pnas.69.6.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Storch KF, Paz C, Signorovitch J, et al. Intrinsic Circadian Clock of the Mammalian Retina: Importance for Retinal Processing of Visual Information. Cell. 2007;130(4):730–41. doi: 10.1016/j.cell.2007.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takahashi J, DeCoursey P, Bauman L, Menaker M. Spectral sensitivity of a novel photoreceptive system mediating entrainment of mammalian circadian rhythms. Nature. 1984;308(5955):186–8. doi: 10.1038/308186a0. [DOI] [PubMed] [Google Scholar]
- 30.Vanselow K, Vanselow JT, Westermark PO, et al. Differential effects of PER2 phosphorylation: molecular basis for the human familial advanced sleep phase syndrome (FASPS) Genes & Development. 2006;20(19):2660–72. doi: 10.1101/gad.397006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wolff G, Esser KA. Scheduled Exercise Phase Shifts the Circadian Clock in SkeletalMuscle. Medicine & Science in Sports & Exercise. 2012;44(9):1663–70. doi: 10.249/MSS.0b013e318255cf4c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoo SH, Ko CH, Lowrey PL, et al. A noncanonical E-box enhancer drives mouse Period2 circadian oscillations in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(7):2608–13. doi: 10.1073/pnas.0409763102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoo S-H, Mohawk Jennifer A, Siepka Sandra M, et al. Competing E3 Ubiquitin Ligases Govern Circadian Periodicity by Degradation of CRY in Nucleus and Cytoplasm. Cell. 2013;152(5):1091–105. doi: 10.1016/j.cell.2013.01.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zambon A, McDearmon E, Salomonis N, et al. Time- and exercise-dependent gene regulation in human skeletal muscle. Genome Biology. 2003;4(10):R61. doi: 10.1186/gb-2003-4-10-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang X, Patel SP, McCarthy JJ, Rabchevsky AG, Goldhamer DJ, Esser KA. A non-canonical E-box within the MyoD core enhancer is necessary for circadian expression in skeletal muscle. Nucleic Acids Research. 2012;40(8):3419–30. doi: 10.1093/nar/gkr1297. [DOI] [PMC free article] [PubMed] [Google Scholar]