Abstract
A prime objective of genomic medicine is the identification of disease-causing mutations and the mechanisms by which such events result in disease. As most disease phenotypes arise not from single genes and proteins but from a complex network of molecular interactions, a priori knowledge about the molecular network serves as a framework for biological inference and data mining. Here we review recent developments at the interface of biological networks and mutation analysis. We examine how mutations may be treated as a perturbation of the molecular interaction network and what insights may be gained from taking this perspective. We review work that aims to transform static networks into rich context-dependent networks and recent attempts to integrate non-coding RNAs into such analysis. Finally, we conclude with an overview of the many challenges and opportunities that lie ahead.
Introduction
Genome-wide association studies (GWAS) have identified numerous risk loci for common complex diseases, and next-generation sequencing (NGS) based association strategies are now emerging to characterize the contribution of rare variants to human genetic disorders [1,2]. While these studies have provided useful insights into the heritability of diseases, prediction of disease risk from genetic information remains challenging. In addition, without a basic understanding of the biological mechanisms by which most of the candidate loci cause disease, it remains difficult to develop therapeutic strategies for countering them.
The phenotypic effects of genetic alterations result from disruptions of biological activities within cells. These activities arise from the coordinated expression and interaction of various molecules such as proteins, nucleic acids and metabolites [3–7]. Networks can provide a framework for visualizing and performing inference on the set of intracellular molecular interactions and are a promising intermediate for studying genotype-phenotype relationships.
In the ideal case, a candidate locus can be linked to phenotype using canonical ‘pathways’ curated from the biomedical literature, i.e., sequences of experimentally characterized molecular interactions that give rise to a common function. For example, Lee et al. identified candidate de novo somatic mutations in cases of hemimegalencephaly (HME) [8] and found an enrichment of mutations in genes encoding key proteins in the canonical PIK3CA-AKT-mTOR pathway in the affected brain tissue. Based on the structure of this well-studied pathway, they applied an assay to detect pathway activity downstream of the mutation events and determined that the de novo mutations were associated with elevated mTOR activity. Their findings further suggest patients with HME may benefit from treatment with mTOR inhibitors.
In most cases, candidate genes implicated by GWAS or NGS-based studies are not well characterized and their products are not included in available canonical signaling pathways; furthermore, canonical pathways are likely to be incomplete and may even be inaccurate [7]. Systematic screens of the proteome suggest that canonical pathways capture only a fraction of the true protein-protein interactions that occur within the cell [9] and many such interactions may depend on tissue and condition-specific factors [10]. In addition, new classes of molecule such as microRNAs and lincRNAs are increasingly implicated in regulating the activity of protein coding genes [7,11–14].
In contrast to canonical pathways, network models are often built from systematic experimental screens, broad surveys of the literature or public databases of molecular interactions. These models can easily be extended to incorporate new molecular species or different types of relationship between molecules and represent essential tools for biological inference. Nonetheless, it is important to be aware that networks are subject to various ascertainment biases including those introduced by measurement technologies, selection of proteins for systematic study or due to variation in the number of experiments or studies performed for particular genes.
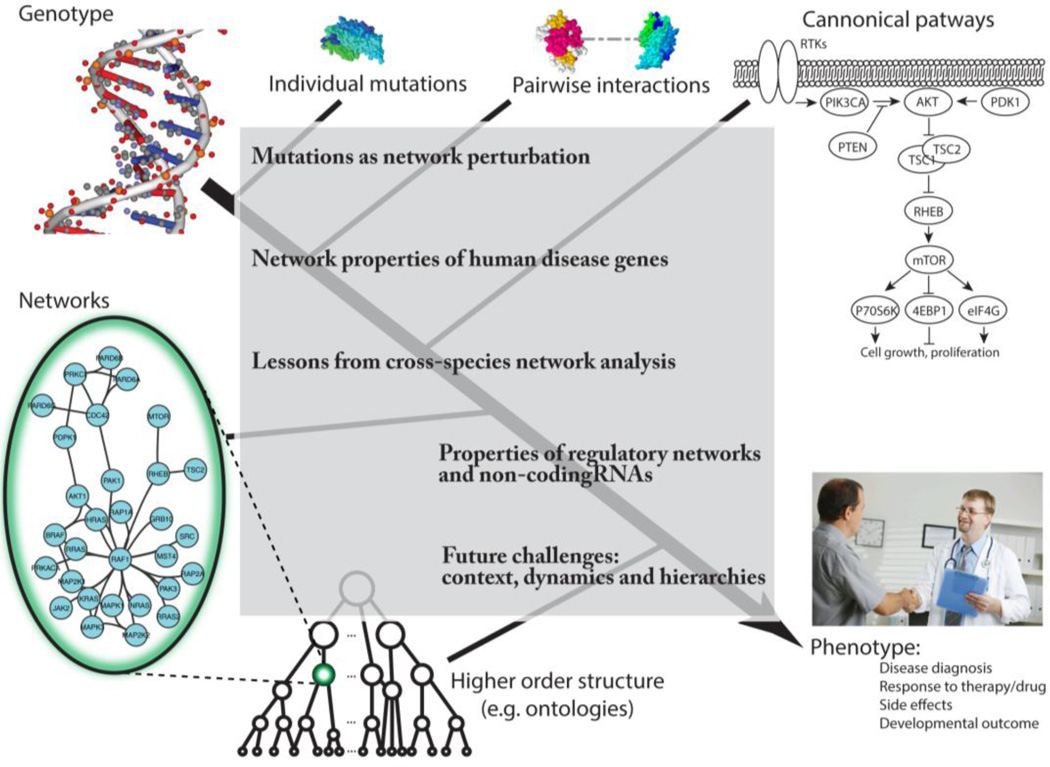
Modeling genotype-phenotype associations will require understanding the consequences of genetic alterations at multiple scales (Figure 1), several of which can be modeled with networks. Genetic alterations impacting the abundance or activity of individual molecules will affect the interactions in which those molecules participate. If the affected interactions are an important component in the larger network mediating a critical biological process or cellular behavior, a disease phenotype is more likely to occur. Here, we review developments in modeling molecular interactions within the cell, how mutations impact molecular interactions and biological processes in disease phenotypes, and how this knowledge can be exploited to elucidate key genotype-phenotype relationships.
Figure 1. A hierarchical perspective of biological interactions mediating genotype-phenotype relationships.
Protein activity is determined by protein amino acid sequence and structure. Proteins contribute to biological processes through interactions with other molecules in the cell. Biological processes arise from coordinated groups of molecular interactions, and in turn can interact to mediate higher order cellular behaviors and responses to environmental cues. Advances in several areas of network research are improving our understanding of how the organization of biological systems mediates genotype-phenotype relationships. This knowledge will be essential for identifying mutations underlying disease associations and their mechanisms of pathogenesis.
Networks for biological inference
Networks provide a framework for deriving information from a set of relationships among biological entities. In models of sub-cellular biological processes, network nodes are typically genes, proteins, nucleic acids or metabolites, and edges represent physical interactions or a rich variety of functional associations (Table 1). Hybrid networks that are mixtures of different types of relationships are prevalent as well.
Table 1.
A summary of several common varieties of biological network and some examples.
| Network type | Nodes | Edges | Example |
|---|---|---|---|
| Protein-protein interaction (PPI) | Proteins | Physical interactions | HPIN[26*] |
| Structurally resolved PPI | Protein | Physical interactions | HSIN[26*], SIN[58], Interactome3D[44], INstruct [45] |
| Protein-DNA interaction | Transcription factors | Transcription factor DNA binding | [91,92] |
| Co-expression | Proteins | Common expression | [93] |
| Genetic interaction (GI) | Genes | Common function | [18,69] |
| Difference | Genes | Differential function | [66] |
| Metabolic | Enzymes, Metabolites | Biochemical reactions | [94] |
| Non-coding RNAs | miRNA, lincRNA, asRNA, target genes | Physical interactions, common function | [95] |
| Integrated | Any | Any | HumanNet[54], BioGrid[96] |
| Hierarchical | Any | Any | Nexo[90**] |
Biological network models can be constructed from systematic genome-wide unbiased screens or focused interrogation of distinct biological functions. For complex disorders that are poorly characterized, mapping candidate genes and mutations implicated by association studies onto holistic network models can implicate underlying biological processes (Table 2). In a recent GWAS of coronary artery disease (CAD), Deloukas et al. identified subnetworks enriched for genes implicated by variable expression with or physical proximity to SNPs in a larger protein-protein interaction (PPI) network [15]. Subsequent gene set analysis to determine functional enrichment of the subnetworks, and analysis of subnetwork overlap with canonical pathways implicated crosstalk between lipid metabolism and inflammatory pathways as underlying the pathogenesis of CAD.
Table 2. Summary of recent network-based strategies for identifying biological mechanisms underlying genetic disorders.
Methods are grouped into two types: Exploratory Methods evaluate biological trends relating genotype to phenotype, while Analytic Methods seek to uncover a specific mutation, gene or biological pathway underlying a specific disorder. (LoF = loss of function, PPI = protein-protein interaction, DE = differential expression, eQTL = expression quantitative trait locus).
| Type | Goal | Data | Strategy | Refs |
|---|---|---|---|---|
| Exploratory | Network for analysis of disorders associated with blood vessels | Protein interactions, protein domains | GeneHits: method based on graph kernel diffusion | [16] |
| Network for analysis of HIV host cell defense evasion mechanisms | Affinity-tagging purification / mass spectrometry | MiST: uses information about protein abundance, reproducibility and specificity across replicate experiments | [17] | |
| Explore network properties of LoF tolerant genes | Gene annotations, interactions from multiple network databases | Use custom Multinet to investigate statistical correlation between genes and network properties, fit a linear model to separate essential and LoF tolerant genes using network properties | [58] | |
| Explore molecular basis of genotype-phenotype relationships | Mutation databases, PPI databases | Build a PPI network with structurally resolved protein interaction interfaces for analysis of mutations in inherited diseases | [25,26*] | |
| Explore the relationship between network state and cellular outcome | TP53 signaling network, condition specific cellular outcome data | Build a Boolean model of simplified TP53 signaling, map model dynamics to cellular outcomes, use the model to simulate how removing genes affects cellular outcome | [28**] | |
| Evaluate how a network motif contributes to cell fate decisions | Cell cycle pathway, yeast response to mating pheromones | Generate hypotheses based on network structure, test experimentally and build differential equation models | [29**] | |
| Explore how drugs rewire biological networks | Time series gene expression, cellular response, growth factor signaling and DNA damage response pathways | Identify candidate genes from pathways or DE after drug exposure, select a subset of genes based on prior knowledge or pathway structure, use time series data for genes model signaling to cellular outcome | [31**] | |
| Explore how SNPs effect gene expression in different tissues | Microarray based SNP and expression measurements | Combine eQTL analysis with a sampling approach to detect tissue specific SNP effects on expression | [64*] | |
| Organize genetic interaction as a hierarchical network | Genetic interaction screens in yeast | A minimum description length criteria is minimized using greedy and local search methods from an initial clustering. | [67] | |
| Organize interaction edges in a hierarchical ontology of terms | Physical and genetic interactions, mRNA co-expression | Combine probabilistic clustering with an ontology alignment method to produce robust hierarchical structure directly from experimental measurements and networks. | [41,90**] | |
| Analytic | Identify biological pathways underlying hemimegalenchephaly | De novo somatic mutations, pathway gene sets | Map onto canonical pathways | [8] |
| Identify disease genes from de novo CNVs in autism cases | De novo CNVs, protein architecture, function, and expression, pathways | Identify genes across CNVs implicated in similar phenotypes | [19] | |
| Identify biological pathways underlying CAD | CAD GWAS loci, commercial network database | Identify subnetworks from GWAS implicated genes, annotate subnetworks, identify overlap with canonical pathways | [15] | |
| Identify genes underlying type 1 diabetes | GWAS loci, protein interaction network, gene expression | Identify subnetworks from GWAS implicated genes, map DE onto subnetworks and test for statistical enrichment of DE genes | [56] | |
| Identify genes underlying autism | De novo somatic mutations, protein interaction network | Identify connected subnetworks based on de novo mutated genes, functionally annotate subnetworks | [57] | |
| Identify genes that regulate plasma insulin levels | Genotypes, clinical traits, transcriptional, hybrid network | Identify subnetworks from eQTLs in different tissues, prioritize genes that participate in inter-subnetwork edges | [59] | |
| Identify cancer related genes and pathways | Gene expression data and SNP data | A set-cover based approach is used to identify subnetworks which explain an eQTL relationship between causal genes and potential targets | [81] |
If the disease is better understood, focused models may enable development of specific biological hypotheses about the mechanisms by which alterations cause disease. For example, Chu et al. constructed a network of protein interactions involved in angiogenesis, which they dub “the angiome”, in order to study diseases related to irregular blood vessel formation [16]. In another example, a network of human-HIV protein complexes constructed by affinity tagging and purification mass spectrometry has provided a near-comprehensive view of how HIV evades host cell defenses [17]. While focused approaches represent only a partial view of the cell, the resulting networks provide an intelligent framework for constraining hypothesis testing to proteins most relevant to a disease. On the other hand, focused screens may miss systems level trends, for example cross-talk between biological processes, that can play a role in disease [18].
Network edges can also represent abstract relationships derived from biological knowledge. Gilman et al. built a network where all pairs of proteins are connected by a weighted edge representing the a priori expectation that the proteins participate in the same phenotype. Edge weights were based on evidence sources such as tissue-specific expression, pathway membership, common functional annotations and similar domain composition [19]. They then searched over this network to identify the most functionally similar genes affected by de novo copy number variants (CNVs) in autism cases.
Mutations as network perturbations
The majority of known disease mutations annotated in the Human Gene Mutation Database (HGMD) cause changes to the amino acid sequence of proteins [20]. These changes can have a spectrum of consequences ranging from completely abrogating protein activity to having no effect at all, and a variety of computational strategies have been developed to predict the functional consequences of mutation at the protein level [21–23]. Changes to a protein’s activity are indirectly linked to altered cellular behaviors by the network of molecular interactions in which it participates. Thus it has been proposed that to understand genotype-phenotype relationships it will be necessary to quantify the effects of mutations on molecular networks [24].
To investigate how interaction networks mediate phenotypic effects of mutations, Zhong et al. experimentally profiled protein interactions for twenty-nine alleles associated with five genetic disorders [25]. This profiling suggested that mutations could have three distinct effects for the PPI network: they could eliminate all interactions, remove a subset of interactions, or have no effect on interactions. To more systematically study how mutations affect physical interactions networks, Wang et al. constructed a high quality PPI network with structurally resolved interaction interfaces [26*]. Using this network, they analyzed disease-associated mutations from OMIM [27] and HGMD and demonstrated enrichment for in-frame mutations such as missense or in-frame insertions and deletions at interaction interfaces. They also found that mutations occurring at distinct interaction interfaces in the same protein could explain many cases where a single gene is involved in multiple disorders (i.e. pleiotropy) or in disorders with multiple distinct modes of inheritance [25,26*].
Models of how PPIs are rewired by mutations, sometimes referred to as “network perturbation models”, may present a useful strategy for functionally prioritizing candidate disease mutations and developing hypotheses about biological processes underlying pathogenesis [4,25]. These models can also be used to analyze the combined effects of multiple mutation and expression changes. For example, TP53 signaling is associated with cell cycle arrest and apoptosis in response to cell damage. Choi et al. used a simplified model of the TP53 signaling network to map combinatorial network perturbations to cellular outcome [28**]. They then used this model to explore how fixing the activation of specific molecules constrained the cellular behaviors available and what parts of the network could be targeted with therapeutics to force the apoptotic state. Relatedly, Doncic et al. recently found that a simple three-gene motif embedded within a more complex network structure was sufficient to explain yeast cellular state decisions in response to mating pheromone, suggesting that it may not be necessary to model the full complexity of biological networks to capture molecular determinants of cellular behaviors [29**].
In addition to the effects on individual edges in the network, downstream processes in the cell may be rewired to maintain homeostasis in the face of perturbations [30]. Intriguingly, Lee et al. showed that deliberate perturbation of networks to achieve specific rewiring could serve as a therapeutic strategy in cancer [31**]. Triple negative breast cancer cells exposed to an EGFR inhibitor prior to chemotherapy showed increased sensitivity to genotoxic therapy. The timing of exposure to EGFR inhibitor greatly influenced sensitivity to subsequent chemotherapy suggesting that temporal dynamics of network rewiring are a determinant of cellular response to environment.
In studies of inherited disease, causal mutations are often buried in a list of candidate variants uncovered by sequencing of risk loci or disease exomes [32], and in cancers, the majority of detected somatic mutations are thought to be neutral “passenger” events [33,34]. It has also been suggested that most post-translational modifications may not affect protein activity [35]. Information about protein sequence and structure provides important clues for discriminating effects of distinct alterations to proteins [21–23]. Thus integrated approaches combining protein sequence and structural information with networks may provide a powerful framework for identifying disease mutations and reasoning about their molecular mechanisms.
The biophysical mechanisms by which mutations alter protein interactions are diverse and are usually not captured in the abstractions provided by simple interaction networks [36,37]. Mutations altering protein conformation or binding affinity can contribute to disease phenotype without removing network edges [38–40]. Furthermore, highly connected proteins in the network are unlikely to interact with all partners simultaneously, as interaction interfaces often overlap [41,42]. Network representations that capture mutually exclusivity of binding may be helpful for predicting the functional consequences of mutations [37,42,43].
Structurally resolved interaction networks are becoming available for several species through databases such as Interactome3D and INstruct [44,45]. Studying candidate disease mutations in the context of these networks may provide important clues as to how mutations affect biological processes. Due to the limited availability of co-crystallization protein structures [46] strategies have been developed to predict structure at protein interfaces using homology models [26*]. Nonetheless, this type of analysis will only be possible for a subset of candidate disease mutations.
Joint study of co-evolution of amino-acid residues at protein interfaces and network structure may provide insights into which residues are essential for maintaining interactions [40,47,48]. Fridman et al. found that affinity-altering mutations in proliferating cell nuclear antigen (PCNA) could have more severe consequences for DNA replication and repair than mutations completely abolishing interactions [40]. Their findings suggest that even within interfaces, mutations are likely to have distinct phenotypic consequences. Thus it may be important to include manipulation of specific interactions as part of mutagenesis studies when experimentally evaluating candidate disease genes. Emerging genome engineering strategies provide exciting opportunities for experimentally characterizing domain specific effects of mutations on network activities [49].
Network properties of human disease genes
The non-random organization of biological networks suggests that their topology may encode information about how molecular interactions contribute to biological phenotypes [50]. Molecular interaction networks within the cell tend to be modular; that is, proteins related to the same biological activities often form connected modules within networks [5–7,50,51]. Goh et al. showed that this phenomenon extends to disease genes as well; genes implicated in the same diseases often cluster within PPI networks [52,53].
The existence of functional and disease modules within interactome networks supports a “guilt-by-association” (GBA) strategy for identifying novel disease-associated genes [5,54]. GBA has been used to intelligently reduce the list of candidate disease genes in association studies [54,55]. Bergholdt et al. combined PPI network overlap with genes located at GWAS risk loci and subnetwork-based enrichment for differential expression to identify new candidate type I diabetes disease genes [56]. Identification of network modules enriched for mutation or variable expression under disease conditions can point to specific biological processes disrupted in disease. For example, analysis of the network distribution of de novo mutations in sporadic cases with autism spectrum disorders implicated a highly interconnected subnetwork of proteins involved in β-catenin/chromatin remodeling [57].
Goh et al. also investigated differences in network connectivity of three classes of genes: essential, inherited and somatic disease genes [52,53]. They reported that essential genes were more likely to have a large number of interaction partners and therefore be central in the network, while inherited disease genes generally had fewer interaction partners and were more peripheral. By contrast, somatic disease genes often looked more like essential genes. Khurana et al. further explored gene essentiality and selection in the context of different types of biological network (PPI, metabolic, post-translational modification, regulatory, etc.) as well as in a pooled network and found that highly connected genes are more likely to show strong signatures of selection [58]. Using topological and selection properties of genes, they built a logistic regression model capable of distinguishing essential genes from genes tolerant to loss-of-function events, suggesting that these properties could be useful for selecting candidate genes for sequencing and follow-up studies. Tu et al. used topological location at the interface between subnetworks with differential expression (DE) mediated by plasma-insulin associated genetic loci to implicate an Alzheimer’s related gene, App, in type 2 diabetes [59].
These applications demonstrate how characteristics of biological networks such as topology and modularity can be used to prioritize candidate disease genes implicated by association studies. Inference based on network architecture may be particularly sensitive to the previously noted ascertainment biases that can affect network models; highly studied genes are more likely to have a large number of edges in the network than less frequently studied genes [4,5,18]. This is less of an issue for networks derived from systematic experimental screens [4,7,60], although technology-specific biases are suspected to exist [61].
Lessons from cross-species network analysis
Mounting evidence from both the study of model organisms [62*,63**] and GWAS [64*,65,4] suggests that much of the ‘missing heritability’ of genetic disease may result from genetic interactions (GIs). GI maps have been widely used to study epistatic phenomena in model organisms [29**,51,66,67] and have more recently been applied to mammalian species and human cell lines.
The most comprehensive GI networks to date have been generated from systematic screens in model organisms. For this reason, it is of interest to determine whether studies of orthologous proteins in model organisms could inform missing interactions in human networks. In a recent attempt to experimentally address this question on a systems level, two evolutionarily diverged yeast species were compared: the budding yeast S. cerevisiae and the fission yeast S. pombe, which are separated by an estimated 400–800 million years of evolution (an evolutionary distance greater than the divergence between humans and fish). Comparison of systematic pairwise genetic interaction screens conducted in both species[18,68,69] showed a hierarchical conservation of network modules, with highest conservation observed for interactions within protein complexes (68–70%), lower conservation of interactions within biological processes (38–58%) and lowest conservation of interactions between distinct biological processes (15–19%)[18]. In some cases, there was functional “repurposing” of complexes between species [69].
Interestingly, although globally only a small fraction of the specific interactions between biological processes were conserved, the total number of interactions was similar, suggesting that coordination of biological processes may be a design principle in eukaryotic systems [18]. Due to the aforementioned divergence between these yeast species, Ryan et al. suggest that these trends will most likely pertain to other eukaryotic species as well. These studies provide compelling evidence that cross-species networks can aid our understanding of human disease proteins and the biological processes in which they participate.
A uniquely informative perspective is afforded by examining ‘difference networks’, which are emerging as an exciting strategy to examine the broader effects of perturbations on biological processes in the cell [30]. Difference networks can be derived from systematic mapping of interactions in cells under different conditions. In these networks, edges represent the interactions that differ between the tested conditions and can capture more dynamic effects of particular (e.g. drug) or environmental (e.g. heat) perturbations on the network [66,70].
Regulatory networks and non-coding DNA
Most GWAS-implicated risk variants occur outside of protein coding genes [71–73]. Recently it has been suggested that the majority of the genome is involved in biochemical and regulatory activities, not just the 1.5% encoding proteins [74]. Non-coding genetic alterations, even those affecting non-coding RNA (ncRNA) sequence, are suspected to mediate phenotypic effects primarily by altering the abundance of proteins in the cell and thus perturbing PPI networks through stoichiometric effects [75–77]. Indeed, many variants detected by GWAS are located at DNA regulatory elements [78**]. An early investigation of the tissue-specific effects of genetic variants on gene expression uncovered surprisingly complex relationships, suggesting that network models may be essential for dissecting phenotypic consequences of non-coding variation [64*].
An analysis conducted as part of the Encyclopedia of DNA Elements (ENCODE) project [79] compared the genome-wide binding patterns of 119 distinct transcription and DNA binding factors (TFs) across five different cell lines [80]. These data were used to construct a hierarchical representation of transcription factor regulation onto which protein and non-coding RNA interaction data as well as post-translational modifications were integrated. The combined network suggested the existence of three tiers of transcriptional regulation with distinct properties and architectures. Kim et al. used an interaction network of similar composition to implicate genes and network paths capable of mediating disease-related expression changes downstream of copy number variants [81].
Increasing evidence points to an important role for ncRNAs in complex disorders. On the level of mutations, microRNAs (miRNAs) have been shown to play a mechanistic role in the effects of often ignored synonymous mutations [14]. A recent work has shown that a network of microRNAs may play a key role in the epithelial to mesenchymal transformation of ovarian cancers [82*]. Recently, the importance of other ncRNA species have also been highlighted, such as the role of anti-sense RNAs on PTEN regulation [83], broad epigenetic effects of HOTAIR a long intergenic ncRNA (lincRNA) in breast cancer [12], and the role of PCAT-1, another lincRNA, on the progression of prostate cancer [13].
Future challenges: context, dynamics and hierarchies
Biological network models still fall short of capturing many important aspects of biological systems. Cells exhibit dynamic responses to environmental stimuli [84] and cells of different tissue types are characterized by distinct gene expression patterns [10,64*]. These properties are key determinants of phenotype but are not captured by the standard static network models that are prevalent in the field. Attempts to estimate the completeness and accuracy of existing protein interaction data suggest that 92% or more of binary human PPIs remain to be uncovered [3,85]. These estimates do not account for the possibility that distinct protein isoforms participate in different interactions. In addition, new molecular species are still being discovered and have not yet been incorporated into network models [7]. Constructing network models that accurately capture the molecular composition and interactions in specific cell types and under distinct conditions will be essential for effectively modeling genotype-phenotype relationships.
New experimental techniques are rapidly emerging that will enable systematic screens of molecular interactions in mammalian cells. Mass spectrometry (MS)-based techniques promise to enable systematic cell type-specific screens of the proteome and protein post-translational modifications [61]. Proteomics may also aid in discovery of as yet undiscovered protein coding genes [86]. Until now, the majority of GI screens have been performed in model organisms, especially yeast, by exhaustively knocking out pairs of genes and measuring the effects on colony size. Novel approaches using RNAi technologies are now enabling systematic mapping of GIs in mammalian cells [87–89].
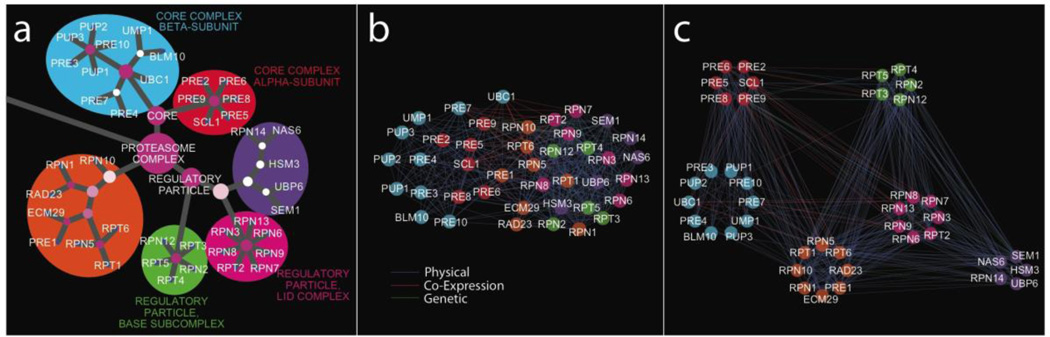
New strategies for network construction and visualization will also aid the search for disease causing genes and mutations. Reformulating interactomes as hierarchies can provide representations of biological information that are easier to interpret than the typical “hairball” that results when thousands of interactions are simultaneously displayed [41,90**] (Figure 2). Mapping molecular measurement data onto such hierarchies will provide novel biological hypotheses about the pathogenesis of complex inherited disease. Furthermore, the hierarchical structure can highlight inconsistent edges likely to be false positives or of lesser importance, and suggest new relationships among distinct biological complexes and processes. Aside from a few pioneering efforts, the space of hierarchical network modeling remains largely unexplored.
Figure 2. Hierarchical representations provide interpretable views of how molecular networks contribute to biological function.
The subnetwork comprising interactions among proteins of the S. cerevisiae proteasome is depicted using different network layouts: (a) hierarchical (b) force-directed and (c) a layout showing within-module edges and between-module edges. In the hierarchical representation, distal nodes are included in proximal nodes (for example the node labeled “core” encapsulates the alpha and beta subunits depicted in red and cyan). Node size corresponds to the number of genes participating in the term and node color gives degree of correspondence to annotated biological activities. Branches and nodes corresponding to physical complexes with known biological function are labeled. Node colors in panels (b) and (c) match the complexes highlighted in panel (a). Reproduced with permission from Dutkowski et al. [90**].
Conclusions
Biological networks are increasingly being applied to study the mechanisms by which genetic alterations cause phenotypic changes at the cellular level. Network organization and structure can help explain many disease phenomena such as locus heterogeneity, variable penetrance, pleiotropy, inheritance models and comorbidity. We believe these efforts are in their infancy. Limited knowledge of the dynamic and context-specific interplay of molecules within cell and our incomplete understanding of the makeup of the human genome has prevented effective modeling of the heritable contributions to human disease. Advances in experimental measurement technologies will soon enable large-scale screens to fill in much of our missing knowledge.
Acknowledgements
This work was supported by NIH grants P41 GM103504 and P50 GM085764.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Singleton AB, Hardy J, Traynor BJ, Houlden H. Towards a complete resolution of the genetic architecture of disease. Trends Genet. 2010;26:438–442. doi: 10.1016/j.tig.2010.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Luo L, Boerwinkle E, Xiong M. Association studies for next-generation sequencing. Genome Res. 2011;21:1099–1108. doi: 10.1101/gr.115998.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vidal M, Cusick ME, Barabasi AL. Interactome networks and human disease. Cell. 2011;144:986–998. doi: 10.1016/j.cell.2011.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.del Sol A, Balling R, Hood L, Galas D. Diseases as network perturbations. Curr Opin Biotechnol. 2010;21:566–571. doi: 10.1016/j.copbio.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 5.Ideker T, Sharan R. Protein networks in disease. Genome Res. 2008;18:644–652. doi: 10.1101/gr.071852.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Furlong LI. Human diseases through the lens of network biology. Trends Genet. 2013;29:150–159. doi: 10.1016/j.tig.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 7.Califano A, Butte AJ, Friend S, Ideker T, Schadt E. Leveraging models of cell regulation and GWAS data in integrative network-based association studies. Nat Genet. 2012;44:841–847. doi: 10.1038/ng.2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee JH, Huynh M, Silhavy JL, Kim S, Dixon-Salazar T, Heiberg A, Scott E, Bafna V, Hill KJ, Collazo A, et al. De novo somatic mutations in components of the PI3K-AKT3-mTOR pathway cause hemimegalencephaly. Nat Genet. 2012;44:941–945. doi: 10.1038/ng.2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guruharsha KG, Kankel MW, Artavanis-Tsakonas S. The Notch signalling system: recent insights into the complexity of a conserved pathway. Nat Rev Genet. 2012;13:654–666. doi: 10.1038/nrg3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bossi A, Lehner B. Tissue specificity and the human protein interaction network. Mol Syst Biol. 2009;5:260. doi: 10.1038/msb.2009.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prensner JR, Iyer MK, Balbin OA, Dhanasekaran SM, Cao Q, Brenner JC, Laxman B, Asangani IA, Grasso CS, Kominsky HD, et al. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat Biotechnol. 2011;29:742–749. doi: 10.1038/nbt.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salzman DW, Weidhaas JB. miRNAs in the spotlight: Making 'silent' mutations speak up. Nat Med. 2011;17:934–935. doi: 10.1038/nm0811-934. [DOI] [PubMed] [Google Scholar]
- 15.Deloukas P, Kanoni S, Willenborg C, Farrall M, Assimes TL, Thompson JR, Ingelsson E, Saleheen D, Erdmann J, Goldstein BA, et al. Large-scale association analysis identifies new risk loci for coronary artery disease. Nat Genet. 2013;45:25–33. doi: 10.1038/ng.2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chu LH, Rivera CG, Popel AS, Bader JS. Constructing the angiome: a global angiogenesis protein interaction network. Physiol Genomics. 2012;44:915–924. doi: 10.1152/physiolgenomics.00181.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jager S, Cimermancic P, Gulbahce N, Johnson JR, McGovern KE, Clarke SC, Shales M, Mercenne G, Pache L, Li K, et al. Global landscape of HIV-human protein complexes. Nature. 2012;481:365–370. doi: 10.1038/nature10719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ryan CJ, Roguev A, Patrick K, Xu J, Jahari H, Tong Z, Beltrao P, Shales M, Qu H, Collins SR, et al. Hierarchical modularity and the evolution of genetic interactomes across species. Mol Cell. 2012;46:691–704. doi: 10.1016/j.molcel.2012.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gilman SR, Iossifov I, Levy D, Ronemus M, Wigler M, Vitkup D. Rare de novo variants associated with autism implicate a large functional network of genes involved in formation and function of synapses. Neuron. 2011;70:898–907. doi: 10.1016/j.neuron.2011.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stenson PD, Ball EV, Mort M, Phillips AD, Shaw K, Cooper DN. The Human Gene Mutation Database (HGMD) and its exploitation in the fields of personalized genomics and molecular evolution. Curr Protoc Bioinformatics. 2012;Chapter 1(Unit1 13) doi: 10.1002/0471250953.bi0113s39. [DOI] [PubMed] [Google Scholar]
- 21.Cline MS, Karchin R. Using bioinformatics to predict the functional impact of SNVs. Bioinformatics. 2011;27:441–448. doi: 10.1093/bioinformatics/btq695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jordan DM, Ramensky VE, Sunyaev SR. Human allelic variation: perspective from protein function, structure, and evolution. Curr Opin Struct Biol. 2010;20:342–350. doi: 10.1016/j.sbi.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sunyaev SR. Inferring causality and functional significance of human coding DNA variants. Hum Mol Genet. 2012;21:R10–R17. doi: 10.1093/hmg/dds385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang X, Gulbahce N, Yu H. Network-based methods for human disease gene prediction. Brief Funct Genomics. 2011;10:280–293. doi: 10.1093/bfgp/elr024. [DOI] [PubMed] [Google Scholar]
- 25.Zhong Q, Simonis N, Li QR, Charloteaux B, Heuze F, Klitgord N, Tam S, Yu H, Venkatesan K, Mou D, et al. Edgetic perturbation models of human inherited disorders. Mol Syst Biol. 2009;5:321. doi: 10.1038/msb.2009.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang X, Wei X, Thijssen B, Das J, Lipkin SM, Yu H. Three-dimensional reconstruction of protein networks provides insight into human genetic disease. Nat Biotechnol. 2012;30:159–164. doi: 10.1038/nbt.2106. The authors use a structurally resolved PPI network to explore how mutations contribute to disease characteristics such as pleiotropy and inheritance models
- 27.Amberger J, Bocchini C, Hamosh A. A new face and new challenges for Online Mendelian Inheritance in Man (OMIM(R)) Hum Mutat. 2011;32:564–567. doi: 10.1002/humu.21466. [DOI] [PubMed] [Google Scholar]
- 28. Choi M, Shi J, Jung SH, Chen X, Cho KH. Attractor landscape analysis reveals feedback loops in the p53 network that control the cellular response to DNA damage. Sci Signal. 2012;5:ra83. doi: 10.1126/scisignal.2003363. The authors develop a framework for mapping the joint states of proteins in a pathway to cellular phenotypes and use this mapping to predict how to intervene in the pathway to achieve a specific outcome.
- 29. Doncic A, Skotheim JM. Feedforward Regulation Ensures Stability and Rapid Reversibility of a Cellular State. Mol Cell. 2013 doi: 10.1016/j.molcel.2013.04.014. The authors establish how a 3-gene feed forward regulatory motif can enable reversible cellular state transitions.
- 30.Ideker T, Krogan NJ. Differential network biology. Mol Syst Biol. 2012;8:565. doi: 10.1038/msb.2011.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lee MJ, Ye AS, Gardino AK, Heijink AM, Sorger PK, MacBeath G, Yaffe MB. Sequential application of anticancer drugs enhances cell death by rewiring apoptotic signaling networks. Cell. 2012;149:780–794. doi: 10.1016/j.cell.2012.03.031. The authors demonstrate network rewiring in response to continuous drug exposure over time and how this rewiring could be used to identify therapeutic opportunities.
- 32.Cooper GM, Shendure J. Needles in stacks of needles: finding disease-causal variants in a wealth of genomic data. Nat Rev Genet. 2011;12:628–640. doi: 10.1038/nrg3046. [DOI] [PubMed] [Google Scholar]
- 33.Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, Davies H, Teague J, Butler A, Stevens C, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beltrao P, Albanese V, Kenner LR, Swaney DL, Burlingame A, Villen J, Lim WA, Fraser JS, Frydman J, Krogan NJ. Systematic functional prioritization of protein posttranslational modifications. Cell. 2012;150:413–425. doi: 10.1016/j.cell.2012.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Teyra J, Kim PM. Interpreting protein networks with three-dimensional structures. Nat Methods. 2013;10:43–44. doi: 10.1038/nmeth.2300. [DOI] [PubMed] [Google Scholar]
- 37.Clarke D, Bhardwaj N, Gerstein MB. Novel insights through the integration of structural and functional genomics data with protein networks. J Struct Biol. 2012;179:320–326. doi: 10.1016/j.jsb.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Szilagyi A, Nussinov R, Csermely P. Allo-network drugs: extension of the allosteric drug concept to protein- protein interaction and signaling networks. Curr Top Med Chem. 2013;13:64–77. doi: 10.2174/1568026611313010007. [DOI] [PubMed] [Google Scholar]
- 39.Kiel C, Serrano L. Cell type-specific importance of ras-c-raf complex association rate constants for MAPK signaling. Sci Signal. 2009;2:ra38. doi: 10.1126/scisignal.2000397. [DOI] [PubMed] [Google Scholar]
- 40.Fridman Y, Palgi N, Dovrat D, Ben-Aroya S, Hieter P, Aharoni A. Subtle alterations in PCNA-partner interactions severely impair DNA replication and repair. PLoS Biol. 2010;8:e1000507. doi: 10.1371/journal.pbio.1000507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van Roey K, Gibson TJ, Davey NE. Motif switches: decision-making in cell regulation. Curr Opin Struct Biol. 2012;22:378–385. doi: 10.1016/j.sbi.2012.03.004. [DOI] [PubMed] [Google Scholar]
- 42.Kim PM, Lu LJ, Xia Y, Gerstein MB. Relating three-dimensional structures to protein networks provides evolutionary insights. Science. 2006;314:1938–1941. doi: 10.1126/science.1136174. [DOI] [PubMed] [Google Scholar]
- 43.Engin HB, Keskin O, Nussinov R, Gursoy A. A strategy based on protein-protein interface motifs may help in identifying drug off-targets. J Chem Inf Model. 2012;52:2273–2286. doi: 10.1021/ci300072q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mosca R, Ceol A, Aloy P. Interactome3D: adding structural details to protein networks. Nat Methods. 2013;10:47–53. doi: 10.1038/nmeth.2289. [DOI] [PubMed] [Google Scholar]
- 45.Meyer MJ, Das J, Wang X, Yu H. INstruct: a database of high quality three-dimensional structurally resolved protein interactome networks. Bioinformatics. 2013 doi: 10.1093/bioinformatics/btt181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Havugimana PC, Hart GT, Nepusz T, Yang H, Turinsky AL, Li Z, Wang PI, Boutz DR, Fong V, Phanse S, et al. A census of human soluble protein complexes. Cell. 2012;150:1068–1081. doi: 10.1016/j.cell.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guharoy M, Chakrabarti P. Conserved residue clusters at protein-protein interfaces and their use in binding site identification. BMC Bioinformatics. 2010;11:286. doi: 10.1186/1471-2105-11-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fraser JS, Gross JD, Krogan NJ. From systems to structure: bridging networks and mechanism. Mol Cell. 2013;49:222–231. doi: 10.1016/j.molcel.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Levin M, Amar D, Aharoni A. Employing directed evolution for the functional analysis of multi-specific proteins. Bioorg Med Chem. 2013 doi: 10.1016/j.bmc.2013.04.052. [DOI] [PubMed] [Google Scholar]
- 50.Taylor IW, Wrana JL. Protein interaction networks in medicine and disease. Proteomics. 2012;12:1706–1716. doi: 10.1002/pmic.201100594. [DOI] [PubMed] [Google Scholar]
- 51.Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, Ding H, Koh JL, Toufighi K, Mostafavi S, et al. The genetic landscape of a cell. Science. 2010;327:425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Goh KI, Choi IG. Exploring the human diseasome: the human disease network. Brief Funct Genomics. 2012;11:533–542. doi: 10.1093/bfgp/els032. [DOI] [PubMed] [Google Scholar]
- 53.Goh KI, Cusick ME, Valle D, Childs B, Vidal M, Barabasi AL. The human disease network. Proc Natl Acad Sci U S A. 2007;104:8685–8690. doi: 10.1073/pnas.0701361104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee I, Blom UM, Wang PI, Shim JE, Marcotte EM. Prioritizing candidate disease genes by network-based boosting of genome-wide association data. Genome Res. 2011;21:1109–1121. doi: 10.1101/gr.118992.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moreau Y, Tranchevent LC. Computational tools for prioritizing candidate genes: boosting disease gene discovery. Nat Rev Genet. 2012;13:523–536. doi: 10.1038/nrg3253. [DOI] [PubMed] [Google Scholar]
- 56.Bergholdt R, Brorsson C, Palleja A, Berchtold LA, Floyel T, Bang-Berthelsen CH, Frederiksen KS, Jensen LJ, Storling J, Pociot F. Identification of novel type 1 diabetes candidate genes by integrating genome-wide association data, protein-protein interactions, and human pancreatic islet gene expression. Diabetes. 2012;61:954–962. doi: 10.2337/db11-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.O'Roak BJ, Vives L, Girirajan S, Karakoc E, Krumm N, Coe BP, Levy R, Ko A, Lee C, Smith JD, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485:246–250. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Khurana E, Fu Y, Chen J, Gerstein M. Interpretation of genomic variants using a unified biological network approach. PLoS Comput Biol. 2013;9:e1002886. doi: 10.1371/journal.pcbi.1002886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tu Z, Keller MP, Zhang C, Rabaglia ME, Greenawalt DM, Yang X, Wang IM, Dai H, Bruss MD, Lum PY, et al. Integrative analysis of a cross-loci regulation network identifies App as a gene regulating insulin secretion from pancreatic islets. PLoS Genet. 2012;8:e1003107. doi: 10.1371/journal.pgen.1003107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dixon SJ, Costanzo M, Baryshnikova A, Andrews B, Boone C. Systematic mapping of genetic interaction networks. Annu Rev Genet. 2009;43:601–625. doi: 10.1146/annurev.genet.39.073003.114751. [DOI] [PubMed] [Google Scholar]
- 61.Altelaar AF, Munoz J, Heck AJ. Next-generation proteomics: towards an integrative view of proteome dynamics. Nat Rev Genet. 2013;14:35–48. doi: 10.1038/nrg3356. [DOI] [PubMed] [Google Scholar]
- 62. Dowell RD, Ryan O, Jansen A, Cheung D, Agarwala S, Danford T, Bernstein DA, Rolfe PA, Heisler LE, Chin B, et al. Genotype to phenotype: a complex problem. Science. 2010;328:469. doi: 10.1126/science.1189015. In an elegant experiment exploiting properties of yeast genetics the authors demonstrate how even in this simple eukaryote gene essentiality may depend on complex interaction effects.
- 63. Lehner B. Genotype to phenotype: lessons from model organisms for human genetics. Nat Rev Genet. 2013;14:168–178. doi: 10.1038/nrg3404. In this landmark review the authors show that one explanation for the ‘missing heritability’ is the existence complex interaction effects.
- 64. Fu J, Wolfs MG, Deelen P, Westra HJ, Fehrmann RS, Te Meerman GJ, Buurman WA, Rensen SS, Groen HJ, Weersma RK, et al. Unraveling the regulatory mechanisms underlying tissue-dependent genetic variation of gene expression. PLoS Genet. 2012;8:e1002431. doi: 10.1371/journal.pgen.1002431. The authors examine tissue specific effects of single nucleotide polymorphisms and describe a complex interplay between heritable variants and specific tissues.
- 65.Zuk O, Hechter E, Sunyaev SR, Lander ES. The mystery of missing heritability: Genetic interactions create phantom heritability. Proc Natl Acad Sci U S A. 2012;109:1193–1198. doi: 10.1073/pnas.1119675109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bandyopadhyay S, Mehta M, Kuo D, Sung MK, Chuang R, Jaehnig EJ, Bodenmiller B, Licon K, Copeland W, Shales M, et al. Rewiring of genetic networks in response to DNA damage. Science. 2010;330:1385–1389. doi: 10.1126/science.1195618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jaimovich A, Rinott R, Schuldiner M, Margalit H, Friedman N. Modularity and directionality in genetic interaction maps. Bioinformatics. 2010;26:i228–i236. doi: 10.1093/bioinformatics/btq197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dixon SJ, Fedyshyn Y, Koh JL, Prasad TK, Chahwan C, Chua G, Toufighi K, Baryshnikova A, Hayles J, Hoe K-L. Significant conservation of synthetic lethal genetic interaction networks between distantly related eukaryotes. Proceedings of the National Academy of Sciences. 2008;105:16653–16658. doi: 10.1073/pnas.0806261105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Frost A, Elgort MG, Brandman O, Ives C, Collins SR, Miller-Vedam L, Weibezahn J, Hein MY, Poser I, Mann M, et al. Functional repurposing revealed by comparing S. pombe and S. cerevisiae genetic interactions. Cell. 2012;149:1339–1352. doi: 10.1016/j.cell.2012.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Casanueva MO, Burga A, Lehner B. Fitness trade-offs and environmentally induced mutation buffering in isogenic C. elegans. Science. 2012;335:82–85. doi: 10.1126/science.1213491. [DOI] [PubMed] [Google Scholar]
- 71.Kumar V, Westra HJ, Karjalainen J, Zhernakova DV, Esko T, Hrdlickova B, Almeida R, Zhernakova A, Reinmaa E, Vosa U, et al. Human disease-associated genetic variation impacts large intergenic non-coding RNA expression. PLoS Genet. 2013;9:e1003201. doi: 10.1371/journal.pgen.1003201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hindorff LA, Gillanders EM, Manolio TA. Genetic architecture of cancer and other complex diseases: lessons learned and future directions. Carcinogenesis. 2011;32:945–954. doi: 10.1093/carcin/bgr056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Freedman ML, Monteiro AN, Gayther SA, Coetzee GA, Risch A, Plass C, Casey G, De Biasi M, Carlson C, Duggan D, et al. Principles for the post-GWAS functional characterization of cancer risk loci. Nat Genet. 2011;43:513–518. doi: 10.1038/ng.840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dunham I, Kundaje A, Aldred SF, Collins PJ, Davis CA, Doyle F, Epstein CB, Frietze S, Harrow J, Kaul R, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 76.Kaikkonen MU, Lam MT, Glass CK. Non-coding RNAs as regulators of gene expression and epigenetics. Cardiovasc Res. 2011;90:430–440. doi: 10.1093/cvr/cvr097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kowalczyk MS, Higgs DR, Gingeras TR. Molecular biology: RNA discrimination. Nature. 2012;482:310–311. doi: 10.1038/482310a. [DOI] [PubMed] [Google Scholar]
- 78. Maurano MT, Humbert R, Rynes E, Thurman RE, Haugen E, Wang H, Reynolds AP, Sandstrom R, Qu H, Brody J, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. The authors explore the effect of disease-associated variants on regulatory regions of the genome. Among their findings is the observation that variants appear to cluster in transcriptional regulatory pathways.
- 79.Rosenbloom KR, Sloan CA, Malladi VS, Dreszer TR, Learned K, Kirkup VM, Wong MC, Maddren M, Fang R, Heitner SG, et al. ENCODE data in the UCSC Genome Browser: year 5 update. Nucleic Acids Res. 2013;41:D56–D63. doi: 10.1093/nar/gks1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gerstein MB, Kundaje A, Hariharan M, Landt SG, Yan KK, Cheng C, Mu XJ, Khurana E, Rozowsky J, Alexander R, et al. Architecture of the human regulatory network derived from ENCODE data. Nature. 2012;489:91–100. doi: 10.1038/nature11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kim YA, Wuchty S, Przytycka TM. Identifying causal genes and dysregulated pathways in complex diseases. PLoS Comput Biol. 2011;7:e1001095. doi: 10.1371/journal.pcbi.1001095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Yang D, Sun Y, Hu L, Zheng H, Ji P, Pecot CV, Zhao Y, Reynolds S, Cheng H, Rupaimoole R, et al. Integrated analyses identify a master microRNA regulatory network for the mesenchymal subtype in serous ovarian cancer. Cancer Cell. 2013;23:186–199. doi: 10.1016/j.ccr.2012.12.020. The authors analyze patient matched microRNA, gene expression and copy number alteration data. They uncover a microRNA signature that appears to regulate aspects of the epithelial-mesenchymal transition.
- 83.Johnsson P, Ackley A, Vidarsdottir L, Lui WO, Corcoran M, Grander D, Morris KV. A pseudogene long-noncoding-RNA network regulates PTEN transcription and translation in human cells. Nat Struct Mol Biol. 2013;20:440–446. doi: 10.1038/nsmb.2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Park Y, Bader JS. How networks change with time. Bioinformatics. 2012;28:i40–i48. doi: 10.1093/bioinformatics/bts211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Venkatesan K, Rual JF, Vazquez A, Stelzl U, Lemmens I, Hirozane-Kishikawa T, Hao T, Zenkner M, Xin X, Goh KI, et al. An empirical framework for binary interactome mapping. Nat Methods. 2009;6:83–90. doi: 10.1038/nmeth.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Brosch M, Saunders GI, Frankish A, Collins MO, Yu L, Wright J, Verstraten R, Adams DJ, Harrow J, Choudhary JS, et al. Shotgun proteomics aids discovery of novel protein-coding genes, alternative splicing, and "resurrected" pseudogenes in the mouse genome. Genome Res. 2011;21:756–767. doi: 10.1101/gr.114272.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bassik MC, Kampmann M, Lebbink RJ, Wang S, Hein MY, Poser I, Weibezahn J, Horlbeck MA, Chen S, Mann M, et al. A systematic mammalian genetic interaction map reveals pathways underlying ricin susceptibility. Cell. 2013;152:909–922. doi: 10.1016/j.cell.2013.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Roguev A, Talbot D, Negri GL, Shales M, Cagney G, Bandyopadhyay S, Panning B, Krogan NJ. Quantitative genetic-interaction mapping in mammalian cells. Nat Methods. 2013;10:432–437. doi: 10.1038/nmeth.2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Laufer C, Fischer B, Billmann M, Huber W, Boutros M. Mapping genetic interactions in human cancer cells with RNAi and multiparametric phenotyping. Nat Methods. 2013;10:427–431. doi: 10.1038/nmeth.2436. [DOI] [PubMed] [Google Scholar]
- 90. Dutkowski J, Kramer M, Surma MA, Balakrishnan R, Cherry JM, Krogan NJ, Ideker T. A gene ontology inferred from molecular networks. Nat Biotechnol. 2013;31:38–45. doi: 10.1038/nbt.2463. The authors demonstrate a clustering paradigm that uncovers an ontology structure from systematic experimental data in yeast and demonstrate that the resulting ontology shares a significant similarity to the manually curated Gene Ontology, while also uncovering hierarchical structure poorly annotated in the GO.
- 91.Moignard V, Macaulay IC, Swiers G, Buettner F, Schutte J, Calero-Nieto FJ, Kinston S, Joshi A, Hannah R, Theis FJ, et al. Characterization of transcriptional networks in blood stem and progenitor cells using high-throughput single-cell gene expression analysis. Nat Cell Biol. 2013;15:363–372. doi: 10.1038/ncb2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Amit I, Garber M, Chevrier N, Leite AP, Donner Y, Eisenhaure T, Guttman M, Grenier JK, Li W, Zuk O, et al. Unbiased reconstruction of a mammalian transcriptional network mediating pathogen responses. Science. 2009;326:257–263. doi: 10.1126/science.1179050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nayak RR, Kearns M, Spielman RS, Cheung VG. Coexpression network based on natural variation in human gene expression reveals gene interactions and functions. Genome Res. 2009;19:1953–1962. doi: 10.1101/gr.097600.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pah AR, Guimera R, Mustoe AM, Amaral LA. Use of a global metabolic network to curate organismal metabolic networks. Sci Rep. 2013;3:1695. doi: 10.1038/srep01695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gennarino VA, D'Angelo G, Dharmalingam G, Fernandez S, Russolillo G, Sanges R, Mutarelli M, Belcastro V, Ballabio A, Verde P, et al. Identification of microRNA-regulated gene networks by expression analysis of target genes. Genome Res. 2012;22:1163–1172. doi: 10.1101/gr.130435.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Stark C, Breitkreutz BJ, Chatr-Aryamontri A, Boucher L, Oughtred R, Livstone MS, Nixon J, Van Auken K, Wang X, Shi X, et al. The BioGRID Interaction Database: 2011 update. Nucleic Acids Res. 2011;39:D698–D704. doi: 10.1093/nar/gkq1116. [DOI] [PMC free article] [PubMed] [Google Scholar]