Abstract
Lack of an objective, accurate, and noninvasive embryo assessment strategy remains one of the major challenges encountered in in vitro fertilization. Cumulus and mural granulosa cells reflect the characteristics of the oocyte, providing a noninvasive means to assess oocyte quality. Specifically, transcriptomic profiling of follicular cells may help identify biomarkers of oocyte and embryo competence. Current transcriptomics technologies include quantitative reverse transcriptase–polymerase chain reaction (qRT-PCR) for analysis of individual genes and microarrays and high-throughput deep sequencing for whole genome expression profiling. Recently, using qRT-PCR and microarray technologies, a multitude of studies correlated changes in cumulus or granulosa cell gene expression with clinically relevant outcome parameters, including in vitro embryo development and pregnancy. While the initial findings are promising, a clinical benefit from the use of identified biomarker genes remains to be demonstrated in randomized controlled trials.
Infertility affects approximately 15% of reproductive-age couples (1) and in addition to medical consequences, has significant social and financial implications. Among the treatment modalities available to infertile couples, IVF offers the highest success rates of pregnancy and live-birth outcomes. A critical step in IVF treatment is assessment of oocyte and embryo competence to determine the most viable embryo(s) to be transferred. Currently, embryo assessment strategies rely primarily on embryo morphology and cleavage rate. While these methodologies have been successful in improving pregnancy rates and reducing multiple gestations, their precision is less than what is desired (2). Consequently, many centers perform multiple embryo transfers (ETs) to increase the chances of success for a given cycle, at the expense of a significantly higher risk of multiple gestations. Multiple gestations, in turn, result in an increased risk of preterm birth and its complications, such as cerebral palsy, and infant death (reviewed in reference 2). Therefore, the development of an objective and accurate test to assess oocyte and embryo viability remains one of the most significant contemporary goals of reproductive medicine.
In an attempt to develop novel embryo assessment strategies that can be used alone or in combination with morphologic criteria, invasive and noninvasive methods have been applied (reviewed in reference 3). These include the assessment of the genome using comparative genomic hybridization (CGH) arrays, single nucleotide polymorphism (SNP) arrays, quantitative real time–polymerase chain reaction (qPCR), transcriptomic analysis of cumulus/granulosa cells, and proteomic and metabolomic analysis of embryo culture media (reviewed in reference 3).
Among these approaches, transcriptomic analysis of cumulus/granulosa cells has been proposed as a noninvasive tool to assess oocyte quality and viability as a surrogate for the reproductive potential of embryos. In this review, we first describe the basic aspects of folliculogenesis and the interactions between the oocyte and the stromal cells of the follicle. Methods used to study cumulus/granulosa cell gene expression are then outlined followed by the review of the studies assessing transcriptional analysis of cumulus/granulosa cells in correlation with oocyte and embryo competence. Lastly, the variations in the findings of these studies are explored from a methodological perspective.
FOLLICULOGENESIS: BASIC ASPECTS
Folliculogenesis requires a carefully orchestrated cross talk between the oocyte and the surrounding somatic cells. During fetal life, primordial germ cells (PGCs) migrate to the future gonad, undergo mitosis, and give rise to oogonia (4). The oogonia are then transformed into oocytes as they enter the first meiotic division. Primordial follicles are formed perinatally as the oocytes arrested in prophase of the first meiotic division become enveloped by a single layer of flattened granulosa cells that are surrounded by a basement membrane (5) (Fig. 1). Thereafter, through an unknown selection mechanism, individual primordial follicles are recruited from this resting pool to undergo growth and differentiation (6). During this process, granulosa cells surrounding the oocyte become cuboidal and form the primary follicle (7). Then granulosa cells proliferate and form multiple layers of somatic cells that surround the oocyte, resulting in the formation of a secondary follicle. This is followed by the formation of small, fluid-filled cavities within the follicle that coalesce to form the early antral (or tertiary) follicle (8). In the absence of gonadotropin stimulation, these follicles become atretic and disappear from the ovary. However, once puberty ensues, pituitary follicle stimulating hormone (FSH) stimulates further follicular growth (5). Under the influence of FSH, the antrum continues to enlarge, resulting in the formation of a preovulatory (also called antral or late antral) follicle (Fig. 1). In the preovulatory follicle, the oocyte is surrounded by cumulus cells, a specialized type of granulosa cell, distinct from the mural granulosa cells that line the antrum (9).
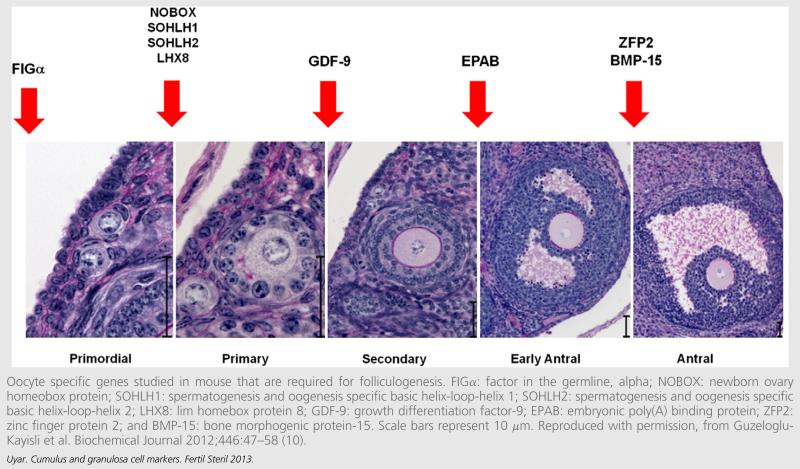
FIGURE 1.
Oocyte specific genes studied in mouse that are required for folliculogenesis. FIGa: factor in the germline, alpha; NOBOX: newborn ovary homeobox protein; SOHLH1: spermatogenesis and oogenesis specific basic helix-loop-helix 1; SOHLH2: spermatogenesis and oogenesis specific basic helix-loop-helix 2; LHX8: lim homebox protein 8; GDF-9: growth differentiation factor-9; EPAB: embryonic poly(A) binding protein; ZFP2: zinc finger protein 2; and BMP-15: bone morphogenic protein-15. Scale bars represent 10 μm. Reproduced with permission, from Guzeloglu-Kayisli et al. Biochemical Journal 2012;446:47–58 (10).
The oocyte within the preovulatory follicle will remain arrested in prophase I until after the LH surge, which precedes ovulation (10). Several critical steps are necessary to activate and mature the preovulatory follicle. It is during this period that the cumulus cells undergo “cumulus expansion,” a process that requires cumulus cells to produce hyaluronic acid that is deposited into the extracellular space and further stabilized by secreted proteins (11, 12). This newly formed extracellular matrix binds the oocyte and cumulus cells together (11, 12). In the meantime, the oocyte resumes meiotic division and begins the process of maturation. The culmination of these processes is the formation of a mature cumulus-oocyte complex (COC) containing an oocyte arrested at the metaphase of the second meiotic division (MII) and ready for ovulation and subsequent fertilization (10).
INTERACTIONS BETWEEN THE OOCYTE AND THE STROMAL CELLS OF THE FOLLICLE
Follicular development, oocyte maturation, cumulus expansion, and ovulation rely on continuing cross talk between the oocyte and the somatic follicular cells. In this section, we will briefly review key aspects of these processes as they have implications for studies investigating cumulus/granulosa cell gene expression as a proxy for oocyte viability.
Implications of Oocyte-Somatic Cell Interactions for Follicular Development
Studies in murine models have shown that rodents deficient in oocytes, secondary to mutations or chemical ablation, are also deficient in follicles, suggesting that the oocyte plays a critical role in the growth and development of the surrounding follicle (13). During folliculogenesis, factors exclusively or predominantly expressed by the oocyte are required for the progression of the follicle through different developmental stages.
Among these, one of the earliest transcription factors implicated in postnatal oocyte-specific gene expression is FIGα (8). FIGα is required for primordial follicle formation, and Figα-knockout mice are devoid of primordial follicles (8). Another key factor expressed in the oocyte is NOBOX, an oocyte-specific homeobox gene product expressed in germ cell cysts and in the oocytes of primordial and growing follicles (14). Nobox-knockout female mice are infertile and have atrophic ovaries that are devoid of oocytes at 6 weeks of age (15). Interestingly, unlike the findings seen in Figα-knockout mice, germ cell proliferation and initial primordial follicle formation occur in the absence of NOBOX (15). Thus the lack of NOBOX inhibits follicle growth beyond the primordial follicle stage and accelerates the loss of oocytes (15).
Another key mediator for folliculogenesis, growth differentiation factor-9 (GDF-9), is produced by the oocyte from the primary follicle stage until the time of ovulation (16, 17). In Gdf-9-knockout mice, follicular development is arrested at the primary follicle stage, despite the presence of numerous primordial and early primary follicles (18). Additionally, an oocyte-specific homolog of GDF-9 called bone morphogenetic protein 15 (BMP-15) has been cloned on the X-chromosome (19). Yan et al. showed a synergistic role for BMP-15 and GDF-9 in ovarian folliculogenesis (20). Despite having relatively normal ovarian histology, Bmp-15-knockout mice are subfertile, secondary to decreased ovulation and fertilization rates (20). Double homozygous knockout females for BMP-15 and GDF-9 (Bmp-15–/–; Gdf-9–/–) experience oocyte loss resembling the phenotype seen in the GDF-9-knockout mice. Interestingly, homozygous Bmp-15 and heterozygous Gdf-9-knockout (Bmp-15–/–; Gdf-9+/–) females have oocytes that fail to adhere to cumulus cells and often remain trapped in the corpus luteum (20).
Gene expression during oocyte maturation, fertilization, and early embryo development until zygotic gene activation relies on translational activation of specific maternally derived mRNA that was attained during the first meiotic arrest (reviewed in reference 21). Embryonic poly (A)-binding protein (EPAB), which is exclusively expressed in gametes and early embryos (22, 23), is the predominant poly (A)-binding protein that stabilizes maternal RNAs and promotes their translation (10). Epab-knockout female mice are infertile owing to the inability of the oocytes to undergo maturation. Interestingly, although EPAB is not expressed in the somatic cells of the follicle (22), follicular development beyond the secondary follicle stage is impaired. Furthermore, the follicles that reach the preovulatory stage fail to undergo cumulus expansion in Epab-knockout mice (10).
The above-mentioned findings establish that follicular development is adversely affected by the absence of normal oogenesis. The oocyte-specific genes that are required for follicular development, and the stages at which follicular development is arrested in knockout mouse models of these genes are shown in Figure 1 (8, 10, 15, 18, 20, 24–26).
Implications of Oocyte-Somatic Cell Interactions for Oocyte Maturation and Cumulus Expansion
The growing oocyte derives most of its substrates for energy metabolism and biosynthesis from the surrounding somatic cells, stressing the importance of the intercommunication between these cells (27, 28). Cumulus cells communicate with each other and with the oocyte through specialized gap junctions that allow metabolic exchange and transport of signaling molecules (29). The fundamental unit of the gap junctions is the connexon, which is a hexamer of proteins called connexins (Cxs) (30). Cx43, is a major contributor to gap junctions in human cumulus cells and is essential for the developmental competence of human oocytes (30). Reduction of Cx43 expression in the COC has been shown to initiate the resumption of meiosis by disrupting the gap junctions (30). Other genes involved in gap junctions, such as Cx37 and Cx40, have also been detected on microarray studies and may play an additional role in this process (31).
Cumulus cells play a vital role in regulating oocyte maturation (32, 33). During the meiotic arrest, cyclic guanosine monophosphate (cGMP) from cumulus cells passes into the oocyte through gap junctions and inhibits the hydrolysis of cyclic adenosine monophosphate (cAMP) by the phosphodiesterase PDE3A. This inhibition maintains a high concentration of cAMP in the oocyte and blocks meiotic progression. LH reverses the inhibitory signal by lowering cGMP levels in the somatic cells and by closing gap junctions between the oocyte and the somatic cells. The resulting decrease in oocyte cGMP relieves the inhibition of PDE3A, causes a decrease in oocyte cAMP and leads to the resumption of meiosis (34). It is noteworthy that while the development of the follicle is regulated by bidirectional signals between the oocyte and surrounding somatic cells, the LH-induced maturation of the oocyte is under the control of signals from the follicular somatic cells (reviewed in reference 35).
Cumulus expansion is another critical aspect in the final stages of follicular development. Oocytes obtained from follicles with impaired cumulus expansion have limited potential for implantation (36). GDF-9 secreted by the oocyte has been shown to play a key role in cumulus expansion (37). GDF-9 functions as an oocyte-secreted paracrine factor that regulates several key granulosa cell enzymes involved in cumulus cell expansion, thus creating a microenvironment optimal for oocyte developmental competence (38). Cyclooxygenase 2 (COX2), gremlin1 (GREM1), hyaluronic acid synthase 2 (HAS2), and pentraxin 3 (PTX3) are all downstream GDF-9 target genes found in cumulus cells and have been evaluated as markers for oocyte developmental competence (39, 40).
Evidence accumulating from several elegant studies using the model organisms described above strongly suggests that key events during oogenesis and folliculogenesis rely on interactions between the oocyte and the somatic cells. These data strongly suggest that cumulus and granulosa cells may be used to gain insight into the viability and reproductive potential of the oocytes. In the following sections we will introduce analytical methods that are applied for cumulus and granulosa cell analysis and review the findings from clinical studies.
MOLECULAR BIOLOGY AND BIOINFORMATICS METHODS APPLIED TO THE ANALYSIS OF CUMULUS AND GRANULOSA CELLS
The survival and function of a cell relies on complex molecular pathways that are activated in a timely manner in response to changing biological needs. Since gene expression proceeds through transcription of the genetic code into messenger RNA (mRNA), relative quantities of individual mRNAs can be assessed as an approximation of the expression levels of corresponding genes and provide information on the well-being of a cell or tissue.
The total RNA content of a cell is termed the transcriptome and includes mRNAs, ribosomal RNAs (rRNAs), transfer RNAs (tRNAs), and microRNAs (miRNAs). Transcriptomics studies enable qualitative and quantitative characterization of gene expression in a cell or tissue under physiologic or pathologic conditions.
Within the context of IVF (3), transcriptomics analysis enables monitoring of gene expression in somatic cells of the follicle, gametes, and embryos. Transcriptomics technology offers quantitative reverse transcription polymerase chain reaction (qRT-PCR), for analysis of individual genes, and microarrays and high-throughput deep sequencing techniques for whole genome transcriptomic profiling. The result of genome-wide transcriptome analysis is the generation of a list of genes that are differentially expressed between the experimental conditions. Analysis of cumulus and/or granulosa cells transcriptomics in association with oocyte maturation, fertilization, embryo competence, and pregnancy outcome may help identify novel diagnostic biomarkers as an alternative to conventional morphological criteria. Here we provide brief definitions of the transcriptomic technologies and related cumulus cell–specific procedures.
Sample Collection, RNA Isolation, and Quality Assessment
The common initial step in transcriptomic experiments is sample collection and RNA isolation, where high-quality starting material is a prerequisite for reliable analysis. Sampling of somatic components of the follicle poses specific challenges. Compared with granulosa cells, mechanical separation of COCs from contaminating cells is easier. In addition, laser capture technology can be quite useful in minimizing the effect of regional differences in the cumulus physiology. Cumulus cells must also be separated from the attached mural granulosa cells, if any (3). In the context of granulosa cells, potential contamination with blood during follicular aspiration may affect the gene-expression profile. Interestingly, as demonstrated by principal component analysis (PCA) and flow cytometry analysis, the granulosa cell gene-expression profile is closer to that of lymphocytes than of cumulus cells (31). Therefore, granulosa cells need to be carefully separated from leukocytes.
RNA must be purifiedimmediatelyafter tissue/cell isolation as RNA degradation results in a rapid change in transcript profile. If the samples need to be stored before RNA isolation, RNAses must be inactivated either by using inhibitors or by rapid cooling. RNA stabilization is an absolute prerequisite for reliable gene-expression analysis. Immediate stabilization of RNA in cumulus and granulosa cell samples is necessary because, directly after harvesting the biological sample, changes in the gene-expression pattern occur owing to specific and nonspecific RNA degradation as well as removal of the oocyte. Such changes in gene-expression pattern need to be avoided for all reliable quantitative gene-expression analyses, such as biochip and array analyses, qRT-PCR, or other nucleic acid–based technologies (41).
The efficiency of downstream processes strongly depends on the concentration, purity, and integrity of starting RNA molecules. According to UV spectroscopy–based analyses, absorbance at 260 nm (A260) is considered a measure of RNA concentration and the ratio A260/A280 indicates the purity of RNA content with respect to protein contamination (42). On the other hand, all nucleic acids including DNA, RNA, and free nucleotides have a peak absorbance at ~260 nm, which affects the accurate determination of RNA concentration. RNA isolation techniques are usually coupled with a subsequent DNase treatment to eliminate DNA contamination. However, it is almost impossible to completely eliminate genomic DNA from RNA samples. Assessment of potential residual DNA contamination in an RNA sample requires further analysis. The acceptable level of contamination may vary depending on the requirements of the downstream application.
Typically, a minus reverse transcriptase (—RT) control (no amplification control, or NAC) reaction is included in RT-PCR experiments to check for the presence of contaminating DNA. A —RT sample contains all the PCR reagents and total RNA except for the RT. In a —RT sample, the cDNA would not be synthesized. Therefore, a visible band in the —RT reaction on agarose gel would indicate DNA contamination. Also, it might be noted that RT-PCR using primers that span an intron (i.e., primers are on two different exons) can prevent DNA contamination from affecting the results.
Alternatively, DNA and RNA concentrations in an RNA sample can be measured separately using a Qubit Fluorometer (Invitrogen), which uses fluorescent dyes to determine the concentration of nucleic acids and proteins where each dye is specific for one type of molecule.
RNA integrity is usually determined by agarose gel quantification of 28S and 18S in human ribosomal RNAs (43). Intact total RNA ran on a gel should demonstrate clear 28S and 18S bands with a 28S/18S ratio between 1.8 and 2. The major drawback of this method is the amount of required RNA needed for analysis. In case of a limited amount of samples, such as with oocyte and cumulus cells, a sufficient amount of RNA (~200 ng) may not be available for integrity assessment. SYBR Gold and SYBR Green II stains allow smaller amount of samples (1–2 ng) to be used for preexperimental analysis.
As an alternative to traditional gel–based methods, microfluidics technology provides electrophoretic RNA separation enabling automated high-throughput integrity estimation. Currently, Agilent 2100 Bioanalyzer (Agilent Technologies) and Experion automated electrophoresis system (Bio-Rad Laboratories, Inc.) are the two most popular commercial biodevices that integrate the quantitation and quality assessment of RNA samples in a single step. The resulting data are usually displayed as an electropherogram where the 28S and 18S rRNAs generate two separate peaks and the 28S/18S ratio is calculated as the ratio of the areas under the peak signals. With the advent of these technologies, novel computerized methods have been proposed for estimation of RNA integrity such as Degradometer for Agilent Bioanalyzer by Auer et al. (44) and RNA Quality Indicator (RQI) for Experion system by Denisov et al. (45). The most robust and common method for RNA quality assessment, the RNA Integrity Number (RIN), was introduced by Schroeder et al. which was constructed based on eight features extracted from the electropherogram signal measurements and a Bayesian learning technique (46) (Agilent Technologies). The method produces RIN, a number in the range of 1–10, with 10 being the highest quality samples showing the least degradation.
The accepted level of RNA integrity is usually specific to procedures, tissues, and protocols. In general, cDNA labeling for microarray analysis requires RNA of high integrity. Although there is no universal RIN for all experiments, most microarray procedures require a RIN >7. On the other hand, RT-PCR involves analysis of smaller regions of RNA and therefore is more tolerant to partially degraded RNA. For qRT-PCR experiments, the RIN >5 and RIN >8 have been recommended as good and perfect total RNA quality, respectively (47).
qRT-PCR
Once the sample integrity is confirmed, qRT-PCR analysis may be used to provide reliable quantification of gene expression. PCR has been the most widely used tool to identify the presence or absence of a particular DNA fragment in a biological sample. PCR may also be used to amplify the input material to reach the micrograms of RNA required for microarray and deep sequencing experiments from the picograms of RNA obtained from follicular cells.
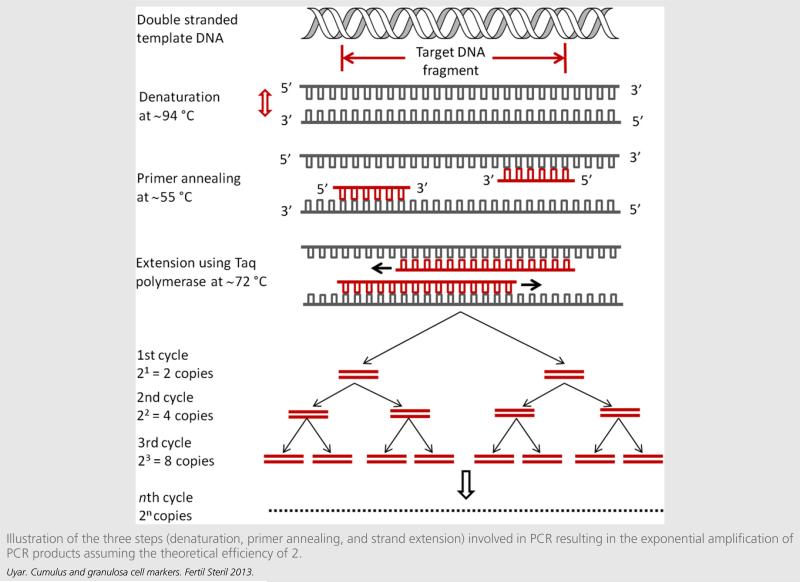
The main idea behind the PCR is the exponential amplification of the target sequence using a pair of primers, deoxynucleotides, and a polymerase enzyme that catalyzes the reaction. Each amplification cycle involves three steps performed at specific temperatures: denaturation of double strands (~94°C), primer annealing to complementary parts on both strands (~65 to 55°C) and primer extension (~72°C), which in turn produces double-stranded molecules twice the amount of the starting material (Fig. 2). The annealing and extension steps of a PCR cycle can also be performed as a single step either by using polymerases that are highly active in the annealing temperature range or by designing longer primers (≥23 bases) that would increase the annealing temperature. A two-step PCR would be a time-saving procedure reducing the cycle period compared to three-step PCR.
FIGURE 2.
Illustration of the three steps (denaturation, primer annealing, and strand extension) involved in PCR resulting in the exponential amplification of PCR products assuming the theoretical efficiency of 2.
The major landmark in the history of PCR was the identification of a thermostable polymerase for DNA synthesis (48). The enzyme was isolated from the bacterium Thermophilus aquaticus that grows in hot springs and hence is resistant to the high temperatures required for PCR denaturation. This greatly simplified the procedure, eliminating the necessity of adding the enzyme at the beginning of each cycle. After ~25 to 40 rounds of amplification, the resulting PCR product is analyzed on an agarose gel containing a fluorescent DNA stain. Ethidium bromide (EtBr) has been used as a common DNA stain in biology labs for a long time. However, owing to its mutagenic nature, the newer trend is to replace EtBr by less harmful and as powerful fluorescent dyes (49). PCR and the use of Thermophilus aquaticus polymerase (Taq polymerase) were discovered by Kary Mullis (48, 50, 51); Kary Mullis was subsequently awarded the Nobel Prize for Chemistry in 1993 for these achievements.
Transcriptomics studies make use of PCR after reverse transcription of mRNA molecules to complementary DNA (cDNA). The combined method is called reverse transcription PCR (RT-PCR) and is widely used for gene-expression analysis. Total RNA is primed with either one of the oligo(dT), random hexamers or gene-specific primers, or a blend of these primers, and cDNA is synthesized. The choice of primers used for RT [oligo d(T) vs. random hexamers] is especially important when the sample may contain inactive transcripts with short poly(A) tails [that may not be reverse transcribed by oligo d(T)] as in the case of oocytes. Once RT is complete, PCR is performed as usual using the cDNA as the template for amplification. The choice of priming strategy has been shown to be an important factor affecting the results of mRNA quantification (52).
In theory, assuming 100% efficiency, the amount of input cDNA is expected to double at the end of each PCR cycle. Conveniently, the resulting products are expected to be proportional to the amount of the starting material. However, the exponential increase cannot go on until the end of the reaction. The amplification reaction reaches a plateau phase, and quantification of the PCR products at this stage may not reflect the actual relative quantities of the input material. Consequently, conventional RT-PCR should not be considered a quantitative strategy and can only be efficiently used to identify the presence or absence of a particular mRNA within a biological sample.
To analyze the variations between different phenotypes, identification of the genes that are expressed in the samples of interest is not sufficient; accurate measurement of gene-expression levels is also crucial. Since agarose gel–based analysis of RT-PCR products does not provide precise quantitative information, researchers sought for a way to accurately quantify the mRNA of a specific gene within a sample. For this purpose, Higuchi et al. pioneered a system for continuous measurement of PCR amplification results (53, 54). This system provided quantitative analysis of RNA transcript levels, and the new approach evolved into qRT-PCR (or so called real-time RT-PCR) technology, which is now widely used for quantification of mRNAs for a number of genes within biological samples. Note that the abbreviation used for real-time PCR is qPCR. Real-time reverse-transcription PCR is denoted as qRT-PCR. The acronym RT-PCR commonly denotes RT polymerase chain reaction and not real-time PCR, but not all investigators adhere to this convention (55, 56).
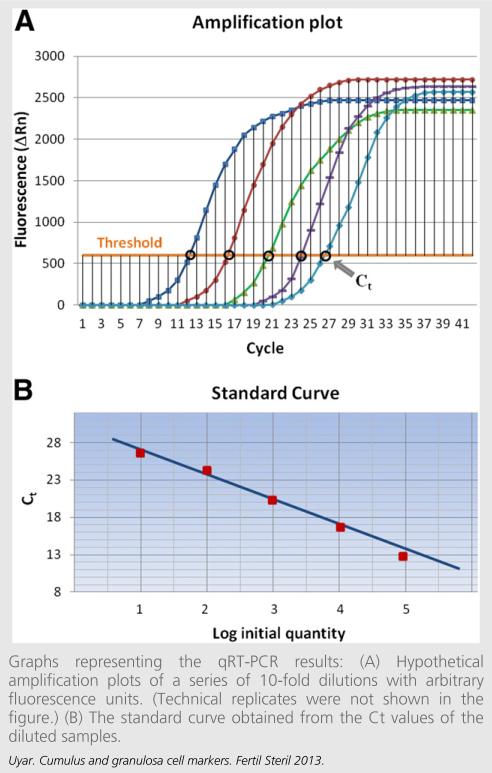
To achieve accurate quantification, there should be a linear relationship between the amount of input material and the resulting product, that is, if a number of samples with known concentrations of cDNA are amplified, the final quantities should be proportional to the initial concentrations. However, this linear relationship is usually observed only in the earlier cycles of PCR, where the later measurements may result in erroneous quantification (Fig. 3A). QRT-PCR is a kinetic approach in which the entire reaction can be observed at all stages; SYBR green is commonly used as the fluorescence source, and the amount of fluorescence included in the newly synthesized cDNA is measured. First the cycle threshold value (Ct) is determined as the number of cycles where the fluorescence signal exceeds the background level. Ct levels are inversely proportional to the amount of input cDNA; a reaction with lower starting material takes more cycles before the amplification is detectable. Then a standard curve is generated from a dilution series of a reference sample. The standard curve plots Ct values versus relative concentrations of the diluted sample in an experiment as shown in Figure 3B. Finally, relative quantities for target abundance in each experimental sample are determined using the standard curve method. It is noteworthy that, Ct, crossing point (Cp), and take-off point (Top) are all used to refer to the same quantification cycle value (Cq) depending on the real-time instrument, and the term Ct is mostly replaced by Cq, which is the official abbreviation in the Real-Time PCR Data Markup Language (RDML) (57).
FIGURE 3.
Graphs representing the qRT-PCR results: (A) Hypothetical amplification plots of a series of 10-fold dilutions with arbitrary fluorescence units. (Technical replicates were not shown in the figure.) (B) The standard curve obtained from the Ct values of the diluted samples.
In a qRT-PCR experiment, in addition to RT control, which is used as a NAC, a no template control (NTC) should be run in parallel to check for contamination of reagents. The NTC reaction includes all of the RT-PCR reagents except the RNA template. Therefore, any amplification product in the NTC indicates contamination by reagents or the environment.
In some cases, nonspecific amplification may occur owing to contamination, mispriming, or primer-dimer artifacts. Mispriming refers to binding of the primers to a nontarget cDNA; primer-dimer artifact is annealing of the primers to themselves and creation of small templates for PCR amplification. To prove that only the target cDNA has been amplified, a melting curve can be generated at the end of the reaction. The melting temperature of a DNA molecule depends on its sequence composition and its length. Hence, unless there is a PCR artifact, all PCR products resulting from the same primer pair should have similar melting curves. Different products can generate different peaks on the melting curve, and PCR artifacts typically have lower melting temperatures. The reactions producing such different peaks must be eliminated from the analysis or the entire experiment must be repeated. Alternatively, TaqMan technology can be used to increase the specificity of quantification in qRT-PCR. The SYBR green dye intercalates to double-stranded PCR products including both specific and nonspecific amplicons. In contrast, the fluorogenic-labeled TaqMan probes, which are synthesized specifically to template sequence, enable detection of only target products. Both TaqMan (40, 58–61) and SYBR green assays (61–67) have been widely used in cumulus studies.
Unless absolute quantification is performed by using controls with a known amount of cDNA, qRT-PCR gene-expression results need to be normalized using endogenous control genes. Normalization to endogenous reference genes does not provide information on the absolute amount of a specific mRNA or its cDNA in a sample. Rather, it provides very reliable information on which sample has a higher (or lower) expression of a trancript (or its cDNA) in a group of samples. These reference genes should be expressed at the same level across all samples independent of the study treatments or the biological conditions being investigated. The expression levels of the putative reference genes, including glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and beta-actin (β-Actin), have been shown to be variant across 16 different human tissues (68). Assessment of unstable reference genes for normalization of expression levels would introduce a bias in the results. Thus, a key aspect of transcriptomics studies is the selection of the most invariant endogenous control according to the tissue type or experimental conditions. It is recommended not to use a single control gene in the qRT-PCR experiments, and mathematical models such as NormFinder (69) and GeNorm (70) have been developed to help with selection of the most suitable normalization genes among a set of candidate genes.
Accuracy of the relative gene-expression measurements in qRT-PCR also correlates with PCR efficiency. The common ΔΔCt method assumes that the samples amplify with comparable efficiencies. To provide precise calculations, the PCR efficiency should be tested at least once, and alternative models adjusting for amplification efficiency should be preferred, if necessary. Moreover, the use of biological and technical replicates is recommended to ascertain the significance of the qRT-PCR results. The PCR reactions are usually run in duplicates, or preferably triplicates, to control for well-to-well variation.
It is noteworthy that, while qRT-PCR is discussed as a transcriptomic methodology, it is not used to assess the entire transcriptome. QRT-PCR is also used for confirmation of the gene-expression levels obtained from high-throughput microarray or RNA-Seq experiments, which will be discussed in the ensuing sections.
Microarrays
Within thepastdecade,microarraystudies havebecomea major component of the transcriptomic investigation, helping to evolve gene-expression research from single-gene analysis to genome-wide transcriptomic profiling. High-throughput microarray experiments provided significant contributions to the scientific literature, with more than 50,000 publications in PubMed matching the keyword “microarray” to date. Microarray technology has also been applied to the study of cumulus and granulosa cell transcriptome within the context of IVF.
Microarray technology relies on hybridization of complementary nucleotide sequences. A DNA microarray consists of single-stranded ~25 base length polynucleotide probes that are synthesized in fixed positions in the form of a two-dimensional array on a solid surface. Each probe is attached to a microscopic spot on the array and corresponds to the base sequence of a specific mRNA. The large number of probes on a single microarray chip enables simultaneous expression analysis for all known genes.
The microarray procedure proceeds through labeling, hybridization, scanning, and data analysis stages (Fig. 4). The procedures presented here are mainly related to the Affymetrix platform, which is the most commonly used microarray system.
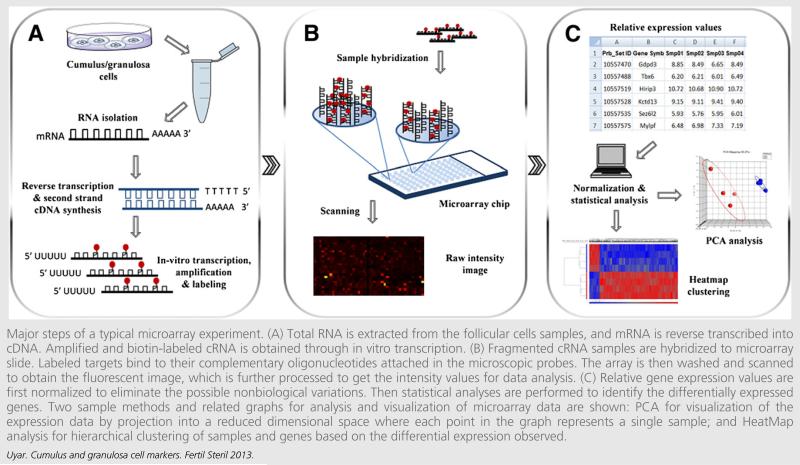
FIGURE 4.
Major steps of a typical microarray experiment. (A) Total RNA is extracted from the follicular cells samples, and mRNA is reverse transcribed into cDNA. Amplified and biotin-labeled cRNA is obtained through in vitro transcription. (B) Fragmented cRNA samples are hybridized to microarray slide. Labeled targets bind to their complementary oligonucleotides attached in the microscopic probes. The array is then washed and scanned to obtain the fluorescent image, which is further processed to get the intensity values for data analysis. (C) Relative gene expression values are first normalized to eliminate the possible nonbiological variations. Then statistical analyses are performed to identify the differentially expressed genes. Two sample methods and related graphs for analysis and visualization of microarray data are shown: PCA for visualization of the expression data by projection into a reduced dimensional space where each point in the graph represents a single sample; and HeatMap analysis for hierarchical clustering of samples and genes based on the differential expression observed.
Labeling, hybridization, and scanning for microarray analysis
First, total RNA is isolated and mRNA is reverse transcribed to cDNA. Amplified and biotin-labeled cRNA is obtained through in vitro transcription. Then fragmented cRNAs are deposited over the array and hybridized overnight. Labeled targets bind to the complementary probes at a quantity that is expected to be proportional to the level of expression of the gene represented by that probe. The array is washed to exclude the targets that have not been hybridized. The micro-array chip is then illuminated by a laser light; labeled molecules emit fluorescence proportional to their quantity. Gene expression is quantified by means of fluorescence intensity, which is captured by the scanners into an image (Fig. 4).
Microarray data analysis
The raw microarray data obtained from the image files need to be processed before differential expression analysis. Considering microarrays, the entire experimental process is more prone to error compared with qRT-PCR since thousands of genes are analyzed simultaneously after a multistage experimental setting where errors might mainly result from the amplification procedure. To reduce possible nonbiological variations and to ensure the integrity of the experiment, a number of controls are performed at each stage.
Once the raw expression values are obtained, it is necessary to ensure the integrity of the experimental procedure at the RNA extraction, amplification, labeling, and hybridization stages. Affymetrix GeneChip Quality Control (71) uses the endogenous housekeeping gene 3’/5’ fluorescence ratio (beta-actin and gapdh), exogenous hybridization control fluorescence intensity (bioB, bioC, bioD and Cre), and polyA control fluorescence intensity (Dap, Lys, Phe, Thr, and Trp). The Microarray Quality Control (MAQC) Consortium (72) provides resources to assess the performance of microarray platforms.
Robust Multichip Average (RMA)
Several statistical methods for preprocessing of microarray data have been developed. Among those methods, there is a consensus on Robust Multi-chip Average (RMA) as the most efficient approach to date. RMA consists of three steps: background adjustment, quantile normalization, and probe set summarization (73–75).
A portion of the observed intensities on a scanned array may be due to background noise, that is, optical noise and/or nonspecific hybridization. RMA background adjustment is a model-based approach using only perfect match values to estimate background density (73). The probe intensities are modeled as the sum of a normal noise component and an exponential signal component.
Among various normalization methods, the widely used Quantile Normalization (74) is based on the assumption that the microarray chips have identical intensity distributions. According to the underlying biological reasoning, a treatment would up-regulate only a number of genes, down-regulate only a number of genes, and leave the rest of the genes unaffected. Moreover, since equal amounts of RNA are processed and loaded to each array, the cumulative expression would be the same among the samples in an experimental setting. When there is a global mRNA change among the samples and the main assumption does not hold, additional external controls should be used to measure actual expression levels (76).
In an Affymetrix GeneChip, probes consist of 25 base oligomers and a gene is represented by a set of probes (n~27). Each probe maps to well-supported exons of known genes. After background adjustment and quantile normalization at the probe level, the intensities of probes within a probe set are summarized to a single value (75). The resulting intensity value is assigned to the gene represented with that probe set. Differential expression analysis is then performed on the background adjusted, normalized, and summarized intensities.
Differential expression analysis: multiple testing issue
The preprocessed gene-expression data are stored in an m × n matrix, where the rows correspond to genes and the columns correspond to samples. Samples are usually categorized as wild type versus knockout, treated versus untreated, diseased versus healthy, or tumor versus normal. In the case of cumulus cell analysis, the samples are categorized according to the outcome measure; that is, oocytes developing to low-quality versus high-quality embryos, oocytes resulting in pregnancy versus failed cycle, and so on. Each category is assumed as a response variable. In a typical microarray experiment, the total number (n) of samples is expected to be around 10 to a few hundreds, and the number (m) of genes is several thousand depending on the species and platforms.
The differential expression analysis can be stated as the simultaneous test for each gene of the null hypothesis of equal mean expression levels in two populations of cells. In this context, a false-positive (type I error) declares that a gene is differentially expressed when it is not. When thousands of hypotheses are tested simultaneously, one for each gene, with a specified type I error probability (e.g., 0.05), the chance of type I errors increases with the number of hypotheses. Statistical literature offers various approaches for multiple hypotheses testing to avoid excessive type I errors (77). The widely used false discovery rate (FDR) is the expected proportion of type I errors among the rejected hypotheses. A common approach to apply FDR in a microarray setting is by calculating the q-value, which is the FDR analog of the P value (78, 79). Commercial tools such as the Partek Genomic Suite and the Bioinformatics toolbox of Matlab exist for differential expression analysis of microarray data.
In a routine microarray data analysis procedure, the expression levels are also investigated in terms of fold change, a parameter that stands for the ratio of mean expression values for each gene among the sample groups, which is expected to be higher than a predefined threshold value, for example, two folds. To conclude that a gene is differentially expressed, the data analysis should consider both FDR and fold change rate.
The results of robust FDR-corrected significance tests may [1] reveal no difference between the populations of interest, [2] point out just a few genes up-regulated or down-regulated in either population, or [3] determine hundreds of differentially expressed genes. In the case of [2], target genes may be further investigated as potential biomarkers of the research interest, especially in case of high fold changes. In the case of [3], more sophisticated analyses are required for biological interpretation, that is, pathway analysis of microarray data.
Pathway analysis
Biological interpretation of the differentially expressed genes is usually explored in the form of gene-ontology (GO) functional categories or metabolic and regulatory pathways. A review of approaches and tools employed for pathway analysis is presented in reference (80). Two of the popular tools are the DAVID, a widely used web-based application focusing on GO classification (81), and Ingenuity Pathway Analysis (IPA), a commercial software with enhanced visualization capabilities (Ingenuity Systems, www.ingenuity.com).
Validation of microarray data
Validation of microarray data is a key component of the transcriptomics approach. Selected transcripts that are found to be significantly different in the experimental group compared with controls should be validated in independent biological replicates using qRT-PCR. In this setting, “independent” signifies that the samples used to validate microarray results should be collected separately from those used for microarray experiments.
Potential pitfalls of microarray analysis
In the last decade, microarrays have been the most popular biotechnological tools for monitoring whole-genome expression levels. Microarray technology has also been a popular research interest for the bioinformatics community as the statistical analysis of microarray data poses significant challenges. The underlying reasons for the complex data analysis procedure may be summarized in three points. First, the experimental procedure consists of multiple stages (RNA extraction, RT, amplification, labeling, and hybridization); each stage results in additional potential for a systematic or random variation in the resulting data. Second, simultaneous multiple hypothesis testing is likely to result in high false-positive rates unless appropriate statistical methods are applied. And third, very large GO databases are searched for identification of biological processes associated with the differentially expressed genes.
Considering such complexities, it is crucial to understand the main assumptions of microarray experiments and reassess the experimental design based on these assumptions. It is also essential to assure reliability, comparability, and reproducibility of microarray experiments. Data obtained from different platforms or different sites may not be consistent (82). Recently, evaluation of reproducibility of the results presented in 18 articles on microarray-based gene-expression profiling revealed that only the data from two studies could be reproduced, while six could be partially reproduced and 10 could not be reproduced (83). Publication of any false discoveries may lead to an undesired domino effect in the scientific literature since the biological databases are extended using the published results.
RNA-Seq
Despite the increasing use of microarrays to explore complex transcriptional mechanisms, the underlying technology has certain limitations. Primarily, existing knowledge about the genomic constitution of the species is a prerequisite for the microarray probe design. This constraint limits detection of alternative splicing patterns or previously unmapped genes. Although tiling arrays are constructed that span exon junctions and overlapping genomic sequences, both expression arrays and tiling arrays suffer from high levels of background noise due to cross-hybridization. Moreover, microarrays have a limited dynamic range of detection that compresses the fold change of highly expressed genes.
Recently, next-generation sequencing technologies have provided an alternative approach called RNA-Seq, which appears to be a more precise characterization of RNA transcripts (84, 85). The RNA-Seq procedure starts with the construction of a cDNA library from a population of RNA. Unlike smaller RNAs, long RNA molecules such as mRNAs first need to be fragmented into smaller pieces (by hydrolysis or nebulization) to be compatible with the sequencing technology. Alternatively, the reverse transcribed cDNA can be fragmented by DNAse I treatment or sonication. Direct single-molecule RNA sequencing is also possible without prior conversion of RNA to cDNA (86).
By using the cDNA library that has been constructed, short sequence reads (30–400 bp) are obtained by high-throughput sequencing of single molecules from one end (single-end sequencing) or both ends (pair-end sequencing). The resulting sequence reads are either mapped to a reference genome or assembled de novo without a priori knowledge of the underlying genome. Each read may correspond to a known exon, a splice variant, or a new candidate gene. Reads are also counted to assess the level of gene expression based on the principle that the number of sequence reads that map to a particular transcript is proportional to its expression level.
Currently, among the existing technologies, the most commonly used platforms for RNA-Seq are the Genome Analyzer (Illumina), SOLiD (ABI), and 454 Sequencing (Life Technologies) for sequencing of DNA and DRS (Helicos) for direct RNA sequencing.
Typical RNA-Seq protocols require at least 1 μg of total RNA. However, this amount of starting material may not be available when studying small tissues such as oocytes. Recently, Antoniou and Taft (87) provided a protocol, specifically for Illumina, to prepare sequencing libraries from mouse oocytes and suggested that this protocol can also be used for follicular cells. Unfortunately, to date there are no publications applying the RNA-Seq approach for analysis of follicular cells in the context of assisted reproduction. While RNA-seq has not yet been applied to the analysis of cumulus/granulosa cell transcriptome, we believe that its use would increase accuracy of testing and may help resolve the inconsistencies between clinical studies discussed in the following sections.
Potential promises and pitfalls of RNA-Seq analysis
An important concern about RNA-Seq is the depth of sequencing since transcriptome coverage strongly depends on the sequencing depth. A small amount of sequencing data would be sufficient for the highly expressed genes where more reads are required for accurate quantification of lower level expressions. Obviously, the cost per sample and the size of the resulting data increase as the sequencing depth increases. The raw data obtained from RNA-Seq experiments are already too large (approximately 20–30 MB sequencing file per sample), requiring parallel processing of high-performance computers for alignment of the reads to an existing genome or assembling the reads into a new genome. Despite these challenges, RNA-Seq would detect tissue (cumulus cell) specific and currently unknown splice variants and polymorphisms.
Reproducibility is also a critical aspect of RNA-Seq experiments. A recent study on the analysis of technical reproducibility of RNA-Seq reported that sequencing data are highly reproducible, with few systematic differences among technical replicates (88). In the same study, the gene-expression differences between two tissues were estimated using the Illumina sequencing platform and compared with data obtained from Affymetrix arrays using the same samples. The results revealed that 81% of genes significantly differentially expressed from the array data were also detected in RNA-Seq. In another study, on the basis of the compatibility of the results obtained from both technologies and with the proven superiority of RNA-Seq, the authors concluded that microarrays remain useful and accurate tools for measuring expression levels and RNA-Seq complements and extend microarray measurements (89). Usually, RNA-Seq is the method of choice for transcript discovery and de novo genome annotation, whereas due to the lower costs and easier data analysis, microarrays are preferable in the experiments involving large number of samples from species with well-annotated genomes. A comparison of the major characteristics of qRT-PCR, microarray, and RNA-Seq is presented in Table 1.
TABLE 1.
Major characteristics of transcriptomics technologies: a comparative representation.
| Feature | qRT-PCR | Microarray | RNA-Seq |
|---|---|---|---|
| Technique | Primer annealing and strand extension | Hybridization | High-throughput sequencing |
| Genome-wide analysis | No | Yes | Yes |
| Reliance on genomic sequence | Yes | Yes | No |
| Throughput | Low | High | High |
| Resolution | Hundreds bp | A few tens of bp | Single base |
| Precision | High | Medium | High |
| Background noise | Low | High | Low |
| Required amount of starting RNA material | Low | High | Low |
| Cost per sample | Low | Medium | High |
| Bioinformatics requirement | Low | Medium | High |
ANALYSIS OF CUMULUS AND GRANULOSA CELLS AS POTENTIAL BIOMARKERS FOR OOCYTE AND EMBRYO QUALITY
Novel embryo assessment strategies take advantage of the rapid developing transcriptomic approaches to explore the gene-expression profile of cumulus and granulosa cells. Recent studies suggest that the transcript levels of the candidate genes in cumulus and granulosa cells are associated with oocyte maturation (40, 62, 63, 90), embryo competence (39, 40, 58–63, 67, 90), or pregnancy (59, 61, 62, 64, 66, 67, 90–92), or live birth outcome (61, 65). The studies are discussed in three groups: qRT-PCR analysis of cumulus cell markers of oocyte and embryo competence (Table 2), microarray analysis of cumulus cell markers of oocyte and embryo competence (Table 3), and granulosa cell markers of oocyte and embryo competence (Table 4).
TABLE 2.
Summary of studies evaluating cumulus cell markers of oocyte and embryo quality using RT-PCR and qRT-PCR.
| Study (ref.) | Outcome measure | Study population | RNA isolation method | RT priming | Endogenous standard | qRT-PCR platform | Genes studied | Results |
|---|---|---|---|---|---|---|---|---|
| McKenzie et al. (40) | Oocyte maturation, fertilization, day 3 embryo morphology | 8(Np), 108 (No) | Picopure RNA isolation kit (Arcturus) | NA | 18s rRNA | Taqman Assay | HAS2, COX2, GREM1, TNFAIP6, PTX3 | ↑ HAS2, ∝ oocyte maturation; ↑ COX2, GREM1 fertilization and day 3 embryo morphology; ↔ TNFAIP, PTX3 not detected |
| Cillo et al. (39) | Fertilization, day 3 embryo morphology | 45 (Np), 90 (No) | Trizol | Random hexamers | 28s rRNA | In this study, RT-PCR was used without real-time analysis | HAS2, GREM1, PTX3 | ↑ HAS2, GREM1 ∝ fertilization and day 3 embryo morphology, ↔ PTX3 |
| Feuerstein et al. (62) | PB formation, day 2 embryo morphology, day 5/6 good or optimal blastocyst development | 47 (Np), 149 (No) | Absolutely RNA Nanoprep kit (Stratagene) | Oligo(dT) and random hexamers | 18s rRNA RPL19 | SYBR green | STAR, COX2, AREG, SCD1, SCD5, CX43 | ↑ STAR, COX2, AREG, SCD1, SCD5 ∝ PB formation; ↓ STAR, COX2, AREG, SCD1, SCD5 ∝; day 5/6 good or optimal blastocyst development; ↔ CX43 |
| Anderson et al. (63) | Cumulus expansion, oocyte maturation, fertilization, day 2/3 embryo morphology, pregnancy (DET) | 75 (Np), 674 (No) | RNeasy micro kit (Qiagen) | Oligo(dT) | GAPDH | SYBR green | GREM1, BDNF, PTGS2, HAS2, TNFAIP6, PTX3 | ↓ BDNF, TNFAIP6, PTX3 ∝ cumulus expansion; ↑ PTGS2 ∝ oocyte maturation; ↓ BDNF ∝ fertilization; ↑ GREM1, ↓ BDNF ∝ day2/3 embryo morphology |
| Gebhardt et al. (65) | Live birth (SET), birth weight | 38 (Np), 38 (No) | RNAqueous micro kit (Ambion) | Random hexamers | GAPDH | SYBR green | ALDOA, LDHA, PFKP, PKM2, AHR, GREM1, PTGS2, STS, HAS2, PTX3, TNFA P6, VCAN | ↑ VCAN, PTGS2 ∝ live birth; ↑ PFKP ∝ birth weight |
| Wathlet et al. (90) | Oocyte maturation, day 3/5 embryo morphology, pregnancy (SET) | 162 (Np), 162 (No) | RNeasy micro kit (Qiagen) | Oligo(dT) and random hexamers; in iScript cDNA synthesis kit (Bio-Rad) | B2M and UBC | SYBR green; Taqman assay (for GREM1); Predictive model development based on eight genes |
SDC4, ALCAM, PTGS2, VCAN, GREM1, TRPM7, CALM2, ITPKA | ↑ PTGS2, ↓VCAN ∝ oocyte maturation; models based on ↑ TRPM7 and ITPKA ∝ day 3 embryo morphology; ↑ SDC4, ITPKA ∝ day 5 embryo morphology; ↑ SDC4, VCAN ∝ pregnancy |
| Wathlet etal. (61) | Day 3/5 embryo morphology, pregnancy, live birth (SET) | 33 (Np), 99 (No) | RNeasy micro kit (Qiagen) | Oligo(dT) and random hexamers; in iScript cDNA synthesis kit (Bio-Rad) | B2M & UBC | SYBR green; Taqman assay (for CAMK1D) Predictive model development based on 11 genes |
TRPM7, SDC4, STC1, STC2, CYP11A1, HSD3B1, CAMK1D, PTHLH, ITPKA, EFNB2, VCAN | Models based on ↑ TRPM7 ∝ day 3 embryo morphology; ↑ CYP11A1 ∝ day 5 embryo morphology; ↑ EFNB2, CAMK1D, STC1 ∝ pregnancy; ↑ EFNB2, CAMK1D, STC2 ∝ live birth |
Note: All studies were of retrospective cohort design, and all collected cumulus cells from individual oocytes. All except one (Cillo et al.) used qRT-PCR. The studies are presented in chronological order in the table. DET = double ET; SET = single ET; Np = number of patients; No = number of oocytes; NA = not available; PB = polar body. Symbols ↑, ↓, ↔, ∝ stand for higher expression level, lower expression level, no difference in expression levels, and associated with, respectively.
TABLE 3.
Summary of selected microarray studies evaluating cumulus cell markers of oocyte and embryo quality.
| Study (ref.) | Microarray platform (no. of arrays performed) | Outcome measure | Study population | RNA isolation method | Endogenous standard | Individual versus pooled samples | Results |
|---|---|---|---|---|---|---|---|
| Zhang et al. (58) | Affymetrix (6) | Fertilization, embryo morphology | 20 (Np), 123 (No) | Trizol (Invitrogen) | 18s rRNA | Pooled | *160 genes were differentially expressed; ** ↑ PTX3 ∝ fertilization and embryo quality |
| Assou et al. (59) | Affymetrix (50) | Day 3 embryo morphology, pregnancy | 30 (Np), 50 (No) | RNeasy Micro kit (Qiagen) | PGK1 | Individual | *630 genes were differentially expressed; ** ↑ BCL2L11, PCK1 & ↓ NF1B; ∝ pregnancy |
| Van Montfoort et al. (60) | Affymetrix (16) | Early cleavage | 6 (Np), 16 + 24 (No), arrays validated in additional 24 samples | Trizol (Invitrogen) | SRP14 and RHOA | Individual | *611 genes were differentially expressed; ** ↓ CCND2, CXCR4, GPX3, CTNND1 DHCR7, DVL3, HSPB1, TRIM28; ∝ early cleavage |
| Assidi et al. (66) | Commercial and custom-made microarrays (8) | Pregnancy | 8 (Np), 14 (No) | Picopure RNA isolation kit (Arcturus) | ACTB and PPIA | Pooled | *491 genes were differentially expressed; ** ↑ DOO8, HIST1H4C, UBQLN1, CALM1, NRP1, PSMD6 & ↓ TOM1 ∝ pregnancy |
| Feuerstein et al. (67) | Whole Human Genome Oligo Microarray (Agilent technologies) (96) | Day 5 embryo morphology, pregnancy | 106 (Np), 197 (No) | RNeasy Micro kit (Qiagen) | RPL19 | Individual | *308 genes were differentially expressed; ** ↑ RGS2 ∝ blastocyst development and pregnancy |
Note: All studies were of retrospective cohort design. The studies are presented in chronological order in the table. No = number of oocytes; Np = number of patients. Symbols ↑, ↓, ↔, ∝ stand for higher expression level, lower expression level, no difference in expression levels, and associated with, respectively.
Results of microarray experiments.
qRT-PCR validation results.
TABLE 4.
Summary of selected studies evaluating granulosa cell markers of oocyte and embryo quality.
| Study (ref.) | Technique | Outcome measure | Study population | RNA isolation method | Endogenous standard | Individual versus pooled samples | Results |
|---|---|---|---|---|---|---|---|
| Hamel et al. (91) | Custom-made and Affymetrix (2×2) | Pregnancy versus embryo developmental failure | 40 (Np), 60 (No) | Trizol (Invitrogen) | ACTB and GAPDH | Pooled |
*115 genes were differentially expressed ** ↑ FDX1, CYP19A1, CDC42, SERPINE2, 3βHSD ∝ pregnancy |
| Hamel et al. (92) | qRT-PCR | Pregnancy versus embryo developmental failure | 34 (Np), 62 (No) | Trizol (Invitrogen) | ACTB, GAPDH and PPIA | Individual | ↑ PGK1, RGS2 ∝ pregnancy |
| Hamel et al. (64) | Custom-made microarray | Pregnancy | 18 (Np), 18 (No) | Trizol (Invitrogen) | ACTB, GAPDH and PPIA | Pooled |
*115 genes were differentially expressed ** ↑ UGP2, PHLDA1 ∝ pregnancy |
Note: All studies were of retrospective cohort design. The studies are presented in chronological order in the table. No = number of oocytes; Np = number of patients. Symbols ↑, ↓, ↔, ∝ stand for higher expression level, lower expression level, no difference in expression levels, and associated with, respectively.
Results of microarray experiments.
qRT-PCR validation results.
qRT-PCR Analysis of Cumulus Cell Markers of Oocyte and Embryo Competence
Human chorionic gonadotropin (hCG) or luteinizing hormone (LH) induces the rapid and transient expression of epidermal growth factor (EGF)-like factors (amphiregulin [AREG], epiregulin [EREG], and betacellulin [BTC]) in mural granulosa and cumulus cells. AREG, EREG, and BTC each induce the expression of prostaglandin synthase-2 (PTGS2, also termed COX2), tumor necrosis factor alpha-induced protein (TNFAIP6), and HAS2, which are necessary for synthesis and stabilization of the extracellular matrix by cumulus cells, cumulus expansion, and subsequent ovulation. These genes’ products and others with similar function were assessed as potential bio-markers by the studies using qRT-PCR for cumulus/granulosa cell transcriptome analysis.
In the first study investigating the association between cumulus cell gene expression and in vitro embryo development, McKenzie et al. assessed the expression levels of HAS2, PTGS2, and GREM1 in 108 human oocytes in correlation to oocyte maturity, fertilization, and embryo quality (40). Using qRT-PCR, they assessed cumulus cells from individual oocytes and found that the expression of PTGS2 and HAS2 was 6-fold higher and that of GREM1 was 15-fold higher in cumulus cells surrounding oocytes that developed into high-grade embryos compared with the cumulus cells of oocytes that resulted in low-grade embryos on day 3. Subsequent studies confirmed the association between GREM1 expression and day 3 embryo development (39, 63). However, Anderson et al. (63) and Gebhardt et al. (65) failed to show a significant association between GREM1 expression and clinical pregnancy, while Wathlet et al. (90) reported GREM1 to be elevated in the cumulus cells of oocytes that resulted in a pregnancy in women treated with GnRH antagonist and recombinant FSH (rFSH) but not with GnRH agonist and highly purified menotropin (HP-hMG). Among the other candidate genes targeted by the initial studies using qRT-PCR, HAS2 or TNFAIP6 have not been consistently associated with embryo development or pregnancy (40, 63, 65) (Table 2).
Feuerstein et al. also used qRT-PCR and focused on target genes that are up-regulated in cumulus cells in response to the preovulatory LH surge that precedes cumulus expansion: steroidogenic acute regulatory protein (STAR), PTGS2, AREG, two Stearoyl-Coenzyme A Desaturases (SCD1 and SCD5), and Cx43 (62). Expression levels of all genes investigated, except Cx43, were increased after resumption of meiosis. Nuclear maturation was thus associated with increased expression of STAR, PTGS2, AREG, SCD1, and SCD5 by cumulus cells. When considering only cumulus associated with metaphase II oocytes, gene expression was independent of morphological status at day 2. In contrast, transcript levels were lower and distributed over a narrower range in cumulus-enclosing oocytes achieving blastocyst development at day 5/6 (62). Based on these results, the authors suggested that the expression levels of the genes investigated might vary in a precise chronological pattern throughout the embryo development.
More recently, Gebhardt et al. reported a significantly higher expression of Versican (VCAN) and PTGS2 in cumulus cells obtained from oocytes that yielded a live birth (65). The same study also identified a correlation between birth weight and expression levels of VCAN and the GREM1 and PKFP genes. It is noteworthy that the studies mentioned above (39, 40, 62, 63, 65) have assessed the expression of individual genes but did not attempt to generate predictive models based on the expression pattern of multiple genes.
In 2011, Wathlet et al. (90) reported the analysis of cumulus cells from intracytoplasmic sperm injection (ICSI) patients stimulated with GnRH antagonist and rFSH or GnRH agonist and HP-hMG. Based on their unpublished microarray results, cumulus cells were analyzed for the expression of eight genes (Syndecan 4 [SDC4], PTGS2, VCAN, activated leukocyte cell adhesion molecule [ALCAM], GREM1, transient receptor potential cation channel, subfamily M member 7 [TRPM7], Calmodulin 2 [CALM2], and inositol 1,4,5-trisphosphate 3-kinase A [ITPKA]) using qRT-PCR. The investigators analyzed their results in relation to the stimulation protocol and correlated within-patient variation in gene expression to oocyte maturity and developmental potential. Models predictive for day 3 or day 5 embryo development and pregnancy in single ET cycles were developed. Wathlet et al. found cumulus cells from mature oocytes to have higher PTGS2 and lower VCAN expression. All genes except VCAN had a positive correlation with day 3 or day 5 morphology and were used to develop predictive models for embryo or blastocyst development. Specific models were obtained for the two stimulation protocols. In both groups, better cleavage-stage embryo prediction relied on TRPM7 and ITPKA expression and pregnancy prediction relied on SDC4 and VCAN expression. The proposed model achieved a pregnancy prediction with a sensitivity of 70% and a specificity of 90%.
In a subsequent study, the same group assessed 11 genes and found no significant difference in the cumulus cell expression of VCAN among the pregnant versus nonpregnant groups, whereas EphrinB2 (EFNB2) and ITPKA were up-regulated and calcium/calmodulin-dependent protein kinase 1D (CAMK1D) showed the same trend in the pregnant group (61). Once again, the investigators developed models for pregnancy prediction. EFNB2, CAMK1D, and Stanniocalcin-2 (STC2) were used for the live-birth prediction model.
Two additional aspects of the studies from Wathlet et al. are noteworthy. First, in addition to generating predictive models based on transcript expression, they provided additional and stronger models combining transcript levels with other clinical parameters such as hormone levels or days of stimulation (61, 90). Second, to identify the appropriate gene for normalization, they assessed the expression of 13 primer pairs: ACTB, GAPDH, ubiquitin C (UBC), beta-2-microglobulin (B2M), tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein (YWHAZ), splicing factor 3a, subunit 1 (SF3A1), 18S, cytochrome c-1 (CYC1), eukaryotic translation initiation factor 4A2 (EIF4A2), succinate dehydrogenase complex, subunit A (SDHA), topoisomerase (DNA) I (TOP1), ATP synthase subunit beta (ATP5B), and AluJ. They found that B2M and UBC were stably expressed in human cumulus cells and used the geometric mean of the B2M and UBC expression as the endogenous normalization factor (90).
Microarray Analysis of Cumulus Cell Markers of Oocyte and Embryo Competence
In the first study applying microarray to investigate cumulus cell transcriptome in women undergoing IVF, Zhang et al. compared pooled cumulus cells from oocytes that failed to fertilize to those derived from oocytes that developed into an 8-cell embryo on day 3 and identified 160 genes differentially expressed between the two groups (58). Among the 160 genes, subsequent qRT-PCR quantification of PTX3 confirmed the association of PTX3 expression with oocyte development. However, this observation was contradictory to the results obtained by Cillo et al. where no difference was observed in PTX3 levels in the cumulus cells isolated from oocytes that resulted in high-quality embryos on day 3 compared with those from oocytes that did not fertilize or developed into poor-quality embryos (39). Similarly, using qRT-PCR, Anderson et al. (63) had not found an association between PTX3 and embryo development or pregnancy.
Van Montfoort et al. conducted a microarray experiment for comparison of gene expression in cumulus cells from eight oocytes resulting in early cleavage embryos and from eight oocytes resulting in non–early cleavage embryos (60). Obtained results revealed that a total of 611 genes were differentially expressed; these genes were involved in a number of biological processes including cell cycle, angiogenesis, and apoptosis. Among the identified genes, cyclin D2 (CCND2), chemokine (C-X-C motif) receptor 4 (CXCR4), glutathione peroxidase 3 (GPX3), catenin delta 1 (CTNND1), 7-dehydrocholesterol reductase (DHCR7), disheveled, dsh homolog 3 (DVL3), heat shock 27 kDa protein 1 (HSBP1), and tripartite motif containing 28 (TRIM28) were reported to be the most significant ones confirmed by subsequent qPCR analysis. The altered expressions of these genes were suggested as an indication of hypoxic conditions or delayed oocyte maturation in non–early cleavage embryos.
Subsequently, using pregnancy as the primary outcome measure in the study, Assou et al. presented a microarray-based comparison of gene-expression profiles in cumulus cells that resulted in the identification of 630 differentially expressed genes (59). The majority of these genes had higher expression levels in the pregnant group, suggesting that transcriptional activation in cumulus cells is essential for the developmental competence of embryos. The up-regulated expression of BCL-like protein 11 (BCL2L11) and phosphoenolpyruvate carboxykinase 1 (PCK1) and the down-regulated expression of nuclear factor IB (NFIB) in the pregnant group were further confirmed by qRT-PCR in that study (59).
It is noteworthy that the number of biological replicates performed in microarray studies has significant implications in determining the potential validity of the data. Increasing the number of biological replicates requires the performance of a higher number of microarrays. This in turn is associated with additional cost and makes the bioinformatics analysis more challenging. In microarray studies investigating cumulus cell biomarkers, a wide range of replicates were used (Table 3). Most recently, Feuerstein et al. (67) collected 197 individual cumulus cell samples from 106 patients undergoing the ICSI procedure and studied cumulus cell gene expression using 96 microarrays to determine the genes associated with oocyte maturation and subsequent blastocyst development. Microarrays were followed by a meta-analysis of the behavior of these genes in other data sets available in the Gene Expression Omnibus, which showed the consistency of this list of genes. Finally, eight genes were selected according to oocyte developmental competence from the 308 differentially expressed genes for further validation by qRT-PCR. Three of these eight selected genes were validated as potential biomarkers (perilipin 2 [PLIN2], regulator of G-protein signaling 2 [RGS2], and angiogenin [ANG]). Experimental parameters including interpatient variability were then assessed, and only the expression level of RGS2 was confirmed to be related to oocyte developmental competence as well as clinical pregnancy.
At the same time, but using a different approach, Fragouli et al. reported up-regulation of SPSB2 in the cumulus cells of oocytes that produced a healthy live birth (93). The main objective of that study was to assess the cumulus cells gene expression as a potential indicator of oocyte aneuploidy. The investigators analyzed the expression of 96 genes using TaqMan low-density array (TLDA) and observed down-regulation of splA/ryanodine receptor domain and SOCS box containing 2 (SPSB2) and TP5313 in cumulus cells from chromosomally abnormal oocytes. These results might suggest that transcriptome analyses of cumulus cells may also help in the identification of novel noninvasive markers of meiotic errors in the oocyte as an alternative to invasive genomic diagnostic tests.
Analysis of Granulosa Cell Markers of Oocyte and Embryo Competence
Three consecutive studies by Hamel et al. reported a correlation between pregnancy outcome and the transcriptome of mural granulosa cells (64, 91, 92). In the first study, the gene expression of granulosa cells has been analyzed using both a custom-made cDNA microarray and an Affymetrix GeneChip (91). The custom-made microarray contained granulosa/cumulus expressed sequence tags (ESTs) from subtracted libraries obtained from pooled samples representing pregnant versus nonpregnant patients. The samples were investigated in two groups: the follicular cells from oocytes that resulted in a positive pregnancy and the follicular cells from oocytes resulting in embryos that represent developmental failure. Among the 115 differentially expressed genes, altered expression of 3-beta-hydroxystreoid dehydrogenase (HSD3β1), ferrodoxin 1 (FDX1), serine (or cysteine) proteinase inhibitor clade E member 2 (SERPINE2), cytochrome P450 aromatase (CYP19A1), and cell division cycle 42 (CDC42) were significantly associated with pregnancy outcome. Using the findings of this initial study, the investigators subsequently identified phosphoglycerate kinase 1 (PGK1), RGS2, regulator of G-protein signaling 3 (RGS3), CDC42 (92), and UDP-glucose pyrophosphorylase 2 (UGP2), and pleckstrin homology-like domain, family A, member 1 (PHLDA1) (64), as potential follicular markers associated with embryo quality resulting in a successful pregnancy.
Methodological Framework for Implementation of Cumulus/Granulosa Cell Transcriptomics as an Embryo Assessment Methodology in the IVF Laboratory
While the application of cutting-edge transcriptomic and bioinformatic tools in the IVF laboratory is exciting, a number of questions remain to be answered before implementing cumulus/granulosa cell transcriptomics as a clinically useful tool. In this section, we will attempt to address some of these questions.
How will the analysis be done in clinical practice?
Microarray analysis requires dedicated equipment and technical staff that would be cost prohibitive in most embryology laboratories and may not produce results quickly enough to allow the information to be used clinically in the limited window of time acceptable for ET. It is therefore unlikely that cumulus/granulosa cells from individual oocytes will be subjected to microarray analysis to obtain a full transcriptomic profile. Instead, a limited number of transcripts identified in microarray studies as predictors of oocyte/embryo viability may be assessed by qRT-PCR. In that case, it will also be important to establish whether an absolute quantification would be necessary or the laboratory would just choose the embryo(s) associated with the highest or lowest expression. In our opinion, as a single gene product is unlikely to predict outcome, multiple genes would need to be used within an algorithm, necessitating absolute quantification.
Are there any concerns about the safety of the procedure?
While the collection of cumulus or granulosa cells is considered noninvasive, it may be associated with additional time for each oocyte outside the incubator. This approach may also result in unindicated removal of cumulus cells and performance of ICSI procedures. In addition, to maintain the identity of each cumulus/granulosa cell sample-embryo relationship, individual culture will be necessary, adding to workload and financial cost in the laboratory. These aspects will need to be studied and a risk-benefit analysis performed before clinical implementation.
What data should be produced before implementation of cumulus or granulosa cell transcriptomic analysis in clinical practice?
The clinical studies summarized above include hypothesis-driven (candidate gene analysis by qRT-PCR) and hypothesis-generating (global analysis by microarray) studies investigating the association between altered cumulus and/or granulosa cell mRNA expression and embryo development and viability. A key second step is to validate the identified predictive transcripts in blinded studies. This has been accomplished for some of the candidates identified by qRT-PCR (GREM1 [39, 40, 63]) and others identified by microarray (PGK1, RGS2, RGS3, CDC42; [92]). Finally, the transcripts or algorithms involving a group of transcripts will need to be tested in randomized clinical trials to determine whether their use alone or in combination with morphology improves IVF outcome parameters such as implantation rate, pregnancy rate, or live-birth rate.
SUMMARY AND CONCLUSIONS
Interactions between the oocyte and the surrounding somatic follicular cells are central to the normal development of the female gamete. Importantly, aberrations in oocyte gene expression have implications for cumulus and granulosa cell proliferation, differentiation, and function. Therefore, it is plausible to assess cumulus or granulosa cells to identify markers of oocyte and embryo viability. On the basis of these observations and assumptions, a number of investigators have studied cumulus and granulosa cell transcriptome to gain insight into the reproductive potential of oocytes and embryos.
Studies published to date demonstrate extensive use of qRT-PCR as a sensitive and efficient method for quantification of individual transcripts. On the other hand, use of microarrays for genome-wide expression profiling has opened a new perspective for researchers, promising identification of the transcriptomic signature of the viable embryos. However, variations in the design and methodology of the microarray experiments and technological limitations complicate the interpretation of the results of microarray studies. Although RNA-Seq technology has been proposed recently as an alternative to microarrays, it has not yet been applied to cumulus/granulosa cell transcriptomic analysis.
The overview of the published results presented in studies assessing cumulus and granulosa cells transcriptomics revealed limited consensus on the identified markers. This may be due to variations in laboratory environment, studied outcome measures, size of study populations, and utilized technologies. In addition, the treatment protocol used and the age of women studied may affect the transcriptomic profile (94). Moreover, the majority of the studies testing the diagnostic power of follicular cells’ gene expression are based on retrospective cohort designs, are limited in extensive performance evaluation, and suffer from lack of clinical validation of the identified markers. We are optimistic that a group of cumulus and/or granulosa cell transcripts will be validated as predictors of oocyte competence. Whether, the use of these markers in the setting of IVF will result in an improvement in pregnancy outcome will need to be tested using a randomized controlled trial design (95).
Acknowledgments
E.S. is supported by award no. R01HD059909 from the National Institutes of Health (NIH). The contents of this manuscript are solely the responsibility of the authors and do not necessarily represent the official view of NIH.
Footnotes
Discuss: You can discuss this article with its authors and with other ASRM members at http://fertstertforum.com/uyara-cumulus-granulosa-cells-biomarkers-embryo-quality/
A.U. has nothing to disclose. S.T. has nothing to disclose. E.S. has nothing to disclose.
REFERENCES
- 1.Report of the Meeting on the Prevention of Infertility at the Primary Health Care Level 12–16 December 1983. Geneva. 1984 [Google Scholar]
- 2.Bromer JG, Seli E. Assessment of embryo viability in assisted reproductive technology: shortcomings of current approaches and the emerging role of metabolomics. Curr Opin Obstet Gynecol. 2008;20:234–41. doi: 10.1097/GCO.0b013e3282fe723d. [DOI] [PubMed] [Google Scholar]
- 3.Seli E, Robert C, Sirard MA. OMICS in assisted reproduction: possibilities and pitfalls. Mol Hum Reprod. 2010;16:513–30. doi: 10.1093/molehr/gaq041. [DOI] [PubMed] [Google Scholar]
- 4.Gondos B, Bhiraleus P, Hobel CJ. Ultrastructural observations on germ cells in human fetal ovaries. Am J Obstet Gynecol. 1971;110:644–52. doi: 10.1016/0002-9378(71)90245-6. [DOI] [PubMed] [Google Scholar]
- 5.Fritz M, Speroff L. The ovary—embryology and development. 8th ed. Lippincott Williams & Wilkins; 2011. [Google Scholar]
- 6.McGee EA, Hsueh AJ. Initial and cyclic recruitment of ovarian follicles. Endocr Rev. 2000;21:200–14. doi: 10.1210/edrv.21.2.0394. [DOI] [PubMed] [Google Scholar]
- 7.Gougeon A, Chainy GB. Morphometric studies of small follicles in ovaries of women at different ages. J Reprod Fertil. 1987;81:433–42. doi: 10.1530/jrf.0.0810433. [DOI] [PubMed] [Google Scholar]
- 8.Soyal SM, Amleh A, Dean J. FIGalpha, a germ cell-specific transcription factor required for ovarian follicle formation. Development. 2000;127:4645–54. doi: 10.1242/dev.127.21.4645. [DOI] [PubMed] [Google Scholar]
- 9.Diaz FJ, Wigglesworth K, Eppig JJ. Oocytes determine cumulus cell lineage in mouse ovarian follicles. J Cell Sci. 2007;120:1330–40. doi: 10.1242/jcs.000968. [DOI] [PubMed] [Google Scholar]
- 10.Guzeloglu-Kayisli O, Lalioti MD, Aydiner F, Sasson I, Ilbay O, Sakkas D, et al. Embryonic poly(A)-binding protein (EPAB) is required for oocyte maturation and female fertility in mice. Biochem J. 2012;446:47–58. doi: 10.1042/BJ20120467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eppig JJ. Intercommunication between mammalian oocytes and companion somatic cells. Bioessays. 1991;13:569–74. doi: 10.1002/bies.950131105. [DOI] [PubMed] [Google Scholar]
- 12.Richards JS, Russell DL, Ochsner S, Espey LL. Ovulation: new dimensions and new regulators of the inflammatory-like response. Annu Rev Physiol. 2002;64:69–92. doi: 10.1146/annurev.physiol.64.081501.131029. [DOI] [PubMed] [Google Scholar]
- 13.Hirshfield AN. Relationship between the supply of primordial follicles and the onset of follicular growth in rats. Biol Reprod. 1994;50:421–8. doi: 10.1095/biolreprod50.2.421. [DOI] [PubMed] [Google Scholar]
- 14.Suzumori N, Yan C, Matzuk MM, Rajkovic A. Nobox is a homeobox-encoding gene preferentially expressed in primordial and growing oocytes. Mech Dev. 2002;111:137–41. doi: 10.1016/s0925-4773(01)00620-7. [DOI] [PubMed] [Google Scholar]
- 15.Rajkovic A, Pangas SA, Ballow D, Suzumori N, Matzuk MM. NOBOX deficiency disrupts early folliculogenesis and oocyte-specific gene expression. Science. 2004;305:1157–9. doi: 10.1126/science.1099755. [DOI] [PubMed] [Google Scholar]
- 16.McPherron AC, Lee SJ. GDF-3 and GDF-9: two new members of the transforming growth factor-beta superfamily containing a novel pattern of cysteines. J Biol Chem. 1993;268:3444–9. [PubMed] [Google Scholar]
- 17.McGrath SA, Esquela AF, Lee SJ. Oocyte-specific expression of growth/differentiation factor-9. Mol Endocrinol. 1995;9:131–6. doi: 10.1210/mend.9.1.7760846. [DOI] [PubMed] [Google Scholar]
- 18.Dong J, Albertini DF, Nishimori K, Kumar TR, Lu N, Matzuk MM. Growth differentiation factor-9 is required during early ovarian folliculogenesis. Nature. 1996;383:531–5. doi: 10.1038/383531a0. [DOI] [PubMed] [Google Scholar]
- 19.Dube JL, Wang P, Elvin J, Lyons KM, Celeste AJ, Matzuk MM. The bone morphogenetic protein 15 gene is X-linked and expressed in oocytes. Mol Endocrinol. 1998;12:1809–17. doi: 10.1210/mend.12.12.0206. [DOI] [PubMed] [Google Scholar]
- 20.Yan C, Wang P, DeMayo J, DeMayo FJ, Elvin JA, Carino C, et al. Synergistic roles of bone morphogenetic protein 15 and growth differentiation factor 9 in ovarian function. Mol Endocrinol. 2001;15:854–66. doi: 10.1210/mend.15.6.0662. [DOI] [PubMed] [Google Scholar]
- 21.Radford HE, Meijer HA, de Moor CH. Translational control by cytoplasmic polyadenylation in Xenopus oocytes. Biochim Biophys Acta. 2008;1779:217–29. doi: 10.1016/j.bbagrm.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seli E, Lalioti MD, Flaherty SM, Sakkas D, Terzi N, Steitz JA. An embryonic poly(A)-binding protein (ePAB) is expressed in mouse oocytes and early preimplantation embryos. Proc Natl Acad Sci U S A. 2005;102:367–72. doi: 10.1073/pnas.0408378102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guzeloglu-Kayisli O, Pauli S, Demir H, Lalioti MD, Sakkas D, Seli E. Identification and characterization of human embryonic poly(A) binding protein (EPAB). Mol Hum Reprod. 2008;14:581–8. doi: 10.1093/molehr/gan047. [DOI] [PubMed] [Google Scholar]
- 24.Rankin TL, O'Brien M, Lee E, Wigglesworth K, Eppig J, Dean J. Defective zonae pellucidae in Zp2-null mice disrupt folliculogenesis, fertility and development. Development. 2001;128:1119–26. doi: 10.1242/dev.128.7.1119. [DOI] [PubMed] [Google Scholar]
- 25.Pangas SA, Choi Y, Ballow DJ, Zhao Y, Westphal H, Matzuk MM, et al. Oogenesis requires germ cell-specific transcriptional regulators Sohlh1 and Lhx8. Proc Natl Acad Sci U S A. 2006;103:8090–5. doi: 10.1073/pnas.0601083103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Choi Y, Yuan D, Rajkovic A. Germ cell-specific transcriptional regulator sohlh2 is essential for early mouse folliculogenesis and oocyte-specific gene expression. Biol Reprod. 2008;79:1176–82. doi: 10.1095/biolreprod.108.071217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heller DT, Schultz RM. Ribonucleoside metabolism by mouse oocytes: metabolic cooperativity between the fully grown oocyte and cumulus cells. J Exp Zool. 1980;214:355–64. doi: 10.1002/jez.1402140314. [DOI] [PubMed] [Google Scholar]
- 28.Brower PT, Schultz RM. Intercellular communication between granulosa cells and mouse oocytes: existence and possible nutritional role during oocyte growth. Dev Biol. 1982;90:144–53. doi: 10.1016/0012-1606(82)90219-6. [DOI] [PubMed] [Google Scholar]
- 29.Tanghe S, Van Soom A, Nauwynck H, Coryn M, de Kruif A. Minireview: functions of the cumulus oophorus during oocyte maturation, ovulation, and fertilization. Mol Reprod Dev. 2002;61:414–24. doi: 10.1002/mrd.10102. [DOI] [PubMed] [Google Scholar]
- 30.Hasegawa J, Yanaihara A, Iwasaki S, Mitsukawa K, Negishi M, Okai T. Reduction of connexin 43 in human cumulus cells yields good embryo competence during ICSI. J Assist Reprod Genet. 2007;24:463–6. doi: 10.1007/s10815-007-9155-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Assou S, Haouzi D, De Vos J, Hamamah S. Human cumulus cells as biomarkers for embryo and pregnancy outcomes. Mol Hum Reprod. 2010;16:531–8. doi: 10.1093/molehr/gaq032. [DOI] [PubMed] [Google Scholar]
- 32.Dekel N, Beers WH. Development of the rat oocyte in vitro: inhibition and induction of maturation in the presence or absence of the cumulus oophorus. Dev Biol. 1980;75:247–54. doi: 10.1016/0012-1606(80)90160-8. [DOI] [PubMed] [Google Scholar]
- 33.Larsen WJ, Wert SE, Brunner GD. A dramatic loss of cumulus cell gap junctions is correlated with germinal vesicle breakdown in rat oocytes. Dev Biol. 1986;113:517–21. doi: 10.1016/0012-1606(86)90187-9. [DOI] [PubMed] [Google Scholar]
- 34.Norris RP, Ratzan WJ, Freudzon M, Mehlmann LM, Krall J, Movsesian MA, et al. Cyclic GMP from the surrounding somatic cells regulates cyclic AMP and meiosis in the mouse oocyte. Development. 2009;136:1869–78. doi: 10.1242/dev.035238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hsieh M, Zamah AM, Conti M. Epidermal growth factor-like growth factors in the follicular fluid: role in oocyte development and maturation. Semin Reprod Med. 2009;27:52–61. doi: 10.1055/s-0028-1108010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Veeck LL. An atlas of human gametes and conceptuses: an illustrated reference of assisted reproductive technology. Parthenon Publishing Group; New York: 1999. [Google Scholar]
- 37.Elvin JA, Clark AT, Wang P, Wolfman NM, Matzuk MM. Paracrine actions of growth differentiation factor-9 in the mammalian ovary. Mol Endocrinol. 1999;13:1035–48. doi: 10.1210/mend.13.6.0310. [DOI] [PubMed] [Google Scholar]
- 38.Pangas SA, Matzuk MM. The art and artifact of GDF9 activity: cumulus expansion and the cumulus expansion-enabling factor. Biol Reprod. 2005;73:582–5. doi: 10.1095/biolreprod.105.042127. [DOI] [PubMed] [Google Scholar]
- 39.Cillo F, Brevini TA, Antonini S, Paffoni A, Ragni G, Gandolfi F. Association between human oocyte developmental competence and expression levels of some cumulus genes. Reproduction. 2007;134:645–50. doi: 10.1530/REP-07-0182. [DOI] [PubMed] [Google Scholar]
- 40.McKenzie LJ, Pangas SA, Carson SA, Kovanci E, Cisneros P, Buster JE, et al. Human cumulus granulosa cell gene expression: a predictor of fertilization and embryo selection in women undergoing IVF. Hum Reprod. 2004;19:2869–74. doi: 10.1093/humrep/deh535. [DOI] [PubMed] [Google Scholar]
- 41.Opitz L, Salinas-Riester G, Grade M, Jung K, Jo P, Emons G, et al. Impact of RNA degradation on gene expression profiling. BMC Med Genomics. 2010;3:36. doi: 10.1186/1755-8794-3-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Manchester KL. Use of UV methods for measurement of protein and nucleic acid concentrations. Biotechniques. 1996;20:968–70. doi: 10.2144/96206bm05. [DOI] [PubMed] [Google Scholar]
- 43.Sambrook J, Russel DW. Molecular cloning: a laboratory manual. 3d ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbord, NY: 2001. [Google Scholar]
- 44.Auer H, Lyianarachchi S, Newsom D, Klisovic MI, Marcucci G, Kornacker K. Chipping away at the chip bias: RNA degradation in microarray analysis. Nat Genet. 2003;35:292–3. doi: 10.1038/ng1203-292. [DOI] [PubMed] [Google Scholar]
- 45.Denisov V, Strong W, Walder M, Gingrich J, Wintz H. Development and validation of RQI: an RNA quality indicator for the Experion automated electrophoresis system. Bio-Rad. 2008 [Google Scholar]
- 46.Schroeder A, Mueller O, Stocker S, Salowsky R, Leiber M, Gassmann M, et al. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol Biol. 2006;7:3. doi: 10.1186/1471-2199-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fleige S, Pfaffl MW. RNA integrity and the effect on the real-time qRT-PCR performance. Mol Aspects Med. 2006;27:126–39. doi: 10.1016/j.mam.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 48.Saiki RK, Gelfand DH, Stoffel S, Scharf SJ, Higuchi R, Horn GT, et al. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988;239:487–91. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- 49.Huang Q, Fu WL. Comparative analysis of the DNA staining efficiencies of different fluorescent dyes in preparative agarose gel electrophoresis. Clin Chem Lab Med. 2005;43:841–2. doi: 10.1515/CCLM.2005.141. [DOI] [PubMed] [Google Scholar]
- 50.Mullis K, Faloona F, Scharf S, Saiki R, Horn G, Erlich H. Specific enzymatic amplification of DNA in vitro: the polymerase chain reaction. Cold Spring Harb Symp Quant Biol. 1986;51(Pt 1):263–73. doi: 10.1101/sqb.1986.051.01.032. [DOI] [PubMed] [Google Scholar]
- 51.Mullis KB, Faloona FA. Specific synthesis of DNA in vitro via a polymerasecatalyzed chain reaction. Methods Enzymol. 1987;155:335–50. doi: 10.1016/0076-6879(87)55023-6. [DOI] [PubMed] [Google Scholar]
- 52.Stahlberg A, Hakansson J, Xian X, Semb H, Kubista M. Properties of the reverse transcription reaction in mRNA quantification. Clin Chem. 2004;50:509–15. doi: 10.1373/clinchem.2003.026161. [DOI] [PubMed] [Google Scholar]
- 53.Higuchi R, Dollinger G, Walsh PS, Griffith R. Simultaneous amplification and detection of specific DNA sequences. Biotechnology (N Y) 1992;10:413–7. doi: 10.1038/nbt0492-413. [DOI] [PubMed] [Google Scholar]
- 54.Higuchi R, Fockler C, Dollinger G, Watson R. Kinetic PCR analysis: real-time monitoring of DNA amplification reactions. Biotechnology (N Y) 1993;11:1026–30. doi: 10.1038/nbt0993-1026. [DOI] [PubMed] [Google Scholar]
- 55.VanGuilder HD, Vrana KE, Freeman WM. Twenty-five years of quantitative PCR for gene expression analysis. Biotechniques. 2008;44:619–26. doi: 10.2144/000112776. [DOI] [PubMed] [Google Scholar]
- 56.Logan J, Edwards K, Saunders N. Real-time PCR: current technology and applications. Caister Academic Press. 2009 [Google Scholar]
- 57.Lefever S, Hellemans J, Pattyn F, Przybylski DR, Taylor C, Geurts R, et al. RDML: structured language and reporting guidelines for real-time quantitative PCR data. Nucleic Acids Res. 2009;37:2065–9. doi: 10.1093/nar/gkp056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang X, Jafari N, Barnes RB, Confino E, Milad M, Kazer RR. Studies of gene expression in human cumulus cells indicate pentraxin 3 as a possible marker for oocyte quality. Fertil Steril. 2005;83(Suppl 1):1169–79. doi: 10.1016/j.fertnstert.2004.11.030. [DOI] [PubMed] [Google Scholar]
- 59.Assou S, Haouzi D, Mahmoud K, Aouacheria A, Guillemin Y, Pantesco V, et al. A non-invasive test for assessing embryo potential by gene expression profiles of human cumulus cells: a proof of concept study. Mol Hum Reprod. 2008;14:711–9. doi: 10.1093/molehr/gan067. [DOI] [PubMed] [Google Scholar]
- 60.van Montfoort AP, Geraedts JP, Dumoulin JC, Stassen AP, Evers JL, Ayoubi TA. Differential gene expression in cumulus cells as a prognostic indicator of embryo viability: a microarray analysis. Mol Hum Reprod. 2008;14:157–68. doi: 10.1093/molehr/gam088. [DOI] [PubMed] [Google Scholar]
- 61.Wathlet S, Adriaenssens T, Segers I, Verheyen G, Janssens R, Coucke W, et al. New candidate genes to predict pregnancy outcome in single embryo transfer cycles when using cumulus cell gene expression. Fertil Steril. 2012;98:432–9. e4. doi: 10.1016/j.fertnstert.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 62.Feuerstein P, Cadoret V, Dalbies-Tran R, Guerif F, Bidault R, Royere D. Gene expression in human cumulus cells: one approach to oocyte competence. Hum Reprod. 2007;22:3069–77. doi: 10.1093/humrep/dem336. [DOI] [PubMed] [Google Scholar]
- 63.Anderson RA, Sciorio R, Kinnell H, Bayne RA, Thong KJ, de Sousa PA, et al. Cumulus gene expression as a predictor of human oocyte fertilisation, embryo development and competence to establish a pregnancy. Reproduction. 2009;138:629–37. doi: 10.1530/REP-09-0144. [DOI] [PubMed] [Google Scholar]
- 64.Hamel M, Dufort I, Robert C, Leveille MC, Leader A, Sirard MA. Identification of follicular marker genes as pregnancy predictors for human IVF: new evidence for the involvement of luteinization process. Mol Hum Reprod. 2010;16:548–56. doi: 10.1093/molehr/gaq051. [DOI] [PubMed] [Google Scholar]
- 65.Gebhardt KM, Feil DK, Dunning KR, Lane M, Russell DL. Human cumulus cell gene expression as a biomarker of pregnancy outcome after single embryo transfer. Fertil Steril. 2011;96:47–52. e2. doi: 10.1016/j.fertnstert.2011.04.033. [DOI] [PubMed] [Google Scholar]
- 66.Assidi M, Montag M, Van der Ven K, Sirard MA. Biomarkers of human oocyte developmental competence expressed in cumulus cells before ICSI: a preliminary study. J Assist Reprod Genet. 2011;28:173–88. doi: 10.1007/s10815-010-9491-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Feuerstein P, Puard V, Chevalier C, Teusan R, Cadoret V, Guerif F, et al. Genomic assessment of human cumulus cell marker genes as predictors of oocyte developmental competence: impact of various experimental factors. PLoS One. 2012;7:e40449. doi: 10.1371/journal.pone.0040449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Radonic A, Thulke S, Mackay IM, Landt O, Siegert W, Nitsche A. Guideline to reference gene selection for quantitative real-time PCR. Biochem Biophys Res Commun. 2004;313:856–62. doi: 10.1016/j.bbrc.2003.11.177. [DOI] [PubMed] [Google Scholar]
- 69.Andersen CL, Jensen JL, Orntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–50. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- 70.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Osorio YFJ, Prina E, Lang T, Milon G, Davory C, Coppee JY, et al. AffyGCQC: a web-based interface to detect outlying genechips with extreme studentized deviate tests. J Bioinform Comput Biol. 2008;6:317–34. doi: 10.1142/s0219720008003400. [DOI] [PubMed] [Google Scholar]
- 72.Shi L, Reid LH, Jones WD, Shippy R, Warrington JA, Baker SC, et al. The MicroArray Quality Control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat Biotechnol. 2006;24:1151–61. doi: 10.1038/nbt1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–64. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 74.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–93. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 75.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hannah MA, Redestig H, Leisse A, Willmitzer L. Global mRNA changes in microarray experiments. Nat Biotechnol. 2008;26:741–2. doi: 10.1038/nbt0708-741. [DOI] [PubMed] [Google Scholar]
- 77.Dudoit S, Shaffer JP, Boldrick JC. Multiple hypothesis testing in microarray experiments. Stat Sci. 2003;18:71–103. [Google Scholar]
- 78.Storey JD. A direct approach to false discovery rates. J Roy Stat Soc Ser B Stat Method. 2002;64:479–98. [Google Scholar]
- 79.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100:9440–5. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Werner T. Bioinformatics applications for pathway analysis of microarray data. Curr Op Biotech. 2008;19:50–4. doi: 10.1016/j.copbio.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 81.Dennis G, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, et al. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 2003:4. [PubMed] [Google Scholar]
- 82.Chen JJ, Hsueh HM, Delongchamp RR, Lin CJ, Tsai CA. Reproducibility of microarray data: a further analysis of microarray quality control (MAQC) data. BMC Bioinforma. 2007;8:412. doi: 10.1186/1471-2105-8-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ioannidis JP, Allison DB, Ball CA, Coulibaly I, Cui X, Culhane AC, et al. Repeatability of published microarray gene expression analyses. Nat Genet. 2009;41:149–55. doi: 10.1038/ng.295. [DOI] [PubMed] [Google Scholar]
- 84.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–8. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 85.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ozsolak F, Platt AR, Jones DR, Reifenberger JG, Sass LE, McInerney P, et al. Direct RNA sequencing. Nature. 2009;461:814–8. doi: 10.1038/nature08390. [DOI] [PubMed] [Google Scholar]
- 87.Antoniou E, Taft R. Gene expression in mouse oocytes by RNA-Seq. Methods Mol Biol. 2012;825:237–51. doi: 10.1007/978-1-61779-436-0_18. [DOI] [PubMed] [Google Scholar]
- 88.Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18:1509–17. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Malone JH, Oliver B. Microarrays, deep sequencing and the true measure of the transcriptome. BMC Biol. 2011;9:34. doi: 10.1186/1741-7007-9-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wathlet S, Adriaenssens T, Segers I, Verheyen G, Van de Velde H, Coucke W, et al. Cumulus cell gene expression predicts better cleavage-stage embryo or blastocyst development and pregnancy for ICSI patients. Hum Reprod. 2011;26:1035–51. doi: 10.1093/humrep/der036. [DOI] [PubMed] [Google Scholar]
- 91.Hamel M, Dufort I, Robert C, Gravel C, Leveille MC, Leader A, et al. Identification of differentially expressed markers in human follicular cells associated with competent oocytes. Hum Reprod. 2008;23:1118–27. doi: 10.1093/humrep/den048. [DOI] [PubMed] [Google Scholar]
- 92.Hamel M, Dufort I, Robert C, Leveille MC, Leader A, Sirard MA. Genomic assessment of follicular marker genes as pregnancy predictors for human IVF. Mol Hum Reprod. 2010;16:87–96. doi: 10.1093/molehr/gap079. [DOI] [PubMed] [Google Scholar]
- 93.Fragouli E, Wells D, Iager AE, Kayisli UA, Patrizio P. Alteration of gene expression in human cumulus cells as a potential indicator of oocyte aneuploidy. Hum Reprod. 2012;27:2559–68. doi: 10.1093/humrep/des170. [DOI] [PubMed] [Google Scholar]
- 94.Adriaenssens T, Wathlet S, Segers I, Verheyen G, De Vos A, Van der Elst J, et al. Cumulus cell gene expression is associated with oocyte developmental quality and influenced by patient and treatment characteristics. Hum Reprod. 2010;25:1259–70. doi: 10.1093/humrep/deq049. [DOI] [PubMed] [Google Scholar]
- 95.Uyar A, Seli E. Embryo assessment strategies and their validation for clinical use: a critical analysis of methodology. Curr Opin Obstet Gynecol. 2012;24:141–50. doi: 10.1097/GCO.0b013e328352cd17. [DOI] [PubMed] [Google Scholar]