Abstract
We tested the hypothesis that an altered community of gut microbes is associated with risk of colorectal cancer (CRC) in a study of 47 CRC case subjects and 94 control subjects. 16S rRNA genes in fecal bacterial DNA were amplified by universal primers, sequenced by 454 FLX technology, and aligned for taxonomic classification to microbial genomes using the QIIME pipeline. Taxonomic differences were confirmed with quantitative polymerase chain reaction and adjusted for false discovery rate. All statistical tests were two-sided. From 794217 16S rRNA gene sequences, we found that CRC case subjects had decreased overall microbial community diversity (P = .02). In taxonomy-based analyses, lower relative abundance of Clostridia (68.6% vs 77.8%) and increased carriage of Fusobacterium (multivariable odds ratio [OR] = 4.11; 95% confidence interval [CI] = 1.62 to 10.47) and Porphyromonas (OR = 5.17; 95% CI = 1.75 to 15.25) were found in case subjects compared with control subjects. Because of the potentially modifiable nature of the gut bacteria, our findings may have implications for CRC prevention.
The human gut hosts a diverse community of bacteria that play key roles in modulating host metabolism and immunity (1) and in the digestion and conversion of dietary constituents into active forms (2). Although a role for this gut microbiota in colorectal cancer (CRC) in humans is suspected (3–6), particularly from comparisons of CRC tumor and adjacent normal tissue (7,8), systematic epidemiologic comparisons between CRC patients and control subjects, considering comprehensive confounders and multiple comparisons, are lacking. From stool samples, we comprehensively surveyed the distal gut microbiota by 16S rRNA gene sequencing and compared the fecal microbial profiles between CRC case subjects and matched control subjects.
We used specimens and data from a case–control study that tested whether fecal mutagens were associated with CRC (9,10). Briefly, case subjects with newly diagnosed, histologically confirmed adenocarcinoma of the colon or rectum were recruited before initiation of treatment during the period from 1985 to 1989 at three Washington, DC, area hospitals. Control subjects were recruited from contemporaneous patients awaiting elective surgery for nononcologic, nongastrointestinal conditions at these hospitals. Before hospitalization and treatment, participants completed written informed consent and diet and demographic questionnaires and provided 2-day fecal samples that were freeze-dried. The lyophilates were pooled, mixed, and stored at −40°C. Among 69 case subjects and 114 control subject, we included for study 47 colorectal cancer case subjects and 94 control subjects for whom at least 100mg of lyophilized feces was available. Case and control subjects were frequency matched by sex and body mass index (Supplementary Table 1, available online). One subject in the CRC case subject group used antibiotics within the past year; results remained unchanged after exclusion of this subject. This study was approved by the National Cancer Institute and the New York Univeristy Institutional Review Board.
We extracted DNA from fecal samples using the Mobio PowerSoil DNA Isolation Kit (Carlsbad, CA) with bead-beating. As we reported previously (11),16S rRNA amplicons covering variable regions V3 to V4 were generated using primers (347F-5′GGAGGCAGCAGTRRGGAAT′-3′ and 803R 5′-CTACCRGGGTATCTAATCC-3′) incorporating Roche 454 FLX Titanium adapters (Branford, CT) and a sample barcode sequence (12). Amplicons were sequenced with the 454 Roche FLX Titanium pyrosequencing system following the manufacturer’s specifications. Laboratory personnel were blinded to case–control status.
Multiplexed, barcorded sequencing data were deconvoluted. Poor-quality sequences were filtered based on sequence less than 200 or more than 600 base pairs, missing or mean quality score less than 25, or mismatched barcode and primer sequences. Chimeric sequences were removed with ChimeraSlayer (13). Filtered sequences were binned into operational taxonomic units with 97% identity and aligned to fully-sequenced microbial genomes (IMG/GG GreenGenes) using the QIIME pipeline (14). Blinded quality control specimens in all sequencing batches (38 aliquots from 9 unmatched parent study control subjects) had good reproducibility. Intraclass correlation coefficients were 0.84 for Shannon diversity index, and 0.43 to 0.59 for relative abundances of major phyla (Supplementary Table 2, available online). To confirm sequencing associations, we performed quantitative polymerase chain reaction for genera Fusobacterium and Porphyromonas with the SYBR Green method (15) using genus-specific primer sets (16,17).
Rarefaction curves were estimated by bootstrapping of 500 random samples at 500 sequence increments. Alpha diversity (Shannon’s diversity and evenness indices) differences between case and control subjects were compared with t tests with Monte Carlo permutations using compare_alpha_diversity.py, a built-in function in the QIIME pipeline (14). Carriage (presence or absence; ie, prevalence) of specific taxa was compared by χ2 analysis, and relative abundances were compared using the nonparametric Wilcoxon test. Odds ratios (ORs) were calculated for taxa, based on logistic regression, adjusting for age and, additionally, for sex, body mass index, race, smoking, and sequencing batch. We report nominal P values and highlight associations that meet a false discovery rate (FDR) adjusted P less than or equal to .05 by the Benjamini and Hochberg method (18). All statistical tests were two-sided, and a P value of less than .05 was considered statistically significant.
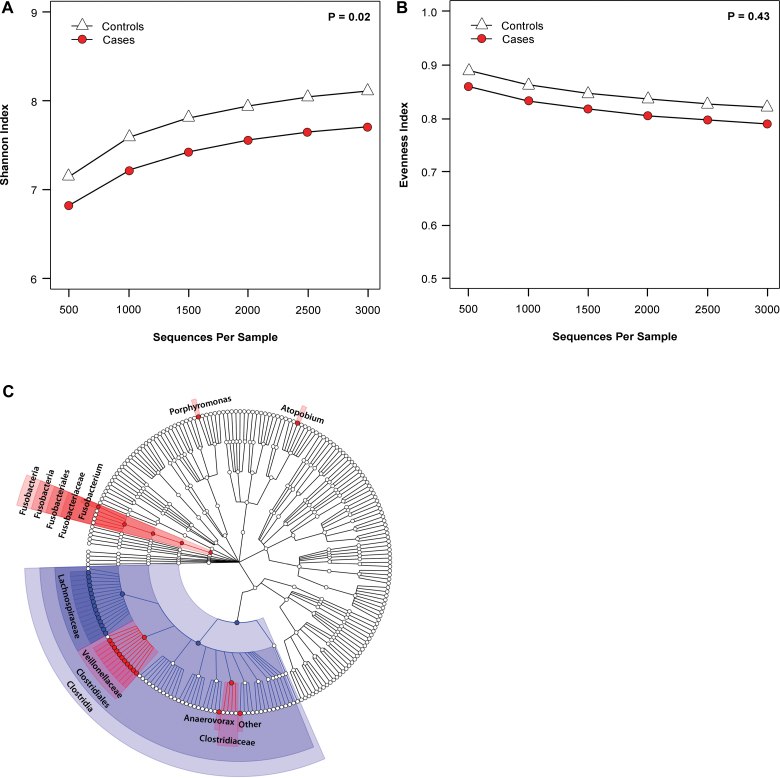
From the 141 fecal study samples (n = 47 CRC case subjects and 94 control subjects), we obtained 794 217 16S rRNA filtered gene sequences (mean ± standard deviation = 4919±2942 reads per sample in control subjects and 4863±2784 per sample in case subjects; P = .91). We assessed sample gut microbial community structure by diversity (ie, how many different taxa are present) and evenness (ie, how evenly distributed are the taxa in a sample) and found that CRC case subjects had decreased community diversity (P = .02) (Figure 1A) but did not differ from control subjects on community evenness (P = .43) (Figure 1B).
Figure 1.
Human gut microbiome in relation to colorectal cancer case-control status. A) Shannon diversity index in 47 colorectal cancer case subjects and 94 control subjects. B) Evenness index in 47 colorectal cancer case subjects and 94 control subjects. Rarefaction curves were estimated by bootstrapping of 500 random samples at 500 sequence increments. Alpha diversity (Shannon’s diversity and evenness indices) differences between case and control subjects were compared with t tests with Monte Carlo permutations using compare_alpha_diversity.py, a built-in function in the QIIME pipeline. C) Cladogram representation of gut microbiome taxa associated with colorectal cancer. Red indicates taxa enriched in colorectal cancer case subjects, and blue indicates taxa enriched in control subjects. Only taxa with nominal P less than .05 based on χ2 test (dichotomized) or Wilcoxon test (continuous) are labeled. The tests were two-sided. Figure was constructed using data presented in in Table 1.
We compared case and control subjects for presence and relative abundance of taxa. Case subjects tended to have enrichment of phylum Bacteroidetes (16.2% vs 9.9% relative abundance for case and control subjects, respectively) and depletion of Firmicutes (74.0% vs 80.3% for case and control subjects, respectively) (Supplementary Figure 1, available online). Within Firmicutes, the relative depletion was most prominent for the class Clostridia (68.6% vs 77.8%; P = .005; FDR-adjusted P ≤ .05), including Coprococcus and other taxa in the family Lachnospiraceae (Table 1; Figure 1C). Gram-positive Clostridia, especially Coprococcus, efficiently ferment dietary fiber and other complex carbohydrates to butyrate, a major colonic metabolite that may inhibit colonic inflammation and carcinogenesis (2,19). Consistent with our result, Clostridia have also been reported to be less abundant in colon tumors than in adjacent normal tissue (7).
Table 1.
Prevalence and relative abundance of selected fecal microbial taxa in 47 colorectal cancer case subjects and 94 control subjects
| Taxa (Phylum; class; order; family; genus) | % positive (carriage)* | Abundance %† | ||||||
|---|---|---|---|---|---|---|---|---|
| Case | Control | P‡ | Age-adjusted OR (95% CI) | Multivariable OR (95% CI)# | Case | Control | P§ | |
| Firmicutes;Clostridia (class) | 100.0 | 100.0 | NA | NA | NA | 68.6 | 77.8 | .005|| |
| Firmicutes;Clostridia;Clostridiales (order) | 100.0 | 100.0 | NA | NA | NA | 68.5 | 77.6 | .006 |
| Firmicutes;Clostridia;Clostridiales; Lachnospiraceae;Ruminococcus | 89.4 | 97.9 | .02 | 0.18 (0.032 to 0.95) | 0.14 (0.024 to 0.85) | 1.5 | 1.5 | .49 |
| Firmicutes;Clostridia;Clostridiales; Lachnospiraceae;Coprococcus | 93.6 | 97.9 | .20 | 0.34 (0.054 to 2.14) | 0.33 (0.055 to 2.42) | 1.7 | 3.7 | .002|| |
| Firmicutes;Clostridia;Clostridiales; Lachnospiraceae;Other | 100.0 | 98.9 | .48 | NA | NA | 16.1 | 21.2 | .005|| |
| Firmicutes;Clostridia;Clostridiales; Clostridiaceae (family) | 4.3 | 0.0 | .04 | NA | NA | 0.04 | 0.0 | NA |
| Firmicutes; Clostridia;Clostridiales; ClostridialesFamilyXIII. IncertaeSedis;Anaerovorax | 4.3 | 0.0 | .04 | NA | NA | 0.3 | 0.0 | NA |
| Firmicutes;Clostridia;Clostridiales; ClostridialesFamilyXIII. IncertaeSedis;Other | 44.7 | 26.6 | .03 | 2.31 (1.10 to 4.86) | 2.38 (1.12 to 5.06) | 0.06 | 0.1 | .49 |
| Firmicutes;Clostridia;Clostridiales; Peptostreptococcaceae; Peptostreptococcus | 10.6 | 2.1 | .03 | 5.26 (0.97 to 28.46) | 4.98 (0.87 to 28.53) | 0.03 | 0.0 | .33 |
| Firmicutes;Clostridia;Clostridiales; Veillonellaceae (family) | 25.5 | 10.6 | .02 | 2.79 (1.10 to 7.09) | 3.02 (1.16 to 7.86) | 0.08 | 0.2 | .37 |
| Firmicutes;Clostridia;Clostridiales; Veillonellaceae;Megasphaera | 14.9 | 4.3 | .03 | 4.14 (1.13 to 15.19) | 4.02 (1.03 to 15.70) | 0.4 | 0.3 | .78 |
| Firmicutes;Clostridia;Clostridiales; Veillonellaceae;Selenomonas | 4.3 | 0.0 | .04 | NA | NA | 0.06 | 0.0 | NA |
| Fusobacteria (phylum) | 36.2 | 16.0 | .007||¶ | 3.71(1.85 to 7.46) | 4.37 (2.10 to 9.09) | 0.06 | 0.1 | .13 |
| Fusobacteria;Fusobacteria;Fusobacteriales; Fusobacteriaceae;Fusobacterium | 31.9 | 11.7 | .004||¶ | 3.74 (1.53 to 9.12) | 4.11 (1.62 to 10.47) | 0.07 | 0.1 | .32 |
| Actinobacteria;Actinobacteria; Coriobacteriales (family) | 46.8 | 69.2 | .01 | 0.41 (0.20 to 0.86) | 0.42 (0.20 to 0.90) | 0.2 | 0.2 | .99 |
| Actinobacteria;Actinobacteria; Coriobacteriales;Coriobacteriaceae; Atopobium | 19.2 | 2.1 | <.001||¶ | 11.15 (2.28 to 54.50) | 14.36 (2.78 to 74.30) | 0.1 | 0.2 | .13 |
| Bacteroidetes;Bacteroidia;Bacteroidales; Porphyromonadaceae;Porphyromonas | 27.7 | 7.5 | .001 ||¶ | 4.58 (1.66 to 12.60) | 5.17 (1.75 to 15.25) | 0.1 | 0.2 | .58 |
* Percentage of case and control subjects who carry the specific taxon. NA = not assessed.
† Median relative abundance of the specific taxon in people who carry the taxon. NA = not assessed.
‡ P values were based on χ2 test (two-sided).
§ P values were based on nonparametric Wilcoxon test (two-sided).
|| False discovery rate–adjusted P values were P less than or equal to .05.
¶ False discover rate tested limited to taxa with at least 0.05% relative abundance for Wilcoxon test (62 tests for genera).
# Multivariable odds ratios (ORs) and 95% confidence intervals (CIs) were calculated for taxa, based on logistic regression with noncarriers as the referent, adjusting for age, sex, body mass index, race, smoking, and sequencing batch.
Carriage of the genus Fusobacterium was statistically significantly greater in case subjects (31.9% vs 11.7% in control subjects) (Table 1; Figures 1C) and was associated with increased CRC risk (multivariable-adjusted OR = 4.11; 95% confidence interval [CI] = 1.62 to 10.47; P = .004; FDR-adjusted P ≤ .05). Relative abundance of Fusobacterium taxa in carriers did not differ (case subject range = 0.009%–28.9%; control subject range = 0.01%–1.3%; P = .32) (Table 1).
Gram-negative, anaerobic Fusobacterium contributes to colitis (20) and to periodontal disease (21), which itself may be related to colon cancer (22). Consistent with our findings, two studies recently reported that Fusobacterium was enriched in human CRC tissue compared with adjacent normal tissue (7,8), and another study reported enrichment of Fusobacterium in rectal swabs from CRC case subjects compared with control subjects (23).
In our study, increased carriage of genera Atopobium and Porphyromonas was also associated with CRC (OR = 14.36, 95% CI = 2.78 to 74.30, P < .001; and OR = 5.17, 95%CI = 1.75 to 15.25, P = .001, respectively) (Table 1). Atopobium, a Gram-positive anaerobic bacterium, is associated with Crohn’s disease (24) and reported to inhibit colon cancer apoptosis in vitro (25). Porphyromonas, commonly found in the mouth and gastrointestinal track, is associated with oral periodontal disease (26). Increased risks of CRC with carriage of Porphyromonas (P = .05; OR = 1.44; 32.1% vs 16.2% in case subjects vs control subjects, respectively) and of Fusobacterium (P = .01; OR = 1.44; 34.3% vs 28.1% in case subjects vs control subjects, respectively) were confirmed by quantitative polymerase chain reaction.
This is the first epidemiologic study comparing the gut microbiome of CRC patients and noncancer control subjects while controlling for potential confounders and taking into account the multiple comparisons involved in microbiome analysis. Other strengths of this study include nonculture-dependent sequencing-based microbiome assessment, which provided a comprehensive survey of the human fecal microbiome.
We did not examine mucosal adherent gut bacteria, which is a limitation because these might be more closely linked to colon carcinogenesis than are bacteria in feces. Possible effects of lyophylization and long-term frozen storage are unknown. However, lyophilization is an excellent method to preserve DNA for long-term storage (27); reproducibility in our masked replicates was good; and taxon distributions of our data are comparable with those of other published fecal microbiome data (28,29). Results from these analyses could be affected by selection bias and other biases that are common to case–control studies. Large prospective studies are warranted to confirm our findings.
In conclusion, this survey of the gut microbiota found that CRC risk was associated with decreased bacterial diversity in feces; depletion of Gram-positive, fiber-fermenting Clostridia; and increased presence of Gram-negative, proinflammatory genera Fusobacterium and Porphyromonas. Because of the potentially modifiable nature of the gut bacteria, our findings may have implications for CRC prevention.
Funding:
This work was supported by the National Cancer Institute (R03CA159414 and R01CA159036); a 2012 Pancreatic Cancer Action Network–AACR Career Development Award, supported by the Daniel and Janet Mordecai Foundation (grant 12-20-25-AHN); and the Intramural Research Program of the National Institutes of Health/National Cancer Institute, Division of Cancer Epidemiology and Genetics.
Supplementary Material
The study sponsor had no role in the design of the study; the collection, analysis, and interpretation of the data; the writing of the manuscript; and the decision to submit the manuscript for publication.
Sequencing was performed in the Next Generation Sequencing Core of the Nucleic Acids Research Facilities at Virginia Commonwealth University.
References
- 1. O’Keefe SJ. Nutrition and colonic health: the critical role of the microbiota. Curr Opin Gastroenterol. 2008;24(1):51–58 [DOI] [PubMed] [Google Scholar]
- 2. Koropatkin NM, Cameron EA, Martens EC. How glycan metabolism shapes the human gut microbiota. Nat Rev Microbiol. 2012;10(5):323–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yang L, Pei Z. Bacteria, inflammation, and colon cancer. World J Gastroenterol. 2006; 12(42):6741–6746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kinross JM, von Roon AC, Holmes E, Darzi A, Nicholson JK. The human gut microbiome: implications for future health care. Curr Gastroenterol Rep. 2008;10(4):396–403 [DOI] [PubMed] [Google Scholar]
- 5. Horie H, Kanazawa K, Okada M, Narushima S, Itoh K, Terada A. Effects of intestinal bacteria on the development of colonic neoplasm: an experimental study. Eur J Cancer Prev. 1999;8(3):237–245 [DOI] [PubMed] [Google Scholar]
- 6. Moore WE, Moore LH. Intestinal floras of populations that have a high risk of colon cancer. Appl Environ Microbiol. 1995;61(9):3202–3207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kostic AD, Gevers D, Pedamallu CS, et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012;22(2):292–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Castellarin M, Warren RL, Freeman JD, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22(2):299–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schiffman MH, Van Tassell RL, Robinson A, et al. Case–control study of colorectal cancer and fecapentaene excretion. Cancer Res. 1989;49(5):1322–1326 [PubMed] [Google Scholar]
- 10. Schiffman MH, Andrews AW, Van Tassell RL, et al. Case–control study of colorectal cancer and fecal mutagenicity. Cancer Res. 1989;49(12):3420–3424 [PubMed] [Google Scholar]
- 11. Ahn J, Yang Y, Paster BJ, et al. Oral microbiome profiles: 16S rRNA pyrosequencing and microarray assay comparison. PLoS One. 2011;6(7):e22788–22795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nossa CW, Oberdor WE, Yang L, et al. In silico design of 16S rRNA gene primers for analysis of human foregut microbiome using next generation sequencing technology. World J Gastroenterol. 2010;16(33):4135–4144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Haas BJ, Gevers D, Earl AM, et al. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011;21(3):494–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sanapareddy N, Legge RM, Jovov B, et al. Increased rectal microbial richness is associated with the presence of colorectal adenomas in humans. Isme J. 2012;6(10):1858–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Knoch B, Nones K, Barnett MP, McNabb WC, Roy NC. Diversity of caecal bacteria is altered in interleukin-10 gene-deficient mice before and after colitis onset and when fed polyunsaturated fatty acids. Microbiology. 2010;156(Pt 11):3306–3316 [DOI] [PubMed] [Google Scholar]
- 17. Rinttila T, Kassinen A, Malinen E, Krogius L, Palva A. Development of an extensive set of 16S rDNA-targeted primers for quantification of pathogenic and indigenous bacteria in faecal samples by real-time PCR. J Appl Microbiol. 2004;97(6):1166–1177 [DOI] [PubMed] [Google Scholar]
- 18. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57(1):289–300 [Google Scholar]
- 19. Hamer HM, Jonkers D, Venema K, Vanhoutvin S, Troost FJ, Brummer RJ. Review article: the role of butyrate on colonic function. Aliment Pharmacol Ther. 2008;27(2):104–119 [DOI] [PubMed] [Google Scholar]
- 20. Ohkusa T, Okayasu I, Ogihara T, Morita K, Ogawa M, Sato N. Induction of experimental ulcerative colitis by Fusobacterium varium isolated from colonic mucosa of patients with ulcerative colitis. Gut. Jan 2003;52(1):79–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Signat B, Roques C, Poulet P, Duffaut D. Fusobacterium nucleatum in periodontal health and disease. Curr Issues Mol Biol. 2011;13(2):25–36 [PubMed] [Google Scholar]
- 22. Ahn J, Segars S, Hayes RB. Periodontal Disease, Porphyromonas gingivalis (P. gingivalis) serum antibody levels and orodigestive cancer mortality. Carcinogenesis. 2012;33(5):1055–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chen W, Liu F, Ling Z, Tong X, Xiang C. Human intestinal lumen and mucosa-associated microbiota in patients with colorectal cancer. PLoS One. 2012;7(6):e39743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Manichanh C, Rigottier-Gois L, Bonnaud E, et al. Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut. 2006;55(2):205–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Altonsy MO, Andrews SC, Tuohy KM. Differential induction of apoptosis in human colonic carcinoma cells (Caco-2) by Atopobium, and commensal, probiotic and enteropathogenic bacteria: mediation by the mitochondrial pathway. Int J Food Microbiol. 2010;137(2–3):190–203 [DOI] [PubMed] [Google Scholar]
- 26. Pihlstrom BL, Michalowicz BS, Johnson NW. Periodontal diseases. Lancet. 2005;366(9499): 1809–1820 [DOI] [PubMed] [Google Scholar]
- 27. Frantzen MA, Silk JB, Ferguson JW, Wayne RK, Kohn MH. Empirical evaluation of preservation methods for faecal DNA. Mol Ecol. 1998;7(10):1423–1428 [DOI] [PubMed] [Google Scholar]
- 28. Turnbaugh PJ, Hamady M, Yatsunenko T, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457(7228):480–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Human Microbiome Project: Structure, function and diversity of the healthy human microbiome. Nature. 2012;486(7402):207–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.