Abstract
The distributions of endophytic bacteria in Alopecurus aequalis Sobol and Oxalis corniculata L. grown in soils contaminated with different levels of polycyclic aromatic hydrocarbons (PAHs) were investigated with polymerase chain reaction followed by denaturing gradient gel electrophoresis technology (PCR-DGGE) and cultivation methods. Twelve types of PAHs, at concentrations varying from 0.16 to 180 mg·kg−1, were observed in the roots and shoots of the two plants. The total PAH concentrations in Alopecurus aequalis Sobol obtained from three different PAH-contaminated stations were 184, 197, and 304 mg·kg−1, and the total PAH concentrations in Oxalis corniculata L. were 251, 346, and 600 mg·kg−1, respectively. The PCR-DGGE results showed that the endophytic bacterial communities in the roots and shoots of the two plants were quite different, although most bacteria belonged to Firmicutes, Proteobacteria, Actinobacteria and Bacteroidetes. A total of 68 endophytic bacterial strains were isolated from different tissues of the two plants and classified into three phyla: Firmicutes, Proteobacteria and Bacteroidetes. In both plants, Bacillus spp. and Pseudomonas spp. were the dominant cultivable populations. With an increase in the PAH pollution level, the diversity and distribution of endophytic bacteria in the two plants changed correspondingly, and the number of cultivable endophytic bacterial strains decreased rapidly. Testing of the isolated endophytic bacteria for tolerance to each type of PAH showed that most isolates could grow well on Luria-Bertani media in the presence of different PAHs, and some isolates were able to grow rapidly on a mineral salt medium with a single PAH as the sole carbon and energy source, indicating that these strains may have the potential to degrade PAHs in plants. This research provides the first insight into the characteristics of endophytic bacterial populations under different PAH pollution levels and provides a species resource for the isolation of PAH-degrading endophytic bacteria.
Introduction
Organic contaminants are frequently detected at relatively high concentrations in soils worldwide [1]. Some of these contaminants may be taken up by plants and translocated into shoots, which is the major pathway by which they reach the food chain [2], [3]. Polycyclic aromatic hydrocarbons (PAHs) are a class of environmental organic pollutants that are considered potentially extremely harmful owing to their toxic, mutagenic, and carcinogenic characteristics [4], [5]. A better understanding of how plants take up and metabolize PAHs from the soil could have considerable benefits for plant PAH risk assessments [6]. Therefore, methods for regulating and controlling the uptake and metabolism of PAHs in plants and effective measures for reducing plant PAH contamination risks have attracted much attention [7], [8].
Some chemicals, such as surfactants and ascorbic acid, can be used to regulate and control PAH absorption and metabolism processes in plants [9], [10]. However, most chemicals are not environmentally friendly and may cause secondary pollution. Additionally, the functionality of such chemicals is always limited by environmental conditions [11]. Previous studies have shown that many microorganisms associated with plants, such as arbuscular mycorrhizal fungi, biofilms on root surfaces, and endophytic bacteria, have enormous potential to degrade PAHs [12], and some of these microorganisms can even reduce PAH concentrations in plants [13]. Therefore, researchers have proposed an attractive strategy for reducing plant PAH contamination risks by utilizing plant-associated microecosystems to control the uptake and metabolism of PAHs by plants. This approach has been the focus of considerable interest in recent years [14].
Endophytic bacteria, defined as a class of microbes that reside within the interior tissues of plants without causing harm to host plants or environments [15], form one of the microbial communities most closely associated with plants. They have established harmonious associations with host plants during including symbiotic, mutualistic, commensalistic, and trophobiotic relationships over a long evolutionary process [16]. This assortment of bacteria have a wide variety of functions including the stimulation of plant growth [17], the promotion of biological nitrogen fixation [18], the protection of plants from harsh external environments, and the control of pathogen activities [19].
Previous studies have shown that some endophytic bacterial strains have the ability to degrade organic pollutants in plants and soils [20], [21]. Germaine et al. [22] inoculated pea plants with a 2,4-D-degrading endophytic bacterium (Pseudomonas putida VM1450) and found that this strain can internally colonize plants, maintain their growth, and cause a 24–35% increase in contaminant removal from the plants. Sheng et al. [14] isolated an endophytic pyrene-degrading bacterium Enterobacter sp. 12J1 from Allium macrostemon Bunge grown in PAH-contaminated soils and found that the bacterium increased plant resistance to pyrene by increasing plant biomass (from 13% to 56%) and promoted pyrene removal from pyrene-amended soils. Therefore, the use of endophytic bacteria to regulate the metabolism of organic pollutants and to reduce contamination risks in plants would be significantly advantageous [14].
Assessing the diversity, distribution, physiology, and ecology of endophytic bacteria in plants is prerequisite for isolating organic contaminant-degrading endophytic bacteria and using them to eliminate organic pollution in plants [23], [24]. There have been some reports regarding endophytic bacterial populations in plants grown in soils polluted with different contaminants [25], [26]. Moore et al. [23] investigated endophytic bacterial populations in poplar trees and found that a number of isolates had the ability to degrade BTEX compounds or to grow in the presence of TCE. Ho et al. [27] isolated 188 endophytic strains from three plants and found that among these strains, 29 not only grew well in the presence of naphthalene, catechol, and phenol but were also able to utilize the pollutant as a sole carbon source for growth. However, to our knowledge, little information is available about endophytic bacterial populations in plants from PAH-contaminated sites.
The aim of this study was to utilize PCR-DGGE technology combined with culture-dependent methods to investigate the distribution and diversity of endophytic bacteria in two plants (Alopecurus aequalis Sobol and Oxalis corniculata L.) that are common in China and also the dominant plants in a PAH-contaminated field. This is an indispensable precondition for the future isolation of functional endophytes to aid in the reduction of plant PAH pollution risk.
Materials and Methods
Chemical reagents
Sixteen PAH standards dissolved in acetonitrile were purchased from ShangHai Anpel Scientific Instruments (Shanghai, China) including naphthalene (NAP), acenaphthylene (ANY), acenaphthene (ANE), fluorine (FLU), phenanthrene (PHE), anthracene (ANT), fluoranthene (FLA), pyrene (PYR), benz[a]anthracene (BaA), chrysene (CHR), benzo[b]fluoranthene (BbF), benzo[k]fluoranthene (BkF), benzo[a]pyrene (BaP), indeno[1,2,3-cd] pyrene (InPy), dibenz[a,h]anthracene (DiahA), and benzo[ghi]perylene (BghiP). The concentration of each compound in the mixture was 200 mg·L−1.
The PAHs used in the tolerance test, including NAP, ACE, PHE, FLU, PYR, ANT, FLA, and BaP, were purchased from Aldrich Chemical Co. with purities >98%.
Media
Beef extract peptone medium contained 5.0 g·L−1 beef extract, 5.0 g·L−1 NaCl, and 10.0 g·L−1 tryptone, pH 7.0. Luria-Bertani (LB) medium contained 10.0 g·L−1 tryptone, 5.0 g·L−1 yeast extract, and 10.0 g·L−1 NaCl, pH 7.0. Mineral salt medium (MSM) contained 1.50 g·L−1 (NH4)2SO4, 1.91 g·L−1 K2HPO4·3H2O, 0.50 g·L−1 KH2PO4, 0.20 g·L−1 MgSO4·7H2O,w=[to]?> and 1 mL of trace element solution (0.1 mg·L−1 CoCl2·6H2O, 0.425 mg·L−1 MnCl2·4H2O, 0.05 mg·L−1 ZnCl2, 0.01 mg·L−1 NiCl2·6H2O, 0.015 mg·L−1 CuSO4·5H2O, 0.01 mg·L−1 Na2MoO4·2H2O, 0.01 mg·L−1 Na2SeO4·2H2O). Plates containing solid media were prepared by adding 18 g·L−1 agar into the above-mentioned liquid media.
Sample collection and PAHs analysis
Amur foxtail (Alopecurus aequalis Sobol), creeping oxalis (Oxalis corniculata L.), and soil samples were obtained from an aromatics factory in Nanjing (permission was obtained from the owner of this private land to perform the study on this site). Samples were collected from three stations (A, Z, and Q) at different distances from the aromatics factory. The physicochemical characteristics of the sampled soils were as follows: pH 5.87, 13.0% sand, 60.7% silt, 26.3% clay, and 1.36% organic matter. The total PAH contents of soils from stations A, Z, and Q were 178, 139, and 89.4 mg·kg−1, respectively. The plant samples were removed from the soil, carefully placed into a plastic bag, and immediately transported to the laboratory where the surface soil was scoured off [28].
Some of the soil and plant samples were freeze dried immediately for the determination of PAH contents. The PAHs were exacted from soil and plant samples as described previously by Ling and Gao [29]. The concentrations of PAHs were analyzed using high-performance liquid chromatography (HPLC) with a reverse-phase C18 column (Inertsil ODS-SP, 5 µm, 4.6×150 mm, GL Sciences Inc., Japan) using gradient elution. The recoveries of PAHs in the soil and plant samples that were investigated averaged between 85%–105% (n = 5, RSD ≤2.52%) after the entire procedure. Table 1 shows the concentrations of PAHs in soils.
Table 1. Concentrations of PAHs in soils.
| PAHs | The concentrations of PAHs in soil (mg·kg−1 dry weight) | ||
| A | Z | Q | |
| NAP | 34.3±3.80a | 25.5±7.30a | 25.7±4.19a |
| ANE | 42.8±1.77a | 39.9±0.48ab | 36.7±0.64b |
| PHE | 8.62±0.67a | 6.45±0.58a | 3.36±0.78b |
| ANT | 72.1±4.71a | 56.6±4.34a | 16.1±7.92b |
| FLU | 0.89±0.02a | 0.72±0.06b | 0.73±1.44b |
| ANY | 2.09±0.05a | 1.01±0.04b | 1.04±0.06b |
| PYR | 1.68±0.02a | 0.94±0.01b | 0.91±0.03b |
| FLA | 3.19±0.18a | 2.14±0.09ab | 0.82±0.34b |
| CHR | 6.62±0.41a | 3.20±0.04b | 1.22±0.49c |
| BaA | 0.94±0.01a | 0.51±0.17b | 0.52±0.01b |
| BbF | 1.77±2.87a | 0.86±4.41b | 0.69±1.97b |
| BghiP | 3.50±0.03a | 1.64±0.04b | 1.66±0.12b |
| ∑PAHs | 178±4.87a | 139±4.20b | 89.5±5.79c |
Note: different letters in the same row indicate significant differences (P<0.05).
Surface sterilization of plant samples
The plant samples were rinsed three times with deionized water and subsequently sterilized by sequential immersion in 75% (v/v) ethanol for 3–5 min, 2% sodium hypochlorite (v/v) for 3 min, and 70% ethanol for 30 sec. Finally, the plant samples were washed three times with sterilized distilled water to remove surface sterilization agents [28]. To determine whether the surface disinfection process was successful, plants were pressed onto fresh beef extract peptone agar plates to detect any remaining epiphytic bacteria.
PCR-DGGE analysis
Total DNA extraction was performed according to the protocol described by Hung et al. [30]. The 16S rDNA V3 sequences were amplified by PCR using the extracted genomic DNA as a template and the bacterial universal primers 341f (with a GC clamp; 5′- CGCCCGCCGCGCGCGGCGGGGGGCGGGGGCACGGGGGGCCTACGGGAGGCAGCAG-3′) and 534r (5′- ATTACCGCGGCTGCTGG-3′). The PCR mixture (25.0 µL) contained 1 µL of DNA template (5 ng µL−1), 12.5 µL of Premix Taq (TaKaRa, Premix Taq Version 2.0), 0.5 µL of primers (12.5 µg·μL−1), and 1 µL of bovine serum albumin (20 µg·μL−1). The PCR was performed in a DNA Engine Thermal Cycler (TaKaRa, D-8308), and the PCR program consisted of an initial denaturation at 94°C for 5 min, 30 cycles at 94°C for 30 sec, 60°C for 30 sec, and 72°C for 30 sec, followed by a final extension step at 72°C for 10 min.
DGGE analysis was performed with a Dcode Multiple System (Bio-Rad Laboratories Inc., Hercules, CA, USA) using the following protocol: Aliquots (25 µL) of the PCR products were loaded onto an 8% (w/v) polyacrylamide gel with a denaturant gradient ranging from 40% to 65%. Electrophoresis was run for 16 h at 120 V and 60°C in 1× TAE buffer (40 mM Tris, 20 mM acetic acid, and 2 mM EDTA), after which the gels were soaked for 30 min in SYBR Green I nucleic acid stain (1∶10,000 dilution) and immediately photographed under UV light. Specific bands were excised from the DGGE gel and washed twice with sterilized distilled water. Each band was used as a direct template for PCR to recover the DNA fragment separated by DGGE. The PCR conditions were the same as those used for the original PCR. The fragments recovered from the PCR were subjected to DGGE again to confirm the equality of their mobility. If a single band appeared in a DGGE gel for one sample, the PCR products were purified with the PCR Cleanup Kit (Axygen, USA) and used for direct sequencing (Invitrogen). When multiple bands appeared in one sample, the bands were repeatedly electrophoresed and excised until only a single band was detectable on the DGGE gel.
Isolation of cultivable endophytic bacteria
After surface disinfection, the root and shoot tissues of plants were cut into pieces and triturated in 5 mL of sterile distilled water. Subsequently, 100 µL aliquots of the appropriate dilutions (10−1, 10−2, and 10−3) were spread onto beef extract peptone medium and incubated at 30°C for 7 days.
Each bacterial strain with a different colonial morphology was identified by 16S rDNA sequence analysis. Genomic DNA was extracted from each isolated endophytic bacterium with a DNA Extraction Kit (Axygen, USA). The PCR mixture was the same as that used for 16S rDNA V3 sequence amplification except that the primers were 27f (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492r (5′-GGTTACCTTGTTACGACT T-3′) [31], and the annealing temperature was altered to 52°C.
DNA Sequence Analysis and Accession Numbers
Analysis of 16S rDNA sequences was performed using the NCBI database and nucleotide BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The sequences were aligned using the ClustalW program. Sequences were identified as the most closely related species with the highest similarity. A phylogenetic tree was constructed using evolutionary distances based on the 16S rDNA V3 sequences with the neighbor-joining method [32]. Tree topologies were evaluated by performing bootstrap analysis of 1,000 datasets with the MEGA 5.05 package.
The 77 fragments of 16S rDNA V3 sequences determined in this study were deposited in the GenBank database under the accession numbers KF051455–KF051531, and the 68 pieces of 16S rDNA sequences determined in this study were deposited with the accession numbers JX994089–JX994132 and JX994134–JX994157.
Tolerance of isolated endophytic bacteria to each type of PAH
A suspension of each isolated endophytic bacterial strain was plated on MSM or LB agar plates containing one of the following PAHs: NAP (with a final concentration of 100 mg·L−1), PHE, FLU, ANT, ACE, PYR, FLU (with final concentration of 30 mg·L−1), or BaP (with a final concentration of 10 mg·L−1), the concrete method was referring to Yao et al. [33]. The plates were incubated for 3–7 days at 30°C, and bacterial growth was monitored regularly to investigate the tolerance of each endophytic bacterial strain to each type of PAH.
Statistical analyses
All data collected were processed using Microsoft Excel 2007. Each data point is the mean of at least three replicates, and error bars represent standard deviations (SD). The data were statistically analyzed using analysis of variance (ANOVA) with the statistical package SPSS 13.0. Differences were considered significant at p values <0.05, and standard deviations obtained from three parallel samples are shown in the figures as error bars.
Results
PAH concentrations in plants from PAH-contaminated soils
In this study, 12 types of PAHs that have been designated by the US Environmental Protection Agency as priority pollutants were detected in soil and plant samples, including NAP, ANE, PHE, ANT, FLU, ANY, PYR, FLA, CHR, BaA, BbF, and BghiP. As shown in Table 1, soil samples obtained from each sampling station contained different concentrations of PAHs, and the total PAH contents of soils from A, Z, and Q stations were 178, 139, and 89.5 mg·kg−1, respectively. ANOVA revealed significant differences among the total PAH concentrations at the three sampling stations. ANE and ANT were the main components in all three soil samples, accounting for 23.9–40.9% and 18.0–40.6% of the total concentrations, respectively.
The 12 types of PAHs detected in rhizosphere soils were also determined in the roots and shoots of the two plants (Tables 2 and 3), and the overwhelming majority of PAHs accumulated in plant roots. For example, at position A, PAHs were enriched to a concentration of 217 mg·kg−1 in the roots of A. aequalis; however, in the shoots, the total PAH concentration was only 88.0 mg·kg−1. Furthermore, for both plants, as soil PAH concentrations increased, the PAH concentrations in the plants also increased. As shown in Tables 2 and 3, the PAH concentrations in the roots of plants from station A were significantly higher than those in plants from the other two stations. Two- to three-ring PAHs were the main detectable pollutants in plant samples, with proportions of 94.9–96.2% in A. aequalis and 87.8–94.2% in O. corniculata grown in differentially contaminated soils. Conversely, four- to six-ring PAHs accounted for only minor proportions of the total PAHs in the two plants (approximately 3.8–5.1% and 5.8–12.2%, respectively).
Table 2. Concentrations of PAHs in Alopecurus aequalis.
| PAHs | Root (mg·kg−1 dry weight of plant) | Shoot (mg·kg−1 dry weight of plant) | ||||
| A | Z | Q | A | Z | Q | |
| NAP | 120±5.43a | 45.5±8.11b | 59.9±21.05ab | 42.8±5.30a | 26.9±5.34a | 20.2±7.82a |
| ANE | 33.9±7.81a | 28.8±6.26a | 24.9±7.80a | 15.9±3.36a | 12.5±0.83a | 9.32±1.85a |
| PHE | 4.85±0.21a | 4.12±0.01a | 3.7±1.31a | 2.31±0.28a | 2.25±0.03a | 1.98±0.71a |
| ANT | 49.0±2.73a | 44.3±6.64a | 30.5±9.02a | 22.6±3.80a | 20.7±1.39a | 23.2±3.93a |
| FLU | 0.44±0.01a | 0.39±0.01a | 0.36±0.13a | 0.21±0.12a | 0.22±0.01a | 0.19±0.06a |
| ANY | 0.73±0.04a | 0.63±0.05a | 0.56±0.22a | 0.39±0.10a | 0.45±0.10a | 0.34±0.07a |
| PYR | 0.62±0.04a | 0.59±0.07a | 0.48±0.06a | 0.30±0.02a | 0.27±0.01a | 0.30±0.01a |
| FLA | 1.72±0.04a | 1.54±0.22a | 1.28±0.24a | 0.82±0.12a | 0.75±0.07a | 0.81±0.04a |
| CHR | 3.78±0.37a | 2.90±0.15ab | 1.81±0.57b | 1.77±0.05a | 1.66±0.34a | 1.61±0.08a |
| BaA | 0.32±0.01a | 0.29±0.03a | 0.25±0.02a | 0.16±0.01a | 0.17±0.00a | 0.16±0.00a |
| BbF | 0.54±0.10a | 0.51±0.04a | 0.43±0.13a | 0.31±0.03a | 0.34±0.02a | 0.31±0.04a |
| BghiP | 0.77±0.01a | 0.71±0.04a | 0.60±0.10a | 0.38±0.01a | 0.36±0.00a | 0.38±0.01a |
| ∑PAHs | 217±11.7a | 130±12.0b | 124±2.30b | 88.0±3.97a | 66.5±2.41b | 58.8±4.08c |
| CFPAHs | 1.21 | 0.93 | 1.39 | 0.49 | 0.48 | 0.66 |
Note: different letters in the same row indicate significant differences (P<0.05); CFPAHs means the plant concentration factors of total PAHs.
Table 3. Concentrations of PAHs in Oxalis corniculata.
| PAHs | Root (mg·kg−1 dry weight of plant) | Shoot (mg·kg−1 dry weight of plant) | ||||
| A | Z | Q | A | Z | Q | |
| NAP | 109±6.53a | 64.1±12.01b | 63.3±19.57b | 49.2±2.78a | 45.8±15.76a | 19.7±10.91b |
| ANE | 83.2±3.09a | 40.2±2.14b | 38.5±3.56b | 30.2±3.71a | 32.3±10.61a | 12.9±3.71b |
| PHE | 18.2±2.83a | 9.28±0.34b | 8.4±2.47b | 5.25±0.32a | 4.53±1.53ab | 2.26±0.54b |
| ANT | 180.±10.12a | 85.8±0.42b | 68.5±9.88c | 47.2±1.41a | 41.3±3.13a | 19.6±4.13b |
| FLU | 1.69±0.06a | 0.78±0.02b | 0.81±0.21b | 0.48±0.04a | 0.48±0.19a | 0.29±0.02a |
| ANY | 2.22±0.40a | 1.16±0.09b | 1.07±0.30b | 0.71±0.08a | 0.75±0.22a | 0.40±0.01a |
| PYR | 2.32±0.88a | 1.12±0.07b | 1.14±0.27b | 0.68±0.02a | 0.61±0.20a | 0.36±0.02b |
| FLA | 6.01±1.12 a | 2.97±0.28b | 1.67±1.26c | 1.75±0.08a | 1.44±0.63ab | 0.64±0.27b |
| CHR | 50.0±6.58a | 6.18±3.83b | 5.83±1.22b | 3.58±0.30a | 2.78±1.36a | 1.45±0.96a |
| BaA | 1.22±0.24a | 0.60±0.09b | 0.60±0.17b | 0.37±0.02a | 0.31±0.09a | 0.21±0.01a |
| BbF | 2.28±0.49a | 1.13±0.15b | 0.60±0.25c | 0.69±0.06a | 0.50±0.04a | 0.44±0.06a |
| BghiP | 3.21±0.67a | 1.58±0.08b | 1.48±0.35b | 0.89±0.03a | 0.77±0.09ab | 0.45±0.03b |
| ∑PAHs | 459±10.6a | 215±14.2b | 192±23.0b | 141±5.74a | 132±12.4a | 58.7±9.34b |
| CFPAHs | 2.57 | 1.54 | 2.15 | 0.79 | 0.94 | 0.66 |
Note: different letters in the same row indicates a significant differences (P<0.05); CFPAHs means the plant concentration factors of total PAHs.
However, there were obvious differences in the absorption and accumulation of PAHs between the two plants. O. corniculata was better able to accumulate PAHs compared with A. aequalis. For example, the PAH concentrations in O. corniculata grown at positions A, Z, and Q were 251, 346, and 600 mg·kg−1, respectively, which were significantly higher than those in A. aequalis (184, 197, and 304 mg·kg−1, respectively at positions A, Z, and Q). Additionally, the concentration of NAP in A. aequalis was the highest among all detected PAHs, accounting for 36.8–53.4% of the total PAH content of the plants from the different stations. In contrast, the concentration of ANT was highest in O. corniculata, accounting for 60.9–62.5% of the total PAH content.
DGGE analysis of the endophytic bacterial community
The PCR-DGGE profiles are shown in Figure 1. All plant samples from the three sites contained band 1 (Pseudomonas sp.), indicating that one of the dominant bacteria was the same in both plants at different levels of PAH contamination. However, there were also some discrepancies between different plant tissues and different places. For example, band 2 (uncultured bacterium clone) was only existed at site Q, and band 3 (Nesterenkonia sp.) was only existed at sites Q and Z. Pollution intensity could significantly influence the diversity of the endophytic bacterial community. As shown in Figure 2, the plants that grew in lightly polluted soils consistently showed the highest diversity.
Figure 1. Representative DGGE for PCR-amplified 16S rDNA V3 fragments from endophytic bacteria in Alopecurus aequalis and Oxalis corniculata.
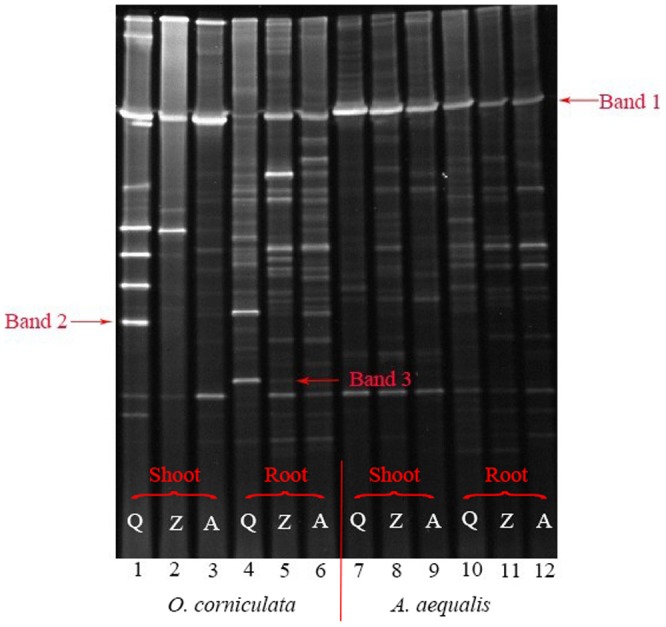
The genus of the band marked in the figure: Band 1 - Pseudomonas sp; Band 2 - uncultured bacterium clone; Band 3 - Nesterenkonia sp.
Figure 2. The similarity of endophytic bacterial community in roots of Alopecurus aequalis (A), shoots of Alopecurus aequalis (B), roots of Oxalis corniculata(C) and shoots of Oxalis corniculata (D).
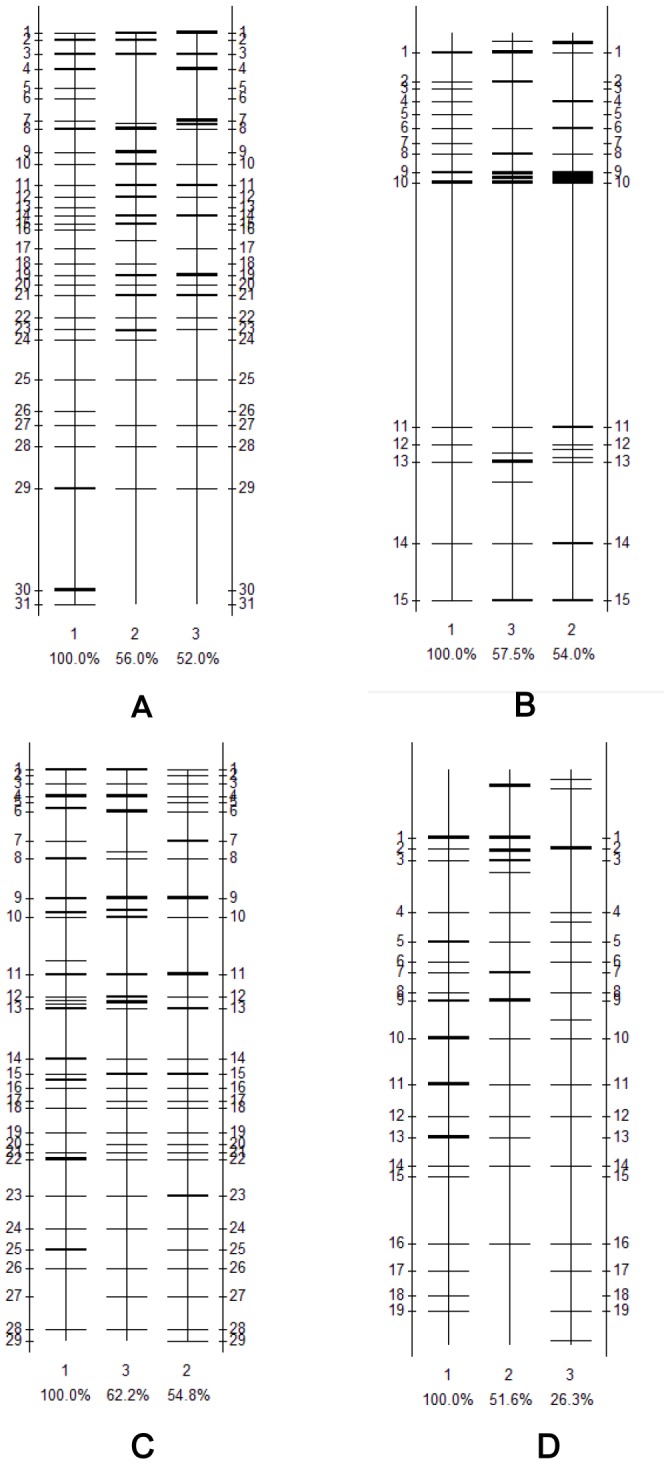
1- Q pollution level (defined as 100%), 2- Z pollution level, 3-A pollution level.
Nearly all the bands found in the DGGE gel were sequenced, and after removing the 16S rDNA and 18S rDNA of mitochondria and chloroplasts, we managed to obtain exactly 77 different bacterial sequences (Tables S1 and S2 in File S1), among which 56 were isolated from A. aequalis (root, 37; shoot, 22; both, 3) and 32 were obtained from O. corniculata (root, 27; shoot, 12; both, 4). Overall, 44.1% of the sequences isolated from A. aequalis were derived from uncultured bacteria, 8.47% were derived from Pseudomonas sp. and 6.78% were derived from Halomonas sp. The remaining 40.7% were derived from approximately 20 different genera of bacteria. In O. corniculata, the highest percentage of sequences (30.6%) was also derived from uncultured bacteria, followed by Pseudomonas sp. (19.4%) and Enterobacter sp. (8.33%). The remaining 41.7% of sequences were derived from 15 different genera of bacteria.
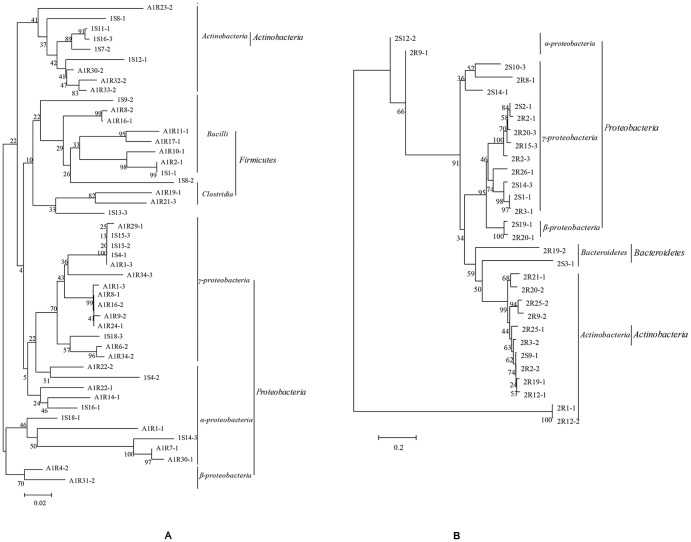
Phylogenetic analysis (Figure 3) of the endophytic bacterial community performed using the results of the DGGE gel indicated that these bacteria belong to four phyla (Firmicutes, Proteobacteria, Actinobacteria and Bacteroidetes), seven classes (Bacilli, Clostridia, α-proteobacteria, β-proteobacteria, γ-proteobacteria, Actinobacteria, Bacteroidetes), and over 30 families. Most of the isolates from A. aequalis belonged to Proteobacteria and Firmicutes, and the remaining belonged to Actinobacteria. Proteobacteria also composed a large percentage of the bacteria isolated from O. corniculata, and the remaining bacteria from O. corniculata belonged to Bacteroidetes and Actinobacteria.
Figure 3. Phylogenetic trees by 16S rDNA V3 sequence analysis of endophytic bacterial community of Alopecurus aequalis (A) and Oxalis corniculata (B).
The codes with characters and numbers indicate the isolated DGGE bands. Bootstrap values are shown for each node in a bootstrap analysis of 1,000 replicates.
Cultivable endophytic bacterial populations in plants from PAH-contaminated soils
A total of 68 species of endophytic bacteria were isolated and identified, among which 39 were isolated from A. aequalis (root, 27; shoot, 20; both, 7), 28 were isolated from O. corniculata (root, 15; shoot, 17; both, 3) and 1 was isolated from both plants. The isolates were identified using 16S rDNA analysis, and these 16S rDNA sequences shared high identities with their most closely related species in the database (≥97%), with most having identities of 99–100% with known bacterial species (Tables S3 and S4 in File S1).
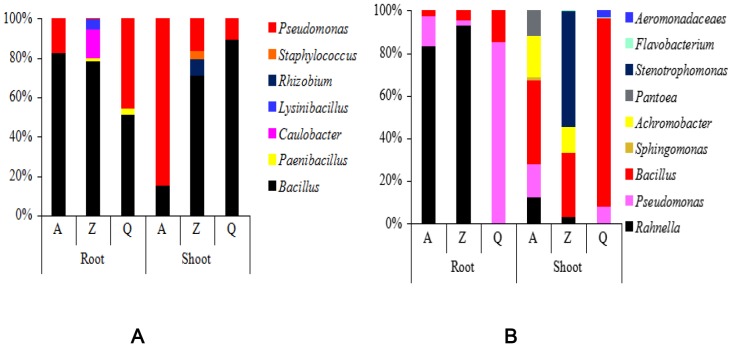
The cultivable endophytic bacterial populations of A. aequalis and O. corniculata grown in PAH-contaminated soils showed low diversity. The 68 endophytic bacterial species were classified into 14 genera belonging to five classes, including Bacilli, α-proteobacteria, β-proteobacteria, γ-proteobacteria, and Flavobacteriaceae. Although more endophytic bacterial species were isolated from A. aequalis than from O. corniculata, the endophytic bacterial population in O. corniculata included more genera than that in A. aequalis. In A. aequalis, Bacillus and Pseudomonas spp. constituted a large proportion, accounting for 65.5% and 24.1%, respectively, of the total bacterial population. Meanwhile, members of Bacillus spp. accounted for the highest proportion of strains in O. corniculata, constituting 43.9% of the total bacterial population, followed by Pseudomonas spp. and Rahnella spp., each constituting 21.1% of the total population.
Amounts of cultivable endophytic bacteria in plants from PAH-contaminated soils
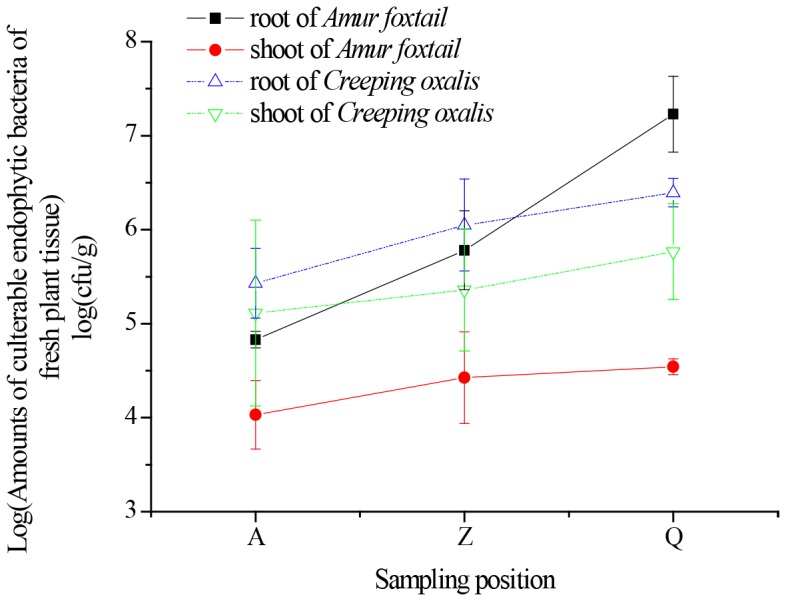
As shown in Figure 4, the cultivable endophytic bacteria detected in the two studied plants were on the order of 104 to 107 CFU per gram of fresh plant tissue in most cases, and the number of endophytic bacteria in roots was higher than that in shoots. As the PAH concentration increased, the total number of cultivable endophytic bacterial strains was reduced. For example, the number of endophytic bacterial strains in A. aequalis from station Q was 251 and 27 times larger than those from stations A and Z, respectively, but the concentration of PAHs was only 0.5 and 0.8 times of those from stations A and Z, respectively. Furthermore, as PAH concentrations increased, the total number of endophytic bacterial cells in plant tissues decreased by different degrees, and the range varied more in plant roots compared with shoots. For example, the number of endophytic bacteria in A. aequalis roots from station Q was 351 times that of roots from station A, while in shoots, the difference was only 3.23 times greater at station Q compared with station A.
Figure 4. Amounts of cultivable endophytic bacterial strains in Alopecurus aequalis and Oxalis corniculata under different PAH pollution levels.
PAH pollution levels also had impacts on the dominant populations of cultivable endophytic bacteria in the two plants (Figure 5). As PAH concentrations increased, the proportion of Bacillus spp. and Pseudomonas spp. showed opposite tendencies in the roots and shoots of A. aequalis. For example, under higher PAH concentrations (station A), Bacillus spp. accounted for 82.4% of the total endophytic bacteria in roots, and at the other two stations, Bacillus spp. accounted for 78.5% (Z) and 51.4% (Q). However, Bacillus spp. in the shoots of A. aequalis showed the opposite trend, with proportions of 15.4%, 70.8%, and 89.4% in samples from stations A, Z, and Q, respectively. Meanwhile, with increasing pollution levels, the proportion of Pseudomonas spp. decreased in plant roots and increased in shoots. The dominant populations in O. corniculata from different stations were quite different. The dominant genus in roots was Rahnella spp. when the pollution level was relatively high (A, Z); however, under a lower level of pollution, the dominant genus became Pseudomonas spp.
Figure 5. Amounts of cultivable endophytic bacterial strains belonging to each genus in Alopecurus aequalis (A) and Oxalis corniculata (B) under different PAH pollution levels.
Tolerance of the isolated endophytic bacteria to each type of PAH
Most of the isolates from the two plants were able to grow well on LB medium containing different PAHs, indicating that they were very tolerant to different PAHs. Some of the isolates were even able to grow rapidly on MSM medium with PAHs as the sole carbon and energy source, illustrating their potential for the degradation of various PAHs (Table S5–S8 in File S1).
Nine bacterial isolates obtained from the roots of A. aequalis grew well on MSM medium containing NAP. Eleven, seven, eight, and ten strains grew normally on MSM medium in the presence of tricyclic ANE, FLU, PHE and ANT, respectively, and ten and five strains grew well on MSM medium containing tetracyclic PYR and FLA, respectively. Only three bacterial strains grew normally on MSM medium with 10 ppm BaP (AF10, AF13, and AF25), and AF13 was the only strain that grew normally on MSM medium with each tested PAH. Many of the isolates from A. aequalis stems grew well on MSM medium with PAHs, and four strains even showed high tolerance to six or seven types of PAHs; however, none of the strains were tolerant to all tested PAHs.
Among all the strains isolated from O. corniculata roots, only CO3 grew well on MSM medium with five types of PAHs. Five isolates grew normally on MSM medium containing NAP, and two, six, two, and three strains grew on MSM medium in the presence of tricyclic ANE, FLU, PHE, and ANT, respectively. One and two strains grew well on MSM medium containing tetracyclic PYR and FLA, respectively. Only one bacterial strain grew normally on MSM medium with 10 mg·L−1 BaP. Some of the isolates from O. corniculata shoots grew well on three or four types of PAHs, but only CO26 grew normally on MSM medium with more than five types of PAHs.
Discussion
Previous studies have shown that many plants have the ability to uptake and accumulate organic pollutants [2], [3], [7]. Gao and Zhu [34] investigated the capacity of 12 plant species to absorb and accumulate PHE and PYR and found that the RCFs (defined as the ratio of the PAH concentration in roots to that in soils on a dry weight basis [35]) of PHE and PYR for plants grown in contaminated soils were 0.05–0.67 and 0.23–4.44, respectively, whereas the SCFs (defined as the ratio of the PAH concentration in shoots to that in soil on a dry weight basis) of PHE and PYR were 0.006–0.12 and 0.004–0.12, respectively. In this study, we also found that both A. aequalis and O. corniculata showed strong abilities to absorb and enrich various types of PAHs (Tables 2 and 3). The RCFs of total PAHs for A. aequalis and O. corniculata were 0.93–1.39 and 1.54–2.57, respectively, and the SCFs of the two plants were 0.48–0.66 and 0.66–0.94, respectively.
Interestingly, the total PAH concentration in O. corniculata was significantly higher than that in A. aequalis. This may be attributable to the different growing seasons (O. corniculata is a perennial herb whereas A. aequalis is an annual herb) or the different lipid contents of the two plants [36], [37]. Chiou et al. [38] built a partition-limited model indicating that water-insoluble contaminants, even in small amounts, were the major compounds present in the plant-lipid phase. Zhu and Gao [39] also confirmed a significant positive correlation between the root concentration of phenanthrene and root lipid content. Interestingly, both A. aequalis and O. corniculata absorbed few high molecular weight PAHs with larger numbers of benzene rings (pentacyclic and hexacyclic), which could be due to the poor bioavailability of these types of PAHs [40].
It is well known to use the surface sterilization methods to remove the epiphytic bacteria before the isolation of endophytic bacteria [28], [41], [42]. In this study, to determine whether the surface disinfection process was successful, plants and water from the final rinse were both pressed onto fresh beef extract peptone agar plates to detect any remaining epiphytic bacteria. No epiphytic bacteria were detected, indicating that the surface disinfection was successful, and the 68 isolated bacterial strains associated to the plants in this study were all endophytic bacteria. The several washes that have been done after sterilization should also ensure the absence of epiphytic dead bacterial cells, which could be detected by PCR-DGGE method. However, additional experiments such as fluorescence in situ hybridization (FISH) or scanning electronic microscopy (SEM) observations could be needed to exclude the presence of epiphytic bacteria.
Although endophytic bacteria have been previously reported in many plants including sugarcane [43], ginseng [44], and aquatic plants [45], among others, little information is available regarding endophytic bacteria in plants grown in PAH-contaminated soils. In this study, a total of 68 endophytic bacterial strains were isolated in A. aequalis and O. corniculata growing in PAH-contaminated stations. As known to us, Bacilli are a group of the commonest bacteria associated with plants and take a certain proportion of the cultivable endophytic bacteria in lots of plants [45]–[47]. In our study, members of Bacillus spp. account for the highest proportion of cultivable endophytic bacterial strains in both Alopecurus aequalis Sobol and Oxalis corniculata L, followed by Pseudomonas spp.. Many bacterial strains belonging to these two genera are known to be capable of degrading various types of organic pollutants, which accounts for the frequent occurrence of these strains in organically contaminated soils in relatively high amounts [23]. In addition, the numbers of endophytic bacteria in the two plants were quite high: 7.84×104−1.70×107 and 4.40×105−3.06×106 cfu·g−1 in A. aequalis and O. corniculata, respectively. In contrast, Garbeva et al. [48] and Araújo et al. [49] obtained only 1.0×103−1.0×105 cfu·g−1 and 103−104 cfu·g−1 of endophytic bacterial cells from the potato and citrus plants, respectively. Some endophytic bacterial strains, such as strains AF14, AF19, and CO4, occurred in both the roots and shoots of a given plant, indicating that endophytic bacterial strains can be transported and spread throughout plant systems[50], [51].
Compared with culture-dependent method, using PCR-DGGE technology we found that bacterial community profiles showed extensive variability depending on the plant origin, tissue, and PAH levels in soil. To further explore this dissimilarity, the Shannon diversity index was calculated for each sample using Quantity One and Bio-Dap software. The results show that from Q to A, the Shannon diversity indices of A. Aequalis were 3.38, 3.2, and 2.93 in roots and 2.55, 3.03, and 2.74 in shoots, respectively. In O. Corniculata, the Shannon diversity indices were 3.34, 3.27, and 3.29 in roots and 2.78, 2.66, and 2.73 in shoots. The Shannon index includes two basic components: abundance and evenness of the species present [52], and this result indicated that different plant samples and pollution levels could lead to different bacterial distributions and levels of diversity. Qing et al. [53] and Robson et al. [54] also obtained a similar conclusion in their studies. Through phylogenetic analysis (Figure 3), we can see that the most prominent groups in the two plants were both related to Proteobacteria, which agrees with other studies [55], [56].
Plant-associated habitats are dynamic environments in which many factors affect the species compositions of microbial communities [57]. The host plant species is one of the major influencing factors [58]. For instance, Chen et al. [45] studied endophytic bacterial species of four aquatic plants, and their results showed that the dominant endophytic bacterial taxa in the four plants were quite different; Pseudomonas spp. and Staphylococcus spp. were the dominant taxa in Phragmites communis and Potamogeton crispus, whereas in Nymphaea tetragona and Najas marina, the dominant taxa were Aeromonas spp. and Bacillus spp. In this study, at the same pollution level, the dominant endophytic bacteria in A. aequalis and O. corniculata were also different. Pollution stress within a plant growing area is another major environmental factor influencing endophytic bacterial communities [59]. Sobral et al. [28] found that many endophytic bacteria could be cultivated from soybeans grown in soil to which glyphosate had been previously applied (pre-planting); however, when the glyphosate was enriched, the taxa of the cultivable endophytic bacteria changed. Similar results were observed in this study: The proportion of each endophytic bacterial genus in a given plant was found to vary at different pollution levels. Even among different tissues of one plant, when the total PAH concentration varied, the endophytic bacterial population changed correspondingly (Figure 5).
Studying endophytic bacterial populations under different pollution levels is a popular means of examining contaminant-degrading bacterial flora [21], [23]. Siciliano et al. [60] found that the amount of contaminant-degrading bacterial cells increased with exposure to a contaminated environment. Phillips et al. [59] analyzed the relationship between the endophytic bacterial community and the capacity for organic pollutant degradation in alfalfa. These authors found that when the dominant population consisted of Pseudomonas spp., the ability of plants to metabolize alkane pollutants improved, whereas when the dominant bacterial populations shifted to Brevundimonas spp. and Pseudomonas rhodesiae, the capacity of the plant to metabolize aromatic hydrocarbons increased. This suggests that the dominant endophytic bacterial taxa isolated from contaminated plants may have the potential to degrade PAHs in plants [23], [45]. Thus, further studies of the tolerance of isolated endophytic bacterial strains to various PAHs were performed.
Many endophytic bacterial strains have been reported to be tolerant of PAHs, and some of these can even degrade various PAHs by using them as sole carbon and energy sources for cell growth [14], [22], [40]. Furthermore, previous studies have proposed that endophytic bacteria could assist plants in remediating organic pollutants, mainly through direct metabolism of the organic pollutants [61], the promotion of plant growth and reduction of plant disease [40], the regulation of plant enzyme system activity and the promotion of metabolic gene expression [62]. Accordingly, researchers have proposed that identifying endophytic bacteria that degrade PAHs and inoculating them into host plants grown at PAH-contaminated sites would be of great significance for reducing plant PAH pollution risks [63]. In this study, some isolates showing strong tolerance to many types of PAHs were identified. However, much work remains to be done before these bacteria can be used for plant PAH pollution reduction. First, the PAH-degrading abilities of each screened endophyte must be confirmed, and the functional strains should then be inoculated into target plants. Lastly, the impacts and mechanisms of PAH-degrading endophytes on the uptake of PAHs by plants must be clarified.
Conclusions
Alopecurus aequalis and Oxalis corniculata grown in PAH-contaminated sites were selected to investigate the concentrations of various PAHs and the distributions of endophytic bacteria in plant roots and shoots. The results revealed that both plants could absorb and accumulate PAHs in the roots and shoots, with O. corniculata exhibiting the higher enrichment ability. The endophytic bacterial communities in the roots and shoots of the two plants were quite different, although most of these bacteria belonged to Firmicutes and Proteobacteria. As pollution increased, the diversity and distribution of endophytic bacterial strains in both plants changed correspondingly, while Proteobacteria always accounted for a large percentage of the total, and the number of cultivable endophytic bacterial strains decreased rapidly. Most bacterial isolates from the two plants showed strong tolerance to different PAHs, and some of them were able to grow rapidly with PAHs as their sole carbon and energy sources, indicating that these strains may have the potential to degrade PAHs in plants. It's represented a significant opportunity to study how to reduce plant PAH contamination risk and remediate PAH-contaminated soils.
Supporting Information
Contains Tables S1–S8. Table S1. Identified 16S rDNA V3 sequences in Alopecurus aequalis. Table S2. Identified 16S rDNA V3 sequences in Oxalis corniculata. Table S3. Identified 16S rDNA sequences of endophytic bacterial isolates in Alopecurus aequalis. Table S4. Identified 16S rDNA sequences of endophytic bacterial isolates in Oxalis corniculata. Table S5. Tolerance to each PAH of different endophytic bacteria isolated from Alopecurus aequalis roots. Table S6. Tolerance to each PAH of different endophytic bacteria isolated from Alopecurus aequalis shoots. Table S7. Tolerance to each PAH of different endophytic bacteria isolated from Oxalis corniculata roots. Table S8. Tolerance to each PAH of different endophytic bacteria isolated from Oxalis corniculata shoots.
(DOC)
Funding Statement
This work was financially supported by the National Science Foundation of China (41171380, 41201501, 41071212, 21077056, 41171193), the Science Foundation of Jiangsu Province (BK2012370, BE2011780), the China Postdoctoral Science Foundation (2011M501246) and the Doctoral Fund of Ministry of Education of China (20120097120012). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Tao S, Cui YH, Xu FL, Li BG, Cao J, et al. (2004) Polycyclic aromatic hydrocarbons (PAHs) in agricultural soil and vegetables from Tianjin. Sci Total Environ 320(1): 11–24. [DOI] [PubMed] [Google Scholar]
- 2. Franzaring J, Eerden LJM (2000) Accumulation of airborne persistent organic pollutants (POPs) in plants. Basic Appl Ecol 1(1): 25–30. [Google Scholar]
- 3. Mckone TE, Maddalena RL (2007) Plant uptake of organic pollutants from soil: Bioconcentration estimates based on models and experiments. Environ Toxicol Chem 26(12): 2494–2504. [DOI] [PubMed] [Google Scholar]
- 4. Mortelmans K, Hawarth S, Lawlor T, Speck W, Tainer B, et al. (1986) Salmonella mutagenicity tests: II. Results from the testing of 270 chemicals. Environ Mutagen 8: 7–119. [PubMed] [Google Scholar]
- 5. Pahlman R, Pelkonen O (1987) Mutagenicity studies of different polycyclic aromatic hydrocarbons: the significance of enzymatic factors and molecular structures. Carcinogenesis 8(6): 773–778. [DOI] [PubMed] [Google Scholar]
- 6. Collins C, Fryer M, Grosso A (2006) Plant uptake of non-ionic organic chemicals. Environ Sci Technol 40(1): 45–52. [DOI] [PubMed] [Google Scholar]
- 7. Gao YZ, Cao XZ, Kang FX, Cheng ZX (2011) PAHs pass through the cell wall and partition into organelles of arbuscular mycorrhizal roots of ryegrass. J Environ Qual 40(2): 653–656. [DOI] [PubMed] [Google Scholar]
- 8. Wang YX, Yamazoe A, Suzuki S, Liu CT, Aono T, et al. (2004) Isolation and characterization of dibenzofuran-degrading Comamonas sp. strains isolated from white clover roots. Curr Microbiol 49(4): 288–294. [DOI] [PubMed] [Google Scholar]
- 9. Gao YZ, Shen Q, Ling WT, Ren LL (2008) Uptake of polycyclic aromatic hydrocarbons by Trifolium pretense L. from water in the presence of a nonionic surfactant. Chemosphere 72(4): 636–643. [DOI] [PubMed] [Google Scholar]
- 10. Gao Y, Li H, Gong S (2012) Ascorbic acid enhances the accumulation of polycyclic aromatic hydrocarbons (PAHs) in roots of tall fescue (Festuca arundinacea Schreb.). PLoS One 7(11): e50467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pulford ID, Watson C (2003) Phytoremediation of heavy metal-contaminated land by trees-a review. Environ Int 29(4): 529–540. [DOI] [PubMed] [Google Scholar]
- 12. Viñas M, Sabaté J, José M, Solanas AM (2005) Bacterial community dynamics and polycyclic aromatic hydrocarbon degradation during bioremediation of heavily creosote-contaminated soil. Appl Environ Microbiol 71(11): 7008–7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dai CC, Tian LS, Zhao YT, Chen Y, Xie H (2010) Degradation of phenanthrene by the endophytic fungus Ceratobasidum stevensii found in Bischofia polycarpa . Biodegradation 21(2): 245–255. [DOI] [PubMed] [Google Scholar]
- 14. Sheng XF, Chen XB, He LY (2008) Characteristics of an endophytic pyrene- degrading bacterium of Enterobacter sp. 12J1 from Allium macrostemon Bunge. Int Biodeterior Biodegrad 62(2): 88–95. [Google Scholar]
- 15. Harish S, Kavino M, Kumar N, Saravanakumara D, Soorianathasundaramb K, et al. (2008) Biohardening with plant growth promoting rhizosphere and endophytic bacteria induces systemic resistance against banana bunchy top virus . Appl Soil Ecol 39 (2): 187–200. [Google Scholar]
- 16. Lodewyckx C, Vangronsveld J, Porteous F, Moorea Edward RB, Taghavi S, et al. (2002) Endophytic bacteria and their potential applications. Crit Rev in Plant Sci 21(6): 583–606. [Google Scholar]
- 17. Shin DS, Park MS, Jung S, Lee KH, Bae KS, et al. (2007) Plant growth-promoting potential of endophytic bacteria isolated from roots of coastal sand dune plants. J Microbiol Biotechnol 17 (8): 1361–1368. [PubMed] [Google Scholar]
- 18. Davison J (1988) Plant beneficial bacteria. Nature Biotechnol 6: 282–286. [Google Scholar]
- 19. Brooksd DS, Gonzalez CF, Appel DN, Filer TH (1994) Evaluation of endophytic bacteria as potential biological control agents for oak wilt. Biol Control 4(4): 373–381. [Google Scholar]
- 20. Rajkumar M, Ae N, Freitas H (2009) Endophytic bacteria and their potential to enhance heavy metal phytoextraction. Chemosphere 77(2): 153–160. [DOI] [PubMed] [Google Scholar]
- 21. Khan Z, Doty S (2011) Endophyte-assisted phytoremediation. Curr Top Plant Biol 12: 97–105. [Google Scholar]
- 22. Germaine KJ, Liu XM, Cabellos GG, Hogan JP, Ryan D, et al. (2006) Bacterial endophyte-enhanced phytoremediation of the organochlorine herbicide 2,4- dichlorophenoxyacetic acid. FEMS Microbiol Ecol 57(2): 302–310. [DOI] [PubMed] [Google Scholar]
- 23. Moore FP, Barac T, Borremans B, Oeyen L, Vangronsveld J, et al. (2006) Endophytic bacterial diversity in poplar trees growing on a BTEX-contaminated site: The characterisation of isolates with potential to enhance phytoremediation. Syst Appl Microbiol 29(7): 539–556. [DOI] [PubMed] [Google Scholar]
- 24. Tang J, Wang R, Niu X, Zhou Q (2010) Enhancement of soil petroleum remediation by using a combination of ryegrass (Lolium perenne) and different microorganisms. Soil Till Res 110: 87–93. [Google Scholar]
- 25. Kaplan CW, Kitts CL (2004) Bacterial succession in a petroleum land treatment unit. Appl Environ Microbiol 70 (3): 1777–1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nogales B, Moore ERB, Llobet-Brossa E, Rossello-Mora R, Amann R, et al. (2001) Combined use of 16S ribosomal DNA and 16S rRNA to study the bacterial community of polychlorinated biphenyl-polluted soil. Appl Environ Microbiol 67(4): 1874–1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ho YN, Shih CH, Hsiao SC, Huang CC (2009) A novel endophytic bacterium, Achromobacter xylosoxidans, helps plants against pollutant stress and improves phytoremediation. J Biosci Bioeng 108: S75–S95. [Google Scholar]
- 28. Sobral JK, Arau˜jo WL, Mendes R, Kleiner AAP, João LA (2005) Isolation and characterization of endophytic bacteria from soybean (Glycine max) grown in soil treated with glyphosate herbicide. Plant Soil 273(1–2): 91–99. [Google Scholar]
- 29. Ling WT, Gao YZ (2004) Promoted dissipation of phenanthrene and pyrene in soils by amaranth (Amaranthus tricolor L.). Environ Geol 46(5): 553–560. [Google Scholar]
- 30. Hung PQ, Annapurna K (2004) Isolation and characterization of endophytic bacteria in soybean (Glycine sp.). Omonrice 12: 92–101. [Google Scholar]
- 31. Byers HK, Stackebrandt E, Hayward C, Blackall LL (1998) Molecular investigation of a microbial mat associated with the great artesian basin. FEMS Microbiol Ecol 25(4): 391–403. [Google Scholar]
- 32. Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, et al. (1997) Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res 25(17): 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yao ZL, Shi RJ, Zhang Y, Zhang ZZ, Xu H, et al.. (2008) Improved solid double-layer flat plate method for quick high-flux sifting motion of polycyclic aromatic hydrocarbon degradation bacterium. CN 101245366.
- 34. Gao YZ, Zhu LZ (2004) Plant uptake, accumulation and translocation of phenanthrene and pyrene in soils. Chemosphere 55(9): 1169–1178. [DOI] [PubMed] [Google Scholar]
- 35. Briggs GG, Bromilow RH, Evans AA, Williams M (1983) Relationship between lipophilicity and the distribution of non-ionised chemicals in barley shoots following uptake by the roots. Pestic Sci 14(5): 492–500. [Google Scholar]
- 36. Editorial Committee of Flora of China (1987) Flora of China. Beijing: Science Press. 9 (3): 261p. [Google Scholar]
- 37. Editorial Committee of Flora of China (1999) Flora of China. Beijing: Science Press. 43(1): 11 p. [Google Scholar]
- 38. Chiou CT, Sheng GY, Manes M (2001) A partition-limited model for the plant uptake of organic contaminants from soil and water. Environ Sci Technol 35 (7): 1437–1444. [DOI] [PubMed] [Google Scholar]
- 39. Zhu LZ, Gao YZ (2004) Prediction of phenanthrene uptake by plants with a partition-limited model. Environ Pollut 131(3): 505–508. [DOI] [PubMed] [Google Scholar]
- 40. Juhasz AL, Naidu R (2000) Bioremediation of high molecular weight polycyclic aromatic hydrocarbons: a review of the microbial degradation of benzo[a]pyrene. Int Biodeter Biodegr 42(1–2): 57–88. [Google Scholar]
- 41. Hoang HL, Dominik DS, Baldwin IT (2008) Native bacterial endophytes promote host growth in a species-specific manner; phytohormone manipulations do not result in common growth responses. PLoS ONE 3(7): e2702 doi:10.1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Brecht V, Daan V, Hester S, Braam VW, Benny L, et al. (2011) Endophytic Bacteria in Toxic South African Plants: Identification, Phylogeny and Possible Involvement in Gousiekte. PLoS ONE 6: e19265 10.1371/journal.Pone.0019265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mendes R, Kleiner AAP, Araujo WL, Raaijmakers JM (2007) Diversity of cultivated endophytic bacteria from sugarcane: genetic and biochemical characterization of Burkholderia cepacia complex isolates. Appl Environ Microbiol 73(22): 7259–7267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Vendan RT, Young JY, Sun HL, Young HR (2010) Diversity of endophytic bacteria in ginseng and their potential for plant growth promotion. J Microbiol 48(5): 559–565. [DOI] [PubMed] [Google Scholar]
- 45. Chen WM, Tang YQ, Mori K, Wu XL (2012) Distribution of culturable endophytic bacteria in aquatic plants and their potential for bioremediation in polluted waters. Aquat Biol 15: 99–110. [Google Scholar]
- 46. West ER, Cother EJ, Steel CC, Ash GJ (2010) The characterization and diversity of bacterial endophytes of grapevine. Can J Microbiol. 56 (3): 209–16 10.1139/w10-004 [DOI] [PubMed] [Google Scholar]
- 47. Miliūtė I, Buzaitė O (2011) IAA production and other plant growth promoting traits of endophytic bacteria from apple tree. Biologija 57(2): 98–102. [Google Scholar]
- 48. Garbeva P, Overbeek VL, Vuurde VJ, Elsas VJ (2001) Analysis of endophytic bacterial communities of potato by plating and denaturing gradient gel electrophoresis (DGGE) of 16S rDNA based PCR fragments. Microbial Eco 41: 369–383. [DOI] [PubMed] [Google Scholar]
- 49. Araújo WL, Marcon J, Maccheroni W, Elsas JD, Vuurde JW, et al. (2002) Diversity of endophytic bacterial populations and their interaction with Xylella fastidiosa in citrus plants. Appl Environ Microbiol 68: 4906–4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Compant S, Clement C, Sessitsch A (2010) Plant growth-promoting bacteria in the rhizo-and endosphere of plants: Their role, colonization, mechanisms involved and prospects for utilization. Soil Biol Biochem 42(5): 669–678. [Google Scholar]
- 51. McCully ME (2001) Niches for bacterial endophytes in crop plants: a plant biologist's view Aust. Aust J Plant Physiol 28(9): 983–990. [Google Scholar]
- 52. Kapley A, Siddiqui S, Misra K, Ahmad SM, Purohit HJ (2007) Preliminary analysis of bacterial diversity associated with the Porites coral from the Arabian sea. World J Microb Biot 23: 923–930. [Google Scholar]
- 53. Qing W, Zhao X, Zhao SY (2006) Application of PCR-DGGE in research of bacterial diversity in drinking water. Biomed Environ Sci 19(5): 371–374. [PubMed] [Google Scholar]
- 54.Robson Andreazza SP, Bortolon L, Okeke BC, Bento FM, Camargo FAO (2010) 19th World Congress of Soil Science, Soil Solutions for a Changing World 1–6 August 2010, Brisbane, Australia. Published on DVD.
- 55. Chelius M, Triplett E (2001) The Diversity of Archaea and Bacteria in Association with the Roots of Zea mays L. Microbial Eco 41: 252–263. [DOI] [PubMed] [Google Scholar]
- 56. Sun L, Qiu F, Zhang X, Dai X, Dong X, et al. (2008) Endophytic bacterial diversity in rice (Oryza sativa L.) roots estimated by 16S rDNA sequence analysis. Microbial Eco 55: 415–424. [DOI] [PubMed] [Google Scholar]
- 57. Zak DR, Holmes WE, White DC, Peacock AD, Tilman D (2003) Plant diversity, soil microbial communities, and ecosystem function: are there any links? Ecology 84: 2042–2050. [Google Scholar]
- 58. Sessitsch A, Reiter B, Pfeifer U, Wilhelm E (2002) Cultivation-independent population analysis of bacterial endophytes in three potato varieties based on eubacterial and Actinomycetes-specific PCR of 16S rRNA genes. FEMS Microbiol Ecol 39(1): 23–32. [DOI] [PubMed] [Google Scholar]
- 59. Phillips LA, Germida JJ, Farrell RE (2008) Hydrocarbon degradation potential and activity of endophytic bacteria associated with prairie plants. Soil Biol Biochem 40(12): 3054–3064. [Google Scholar]
- 60. Siciliano SD, Fortin N, Mihoc A, Wisse G, Labelle S, et al. (2001) Selection of specific endophytic bacterial genotypes by plants in response to soil contamination. Appl Environ Microbiol 67(6): 2469–2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Compant S, Duffy B, Nowak J, Clément C, Barka EA (2005) Use of plant growth-promoting bacteria for biocontrol of plant diseases: principles, mechanisms of action, and future prospects. Appl Environ Microbiol 71(9): 4951–4959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Taghavi S, Barac T, Greenberg B, Borremans B, Vangronsveld J, et al. (2005) Horizontal gene transfer to endogenous endophytic bacteria from poplar improves phytoremediation of toluene. Appl Environ Microbiol 71 (12): 8500–8505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Newman LA, Reynolds CM (2005) Bacteria and phytoremediation: new uses for endophytic bacteria in plants. Trends Biotechnol 23(1): 6–8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Contains Tables S1–S8. Table S1. Identified 16S rDNA V3 sequences in Alopecurus aequalis. Table S2. Identified 16S rDNA V3 sequences in Oxalis corniculata. Table S3. Identified 16S rDNA sequences of endophytic bacterial isolates in Alopecurus aequalis. Table S4. Identified 16S rDNA sequences of endophytic bacterial isolates in Oxalis corniculata. Table S5. Tolerance to each PAH of different endophytic bacteria isolated from Alopecurus aequalis roots. Table S6. Tolerance to each PAH of different endophytic bacteria isolated from Alopecurus aequalis shoots. Table S7. Tolerance to each PAH of different endophytic bacteria isolated from Oxalis corniculata roots. Table S8. Tolerance to each PAH of different endophytic bacteria isolated from Oxalis corniculata shoots.
(DOC)