Abstract
Glioblastoma (GBM) is the most prevalent primary brain tumor and ranks among the most lethal of human cancers with conventional therapy offering only palliation. Great strides have been made in understanding brain cancer genetics and modeling these tumors with new targeted therapies being tested but these advances have not translated into substantially improved patient outcomes. Multiple chemotherapeutic agents, including temozolomide, the first-line treatment for glioblastoma, have been developed to kill cancer cells. However, the response to temozolomide in GBM is modest. Radiation is also moderately effective but this approach is plagued by limitations due to collateral radiation damage to healthy brain tissue and development of radioresistance. Therapeutic resistance is attributed at least in part to a cell population within the tumor that possesses stem-like characteristics and tumor propagating capabilities, referred to as cancer stem cells. Within GBM, the intratumoral heterogeneity is derived from a combination of regional genetic variance and a cellular hierarchy often regulated by distinct cancer stem cell niches, most notably perivascular and hypoxic regions. With the recent emergence as a key player in tumor biology, cancer stem cells have symbiotic relationships with the tumor microenvironment, oncogenic signaling pathways, and epigenetic modifications. The origins of cancer stem cells and their contributions to brain tumor growth and therapeutic resistance are under active investigation with novel anti-cancer stem cell therapies offering potential new hope for this lethal disease.
Keywords: Glioma stem cells, Microenvironment, Hypoxia, Epigenetics, Therapeutic resistance
1. Introduction
Glioblastoma (GBM; World Health Organization grade IV glioma) is the most prevalent primary malignant brain tumor. GBM is a highly cellular tumor with pleomorphic cells expressing astrocytic lineage markers, nuclear and cytoplasmic atypia, frequent mitoses, and necrosis and endothelial proliferation as common findings. Its resistance to conventional therapies, rapid growth and highly infiltrative nature necessitate the use of highly aggressive therapies including tumor resection, radiation, and chemotherapy (Furnari et al., 2007; Stupp et al., 2009). Standard post-resection treatment includes the oral methylator temozolomide (TMZ) administered concurrently with radiation – which continues to evolve with use of new technologies allowing for precise tumor-specific delivery while sparing the normal brain from intense radiotherapy – followed by adjuvant TMZ. However, even with such therapies, prognosis is dismal with the median patient survival of patients under 70 years old with a resection between 14–16 months (Stupp et al., 2009). Older patients fare far worse and there is no accepted standard-of-care for the elderly. Notably, 80–90% of tumors recur within 2–3 cm of the original resection cavity and often in a nodular pattern suggesting a clonal source of recurrence. Numerous therapies have entered clinical trial for GBM therapy, and while many have shown initial promise, nearly all have had minimal or no efficacy (Adamson et al., 2009; Lima et al., 2012; Tobias et al., 2013). The anti-angiogenic bevacizumab (Avastin) has recently been FDA-approved for use with GBM and improves progression-free survival, but does not offer clear improvement in overall survival in either the recurrent or upfront setting. The minimal improvement in GBM treatment compared to other cancers along with the highly infiltrative nature of GBM stress the need for more effective and less toxic therapies. To develop more effective treatments, it is critical to understand pathways involved in the basic biology of GBM and to identify important targets for therapy. This has been aided by the recent delineation of tumor subgroups by Heidi Phillips (Phillips et al., 2006) and the Cancer Genome Atlas (TCGA), which have provided additional granularity to this disease, although this has not translated into changes in clinical management to date. Brain tumors consist of heterogeneous cell types arising from an equally complex microenvironment making it challenging yet necessary to identify the cells that are at the root of tumor development, progression and therapeutic resistance.
More than 150 years ago, German pathologist Rudolf Virchow proposed that cancers arise from the activation of dormant, embryonic-like cells present in mature tissue. Later, first in leukemias and later in breast cancers, cellular hierarchies were defined with selected cells functionally capable of self-renewal and propagating heterogeneous tumors phenocopying parental tumors (Bonnet and Dick, 1997; Lapidot et al., 1994; Reya et al., 2001). The inability to effectively treat GBM may be due in part to their cellular heterogeneity, which may be governed by at least two interactive models involving stochastic or clonal evolution and hierarchies or cancer stem cells (Reya et al., 2001). The clonal evolution model of cancer posits that selected cancer cells have the ability to proliferate, self-renew and regenerate the tumor based on the random acquisition of mutational events, creating clonally derived subpopulations within the tumor. The cancer stem cell model postulates that there is a subpopulation of cells within the tumor that has stem-like properties, including the ability to self-renew, give rise to the complex cell types within the tumor, and proliferate. Cancer stem cells yield the bulk of the cells in the tumor, which have limited tumorigenic potential and display a more differentiated phenotype. These models are not mutually exclusive, but the debate has become energized with recent studies suggesting that the cancer stem cell subpopulation could contribute to tumor growth, recurrence and therapeutic resistance. Of note, cancer stem cells remain controversial due to unresolved issues in enrichment markers, functional assays, and cellular origin (Rahman et al., 2011), yet the importance of these cells has been supported by several groups demonstrating that several types of brain tumors (gliomas, medulloblastomas, and ependymomas) display a functional cellular heterogeneity with a potential hierarchy of differentiation. Cancer stem cells, when compared to their non-stem cell counterparts, display much greater tumorigenic potential when injected into the brains of immunocompromised mice. Furthermore, studies from our laboratory and others have demonstrated that cancer stem cells are preferentially resistant to radiation through activation of the DNA damage checkpoint (Bao et al., 2006a; Blazek et al., 2007; Hambardzumyan et al., 2008b) while others have revealed chemoresistance (Liu et al., 2006; Todaro et al., 2007; Wulf et al., 2001).
2. Defining glioma stem cells
2.1 Identification
The defining features of cancer stem cells are evolving but for GBM stem cells (GSC), it is relevant to note some shared characteristics with normal neural progenitors including the expression of neural stem cell markers, as well as having the capacity for self-renewal and long term proliferation and the formation of neurospheres. Less certain is the ability to differentiate into multiple nervous system lineages (neurons, astrocytes, and oligodendrocytes). There are clearly important differences between cancer stem cells and their normal counterparts, including genetic lesions and tumor formation with unresolved issues of quiescence (Galli et al., 2004; Hemmati et al., 2003; Rich and Eyler, 2008; Sanai et al., 2005; Singh et al., 2004, 2003; Taylor et al., 2005; Vescovi et al., 2006; Yuan et al., 2004). Prospective functional GSC enrichment poses challenges and disputes across the field due to technical variance and lack of universal markers, suggesting the need for continued efforts in validating GSC markers with improved consistency in enrichment methods. Initial reports by Singh and colleagues demonstrated that as few as 100 brain tumor cells expressing the neural stem cell surface marker, CD133 (prominin-1), were capable of recapitulating tumor growth in vivo upon transplantation into immunocompromised mice (Singh et al., 2004). Over the last decade several additional candidate markers used to enrich for GSCs have emerged, including A2B5 (Ogden et al., 2008; Tchoghandjian et al., 2010), CD44 (Anido et al., 2010), CD171 (L1CAM) (Bao et al., 2008), CD15 (SSEA1) (Read et al., 2009; Son et al., 2009; Ward et al., 2009), CD49f (integrin α6) (Lathia et al., 2010), Musashi (Thon et al., 2010), Nestin (Bexell et al., 2009; Zhang et al., 2008), Nanog (Guo et al., 2011; Mathieu et al., 2011; Niu et al., 2011), Oct4 (Guo et al., 2011; Ikushima et al., 2011; Mathieu et al., 2011) and Sox2 (Ge et al., 2010; Guo et al., 2011; Hägerstrand et al., 2011), yet CD133 has remained as the most popular marker to date. Other functional assays include sphere formation that limits the ability to compare cells and requires substantial time in culture, the side population that has failed to validate, and the Aldefluor assay. There is still no clear universal GSC marker, which is likely due to differences in diversity of cellular hierarchies between tumors (possibly reflected in different TCGA subgroups). Some scientists have dismissed the cancer stem cell hypothesis due to the lack of consistent markers but this is a simplistic view as no two human tumors have identical genotypes so it is likely that the molecular features of cellular hierarchies show variability, with evidence of dynamic marker information within tumors during disease course. Cancer stem cells play integral roles in maintaining and promoting tumor niches (perivascular-proliferative and perinecrotic-hypoxic, Figure 1). A number of molecular targets that regulate GSCs directly influence the cellular microenvironment, further regulating the cancer stem cell state (Bao et al., 2006b; Calabrese et al., 2007; Gangemi et al., 2009; Heddleston et al., 2009; Lathia et al., 2010; Li et al., 2009a; Seidel et al., 2010; Soeda et al., 2009; Wang et al., 2008). It is likely that the GSC state is a reversible phenotype and that no single cellular marker is universally informative for GSCs, necessitating the use of validated functional assays and the use patient derived tumors with no or limited time in culture to prevent potential changes from the original tumor phenotype.
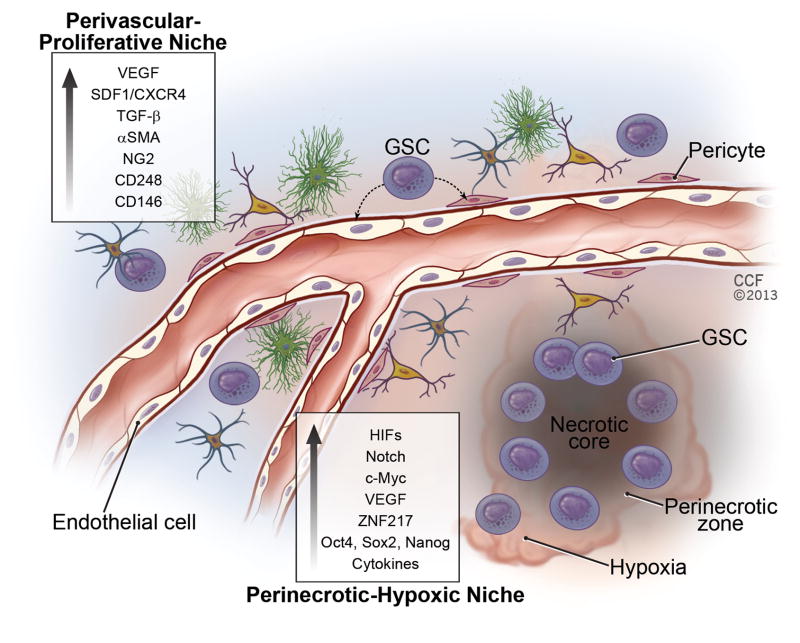
Figure 1.
GBM stem cell (GSC) microenvironment. GSCs often reside and thrive in perivascular and hypoxic niches where growth factors and cytokines promote their maintenance. In turn, GSCs up-regulate the expression, activation, and secretion of a number of niche-dependent signaling molecules and transcription factors involved in pathways such as angiogenesis, proliferation, migration, hypoxia response, etc. GSCs interact with other tumor cells and have the potential to differentiate into other cell types including endothelial cells and pericytes.
2.2 Characterization
Molecular signals that drive tumor formation and maintenance are frequently shared with normal development and wound responses, processes in which normal stem and progenitor cells function (Al-Hajj et al., 2003; Bonnet and Dick, 1997; Galli et al., 2004; Hemmati et al., 2003; Lapidot et al., 1994; Reya et al., 2001; Rich and Eyler, 2008; Rosen and Jordan, 2009; Sanai et al., 2005; Shackleton et al., 2009; Singh et al., 2004, 2003; Taylor et al., 2005; Vescovi et al., 2006; Yuan et al., 2004; Hongwu Zheng et al., 2008). However, GSCs exhibit significant distinctions from normal stem cells in frequency, proliferation, aberrant expression of differentiation markers, chromosomal abnormalities, and tumor formation (Al-Hajj et al., 2003; Bonnet and Dick, 1997; Galli et al., 2004; Hemmati et al., 2003; Lapidot et al., 1994; Reya et al., 2001; Rich and Eyler, 2008; Rosen and Jordan, 2009; Sanai et al., 2005; Shackleton et al., 2009; Singh et al., 2004, 2003; Taylor et al., 2005; Vescovi et al., 2006; Yuan et al., 2004; Hongwu Zheng et al., 2008). Importantly, cancer stem cells need not be derived from normal stem cells but may be subjected to evolutionary pressures that select for the capacity to self-renew (Al-Hajj et al., 2003; Bonnet and Dick, 1997; Galli et al., 2004; Hemmati et al., 2003; Lapidot et al., 1994; Reya et al., 2001; Rich and Eyler, 2008; Rosen and Jordan, 2009; Sanai et al., 2005; Shackleton et al., 2009; Singh et al., 2004, 2003; Taylor et al., 2005; Vescovi et al., 2006; Yuan et al., 2004; Hongwu Zheng et al., 2008). The potent tumorigenic capacity of cancer stem cells coupled with increasing evidence of radioresistance and chemoresistance suggests that cancer stem cells contribute to tumor maintenance and recurrence and that targeting cancer stem cells may offer new avenues of therapeutic intervention (Zhou et al., 2009). This hypothesis has been recently validated in breast cancer clinical trials in which patients undergoing treatment with cytotoxic chemotherapy experienced an increase in breast cancer stem cells in the surviving tumor while the use of a targeted therapeutic against the stem cell population stabilized the cancer stem cell population (Creighton et al., 2009; Li et al., 2008; Schott et al., 2013). Together these data suggest that significant effort should be undertaken to identify potential targets in cancer stem cells that not only promote tumor maintenance but would be amenable to disruption. Overall, GSCs add additional complexity to brain cancer models with GSCs as potential products and regulators of their microenvironment, contributing to the concept of both a cell autonomous and an extrinsically defined cancer stem cell (Rosen and Jordan, 2009).
3. Microenvironmental regulation of GSCs
3.1 Perivascular niche
Several molecular GBM cancer stem cell targets that integrate the cellular microenvironment with the cell state have been defined (Bao et al., 2006b; Calabrese et al., 2007; Gangemi et al., 2009; Guryanova et al., 2011; Heddleston et al., 2009; Lathia et al., 2010; Li et al., 2009a; Seidel et al., 2010; Soeda et al., 2009; Wang et al., 2008). GSCs reside in specific anatomical and functional locations or niches that consist of direct contact with several cell types and as well as access to extracellular matrix and secreted factors that likely play a role in maintaining properties of stem cell self-renewal and proliferation (Figure 1). Neural stem cell maintenance and self-renewal is regulated by adhesion and a vascular niche, so it is not surprising that GSCs are commonly located within close regulatory contact with the tumor vasculature. Medulloblastoma cells positive for both nestin (a filament protein expressed during neural development) and CD133 reside next to capillaries, particularly those with the highest microvessel density. CD133-positive cells receive secreted factors from endothelial cells that foster a self-renewing and undifferentiated state for GSCs (Calabrese et al., 2007). Co-implanting endothelial cells with CD133-positive cells into the brains of immunocompromised mice rapidly accelerates tumor growth, further highlighting the importance of how the endothelium can aid in promoting GSC tumorigenicity. When the contribution of the perivascular niche is inhibited through the use of anti-angiogenics (such as bevacizumab), the percentage of self-renewing GSCs is reduced as is the growth of glioma orthotopic xenografts.
GSCs are not passive recipients of microenvironmental cues as they in turn can stimulate angiogenesis through elaboration of pro-angiogenic growth factors and physical contributions to the vasculature (Bao et al., 2006b; Ricci-Vitiani et al., 2010; Soda et al., 2011; Wang et al., 2010). Patient-derived GSCs produce much higher levels of VEGF (consistently up-regulated 10- to 20-fold) compared to non-GSCs, under normoxia and hypoxia (Bao et al., 2006b). Tumors generated from CD133-positive glioma cells are highly angiogenic with increased tumor vascularity, necrosis, and hemorrhage compared to CD133− glioma cells. GSCs also induce endothelial cell migration and tube formation whereas overexpression of vascular endothelial growth factor (VEGF) in GSCs induces the expansion of vascular-rich GBM tumors (Bao et al., 2006b; Oka et al., 2007). Cultures enriched for GSCs induced higher levels of endothelial progenitor cell proliferation, recruitment, and mobilization compared to low-GSC cultures (Folkins et al., 2009). When VEGF or stromal-derived factor 1 (SDF-1) is blocked in high-GSC cultures, all aspects of angiogenesis are severely reduced (Bao et al., 2006b; Folkins et al., 2009).
To add further complexity to the relationship of GSCs with the endothelium, recent studies have identified endothelial cells of possible tumor origin. A subset of endothelial cells within GBM that contain the same somatic mutations often present in tumors cells such as amplification of EGFR and chromosome 7 (Wang et al., 2010). Between 20–90% of endothelial cells in GBM have the same genomic alterations as tumor cells, further supporting the idea that vascular endothelium have a neoplastic origin (Ricci-Vitiani et al., 2010). Tumor-derived GSCs cultured in endothelial medium express endothelium markers and that these cells have the ability to differentiate in vitro and in vivo into mature endothelial cells. While exposure to the anti-angiogenic bevacizumab or RNA interference against VEGFR2 inhibit the differentiation of tumor endothelial progenitors, neither of these treatments have the same effect of blocking differentiation from GSCs to endothelial progenitors (Wang et al., 2010; Ricci-Vitiani et al., 2010). However, analysis of a large number of human brain tumors has suggested that tumor-derived endothelia are rare (Rodriguez et al., 2012). In contrast, GSCs were shown to differentiate into pericytes to support vessel function and tumor growth in a recent study. Upon differentiation, a fraction of GSCs express multiple pericyte markers in vitro such as α-SMA, NG2, CD248, and CD146 (Cheng et al., 2013). Lineage tracing of GSCs from 21 GBM xenografts reveal that GSCs gave rise to pericytes in vivo (Cheng et al., 2013). Furthermore, GSCs are recruited to endothelial cells via SDF-1/CXCR4 signaling with GSC differentiation into pericytes due in part through endothelial cell-secreted TGF-β (Cheng et al., 2013). Collectively, these studies indicate that the paracrine interaction between GSCs and the perivascular niche is more representative of a two-way street, with both compartments feeding off of one another.
4. Hypoxia and GSCs
4.1 Hypoxia signaling
Tumor-mediated recruitment or at least attraction of vasculature acts a neoplastic feeding source yet often vessels are highly disorganized and diffuse of oxygen likely due to rapid tumor expansion (Carmeliet and Jain, 2000). Solid tumors, including gliomas, contain regions of irregular blood flow that experience fluctuating abnormal hypoxic oxygen tension levels - ranging from less that 1% to 5% (Dewhirst et al., 2008). In GBM patients, hypoxia is a negative prognostic factor and is associated with tumor aggression (Evans et al., 2010; Sathornsumetee et al., 2008). The hypoxic microenvironment is also linked to therapeutic resistance as MGMT (a key player in TMZ resistance) is expressed in hypoxic regions in gliomas (Pistollato et al., 2010). Furthermore, hypoxia increases the expression of the stem cell marker CD133 in brain tumors (Blazek et al., 2007; Platet et al., 2007). Hypoxia mediated responses are carried out predominantly by hypoxia-inducible factor (HIF) family of transcription factors. Under normoxic conditions, HIFs are hydroxylated by HIF prolyl-hydroxylases allowing for ubiquitination by the von Hippel-Lindau (vHL) tumor-suppressor gene followed by proteasomal degradation (Harris, 2002; Keith and Simon, 2007). Yet under hypoxia, HIF prolyl-hydroxylase which requires oxygen is inhibited. HIF proteins become stabilized leading to dimerization and subsequent binding to hypoxia-responsive elements (HREs) on the promoters of target genes often involved in modulating cell survival, motility, metabolism, and angiogenesis (Harris, 2002; Keith and Simon, 2007).
4.2 Regulatory role of hypoxia inducible factors in GSCs
HIF1α is widely expressed in various tissues while HIF2α expression appears to be more restricted. In parallel, HIF1α promotes proliferation and survival of all cancer cells as well as being activated in normal neural progenitors, potentially limiting its therapeutic index. On the other hand, HIF2α is specifically regulated with enhanced transcription in GSCs even in modest hypoxic conditions while non-glioma stem cells express essentially no HIF2α, making HIF2α an appealing GSC target (Li et al., 2009a). Other hypoxia-induced genes that are higher expressed in GSCs compared to matched non-stem glioma cells include Oct4, glucose transporter type 1 (Glut1), SerpinB9, and VEGF. Targeting HIF1α or HIF2α using RNA interference decreases GSC neurosphere formation and proliferation as well as VEGF activity and endothelial cell growth in vitro (Li et al., 2009a). The gold standard assay used to evaluate cancer stem cells is the ability to propagate tumors in vivo. GSCs transduced with either HIF1α or HIF2α shRNA are unable to initiate tumors in an orthotopic xenograft model, supporting an essential role for both HIFs (Li et al., 2009a).
In non-stem glioma cells, over-expression of HIF2α demonstrates a role in cellular reprogramming of cancer. Ectopic delivery of HIF2α or utilization of a non-degradable HIF2α mutant construct results in an increase in the fraction of CD133-positive cells, an upregulation of Oct4, Nanog, and c-Myc mRNA levels and increased tumor formation in vivo (Heddleston et al., 2009). Similarly, hypoxia regulates the side-population gene signatures associated with the perivascular or perinecrotic niches (Seidel et al., 2010). While overexpressing HIF1α has little effect on cancer stem cell genes, overexpressing HIF2α dramatically increases the levels of side population markers (Seidel et al., 2010). HIF2α expression correlates strongly with poor glioma patient survival (Sathornsumetee et al., 2008) and is induced by acidic stress (Hjelmeland et al., 2011). HIF2α may regulate global chromatin structure through activation of epigenetic modifiers such as the histone methyltransferase, mixed lineage leukemia 1 (MLL) (Heddleston et al., 2012), further demonstrating how the microenvironment can have widespread effects on tumor heterogeneity and even patient outcomes.
Studies primarily examining the role of HIF1α further support hypoxia-mediated expansion and self-renewal of CD133-positive cells as well as regulating expression of surface markers CXCR4, CD144low, and A2B5 through cross-talk and possible amplification with the PI3K-Akt and ERK1/2 signaling pathways (Soeda et al., 2009). Hypoxia stimulates numerous pathways and targets hijacked by GSCs, including the Notch pathway, induction of stem genes (such as KLF4 and Sox2) and overexpression of additional oncogenes like ZNF217 (Bar et al., 2010; Mao et al., 2011). In addition to the aforementioned effects on GSC-mediated angiogenesis, hypoxia augments other GSC-microenvironmental interactions including the potentiation of GSC-mediated immunosuppression. Through pSTAT activation, hypoxia increases GSC secretion of immunosuppressive cytokines (CSF1 and CCL2) and enhances the ability of GSCs to inhibit T cell proliferation and macrophage phagocytosis (Wei et al., 2011). Thus, tumor hypoxia and the wide range of effects on GSC intra- and extracellular signaling pathways might explain why a number of the recent anti-angiogenic and immunosuppressive therapies have not yielded more successful responses.
5. GSC signaling pathways
5.1 Notch
Notch family proteins (Notch 1, 2, 3, 4) are transmembrane receptor proteins that mediate cell-cell interactions and function in proliferation, differentiation, and apoptosis (Purow et al., 2005). Notch receptors interact with five ligands (Delta-like 1, 3, 4 and Jagged-1, 2) (Lathia et al., 2008; Ohishi et al., 2002). Activation of Notch-1, the best described Notch receptor, occurs upon binding to Delta-like or Jagged ligands at the epidermal growth factor region of the Notch-1 extracellular domain (Purow et al., 2005). Two sequential enzymatic cleavages by the γ-secretase complex release the Notch intracellular domain (NICD) from the membrane to the nucleus. The NICD translocates into the nucleus where it binds the CBF1/RBPJk/Su(H)/Lag1 (CSL) transcription factor family. The CSL family can either activate or repress transcription via histone acetylases and histone deacetylases, respectively (Kopan, 2002; Lathia et al., 2008). Upon binding of the NICD, CBF-1/RBP-JK becomes a transcriptional activator of beta-helix loop helix (bHLH) transcription factors known as hairy/enhancer of split (HES) genes.
Notch signaling has a role in neural embryological development and within the adult central nervous system (CNS) both physiologically and pathologically (Presente et al., 2001). Notch signaling promotes neural stem cell survival and self-renewal during embryonic CNS development (Mizutani et al., 2007) and may also have a role in adult brain plasticity (Presente et al., 2001). Pathologically, Notch signaling regulates brain tumor progression and cancer stem cells. Inhibition of Notch-1 signaling via a γ-secretase inhibitor induces medulloblastoma stem cell apoptosis and differentiation and reduces tumor progression (Fan et al., 2006). γ-secretase inhibitors enhance temozolomide treatment of human gliomas in vitro and in flank tumors (Gilbert et al., 2010). Notch signaling is activated in human glioma cultures (Shiras et al., 2007) and primary human GBM specimens (Kanamori et al., 2007; Lino et al., 2010; Shih and Holland, 2006). Notch signaling is also a major pathway activated in GSCs, with evidence demonstrating that Notch promotes radioresistance in GSC (J. Wang et al., 2010). Blocking Notch signaling in human GSCs using high concentrations of γ-secretase inhibitors decreases tumor sphere formation, GSC proliferation, and xenograft growth as well as increases differentiation (Fan et al., 2010). Additionally, in a K-ras induced GBM mouse model, Notch activates the stem-cell marker, nestin, further suggesting that Notch contributes to the stem-like phenotype of GSCs (Shih and Holland, 2006). Other proteins that interact with Notch, including the Delta/Notch-like epidermal growth factor-related receptor (DNER) (Sun et al., 2009), Notch ligand Delta-like 4 (DLLR) (Hoey et al., 2009; Li et al., 2011, p. 4, 2007), CXCL4 (Williams et al., 2008), interferon regulatory factor 7 (Jin et al., 2012), ADAM17 (Chen et al., 2013), inhibitor of differentiation-4 (ID4) (Jeon et al., 2008), and miR-18A*(Turchi et al., 2013), all regulate GBM growth and/or GSC activity (Cheng et al., 2010).
While Notch-1 is the most commonly investigated Notch isoform, recent studies identify a similar role for Notch-2, demonstrating that primary GSCs have high Notch-2 expression (Yoon et al., 2012). Additionally, constitutive Notch-2 signaling in neural stem cells produce features similar to GSCs (Tchorz et al., 2012). Collectively, it is apparent that Notch signaling represents a pathway commonly in the midst of regulating the GSC phenotype.
5.2 Phosphotidylinositol 3-kinase (PI3K)/Akt
Receptor tyrosine kinases (RTKs) promote signal transduction after being activated by cytokines or growth factors, including epidermal growth factor (EGF) and basic fibroblast growth factor (bFGF), two mitogens typically used to grow GSCs in culture (Lee et al., 2006). One of the best characterized pathways downstream from RTK is the Akt pathway, an important component of the PI3K/Akt/mTOR pathway that is commonly found in human cancers (Hambardzumyan et al., 2008c). Over-activation of Akt, which is involved in cell survival and growth, can occur through loss of PTEN, oncogenic ras, and mutations of PI3K (Hambardzumyan et al., 2008b, 2008c). Downstream targets of Akt include the BCL-2 family member BAD, caspase-9, HDM2, Forkhead transcription factors, and others, many of which are involved in cell survival (Brunet et al., 1999; Cardone et al., 1998; Datta et al., 1997; Hambardzumyan et al., 2008a; Mayo and Donner, 2001). PTEN deletions activate the PI3K/Akt pathway, possibly involved in the pathogenesis of medulloblastomas (Hartmann et al., 2006). In malignant gliomas, the PI3K/Akt/mTOR pathway is commonly activated by EGFR, which is frequently amplified or constitutively activated in GBM and the GSC subpopulation (Choe et al., 2003; Huang et al., 2010; Moscatello et al., 1998). Overexpression of EGFR in mice induces formation of glioma-like tumors (Holland et al., 1998) and inhibition of EGFR decreases GSC proliferation and increases GSC apoptosis (Soeda et al., 2008). Transducing primary astrocytes with Akt and c-Myc increases tumorigenicity and increases expression of several stem cell markers (Radke et al., 2013). CD133 may directly bind the p85 regulatory subunit of PI3K to activate the pathway (Wei et al., 2013). CD133 knockdown inhibits PI3K/Akt pathway activity while reducing GSC self-renewal and tumorigenicity. CD133 phosphorylation not only overlaps with Akt activation, but also correlates with tumor grade. Directly inhibiting Akt preferentially disrupts GSC maintenance causing increased apoptosis, decreased neurosphere formation, and reduced migration and invasion (Eyler et al., 2008; Gallia et al., 2009). In vivo, targeting Akt delays intracranial tumor development in mice thereby extending survival (Eyler et al., 2008; Gallia et al., 2009).
5.3 Hedgehog
During CNS development, Sonic Hedgehog-Gli signaling is involved in the patterning of cell differentiation and organization of dorsal brain structures (Cayuso et al., 2006; Stecca and Ruiz i Altaba, 2005). In adults, this pathway regulates neural stem cells but also aberrantly activated in cancer to promote tumor progression and metastatic growth (Ruiz i Altaba et al., 2007). Hedgehog-Gli signaling is overactivated in GBM and medulloblastoma, and to a lesser extent, in neuroblastoma, as compared with normal tissues (Shahi et al., 2008). In GSCs, Hedgehog-Gli signaling increases expression of stem genes (i.e. CD133, Olig2, Oct4, Nanog, Sox2, etc.), promotes self-renewal, and supports glioma growth and survival (Bar et al., 2007; Clement et al., 2007; Ehtesham et al., 2007; Xu et al., 2008). Inhibiting the hedgehog pathway has a number of effects on GSCs, including suppression of self-renewal and migration, induction of apoptosis, enhancement of TMZ efficacy, and delay of tumor development in mice (Clement et al., 2007; Uchida et al., 2011; Ulasov et al., 2011). While hedgehog antagonists have shown limited activity in early clinical trials for glioma patients, the use of a hedgehog pathway inhibitor (GDC-0449) in a patient with metastatic medulloblastoma led to rapid, albeit transient, regression of the tumor and associated symptoms (Rudin et al., 2009).
5.4 Bone morphogenetic protein
Bone morphogenetic proteins (BMPs) are a family of growth factors that bind to their cell-surface receptor kinases to regulate proliferation, apoptosis, and differentiation in neural stem cells (Panchision and McKay, 2002). The canonical effectors are the Smad proteins (Receptor Smads: Smad 1/5/8, Co-Smad: Smad 4), which translocate into the nucleus to regulate transcription (Li et al., 2009b). In GSCs, BMPs induce differentiation, determined by an increase in the number of GFAP-positive cells, and delay tumor growth (Lee et al., 2008). In vivo delivery of BMP4 blocks GBM growth in some models (Piccirillo et al., 2006). BMP4 inhibits GSC proliferation through down-regulation of cyclin D1 and induces GSC apoptosis in GSCs via increasing Bax expression and inhibiting Bcl-2 and Bcl-xL (Zhou et al., 2011). GSC tumorigenicity increases upon epigenetic silencing of the BMP receptor, BMPR1B (Lee et al., 2008). It is important to note that in this study, not all GSCs were responsive to BMP-mediated glial differentiation. Specifically, the GSCs isolated from a particular patient had defective BMP signaling and hypermethylation of the BMPR1B promoter, which could be restored by demethylation or knockdown of EZH2 (a facilitator of CpG methylation). Accordingly, as treatment options become more personalized it is important to consider the individual tumor’s genomic and epigenetic characteristics when determining optimal treatment.
5.5 Wnt β-catenin
Highly conserved in numerous organisms, the canonical Wnt β-catenin pathway is involved in embryonic development, as well as controls homeostatic self-renewal and regulates normal and cancer stem cell maintenance (Cheng et al., 2010; Clevers, 2006; Grigoryan et al., 2008). The signal cascade is initiated by Wnt ligands, causing binding of Wnt to specific receptors in the Frizzled and low density lipoprotein receptor-related protein/alpha2-macroglobulin receptor (LRP) families. In the presence of Wnt ligands, the destruction complex that usually captures β-catenin is disassembled, and the free β-catenin is able to translocate to the nucleus where it binds the T-cell factor/lymphoid enhancer factor (TCF/LEF) family of transcription factors thereby transcribing Wnt target genes (Grigoryan et al., 2008). Both gain- and loss-of-function mutations of Wnt are associated with CNS cancers. Genomic alterations in the Wnt pathway are present in medulloblastomas (Rogers et al., 2012; Thompson et al., 2006) and high-grade gliomas (Schüle et al., 2012). β-catenin is a negative prognostic factor for GBM, more predictive than Ki67, p53, and EGFR (Rossi et al., 2011). Targeting Wnt in glioma cells in vivo reduces intracranial tumor formation (Kaur et al., 2013). The role of Wnt β-catenin signaling is becoming more clear in cancer stem cells, particularly in glioma and medulloblastoma stem cells (Gong and Huang, 2012; Manoranjan et al., 2013). FoxG1 and FoxM1, effector proteins in the Wnt β-catenin pathway, correlate with tumor grade and invasiveness. Deleting FoxM1 disrupts the Wnt/FoxM1/β-catenin complex and nuclear localization of β-catenin, which regulates self-renewal and tumorigenicity of GSCs (Gong and Huang, 2012; Zhang et al., 2011). Wnt3a and Wnt1 ligands are overexpressed in GSCs and their inhibition decreases cell proliferation, cell migration, and drug resistance (Kaur et al., 2013). Others wnt targets, such as LGR5 (Nakata et al., 2013), Dishevelled 2 (Pulvirenti et al., 2011) and Frizzled 4 (Jin et al., 2011) promote self-renewal, tumorigenicity, and invasiveness of GSCs.
5.6 STAT3
Signal transducer and activator of transcription 3 (STAT3) is involved in neural stem cell and astrocyte development (de la Iglesia et al., 2009) and has oncogenic or tumor suppressive roles in GBM depending on the tumor genotype (de la Iglesia et al., 2008). STAT3 can be activated by the ciliary neurotrophic factor (CNTF) family cytokine receptors in neural cells to, in turn, activate JAK cytokine-receptor associated tyrosine kinases. STAT3 is then recruited to the cytokine receptor, and activated when the tyrosine residue, Tyr705, is phosphorylated (de la Iglesia et al., 2009). STAT3 can also be directly activated by the tyrosine kinase activity of growth factors receptors, such as EGFR. STAT3 mRNA levels in GBMs are associated with patient progression-free survival and overall survival, (Televantou et al., 2013). STAT3 also interacts with inducible nitric oxide synthase (iNOS) and EGFR to promote glioma formation (Puram et al., 2012). Preferential activation of STAT3 in GSC can occur through the activity of the bone marrow X-linked nonreceptor tyrosine kinase, BMX, which is dispensable in normal neural stem cells suggesting a high therapeutic index (Guryanova et al., 2011). Additionally, the STAT3 pathway is required for survival, proliferation, and multipotency of GSCs, and is important in GBM in vivo tumor growth (Cao et al., 2010; Wang et al., 2009; Wei et al., 2010). Inhibiting the STAT3 pathway regulates the cell cycle in GSCs by inducing G1 arrest and downregulation of cyclin D1 and up-regulation of p21WAF1/CIP1. STAT3 inhibition also decreases expression of CD133 and c-Myc in GSCs and causes apoptotic cell death (Sai et al., 2012). Moreover, phosphorylation of STAT3 is a possible mechanism of interferon (IFN) treatment induction of differentiation and reduction of the tumorigenic GSC pool in vivo (Yuki et al., 2009). Further highlighting the range of oncogenic process potentially regulated by STAT3 signaling, STAT3 is necessary for GSC-induced immunosuppressive immunity in GBM (Wu et al., 2010). In this study, inhibiting the STAT3 pathway reversed anti-phagocytic and anti-inflammatory modulation by GSCs.
6. GSC transcription factors
In addition to the signaling pathways that transduce extracellular signals into the cell, transcription factors inside the cells also have an integral role in the regulation of brain tumor stem cells. These factors, which include Sox2, Oct4, Nanog, c-Myc, Olig2, and Bmi1 among others, are involved in GSC survival, maintenance, self-renewal, and proliferation.
6.1 Oct4, Sox2, and Nanog
Oct4 interacts with Sox2 and Nanog to regulate patterns of self-renewal and differentiation in embryonic stem cells (Li et al., 2009b). Takahashi and Yamanaka transduced Oct4 and Sox2 (with Klf4 and c-myc) into mouse embryonic fibroblasts to induce pluripotent stem cells (Takahashi and Yamanaka, 2006). Oct4 is highly expressed in human gliomas and correlates with tumor grade (Du et al., 2009). Oct4 also promotes colony formation and inhibits differentiation in glioma cells, potentially through upregulation of phosphorylated STAT3. Oct4 and Sox2 are increased in GSC and promote tumorigenic activity as validated by tumorsphere formation and intracerebral tumor formation (Ikushima et al., 2011, 2009). ID4 represses miR-9*-mediated suppression of Sox2, thereby increasing activity of Sox2 leading to maintenance of GSC and promoting GSC chemoresistance (Jeon et al., 2011). Inhibiting Nanog via siRNA or resveratrol reduces GSC self-renewal and tumorigenicity (Sato et al., 2013). Nanog expression is higher in GSC, co-expressed with CD133-positive glioma cells, and less expressed in regions enriched for the differentiation marker, GFAP (Niu et al., 2011). Nanog interacts with the hedgehog-gli1 pathway to modulate GSC proliferation, neurosphere formation, and tumor promotion in orthotopic xenografts (Zbinden et al., 2010).
6.2 c-Myc
Discovered more than 30 years ago, the high frequency of c-myc alterations has contributed to a large number of cancer deaths, making it one of the best studied transcription factors and oncoproteins in normal and cancer stem cells (Bishop, 1982; Sheiness et al., 1978). c-Myc increases the efficiency of cellular reprogramming of adult fibroblasts with Oct4, Sox2, and Klf4 to induce pluripotency (Takahashi and Yamanaka, 2006). c-Myc levels correlate with glioma tumor grade (Cheng et al., 2010; Yoshida et al., 2009) and c-Myc is highly expressed in GSCs relative to non-GSCs. Not only does c-Myc promote proliferation, it may represent a GSC-specific survival factor (Wang et al., 2008). Based on flow-cytometry, c-Myc is highly expressed in approximately half of CD133-positive cells acutely isolated from primary human GBM specimens whereas expression is considerably lower in the CD133-negative fraction. In acutely frozen human glioma surgical specimens, 90% of nestin-positive cells co-express c-Myc. Knockdown of c-Myc in GSCs reduces proliferation causing cell cycle arrest in the G(0)/G(1) phase with upregulation of cell cycle regulators p21WAF1/CIP1 and down regulation of cyclin D1. Furthermore, injecting GSCs with limited c-Myc activity into the brains of immunocompromised mice do not form tumors, demonstrating the importance of c-Myc for maintaining the tumorigenic potential of GSCs. Supporting this role for c-Myc in brain tumors, the DePinho lab demonstrated that p53 and Pten deficiencies in their murine glioma models stimulates c-Myc, which upon knockdown, causes GSCs to differentiate, reducing their capability to form tumors in vivo (H Zheng et al., 2008). Along these lines, primary astrocytes on a p53−/− background transduced to express c-Myc and Akt increase their expression of the stem cell markers CD133, Musashi-1, Nestin, and Olig2 with decrease immunopositivity for the differentiation marker, GFAP. Additionally, p53−/− astrocytes expressing c-Myc and Akt are tumorigenic in vivo (Radke et al., 2013).
6.3 Olig2
Olig2 is a basic helix-loop-helix (bHLH) transcription factor that is specific to the CNS (Takebayashi et al., 2000) and functions in the oligodendrocyte lineage as well as other multipotential neuron/glia progenitors (Zhou and Anderson, 2002). Among human brain tumors, Olig2 is highly expressed in diffuse gliomas including the astrocytomas, oligodendrogliomas, and oligoastrocytomas (Ligon et al., 2004). Mice intracranially implanted with neurospheres derived from neural stem/progenitor cells from p16Ink4a/p19Arf null mouse embryos that also contain active mutation of epidermal growth factor receptor (EGFRvIII) have 100% glioma formation (Ligon et al., 2007). These mice do not get tumors when the neurospheres have loss of Olig2 (Ligon et al., 2007). Moreover, Olig2 (but not Olig1) is critical for proliferation of brain tumor stem cells (Ligon et al., 2007). In human GBM sections and quantitative flow cytometry of fresh human GBM specimens, Olig2 is expressed in at least 85% of the Ki67-positive glioma progenitor cells. Of the CD133-positive fraction, nearly all cells (98%) are positive for Olig2. Additionally, Olig2 is expressed in the majority of cycling cells, based on co-localization with BrdU labeling and directly interacts with the p21 gene. Specifically in GSCs, knockdown of L1CAM reduces Olig2 and upregulates p21WAF1/CIP1 to induce apoptosis and reduce GSC growth and neurosphere formation. Similar effects are observed in vivo (Bao et al., 2008). This suggests that Olig2 can control GSC proliferation through multiple avenues, including cell adhesion and cell cycle progression. Furthermore, Olig2 expression may not be restricted to only a stem-like glioma cell phenotype but may also represent a multipotent progenitor cell phenotype still able to contribute to tumor growth as in the cease with PDGF-driven gliomas (Barrett et al., 2012). Forced expression of Neurogenin-2 (Ngn2), which functions in opposition to Olig2, causes sharp down-regulation of Olig1/2, as well as Shh and Myc, in GBM stem-like cells which is accompanied by cell death, inhibition of proliferation, and neuronal differentiation (Guichet et al., 2013). These studies support the role of Olig2 as being important in maintaining glioma stemness and tumor growth forming capabilities.
6.4 Bmi1
Bmi1 is one of the Polycomb group proteins, which act as epigenetic silencers to regulate stem cell function during embryonic development (Acquati et al., 2013). Bmi1 is a component of the Polycomb Repressive Complex 1 found in undifferentiated neural stem cells and high grade gliomas, with higher expression correlating to poor glioma patient survival (Acquati et al., 2013; Häyry et al., 2008; Li et al., 2009b). Bmi1 is enriched in GSCs and is required for their self-renewal (Facchino et al., 2010). Bmi1 is also enriched in chromatin after irradiation and in DNA damage response proteins. By knocking down Bmi1, the DNA damage response is impaired, thereby increasing GSC sensitivity to radiation. GSCs and normal neural stem cells may depend on the same epigenetic mechanism to survive the hyperproliferative state caused by upregulated Bmi1 expression (Acquati et al., 2013). As discussed in more detail below, miR-128 down regulates Bmi1, which blocks GSC self-renewal (Godlewski et al., 2008). Consistent with a reduction in Bmi1 is a decrease in H3K27 methylation and Akt activation along with overexpression of p21WAF/CIP1, a regulator of cell cycle progression. In addition to these findings in glioma stem cells, Bmi1 is integral in the maintenance of medulloblastoma stem cells (Wang et al., 2012).
7. Epigenetic regulation of GSCs
7.1 DNA methylation
Epigenetic regulation controls gene expression through mechanisms other than changes in the underlying genomic sequence. Increasing evidence points to the critical role epigenetics have in defining cellular state and that epigenetic mechanisms help regulate the cellular hierarchy seen in both normal and neoplastic tissue (Carén et al., 2013; Smith and Meissner, 2013; Suvà et al., 2013). The embryonic and induced pluripotent stem cell fields have shown that the epigenetic state of a cell is critically important in determining both the reprogramming and differentiation potential of a cell (Leonardo et al., 2012; Papp and Plath, 2013) and studies have shown this may also be true in cancer stem cells, including GSCs (Stricker et al., 2013; Suvà et al., 2013). The most studied epigenetic mechanisms in GSCs are DNA methylation, chromatin remodeling through histone methylation and regulatory non-coding RNAs (Figure 2) (Carén et al., 2013; Heddleston et al., 2011).
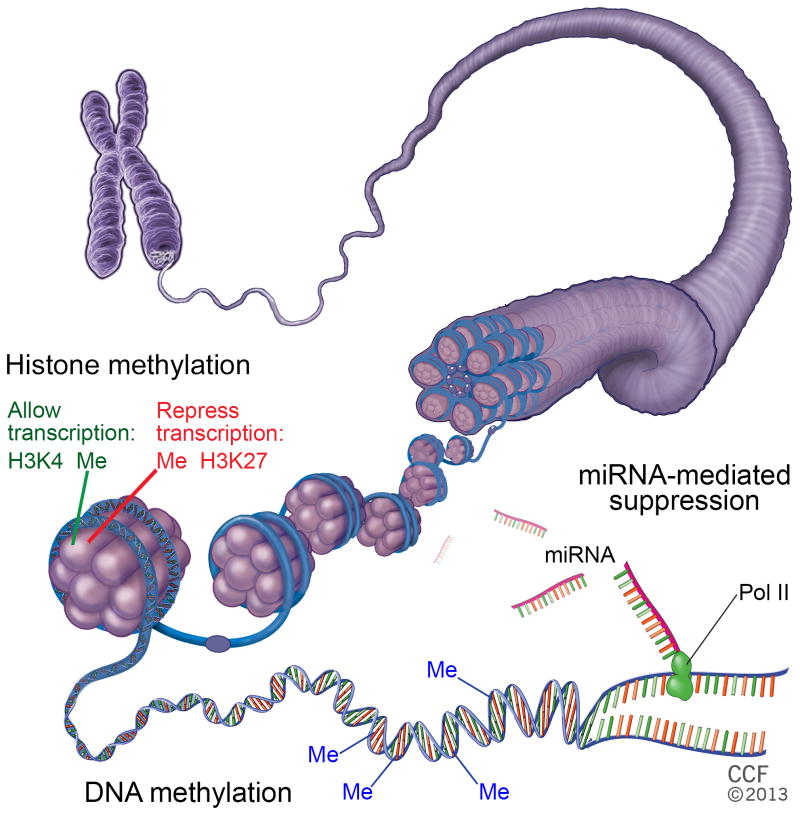
Figure 2.
Epigenetic regulation in GSCs. Epigenetic regulation important in GSC biology includes histone methylation, DNA methylation and miRNA-mediated suppression of target gene translation. Histone methylation in GSCs is mediated by MLL1 (H3K4 methylation), Bmi1 and EZH2 (H3K27 methylation). Demethylation of these marks has been shown to be accomplished through JMJD3. Promoter DNA methylation can suppress the transcription of targets such as CD133, miRNA-211 and TAZ. MicroRNAs, such as miRNAs-124,-125b, -128, -146a, -34a, -9* and -211 regulate various aspects of GSC biology by inhibiting translation of their target genes.
The role of DNA methylation in GBM was initially established by a wide range of reports of individual loci that were transcriptionally controlled by hyper- or hypomethylation, but the most significant advance in our understand of DNA methylation in GBM was the identification that isocitrate dehydrogenase 1 (IDH1) mutations, which are associated with a proneural GBM subgroup, are sufficient to establish the glioma-CpG island methylator phenotype (G-CIMP) (Noushmehr et al., 2010; Turcan et al., 2012; Verhaak et al., 2010). This hypermethylation phenotype leads to the stable modification of DNA methylation patterns on a genome-wide scale, which changes the transcriptional profile and may alter the differentiation state. Specifically in GSCs, DNA methylation of the CD133 promoter may repress transcription of CD133 in the CD133-negative population (Gopisetty et al., 2012) and aberrant DNA methylation of microRNA-211 up-regulates MMP-9 expression and GSC resistance to radiation and chemotherapy (Asuthkar et al., 2012). Expression of the transcriptional co-activator with PDZ-binding motif (TAZ) correlates with the mesenchymal gene expression signature in GBM and that silencing of TAZ by promoter hypermethylation correlated with the proneural signature in GBM (Bhat et al., 2011). Silencing of TAZ in mesenchymal GSCs suppressed the mesenchymal gene expression signature while expressing TAZ in proneural GSCs or NSCs led to increased expression of mesenchymal signature genes (Bhat et al., 2011). Recent evidence also demonstrated that CpG island methylation status, not expression levels, of the two key genes involved in arginine biosynthesis predicts sensitivity to arginine deprivation by pegylated arginine deiminase (ADI-PEG20) in GBM cells including GSCs (Syed et al., 2013).
7.2 Histone methylation
Histone methylation controls cell fate decisions through the interplay of histone methyltransferases and histone demethylases and has been relatively well studied in GSCs. Histone methylation can open chromatin to allow transcription, mediated through the methylation of H3K4 (histone 3 at lysine 4) by the trithorax methyltransferase group, or histone methylation can close chromatin and repress transcription, which is mediated by H3K27 (histone 3 at lysine 27) methylation by a family of polycomb methyltransferase genes. In GSC, hypoxia induces mixed-lineage leukemia 1 (MLL1), a trithorax histone methyltransferase, which then regulates HIF2a expression and tumorigenic potential of GSCs (Heddleston et al., 2012). In a subsequent report, MLL also directly activates the Homeobox gene HOXA10 in GSCs and the HOXA10 enacts a gene expression program that may additionally contribute to the tumorigenic potential of GSCs (Gallo et al., 2013). Transcriptional repression by histone methylation is facilitated through polycomb genes, such as EZH2 and Bmi1. EZH2 silencing of the BMP pathway in GSCs inhibits their ability to differentiate, and inhibition of EZH2 or forced expression of the BMP pathway causes loss of self-renewal and decreased tumorigenicity of GSCs (Lee et al., 2008; Suvà et al., 2009). Bmi1, a key component of the polycomb repressive complex 1 (PRC1) discussed above, is upregulated in GBM and significantly enriched in the GSC population, but is interestingly not expressed in normal astrocytes (Abdouh et al., 2009). Suppression of Bmi1 in human GSCs inhibits their growth in vitro and in vivo (Abdouh et al., 2009) and Bmi1 knockdown in a glioma mouse model suppresses malignant tumor formation (Bruggeman et al., 2007). MicroRNA-128 targets Bmi1, but is downregulated in GBM allowing for the overexpression of Bmi1. However, overexpression of microRNA-128 in neurosphere cultures decreases Bmi1 and blocks self-renewal in these cells (Godlewski et al., 2008). The counterpart to histone methylation is histone demethylation, which is carried out by a family of histone demethylases. While histone demethylation has been less well studied, JMJD3, a H3K27 demethylase, is induced during differentiation of GSCs (Ene et al., 2012). JMJD3 promotes differentiation through both conventional demethylase activity at the INK4A/ARF locus and chromatin independent stabilization of p53. They also find JMJD3 is downregulated in a subset of GBMs due to mutations or DNA hypermethylation and this deregulation may contribute to gliomagenesis (Ene et al., 2012).
7.3 microRNAs
Regulatory non-coding RNAs play a critical role in a wide range of biological processes, including neural development and GBM tumorigenesis. While there are many classes of regulatory non-coding RNA (e.g. lncRNA, siRNA, piRNA, etc.), microRNA (miRNA) is the only class this far to have a clear role in regulating GSC biology. MiRNAs are approximately 22 nucleotide non-coding RNAs that down regulate gene expression by binding to the 3′UTR of their target genes and inhibiting translation (Bartel, 2004). There has been an explosion of research on miRNAs in the past decade, which has touched nearly every field in biology including the GSC field. It is now appreciated that miRNAs are involved initiation, progression and resistance of GBM and some of the miRNAs identified were substantiated in GSCs. This is not surprising as miRNAs have well-established roles in pluripotency and reprogramming (Leonardo et al., 2012), and in GBM it has also been shown that miRNAs regulated neural stem cell pathways (Godlewski et al., 2010; González-Gómez et al., 2011). It has been shown that miRNAs such as miRNA-128 (Godlewski et al., 2008), miRNA-124 (Xia et al., 2012), miRNA-146a (Mei et al., 2011) and miRNA-34a (Guessous et al., 2010) regulate the stem cell phenotype in GSCs and may contribute to gliomagenesis (reviewed in (Zhang et al., 2012)). As GSCs are known to be involved in chemo- and radiotherapy, several reports have analyzed the effects of miRNAs on this process and found that, among others, miRNA-124 (Yang et al., 2012), miRNA-125b (Shi et al., 2012) and miR-9* (Jeon et al., 2011) contribute by regulating mostly known mediators of chemo- or radiotherapy resistance (reviewed in (Chistiakov and Chekhonin, 2012)). As it turns out in GSCs, epigenetic mechanisms such as DNA methylation (Asuthkar et al., 2012) and histone modifications (Katsushima et al., 2012) can also regulate miRNA expression, and in turn miRNAs can regulate epigenetic modifying genes such as Bmi1 (Godlewski et al., 2008), adding to the complexity of epigenetic regulation in GSCs.
While most of the previous studies look at the contribution of individual epigenetic modifiers to GSC biology, a recent report by Stricker and colleagues aimed to get a better appreciation of the overall contribution of the epigenetic landscape to GSC traits (Stricker et al., 2013). They demonstrated that resetting the epigenetic landscape of GSCs through induced pluripotent stem cell reprogramming techniques they could suppress the malignant behavior of these cells only if they were re-differentiated into a non-neural cell types, suggesting that while cancer genome plays a significant role in maintaining a malignant state it is possible to override genetic influence through epigenetic mechanisms. However, this requires substantial manipulation of the cells (Stricker et al., 2013).
As is evident from the many studies above, the epigenetic state of a cell is intimately intertwined with both its microenvironment and its genetic landscape and this interplay determines the cellular fate and therapeutic response of the cell. With the identification of several epigenetic regulators as potential therapeutic targets in GSCs (e.g. Bmi1 (Acquati et al., 2013), MLL1 (Heddleston et al., 2012)) and evidence that epigenetic mechanisms contribute to the cellular hierarchy and therapeutic response in GBM, developing therapies against epigenetic targets may provide significant benefit to patients.
8. The contribution of GSCs to therapeutic resistance
8.1 Radioresistance
Current therapies for high grade brain tumors are only palliative which suggests a resistant population. Increasing evidence suggests that brain tumor recurrence and progression to more aggressive tumor phenotypes is due in part to GSC resistance towards radiation, chemotherapy, and anti-angiogenics. In primary human glioma specimens, radiation dramatically increases cell populations enriched for GSCs and minimally affects GSC tumorigenic potential (Bao et al., 2006a). Furthermore, GSCs are more resistant to radiation-induced apoptosis via activation of DNA damage repair mechanisms. Compared to non-stem tumor cells, GSCs recover more rapidly and are able to more efficiently repair DNA damage. This is likely due to the activation of several DNA damage checkpoint proteins including ataxia telangiectasia mutated (ATM), the cell cycle checkpoint protein Rad17, and the checkpoint kinases Chk1 and Chk2 (Bao et al., 2006a). Inhibition of Chk1 and Chk2 kinases dampens GSC tumor growth ability, suggesting a potential target for sensitizing GSCs to radiation. The cell surface adhesion protein and GSC marker, L1CAM (CD171), enhances DNA checkpoint activation and DNA repair by activating the phosphorylation of ATM, Rad17, Chk1 and Chk2 in response to radiation-induced DNA damage (Cheng et al., 2011). L1CAM directly regulates NBS1, a core component of the MRN complex that is important for activating the DNA damage checkpoint response following double stranded breaks (Cheng et al., 2011). Considering its preferential expression in GSCs, L1CAM represents a potential molecular target that may be beneficial in attenuating radioresistance observed in GBM. An additional mechanism of radioresistance through the DNA damage response may involve the polycomb group protein, BMI1, which has been implicated as an oncogene in glioma regulation and is further enriched in GSCs. Following radiation, BMI1 is redistributed to the chromatin, colocalizing with proteins necessary for the DNA double-stranded break (DSB) response (Facchino et al., 2010). Interestingly, BMI1 deficiency impairs this recruitment, sensitizing GSCs to radiation. Other studies supporting the GSC radioresistance have shown enhanced survival of GSCs following radiation as well as preferential activation of Chk1 and Chk2 without signs of elevated DNA repair (Ropolo et al., 2009).
It is likely that there are multiple, non-mutually exclusive pathways implicated in contributing to the radioresistance of GSCs. Autophagy, the protein degradation system with both cell death and protective roles across different cancers, has recently emerged as a means by which glioma cells respond to radiation. While the benefit of targeting autophagy is controversial in glioma, radiation preferentially induces autophagy in GSC compared to non-stem tumor cells within 24 hours (Lomonaco et al., 2009). GSCs express higher levels of autophagy-related proteins (LC3, ATG5, and ATG12) and autophagy inhibitors or silencing autophagy genes sensitize GSC to radiation, decreasing their viability and sphere-forming capabilities (Lomonaco et al., 2009). Yet the role of autophagy and radiation in brain tumor GSCs is less than straightforward given that other studies demonstrate that activation of autophagy pathways via the inhibition of mTOR signaling in conjunction with radiation increases radiosensitivity, increases neural differentiation and reduces self-renewal, proliferation, and clonogenic ability in GSCs (Wang et al., 2013; Zhuang et al., 2011). Likewise, the combinatorial treatment of cilengitide, an αv integrin inhibitor currently in clinical trials, and radiation induce autophagy in GSC and enhance cytotoxicity, decreasing cell survival (Lomonaco et al., 2011).
Stem cell maintenance pathways are also implicated in promoting radioresistance in GSCs. Constitutively activating the notch pathway in GSCs offers protection from radiotherapy while blocking notch activation with γ-secretase inhibitors in combination with radiation impairs GSC survival and enhances radiation-induced cell death (J. Wang et al., 2010). Notch promotes radioresistance through activation of the PI3K/Akt pathway and the upregulation of the pro-survival protein, Mcl-1. Interestingly, this effect, as well as response to notch inhibition is not observed in non-GSCs (J. Wang et al., 2010). Recently, the radioprotective effect of Akt signaling has also been implicated in response to activation of IGF1 receptors on GSCs as well as through their increased IGF1 secretion (Osuka et al., 2012). Furthermore, treatment of tumors with IGF receptor blockers further increases GSC radiosensitivity. Wnt/β-catenin signaling is another pathway highly involved in embryonic and cancer development which is associated with radioresistance. While identified earlier to mediate radioresistance in mammary progenitor cells and to enrich for breast cancer stem cells (Woodward et al., 2007), more recently Kim and colleagues used transciptomic analysis of orthotopic GBM xenografts with or without in vivo irradiation and revealed that Wnt proteins, including WISP, an effector of activated Wnt, and active β-catenin showed the highest level increase in post-irradiated GBM cells which were also positive for Sox2, a putative stem cell marker (Kim et al., 2012).
8.2 Chemoresistance
GSC display preferential resistance to many cytotoxic agents, including a number of chemotherapeutic agents. Temozolomide (TMZ) is a methylating agent that is the current standard of care for treatment of high grade brain tumors (Hegi et al., 2005; Liu et al., 2006; Shervington and Lu, 2008). TMZ induces DNA alterations by methylating the O6 position of guanine in DNA thereby causing cell damage, triggering cell death. The relative sensitivity of GSC to TMZ is unresolved as some reports have suggested that TMZ depletes GSC (Beier et al., 2008) while others suggest a relative resistance (Chen et al., 2012; Gaspar et al., 2010; Liu et al., 2006; Murat et al., 2008). GSCs express higher levels of the DNA repair enzyme, O6-methylguanine-DNA-methyltransferase (MGMT), which limits TMZ therapeutic efficacy (Liu et al., 2006; Bleau et al., 2009). In a genetically engineered mouse model, TMZ treatment not only fails to inhibit GSC self-renewal but also increases the GSC compartment (Bleau et al., 2009). GSCs are less sensitive to TMZ-induced cell death compared to other tissue and tumor types with TMZ treatment in GBM cells resulting in a stem-like gene signature (Bleau et al., 2009; Ulasov et al., 2011). While therapy with TMZ may slow GBM growth, slightly prolonging survival, tumor recurrence is often inevitable which further indicates a resistant cell population in GBM.
In addition to TMZ, glioma-derived GSCs are more resistant to several other chemotherapeutic agents, including carboplatin, paclitaxel, and etoposide (Liu et al., 2006). In this study, recurrent GBM tissue obtained from multiple patients contain increased mRNA expression of CD133 compared to newly diagnosed tumors, suggesting that GSCs may be responsible for GBM relapse (Liu et al., 2006). Enrichment of GSC surface markers is observed on proliferating glioma cells that withstand lethal exposure of the alkylating agent, BCNU. Tumors develop when these BCNU-resistant cells are subsequently injected into the brains of immunocompromised mice (Kang and Kang, 2007), further demonstrating that identifying new ways of sensitizing GSCs to therapy may improve GBM growth control. This importance is fortified by an elegant study in which a genetically-engineered glioma model was used to lineage trace cells that are responsible for re-growth after TMZ treatment. Cells of strong resemblance to GSCs are indeed the subset that has the capacity to refortify the tumor (Chen et al., 2012).
A potential mechanism for brain GSC drug resistance is increased expression of drug transporters that pump out chemotherapeutic agents. These include ABC (ATP-binding cassette) drug transporters which use the energy from ATP hydrolysis to efflux various substrates, including drugs, across extra- and intracellular membranes. The side population is a functional cellular phenotype associated with cancer stem cells in some cancers but with mixed evidence in gliomas (Broadley et al., 2011; Golebiewska et al., 2013; Harris et al., 2008). In a glioma model in which the side population was highly tumorigenic and enriched following TMZ treatment, Akt regulates ABC transporter activity (Bleau et al., 2009). Other studies suggest that the stem cell surface marker CD133 might have a functional role in promoting drug resistance. Upon exposure to the anti-cancer drugs, camptothecin and doxorubicin, ectopic overexpression of CD133 results in an upregulation of ABC transporters concomitant with a reduction in apoptosis (Angelastro and Lamé, 2010). Recently, melatonin has been shown to enhance the methylation of a class of ABC transporters and when combined with TMZ, causes a synergistic toxic effect on GSCs (Martín et al., 2013).
Additional pathways implicated in brain tumor radio- and chemoresistance include the Notch and Shh signaling pathways. Notch and Shh pathways have many functions yet are most notably involved in promoting proliferation during neurogenesis. In combination with TMZ, targeting Notch and Shh may be an additional strategy to sensitizing GSCs to chemotherapy (Ulasov et al., 2011). Following exposure to TMZ, Notch and Shh pathway activity is enhanced with upregulation of Notch1, Hes1, Hes5 and GLI1 in CD133-positive cells. However, simultaneous treatment of TMZ with Notch and Shh antagonists (GSI-1 and cyclopamine, respectively) more than double the level of toxicity in CD133-positive cells (Ulasov et al., 2011). These findings suggest a possible interaction of Notch and Shh pathways with DNA repair machinery given that TMZ elicits its toxic effects by direct DNA damage.
9. Conclusion and future perspectives
Recent advances in our understanding of brain tumor stem cell specific signaling pathways, transcription factors, niches, and other modes of initiating and propagating malignant brain tumors have led to the development of novel targeted therapies. For example, phase I and phase II clinical trials using γ-secretase inhibitors (RO4929097) to interfere with Notch signaling thereby interfering with GSC maintenance are ongoing (Tanaka et al., 2013). Similarly, vismodegib, an oral medication hedgehog antagonist, is currently in clinical trials for patients with recurrent GBM (Tanaka et al., 2013). The aforementioned signaling pathways, including Hedgehog, Wnt, Notch, and BMP, are all being selectively targeted in tumor initiating cells in various clinical trials for various cancer types (Zhou et al., 2009).
In addition to targeting specific signaling pathways and transcription factors, the cancer stem cell niche, which protects cancer stem cells from therapies, can be targeted as well. For example, factors within the vascular niche promote radioresistance in GSCs (Lathia et al., 2012). Therefore, despite maximal surgical resection and effective radiation therapy, resilient GSCs can recreate a tumor that even more challenging to treat. Accordingly, there are clinical trials using anti-angiogenic drugs such as bevacizumab (Desjardins et al., 2008; Vredenburgh et al., 2007) and cediranib (Batchelor et al., 2010, 2007), to disrupt this protective niche (Gilbertson and Rich, 2007).
To achieve brain cancer cure, all tumor cells, particularly the stem cells, must be eliminated. Cancer stem cells represent an extraordinary population of cells that are able to evade therapy and even change the tumor landscape to promote a more aggressive, ultimately lethal disease. Unfortunately, the current pharmaceutical drug discovery methods fail to recapitulate the hierarchical cancer stem cell phenotype and accordingly high throughput screens fail to identify the brain tumor stem cell targets (Rich, 2008). New methods are being discovered and reported to address these limitations in drug discovery (Diamandis et al., 2007). An added complexity in treating brain tumors is the heterogeneity that exists within each tumor as well as between patients which is becoming increasingly apparent with defined GBM subgroups. Thus, understanding the interplay between GSCs, which likely continue to evolve through disease progression, and their microenvironment will be important as we move towards identifying better biomarkers and delivering more personalized treatment options. An additional obstacle in discovering novel cancer stem cell therapeutic targets is ensuring that there are limited toxicities to the normal brain. With increasing studies identifying brain tumor stem cell targets that are differentially expressed or regulated, validation in pre-clinical and Phase-I clinical trials may prove to be more fruitful. Novel targeting modalities are also being considered including vaccine and viral therapies (Alonso et al., 2012; Jiang and Fueyo, 2010; Jiang et al., 2009, 2007; Pellegatta et al., 2006; Wakimoto et al., 2009). The Neuro-Oncology community must collectively embark on efforts to use optimal human and rodent models informed by genetic and epigenetic information to improve the outcome for brain cancer patients.
Acknowledgments
We sincerely apologize to those whose work we were unable to discuss in this review. We would like to thank members of the Rich and Bao labs for stimulating discussion and critical review of this manuscript. This work was supported by the National Institutes of Health grants CA154130, CA1129958, and GM007250, the American Brain Tumor Association, the James S. McDonnell Foundation, and the Howard Hughes Medical Institute Medical Fellowship.
Biographies
David L. Schonberg, PhD is a Postdoctoral Research Fellow in the laboratory of Dr. Jeremy Rich at the Cleveland Clinic Lerner Research Institute (LRI). He received his undergraduate degree from Miami University followed by receiving a Master in Public Health (MPH) with a focus in Epidemiology and a PhD in Neuroscience from The Ohio State University. After a postdoctoral fellowship with Dr. Robert Wechsler-Reya at Duke University, David joined the Rich laboratory in 2011.
Daniel Lubelski is a MD student at the Cleveland Clinic Lerner College of Medicine (CCLCM) and a Howard Hughes Medical Fellow at the Cleveland Clinic Lerner Research Institute in the laboratory of Dr. Jeremy Rich. He received his undergraduate degree in Neuroscience & Biology from Queens College, City University of New York and did a post-baccalaureate intramural research fellowship at the National Institutes of Health.
Tyler E. Miller is a MD/PhD student in the Case Western Reserve University’s (CWRU) Medical Scientist Training Program. He is pursuing his PhD in the laboratories of Dr. Jeremy Rich at the Cleveland Clinic Lerner Research Institute and Dr. Paul Tesar at Case Western Reserve University. Tyler received his B.S. in Biomedical Science from The Ohio State University.
Jeremy N. Rich, MD is chair of the Department of Stem Cell Biology and Regenerative Medicine at the Cleveland Clinic LRI, professor in the Department of Molecular Medicine CCLCM at CWRU, staff at the Brain Tumor and Neuro-Oncology Center in the Department of Neurology and Taussig Cancer Institute Cleveland Clinic, and Co-Director for the Center for Stem Cell and Regenerative Medicine at the National Center for Regenerative Medicine. Dr. Rich received his undergraduate degree from Washington University, MD from Duke University, Residency in Neurology at Johns Hopkins Hospital, and Neuro-Oncology fellowship at Duke University.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abdouh M, Facchino S, Chatoo W, Balasingam V, Ferreira J, Bernier G. BMI1 sustains human glioblastoma multiforme stem cell renewal. J Neurosci Off J Soc Neurosci. 2009;29:8884–8896. doi: 10.1523/JNEUROSCI.0968-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acquati S, Greco A, Licastro D, Bhagat H, Ceric D, Rossini Z, Grieve J, Shaked-Rabi M, Henriquez NV, Brandner S, Stupka E, Marino S. Epigenetic regulation of survivin by Bmi1 is cell type specific during corticogenesis and in gliomas. Stem Cells Dayt Ohio. 2013;31:190–202. doi: 10.1002/stem.1274. [DOI] [PubMed] [Google Scholar]
- Adamson C, Kanu OO, Mehta AI, Di C, Lin N, Mattox AK, Bigner DD. Glioblastoma multiforme: a review of where we have been and where we are going. Expert Opin Investig Drugs. 2009;18:1061–1083. doi: 10.1517/13543780903052764. [DOI] [PubMed] [Google Scholar]
- Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso MM, Jiang H, Gomez-Manzano C, Fueyo J. Targeting brain tumor stem cells with oncolytic adenoviruses. Methods Mol Biol Clifton Nj. 2012;797:111–125. doi: 10.1007/978-1-61779-340-0_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelastro JM, Lamé MW. Overexpression of CD133 promotes drug resistance in C6 glioma cells. Mol Cancer Res Mcr. 2010;8:1105–1115. doi: 10.1158/1541-7786.MCR-09-0383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anido J, Sáez-Borderías A, Gonzàlez-Juncà A, Rodón L, Folch G, Carmona MA, Prieto-Sánchez RM, Barba I, Martínez-Sáez E, Prudkin L, Cuartas I, Raventós C, Martínez-Ricarte F, Poca MA, García-Dorado D, Lahn MM, Yingling JM, Rodón J, Sahuquillo J, Baselga J, Seoane J. TGF-β Receptor Inhibitors Target the CD44(high)/Id1(high) Glioma-Initiating Cell Population in Human Glioblastoma. Cancer Cell. 2010;18:655–668. doi: 10.1016/j.ccr.2010.10.023. [DOI] [PubMed] [Google Scholar]
- Asuthkar S, Velpula KK, Chetty C, Gorantla B, Rao JS. Epigenetic regulation of miRNA-211 by MMP-9 governs glioma cell apoptosis, chemosensitivity and radiosensitivity. Oncotarget. 2012;3:1439–1454. doi: 10.18632/oncotarget.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao S, Wu Q, Li Z, Sathornsumetee S, Wang H, McLendon RE, Hjelmeland AB, Rich JN. Targeting cancer stem cells through L1CAM suppresses glioma growth. Cancer Res. 2008;68:6043–6048. doi: 10.1158/0008-5472.CAN-08-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, Dewhirst MW, Bigner DD, Rich JN. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006a;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- Bao S, Wu Q, Sathornsumetee S, Hao Y, Li Z, Hjelmeland AB, Shi Q, McLendon RE, Bigner DD, Rich JN. Stem cell-like glioma cells promote tumor angiogenesis through vascular endothelial growth factor. Cancer Res. 2006b;66:7843–7848. doi: 10.1158/0008-5472.CAN-06-1010. [DOI] [PubMed] [Google Scholar]
- Bar EE, Chaudhry A, Lin A, Fan X, Schreck K, Matsui W, Piccirillo S, Vescovi AL, DiMeco F, Olivi A, Eberhart CG. Cyclopamine-mediated hedgehog pathway inhibition depletes stem-like cancer cells in glioblastoma. Stem Cells Dayt Ohio. 2007;25:2524–2533. doi: 10.1634/stemcells.2007-0166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar EE, Lin A, Mahairaki V, Matsui W, Eberhart CG. Hypoxia increases the expression of stem-cell markers and promotes clonogenicity in glioblastoma neurospheres. Am J Pathol. 2010;177:1491–1502. doi: 10.2353/ajpath.2010.091021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett LE, Granot Z, Coker C, Iavarone A, Hambardzumyan D, Holland EC, Nam H, Benezra R. Self-renewal does not predict tumor growth potential in mouse models of high-grade glioma. Cancer Cell. 2012;21:11–24. doi: 10.1016/j.ccr.2011.11.025. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Batchelor TT, Duda DG, di Tomaso E, Ancukiewicz M, Plotkin SR, Gerstner E, Eichler AF, Drappatz J, Hochberg FH, Benner T, Louis DN, Cohen KS, Chea H, Exarhopoulos A, Loeffler JS, Moses MA, Ivy P, Sorensen AG, Wen PY, Jain RK. Phase II study of cediranib, an oral pan-vascular endothelial growth factor receptor tyrosine kinase inhibitor, in patients with recurrent glioblastoma. J Clin Oncol Off J Am Soc Clin Oncol. 2010;28:2817–2823. doi: 10.1200/JCO.2009.26.3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batchelor TT, Sorensen AG, di Tomaso E, Zhang WT, Duda DG, Cohen KS, Kozak KR, Cahill DP, Chen PJ, Zhu M, Ancukiewicz M, Mrugala MM, Plotkin S, Drappatz J, Louis DN, Ivy P, Scadden DT, Benner T, Loeffler JS, Wen PY, Jain RK. AZD2171, a pan-VEGF receptor tyrosine kinase inhibitor, normalizes tumor vasculature and alleviates edema in glioblastoma patients. Cancer Cell. 2007;11:83–95. doi: 10.1016/j.ccr.2006.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beier D, Röhrl S, Pillai DR, Schwarz S, Kunz-Schughart LA, Leukel P, Proescholdt M, Brawanski A, Bogdahn U, Trampe-Kieslich A, Giebel B, Wischhusen J, Reifenberger G, Hau P, Beier CP. Temozolomide preferentially depletes cancer stem cells in glioblastoma. Cancer Res. 2008;68:5706–5715. doi: 10.1158/0008-5472.CAN-07-6878. [DOI] [PubMed] [Google Scholar]
- Bexell D, Gunnarsson S, Siesjö P, Bengzon J, Darabi A. CD133+ and nestin+ tumor-initiating cells dominate in N29 and N32 experimental gliomas. Int J Cancer J Int Cancer. 2009;125:15–22. doi: 10.1002/ijc.24306. [DOI] [PubMed] [Google Scholar]
- Bhat KPL, Salazar KL, Balasubramaniyan V, Wani K, Heathcock L, Hollingsworth F, James JD, Gumin J, Diefes KL, Kim SH, Turski A, Azodi Y, Yang Y, Doucette T, Colman H, Sulman EP, Lang FF, Rao G, Copray S, Vaillant BD, Aldape KD. The transcriptional coactivator TAZ regulates mesenchymal differentiation in malignant glioma. Genes Dev. 2011;25:2594–2609. doi: 10.1101/gad.176800.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop JM. Retroviruses and cancer genes. Adv Cancer Res. 1982;37:1–32. doi: 10.1016/s0065-230x(08)60880-5. [DOI] [PubMed] [Google Scholar]
- Blazek ER, Foutch JL, Maki G. Daoy medulloblastoma cells that express CD133 are radioresistant relative to CD133− cells, and the CD133+ sector is enlarged by hypoxia. Int J Radiat Oncol Biol Phys. 2007;67:1–5. doi: 10.1016/j.ijrobp.2006.09.037. [DOI] [PubMed] [Google Scholar]
- Bleau AM, Hambardzumyan D, Ozawa T, Fomchenko EI, Huse JT, Brennan CW, Holland EC. PTEN/PI3K/Akt pathway regulates the side population phenotype and ABCG2 activity in glioma tumor stem-like cells. Cell Stem Cell. 2009;4:226–235. doi: 10.1016/j.stem.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3:730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- Broadley KWR, Hunn MK, Farrand KJ, Price KM, Grasso C, Miller RJ, Hermans IF, McConnell MJ. Side population is not necessary or sufficient for a cancer stem cell phenotype in glioblastoma multiforme. Stem Cells Dayt Ohio. 2011;29:452–461. doi: 10.1002/stem.582. [DOI] [PubMed] [Google Scholar]
- Bruggeman SWM, Hulsman D, Tanger E, Buckle T, Blom M, Zevenhoven J, van Tellingen O, van Lohuizen M. Bmi1 controls tumor development in an Ink4a/Arf-independent manner in a mouse model for glioma. Cancer Cell. 2007;12:328–341. doi: 10.1016/j.ccr.2007.08.032. [DOI] [PubMed] [Google Scholar]
- Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS, Anderson MJ, Arden KC, Blenis J, Greenberg ME. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857–868. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- Calabrese C, Poppleton H, Kocak M, Hogg TL, Fuller C, Hamner B, Oh EY, Gaber MW, Finklestein D, Allen M, Frank A, Bayazitov IT, Zakharenko SS, Gajjar A, Davidoff A, Gilbertson RJ. A perivascular niche for brain tumor stem cells. Cancer Cell. 2007;11:69–82. doi: 10.1016/j.ccr.2006.11.020. [DOI] [PubMed] [Google Scholar]
- Cao Y, Lathia JD, Eyler CE, Wu Q, Li Z, Wang H, McLendon RE, Hjelmeland AB, Rich JN. Erythropoietin Receptor Signaling Through STAT3 Is Required For Glioma Stem Cell Maintenance. Genes Cancer. 2010;1:50–61. doi: 10.1177/1947601909356352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardone MH, Roy N, Stennicke HR, Salvesen GS, Franke TF, Stanbridge E, Frisch S, Reed JC. Regulation of cell death protease caspase-9 by phosphorylation. Science. 1998;282:1318–1321. doi: 10.1126/science.282.5392.1318. [DOI] [PubMed] [Google Scholar]
- Carén H, Pollard SM, Beck S. The good, the bad and the ugly: Epigenetic mechanisms in glioblastoma. Mol Aspects Med. 2013;34:849–862. doi: 10.1016/j.mam.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmeliet P, Jain RK. Angiogenesis in cancer and other diseases. Nature. 2000;407:249–257. doi: 10.1038/35025220. [DOI] [PubMed] [Google Scholar]
- Cayuso J, Ulloa F, Cox B, Briscoe J, Martí E. The Sonic hedgehog pathway independently controls the patterning, proliferation and survival of neuroepithelial cells by regulating Gli activity. Dev Camb Engl. 2006;133:517–528. doi: 10.1242/dev.02228. [DOI] [PubMed] [Google Scholar]
- Chen J, Li Y, Yu TS, McKay RM, Burns DK, Kernie SG, Parada LF. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012;488:522–526. doi: 10.1038/nature11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Chen L, Zhang R, Yi Y, Ma Y, Yan K, Jiang X, Wang X. ADAM17 regulates self-renewal and differentiation of U87 glioblastoma stem cells. Neurosci Lett. 2013;537:44–49. doi: 10.1016/j.neulet.2013.01.021. [DOI] [PubMed] [Google Scholar]
- Cheng L, Bao S, Rich JN. Potential therapeutic implications of cancer stem cells in glioblastoma. Biochem Pharmacol. 2010;80:654–665. doi: 10.1016/j.bcp.2010.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Huang Z, Zhou W, Wu Q, Donnola S, Liu JK, Fang X, Sloan AE, Mao Y, Lathia JD, Min W, McLendon RE, Rich JN, Bao S. Glioblastoma stem cells generate vascular pericytes to support vessel function and tumor growth. Cell. 2013;153:139–152. doi: 10.1016/j.cell.2013.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Wu Q, Huang Z, Guryanova OA, Huang Q, Shou W, Rich JN, Bao S. L1CAM regulates DNA damage checkpoint response of glioblastoma stem cells through NBS1. Embo J. 2011;30:800–813. doi: 10.1038/emboj.2011.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chistiakov DA, Chekhonin VP. Contribution of microRNAs to radio- and chemoresistance of brain tumors and their therapeutic potential. Eur J Pharmacol. 2012;684:8–18. doi: 10.1016/j.ejphar.2012.03.031. [DOI] [PubMed] [Google Scholar]
- Choe G, Horvath S, Cloughesy TF, Crosby K, Seligson D, Palotie A, Inge L, Smith BL, Sawyers CL, Mischel PS. Analysis of the phosphatidylinositol 3′-kinase signaling pathway in glioblastoma patients in vivo. Cancer Res. 2003;63:2742–2746. [PubMed] [Google Scholar]
- Clement V, Sanchez P, de Tribolet N, Radovanovic I, Ruiz i Altaba A. HEDGEHOG-GLI1 signaling regulates human glioma growth, cancer stem cell self-renewal, and tumorigenicity. Curr Biol Cb. 2007;17:165–172. doi: 10.1016/j.cub.2006.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- Creighton CJ, Li X, Landis M, Dixon JM, Neumeister VM, Sjolund A, Rimm DL, Wong H, Rodriguez A, Herschkowitz JI, Fan C, Zhang X, He X, Pavlick A, Gutierrez MC, Renshaw L, Larionov AA, Faratian D, Hilsenbeck SG, Perou CM, Lewis MT, Rosen JM, Chang JC. Residual breast cancers after conventional therapy display mesenchymal as well as tumor-initiating features. Proc Natl Acad Sci U S A. 2009;106:13820–13825. doi: 10.1073/pnas.0905718106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta SR, Dudek H, Tao X, Masters S, Fu H, Gotoh Y, Greenberg ME. Akt phosphorylation of BAD couples survival signals to the cell-intrinsic death machinery. Cell. 1997;91:231–241. doi: 10.1016/s0092-8674(00)80405-5. [DOI] [PubMed] [Google Scholar]
- De la Iglesia N, Konopka G, Puram SV, Chan JA, Bachoo RM, You MJ, Levy DE, Depinho RA, Bonni A. Identification of a PTEN-regulated STAT3 brain tumor suppressor pathway. Genes Dev. 2008;22:449–462. doi: 10.1101/gad.1606508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De la Iglesia N, Puram SV, Bonni A. STAT3 regulation of glioblastoma pathogenesis. Curr Mol Med. 2009;9:580–590. doi: 10.2174/156652409788488739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desjardins A, Reardon DA, Herndon JE, 2nd, Marcello J, Quinn JA, Rich JN, Sathornsumetee S, Gururangan S, Sampson J, Bailey L, Bigner DD, Friedman AH, Friedman HS, Vredenburgh JJ. Bevacizumab plus irinotecan in recurrent WHO grade 3 malignant gliomas. Clin Cancer Res Off J Am Assoc Cancer Res. 2008;14:7068–7073. doi: 10.1158/1078-0432.CCR-08-0260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewhirst MW, Cao Y, Moeller B. Cycling hypoxia and free radicals regulate angiogenesis and radiotherapy response. Nat Rev Cancer. 2008;8:425–437. doi: 10.1038/nrc2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamandis P, Wildenhain J, Clarke ID, Sacher AG, Graham J, Bellows DS, Ling EKM, Ward RJ, Jamieson LG, Tyers M, Dirks PB. Chemical genetics reveals a complex functional ground state of neural stem cells. Nat Chem Biol. 2007;3:268–273. doi: 10.1038/nchembio873. [DOI] [PubMed] [Google Scholar]
- Du Z, Jia D, Liu S, Wang F, Li G, Zhang Y, Cao X, Ling EA, Hao A. Oct4 is expressed in human gliomas and promotes colony formation in glioma cells. Glia. 2009;57:724–733. doi: 10.1002/glia.20800. [DOI] [PubMed] [Google Scholar]
- Ehtesham M, Sarangi A, Valadez JG, Chanthaphaychith S, Becher MW, Abel TW, Thompson RC, Cooper MK. Ligand-dependent activation of the hedgehog pathway in glioma progenitor cells. Oncogene. 2007;26:5752–5761. doi: 10.1038/sj.onc.1210359. [DOI] [PubMed] [Google Scholar]
- Ene CI, Edwards L, Riddick G, Baysan M, Woolard K, Kotliarova S, Lai C, Belova G, Cam M, Walling J, Zhou M, Stevenson H, Kim HS, Killian K, Veenstra T, Bailey R, Song H, Zhang W, Fine HA. Histone demethylase Jumonji D3 (JMJD3) as a tumor suppressor by regulating p53 protein nuclear stabilization. Plos One. 2012;7:e51407. doi: 10.1371/journal.pone.0051407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans SM, Jenkins KW, Chen HI, Jenkins WT, Judy KD, Hwang W-T, Lustig RA, Judkins AR, Grady MS, Hahn SM, Koch CJ. The Relationship among Hypoxia, Proliferation, and Outcome in Patients with De Novo Glioblastoma: A Pilot Study. Transl Oncol. 2010;3:160–169. doi: 10.1593/tlo.09265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyler CE, Foo WC, LaFiura KM, McLendon RE, Hjelmeland AB, Rich JN. Brain cancer stem cells display preferential sensitivity to Akt inhibition. Stem Cells Dayt Ohio. 2008;26:3027–3036. doi: 10.1634/stemcells.2007-1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Facchino S, Abdouh M, Chatoo W, Bernier G. BMI1 confers radioresistance to normal and cancerous neural stem cells through recruitment of the DNA damage response machinery. J Neurosci Off J Soc Neurosci. 2010;30:10096–10111. doi: 10.1523/JNEUROSCI.1634-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X, Khaki L, Zhu TS, Soules ME, Talsma CE, Gul N, Koh C, Zhang J, Li YM, Maciaczyk J, Nikkhah G, Dimeco F, Piccirillo S, Vescovi AL, Eberhart CG. NOTCH pathway blockade depletes CD133-positive glioblastoma cells and inhibits growth of tumor neurospheres and xenografts. Stem Cells Dayt Ohio. 2010;28:5–16. doi: 10.1002/stem.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X, Matsui W, Khaki L, Stearns D, Chun J, Li Y-M, Eberhart CG. Notch pathway inhibition depletes stem-like cells and blocks engraftment in embryonal brain tumors. Cancer Res. 2006;66:7445–7452. doi: 10.1158/0008-5472.CAN-06-0858. [DOI] [PubMed] [Google Scholar]
- Folkins C, Shaked Y, Man S, Tang T, Lee CR, Zhu Z, Hoffman RM, Kerbel RS. Glioma tumor stem-like cells promote tumor angiogenesis and vasculogenesis via vascular endothelial growth factor and stromal-derived factor 1. Cancer Res. 2009;69:7243–7251. doi: 10.1158/0008-5472.CAN-09-0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A, Hahn WC, Ligon KL, Louis DN, Brennan C, Chin L, DePinho RA, Cavenee WK. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21:2683–2710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- Galli R, Binda E, Orfanelli U, Cipelletti B, Gritti A, De Vitis S, Fiocco R, Foroni C, Dimeco F, Vescovi A. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res. 2004;64:7011–7021. doi: 10.1158/0008-5472.CAN-04-1364. [DOI] [PubMed] [Google Scholar]
- Gallia GL, Tyler BM, Hann CL, Siu I-M, Giranda VL, Vescovi AL, Brem H, Riggins GJ. Inhibition of Akt inhibits growth of glioblastoma and glioblastoma stem-like cells. Mol Cancer Ther. 2009;8:386–393. doi: 10.1158/1535-7163.MCT-08-0680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo M, Ho J, Coutinho FJ, Vanner R, Lee L, Head R, Ling EKM, Clarke ID, Dirks PB. A tumorigenic MLL-homeobox network in human glioblastoma stem cells. Cancer Res. 2013;73:417–427. doi: 10.1158/0008-5472.CAN-12-1881. [DOI] [PubMed] [Google Scholar]
- Gangemi RMR, Griffero F, Marubbi D, Perera M, Capra MC, Malatesta P, Ravetti GL, Zona GL, Daga A, Corte G. SOX2 silencing in glioblastoma tumor-initiating cells causes stop of proliferation and loss of tumorigenicity. Stem Cells Dayt Ohio. 2009;27:40–48. doi: 10.1634/stemcells.2008-0493. [DOI] [PubMed] [Google Scholar]
- Gaspar N, Marshall L, Perryman L, Bax DA, Little SE, Viana-Pereira M, Sharp SY, Vassal G, Pearson ADJ, Reis RM, Hargrave D, Workman P, Jones C. MGMT-independent temozolomide resistance in pediatric glioblastoma cells associated with a PI3- kinase-mediated HOX/stem cell gene signature. Cancer Res. 2010;70:9243–9252. doi: 10.1158/0008-5472.CAN-10-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Y, Zhou F, Chen H, Cui C, Liu D, Li Q, Yang Z, Wu G, Sun S, Gu J, Wei Y, Jiang J. Sox2 is translationally activated by eukaryotic initiation factor 4E in human glioma-initiating cells. Biochem Biophys Res Commun. 2010;397:711–717. doi: 10.1016/j.bbrc.2010.06.015. [DOI] [PubMed] [Google Scholar]
- Gilbert CA, Daou M-C, Moser RP, Ross AH. Gamma-secretase inhibitors enhance temozolomide treatment of human gliomas by inhibiting neurosphere repopulation and xenograft recurrence. Cancer Res. 2010;70:6870–6879. doi: 10.1158/0008-5472.CAN-10-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbertson RJ, Rich JN. Making a tumour’s bed: glioblastoma stem cells and the vascular niche. Nat Rev Cancer. 2007;7:733–736. doi: 10.1038/nrc2246. [DOI] [PubMed] [Google Scholar]
- Godlewski J, Newton HB, Chiocca EA, Lawler SE. MicroRNAs and glioblastoma; the stem cell connection. Cell Death Differ. 2010;17:221–228. doi: 10.1038/cdd.2009.71. [DOI] [PubMed] [Google Scholar]
- Godlewski J, Nowicki MO, Bronisz A, Williams S, Otsuki A, Nuovo G, Raychaudhury A, Newton HB, Chiocca EA, Lawler S. Targeting of the Bmi-1 oncogene/stem cell renewal factor by microRNA-128 inhibits glioma proliferation and self-renewal. Cancer Res. 2008;68:9125–9130. doi: 10.1158/0008-5472.CAN-08-2629. [DOI] [PubMed] [Google Scholar]
- Golebiewska A, Bougnaud S, Stieber D, Brons NHC, Vallar L, Hertel F, Klink B, Schröck E, Bjerkvig R, Niclou SP. Side population in human glioblastoma is non-tumorigenic and characterizes brain endothelial cells. Brain J Neurol. 2013;136:1462–1475. doi: 10.1093/brain/awt025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong A, Huang S. FoxM1 and Wnt/β-catenin signaling in glioma stem cells. Cancer Res. 2012;72:5658–5662. doi: 10.1158/0008-5472.CAN-12-0953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Gómez P, Sánchez P, Mira H. MicroRNAs as regulators of neural stem cell-related pathways in glioblastoma multiforme. Mol Neurobiol. 2011;44:235–249. doi: 10.1007/s12035-011-8196-y. [DOI] [PubMed] [Google Scholar]
- Gopisetty G, Xu J, Sampath D, Colman H, Puduvalli VK. Epigenetic regulation of CD133/PROM1 expression in glioma stem cells by Sp1/myc and promoter methylation. Oncogene. 2012 doi: 10.1038/onc.2012.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoryan T, Wend P, Klaus A, Birchmeier W. Deciphering the function of canonical Wnt signals in development and disease: conditional loss- and gain-of-function mutations of beta-catenin in mice. Genes Dev. 2008;22:2308–2341. doi: 10.1101/gad.1686208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guessous F, Zhang Y, Kofman A, Catania A, Li Y, Schiff D, Purow B, Abounader R. microRNA-34a is tumor suppressive in brain tumors and glioma stem cells. Cell Cycle Georget Tex. 2010;9:1031–1036. doi: 10.4161/cc.9.6.10987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guichet PO, Bieche I, Teigell M, Serguera C, Rothhut B, Rigau V, Scamps F, Ripoll C, Vacher S, Taviaux S, Chevassus H, Duffau H, Mallet J, Susini A, Joubert D, Bauchet L, Hugnot JP. Cell death and neuronal differentiation of glioblastoma stem-like cells induced by neurogenic transcription factors. Glia. 2013;61:225–239. doi: 10.1002/glia.22429. [DOI] [PubMed] [Google Scholar]
- Guo Y, Liu S, Wang P, Zhao S, Wang F, Bing L, Zhang Y, Ling EA, Gao J, Hao A. Expression profile of embryonic stem cell-associated genes Oct4, Sox2 and Nanog in human gliomas. Histopathology. 2011;59:763–775. doi: 10.1111/j.1365-2559.2011.03993.x. [DOI] [PubMed] [Google Scholar]
- Guryanova OA, Wu Q, Cheng L, Lathia JD, Huang Z, Yang J, MacSwords J, Eyler CE, McLendon RE, Heddleston JM, Shou W, Hambardzumyan D, Lee J, Hjelmeland AB, Sloan AE, Bredel M, Stark GR, Rich JN, Bao S. Nonreceptor tyrosine kinase BMX maintains self-renewal and tumorigenic potential of glioblastoma stem cells by activating STAT3. Cancer Cell. 2011;19:498–511. doi: 10.1016/j.ccr.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hägerstrand D, He X, Bradic Lindh M, Hoefs S, Hesselager G, Ostman A, Nistér M. Identification of a SOX2-dependent subset of tumor- and sphere-forming glioblastoma cells with a distinct tyrosine kinase inhibitor sensitivity profile. Neuro-Oncol. 2011;13:1178–1191. doi: 10.1093/neuonc/nor113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hambardzumyan D, Becher OJ, Holland EC. Cancer stem cells and survival pathways. Cell Cycle Georget Tex. 2008a;7:1371–1378. doi: 10.4161/cc.7.10.5954. [DOI] [PubMed] [Google Scholar]
- Hambardzumyan D, Becher OJ, Rosenblum MK, Pandolfi PP, Manova-Todorova K, Holland EC. PI3K pathway regulates survival of cancer stem cells residing in the perivascular niche following radiation in medulloblastoma in vivo. Genes Dev. 2008b;22:436–448. doi: 10.1101/gad.1627008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hambardzumyan D, Squatrito M, Carbajal E, Holland EC. Glioma formation, cancer stem cells, and akt signaling. Stem Cell Rev. 2008c;4:203–210. doi: 10.1007/s12015-008-9021-5. [DOI] [PubMed] [Google Scholar]
- Harris AL. Hypoxia--a key regulatory factor in tumour growth. Nat Rev Cancer. 2002;2:38–47. doi: 10.1038/nrc704. [DOI] [PubMed] [Google Scholar]
- Harris MA, Yang H, Low BE, Mukherjee J, Mukherje J, Guha A, Bronson RT, Shultz LD, Israel MA, Yun K. Cancer stem cells are enriched in the side population cells in a mouse model of glioma. Cancer Res. 2008;68:10051–10059. doi: 10.1158/0008-5472.CAN-08-0786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann W, Digon-Söntgerath B, Koch A, Waha A, Endl E, Dani I, Denkhaus D, Goodyer CG, Sörensen N, Wiestler OD, Pietsch T. Phosphatidylinositol 3′-kinase/AKT signaling is activated in medulloblastoma cell proliferation and is associated with reduced expression of PTEN. Clin Cancer Res Off J Am Assoc Cancer Res. 2006;12:3019–3027. doi: 10.1158/1078-0432.CCR-05-2187. [DOI] [PubMed] [Google Scholar]
- Häyry V, Tynninen O, Haapasalo HK, Wölfer J, Paulus W, Hasselblatt M, Sariola H, Paetau A, Sarna S, Niemelä M, Wartiovaara K, Nupponen NN. Stem cell protein BMI-1 is an independent marker for poor prognosis in oligodendroglial tumours. Neuropathol Appl Neurobiol. 2008;34:555–563. doi: 10.1111/j.1365-2990.2008.00949.x. [DOI] [PubMed] [Google Scholar]
- Heddleston JM, Hitomi M, Venere M, Flavahan WA, Yang K, Kim Y, Minhas S, Rich JN, Hjelmeland AB. Glioma stem cell maintenance: the role of the microenvironment. Curr Pharm Des. 2011;17:2386–2401. doi: 10.2174/138161211797249260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heddleston JM, Li Z, McLendon RE, Hjelmeland AB, Rich JN. The hypoxic microenvironment maintains glioblastoma stem cells and promotes reprogramming towards a cancer stem cell phenotype. Cell Cycle Georget Tex. 2009;8:3274–3284. doi: 10.4161/cc.8.20.9701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heddleston JM, Wu Q, Rivera M, Minhas S, Lathia JD, Sloan AE, Iliopoulos O, Hjelmeland AB, Rich JN. Hypoxia-induced mixed-lineage leukemia 1 regulates glioma stem cell tumorigenic potential. Cell Death Differ. 2012;19:428–439. doi: 10.1038/cdd.2011.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegi ME, Diserens A-C, Gorlia T, Hamou M-F, de Tribolet N, Weller M, Kros JM, Hainfellner JA, Mason W, Mariani L, Bromberg JEC, Hau P, Mirimanoff RO, Cairncross JG, Janzer RC, Stupp R. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- Hemmati HD, Nakano I, Lazareff JA, Masterman-Smith M, Geschwind DH, Bronner-Fraser M, Kornblum HI. Cancerous stem cells can arise from pediatric brain tumors. Proc Natl Acad Sci U S A. 2003;100:15178–15183. doi: 10.1073/pnas.2036535100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjelmeland AB, Wu Q, Heddleston JM, Choudhary GS, MacSwords J, Lathia JD, McLendon R, Lindner D, Sloan A, Rich JN. Acidic stress promotes a glioma stem cell phenotype. Cell Death Differ. 2011;18:829–840. doi: 10.1038/cdd.2010.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoey T, Yen WC, Axelrod F, Basi J, Donigian L, Dylla S, Fitch-Bruhns M, Lazetic S, Park IK, Sato A, Satyal S, Wang X, Clarke MF, Lewicki J, Gurney A. DLL4 blockade inhibits tumor growth and reduces tumor-initiating cell frequency. Cell Stem Cell. 2009;5:168–177. doi: 10.1016/j.stem.2009.05.019. [DOI] [PubMed] [Google Scholar]
- Holland EC, Hively WP, DePinho RA, Varmus HE. A constitutively active epidermal growth factor receptor cooperates with disruption of G1 cell-cycle arrest pathways to induce glioma-like lesions in mice. Genes Dev. 1998;12:3675–3685. doi: 10.1101/gad.12.23.3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z, Cheng L, Guryanova OA, Wu Q, Bao S. Cancer stem cells in glioblastoma--molecular signaling and therapeutic targeting. Protein Cell. 2010;1:638–655. doi: 10.1007/s13238-010-0078-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikushima H, Todo T, Ino Y, Takahashi M, Miyazawa K, Miyazono K. Autocrine TGF-beta signaling maintains tumorigenicity of glioma-initiating cells through Sry-related HMG-box factors. Cell Stem Cell. 2009;5:504–514. doi: 10.1016/j.stem.2009.08.018. [DOI] [PubMed] [Google Scholar]
- Ikushima H, Todo T, Ino Y, Takahashi M, Saito N, Miyazawa K, Miyazono K. Glioma-initiating cells retain their tumorigenicity through integration of the Sox axis and Oct4 protein. J Biol Chem. 2011;286:41434–41441. doi: 10.1074/jbc.M111.300863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon H-M, Jin X, Lee J-S, Oh S-Y, Sohn Y-W, Park H-J, Joo KM, Park W-Y, Nam D-H, DePinho RA, Chin L, Kim H. Inhibitor of differentiation 4 drives brain tumor-initiating cell genesis through cyclin E and notch signaling. Genes Dev. 2008;22:2028–2033. doi: 10.1101/gad.1668708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon H-M, Sohn Y-W, Se-Yeong Oh, Se-Young Oh, Kim S-H, Beck S, Kim S, Kim H. ID4 imparts chemoresistance and cancer stemness to glioma cells by derepressing miR-9*-mediated suppression of SOX2. Cancer Res. 2011;71:3410–3421. doi: 10.1158/0008-5472.CAN-10-3340. [DOI] [PubMed] [Google Scholar]
- Jiang H, Fueyo J. Eradication of brain tumor stem cells with an oncolytic adenovirus. Discov Med. 2010;10:24–28. [PubMed] [Google Scholar]
- Jiang H, Gomez-Manzano C, Aoki H, Alonso MM, Kondo S, McCormick F, Xu J, Kondo Y, Bekele BN, Colman H, Lang FF, Fueyo J. Examination of the therapeutic potential of Delta-24-RGD in brain tumor stem cells: role of autophagic cell death. J Natl Cancer Inst. 2007;99:1410–1414. doi: 10.1093/jnci/djm102. [DOI] [PubMed] [Google Scholar]
- Jiang H, Gomez-Manzano C, Lang FF, Alemany R, Fueyo J. Oncolytic adenovirus: preclinical and clinical studies in patients with human malignant gliomas. Curr Gene Ther. 2009;9:422–427. doi: 10.2174/156652309789753356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin X, Jeon H-Y, Joo KM, Kim J-K, Jin J, Kim SH, Kang BG, Beck S, Lee SJ, Kim JK, Park A-K, Park W-Y, Choi Y-J, Nam D-H, Kim H. Frizzled 4 regulates stemness and invasiveness of migrating glioma cells established by serial intracranial transplantation. Cancer Res. 2011;71:3066–3075. doi: 10.1158/0008-5472.CAN-10-1495. [DOI] [PubMed] [Google Scholar]
- Jin X, Kim Sung-Hak, Jeon H-M, Beck S, Sohn Y-W, Yin J, Kim J-K, Lim YC, Lee J-H, Kim Se-Hyuk, Kang S-H, Pian X, Song M-S, Park JB, Chae Y-S, Chung Y-G, Lee S-H, Choi Y-J, Nam D-H, Choi YK, Kim H. Interferon regulatory factor 7 regulates glioma stem cells via interleukin-6 and Notch signalling. Brain J Neurol. 2012;135:1055–1069. doi: 10.1093/brain/aws028. [DOI] [PubMed] [Google Scholar]
- Kanamori M, Kawaguchi T, Nigro JM, Feuerstein BG, Berger MS, Miele L, Pieper RO. Contribution of Notch signaling activation to human glioblastoma multiforme. J Neurosurg. 2007;106:417–427. doi: 10.3171/jns.2007.106.3.417. [DOI] [PubMed] [Google Scholar]
- Kang M-K, Kang S-K. Tumorigenesis of chemotherapeutic drug-resistant cancer stem-like cells in brain glioma. Stem Cells Dev. 2007;16:837–847. doi: 10.1089/scd.2007.0006. [DOI] [PubMed] [Google Scholar]
- Katsushima K, Shinjo K, Natsume A, Ohka F, Fujii M, Osada H, Sekido Y, Kondo Y. Contribution of microRNA-1275 to Claudin11 protein suppression via a polycomb-mediated silencing mechanism in human glioma stem-like cells. J Biol Chem. 2012;287:27396–27406. doi: 10.1074/jbc.M112.359109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur N, Chettiar S, Rathod S, Rath P, Muzumdar D, Shaikh ML, Shiras A. Wnt3a mediated activation of Wnt/β-catenin signaling promotes tumor progression in glioblastoma. Mol Cell Neurosci. 2013;54:44–57. doi: 10.1016/j.mcn.2013.01.001. [DOI] [PubMed] [Google Scholar]
- Keith B, Simon MC. Hypoxia-inducible factors, stem cells, and cancer. Cell. 2007;129:465–472. doi: 10.1016/j.cell.2007.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Kim KH, Lee Jeena, Lee Y-A, Kim M, Lee SJ, Park K, Yang H, Jin J, Joo KM, Lee Jeongwu, Nam D-H. Wnt activation is implicated in glioblastoma radioresistance. Lab Investig J Tech Methods Pathol. 2012;92:466–473. doi: 10.1038/labinvest.2011.161. [DOI] [PubMed] [Google Scholar]
- Kopan R. Notch: a membrane-bound transcription factor. J Cell Sci. 2002;115:1095–1097. doi: 10.1242/jcs.115.6.1095. [DOI] [PubMed] [Google Scholar]
- Lapidot T, Sirard C, Vormoor J, Murdoch B, Hoang T, Caceres-Cortes J, Minden M, Paterson B, Caligiuri MA, Dick JE. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367:645–648. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- Lathia JD, Gallagher J, Heddleston JM, Wang J, Eyler CE, Macswords J, Wu Q, Vasanji A, McLendon RE, Hjelmeland AB, Rich JN. Integrin alpha 6 regulates glioblastoma stem cells. Cell Stem Cell. 2010;6:421–432. doi: 10.1016/j.stem.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lathia JD, Li M, Hall PE, Gallagher J, Hale JS, Wu Q, Venere M, Levy E, Rani MRS, Huang P, Bae E, Selfridge J, Cheng L, Guvenc H, McLendon RE, Nakano I, Sloan AE, Phillips HS, Lai A, Gladson CL, Bredel M, Bao S, Hjelmeland AB, Rich JN. Laminin alpha 2 enables glioblastoma stem cell growth. Ann Neurol. 2012;72:766–778. doi: 10.1002/ana.23674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lathia JD, Mattson MP, Cheng A. Notch: from neural development to neurological disorders. J Neurochem. 2008;107:1471–1481. doi: 10.1111/j.1471-4159.2008.05715.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Kotliarova S, Kotliarov Y, Li A, Su Q, Donin NM, Pastorino S, Purow BW, Christopher N, Zhang W, Park JK, Fine HA. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9:391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- Lee J, Son MJ, Woolard K, Donin NM, Li A, Cheng CH, Kotliarova S, Kotliarov Y, Walling J, Ahn S, Kim M, Totonchy M, Cusack T, Ene C, Ma H, Su Q, Zenklusen JC, Zhang W, Maric D, Fine HA. Epigenetic-mediated dysfunction of the bone morphogenetic protein pathway inhibits differentiation of glioblastoma-initiating cells. Cancer Cell. 2008;13:69–80. doi: 10.1016/j.ccr.2007.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonardo TR, Schultheisz HL, Loring JF, Laurent LC. The functions of microRNAs in pluripotency and reprogramming. Nat Cell Biol. 2012;14:1114–1121. doi: 10.1038/ncb2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J-L, Sainson RCA, Oon CE, Turley H, Leek R, Sheldon H, Bridges E, Shi W, Snell C, Bowden ET, Wu H, Chowdhury PS, Russell AJ, Montgomery CP, Poulsom R, Harris AL. DLL4-Notch signaling mediates tumor resistance to anti-VEGF therapy in vivo. Cancer Res. 2011;71:6073–6083. doi: 10.1158/0008-5472.CAN-11-1704. [DOI] [PubMed] [Google Scholar]
- Li J-L, Sainson RCA, Shi W, Leek R, Harrington LS, Preusser M, Biswas S, Turley H, Heikamp E, Hainfellner JA, Harris AL. Delta-like 4 Notch ligand regulates tumor angiogenesis, improves tumor vascular function, and promotes tumor growth in vivo. Cancer Res. 2007;67:11244–11253. doi: 10.1158/0008-5472.CAN-07-0969. [DOI] [PubMed] [Google Scholar]
- Li X, Lewis MT, Huang J, Gutierrez C, Osborne CK, Wu M-F, Hilsenbeck SG, Pavlick A, Zhang X, Chamness GC, Wong H, Rosen J, Chang JC. Intrinsic resistance of tumorigenic breast cancer cells to chemotherapy. J Natl Cancer Inst. 2008;100:672–679. doi: 10.1093/jnci/djn123. [DOI] [PubMed] [Google Scholar]
- Li Z, Bao S, Wu Q, Wang H, Eyler C, Sathornsumetee S, Shi Q, Cao Y, Lathia J, McLendon RE, Hjelmeland AB, Rich JN. Hypoxia-inducible factors regulate tumorigenic capacity of glioma stem cells. Cancer Cell. 2009a;15:501–513. doi: 10.1016/j.ccr.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Wang H, Eyler CE, Hjelmeland AB, Rich JN. Turning cancer stem cells inside out: an exploration of glioma stem cell signaling pathways. J Biol Chem. 2009b;284:16705–16709. doi: 10.1074/jbc.R900013200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligon KL, Alberta JA, Kho AT, Weiss J, Kwaan MR, Nutt CL, Louis DN, Stiles CD, Rowitch DH. The oligodendroglial lineage marker OLIG2 is universally expressed in diffuse gliomas. J Neuropathol Exp Neurol. 2004;63:499–509. doi: 10.1093/jnen/63.5.499. [DOI] [PubMed] [Google Scholar]
- Ligon KL, Huillard E, Mehta S, Kesari S, Liu H, Alberta JA, Bachoo RM, Kane M, Louis DN, Depinho RA, Anderson DJ, Stiles CD, Rowitch DH. Olig2-regulated lineage-restricted pathway controls replication competence in neural stem cells and malignant glioma. Neuron. 2007;53:503–517. doi: 10.1016/j.neuron.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima FRS, Kahn SA, Soletti RC, Biasoli D, Alves T, da Fonseca ACC, Garcia C, Romão L, Brito J, Holanda-Afonso R, Faria J, Borges H, Moura-Neto V. Glioblastoma: therapeutic challenges, what lies ahead. Biochim Biophys Acta. 2012;1826:338–349. doi: 10.1016/j.bbcan.2012.05.004. [DOI] [PubMed] [Google Scholar]
- Lino MM, Merlo A, Boulay J-L. Notch signaling in glioblastoma: a developmental drug target? Bmc Med. 2010;8:72. doi: 10.1186/1741-7015-8-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Yuan X, Zeng Z, Tunici P, Ng H, Abdulkadir IR, Lu L, Irvin D, Black KL, Yu JS. Analysis of gene expression and chemoresistance of CD133+ cancer stem cells in glioblastoma. Mol Cancer. 2006;5:67. doi: 10.1186/1476-4598-5-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomonaco SL, Finniss S, Xiang C, Decarvalho A, Umansky F, Kalkanis SN, Mikkelsen T, Brodie C. The induction of autophagy by gamma-radiation contributes to the radioresistance of glioma stem cells. Int J Cancer J Int Cancer. 2009;125:717–722. doi: 10.1002/ijc.24402. [DOI] [PubMed] [Google Scholar]
- Lomonaco SL, Finniss S, Xiang C, Lee HK, Jiang W, Lemke N, Rempel SA, Mikkelsen T, Brodie C. Cilengitide induces autophagy-mediated cell death in glioma cells. Neuro- Oncol. 2011;13:857–865. doi: 10.1093/neuonc/nor073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manoranjan B, Wang X, Hallett RM, Venugopal C, Mack SC, McFarlane N, Nolte SM, Scheinemann K, Gunnarsson T, Hassell JA, Taylor MD, Lee C, Triscott J, Foster CM, Dunham C, Hawkins C, Dunn SE, Singh SK. FoxG1 Interacts with Bmi1 to Regulate Self-Renewal and Tumorigenicity of Medulloblastoma Stem Cells. Stem Cells Dayt Ohio. 2013 doi: 10.1002/stem.1401. [DOI] [PubMed] [Google Scholar]
- Mao X, Yan M, Xue X, Zhang X, Ren H, Guo G, Wang P, Zhang W, Huo J. Overexpression of ZNF217 in glioblastoma contributes to the maintenance of glioma stem cells regulated by hypoxia-inducible factors. Lab Investig J Tech Methods Pathol. 2011;91:1068–1078. doi: 10.1038/labinvest.2011.56. [DOI] [PubMed] [Google Scholar]
- Martín V, Sanchez-Sanchez AM, Herrera F, Gomez-Manzano C, Fueyo J, Alvarez-Vega MA, Antolín I, Rodriguez C. Melatonin-induced methylation of the ABCG2/BCRP promoter as a novel mechanism to overcome multidrug resistance in brain tumour stem cells. Br J Cancer. 2013 doi: 10.1038/bjc.2013.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu J, Zhang Z, Zhou W, Wang AJ, Heddleston JM, Pinna CMA, Hubaud A, Stadler B, Choi M, Bar M, Tewari M, Liu A, Vessella R, Rostomily R, Born D, Horwitz M, Ware C, Blau CA, Cleary MA, Rich JN, Ruohola-Baker H. HIF induces human embryonic stem cell markers in cancer cells. Cancer Res. 2011;71:4640–4652. doi: 10.1158/0008-5472.CAN-10-3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayo LD, Donner DB. A phosphatidylinositol 3-kinase/Akt pathway promotes translocation of Mdm2 from the cytoplasm to the nucleus. Proc Natl Acad Sci U S A. 2001;98:11598–11603. doi: 10.1073/pnas.181181198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei J, Bachoo R, Zhang C-L. MicroRNA-146a inhibits glioma development by targeting Notch1. Mol Cell Biol. 2011;31:3584–3592. doi: 10.1128/MCB.05821-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani K, Yoon K, Dang L, Tokunaga A, Gaiano N. Differential Notch signalling distinguishes neural stem cells from intermediate progenitors. Nature. 2007;449:351–355. doi: 10.1038/nature06090. [DOI] [PubMed] [Google Scholar]
- Moscatello DK, Holgado-Madruga M, Emlet DR, Montgomery RB, Wong AJ. Constitutive activation of phosphatidylinositol 3-kinase by a naturally occurring mutant epidermal growth factor receptor. J Biol Chem. 1998;273:200–206. doi: 10.1074/jbc.273.1.200. [DOI] [PubMed] [Google Scholar]
- Murat A, Migliavacca E, Gorlia T, Lambiv WL, Shay T, Hamou M-F, de Tribolet N, Regli L, Wick W, Kouwenhoven MCM, Hainfellner JA, Heppner FL, Dietrich P-Y, Zimmer Y, Cairncross JG, Janzer R-C, Domany E, Delorenzi M, Stupp R, Hegi ME. Stem cell-related “self-renewal” signature and high epidermal growth factor receptor expression associated with resistance to concomitant chemoradiotherapy in glioblastoma. J Clin Oncol Off J Am Soc Clin Oncol. 2008;26:3015–3024. doi: 10.1200/JCO.2007.15.7164. [DOI] [PubMed] [Google Scholar]
- Nakata S, Campos B, Bageritz J, Bermejo JL, Becker N, Engel F, Acker T, Momma S, Herold-Mende C, Lichter P, Radlwimmer B, Goidts V. LGR5 is a marker of poor prognosis in glioblastoma and is required for survival of brain cancer stem-like cells. Brain Pathol Zurich Switz. 2013;23:60–72. doi: 10.1111/j.1750-3639.2012.00618.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu C-S, Li D-X, Liu Y-H, Fu X-M, Tang S-F, Li J. Expression of NANOG in human gliomas and its relationship with undifferentiated glioma cells. Oncol Rep. 2011;26:593–601. doi: 10.3892/or.2011.1308. [DOI] [PubMed] [Google Scholar]
- Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, Pan F, Pelloski CE, Sulman EP, Bhat KP, Verhaak RGW, Hoadley KA, Hayes DN, Perou CM, Schmidt HK, Ding L, Wilson RK, Van Den Berg D, Shen H, Bengtsson H, Neuvial P, Cope LM, Buckley J, Herman JG, Baylin SB, Laird PW, Aldape K Cancer Genome Atlas Research Network. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17:510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogden AT, Waziri AE, Lochhead RA, Fusco D, Lopez K, Ellis JA, Kang J, Assanah M, McKhann GM, Sisti MB, McCormick PC, Canoll P, Bruce JN. Identification of A2B5+CD133− tumor-initiating cells in adult human gliomas. Neurosurgery. 2008;62:505–514. doi: 10.1227/01.neu.0000316019.28421.95. discussion 514–515. [DOI] [PubMed] [Google Scholar]
- Ohishi K, Varnum-Finney B, Bernstein ID. Delta-1 enhances marrow and thymus repopulating ability of human CD34(+)CD38(−) cord blood cells. J Clin Invest. 2002;110:1165–1174. doi: 10.1172/JCI16167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka N, Soeda A, Inagaki A, Onodera M, Maruyama H, Hara A, Kunisada T, Mori H, Iwama T. VEGF promotes tumorigenesis and angiogenesis of human glioblastoma stem cells. Biochem Biophys Res Commun. 2007;360:553–559. doi: 10.1016/j.bbrc.2007.06.094. [DOI] [PubMed] [Google Scholar]
- Osuka S, Sampetrean O, Shimizu T, Saga I, Onishi N, Sugihara E, Okubo J, Fujita S, Takano S, Matsumura A, Saya H. IGF1 receptor signaling regulates adaptive radioprotection in glioma stem cells. Stem Cells Dayt Ohio. 2012;31:627–640. doi: 10.1002/stem.1328. [DOI] [PubMed] [Google Scholar]
- Panchision DM, McKay RDG. The control of neural stem cells by morphogenic signals. Curr Opin Genet Dev. 2002;12:478–487. doi: 10.1016/s0959-437x(02)00329-5. [DOI] [PubMed] [Google Scholar]
- Papp B, Plath K. Epigenetics of reprogramming to induced pluripotency. Cell. 2013;152:1324–1343. doi: 10.1016/j.cell.2013.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegatta S, Poliani PL, Corno D, Menghi F, Ghielmetti F, Suarez-Merino B, Caldera V, Nava S, Ravanini M, Facchetti F, Bruzzone MG, Finocchiaro G. Neurospheres enriched in cancer stem-like cells are highly effective in eliciting a dendritic cell-mediated immune response against malignant gliomas. Cancer Res. 2006;66:10247–10252. doi: 10.1158/0008-5472.CAN-06-2048. [DOI] [PubMed] [Google Scholar]
- Phillips HS, Kharbanda S, Chen R, Forrest WF, Soriano RH, Wu TD, Misra A, Nigro JM, Colman H, Soroceanu L, Williams PM, Modrusan Z, Feuerstein BG, Aldape K. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- Piccirillo SGM, Reynolds BA, Zanetti N, Lamorte G, Binda E, Broggi G, Brem H, Olivi A, Dimeco F, Vescovi AL. Bone morphogenetic proteins inhibit the tumorigenic potential of human brain tumour-initiating cells. Nature. 2006;444:761–765. doi: 10.1038/nature05349. [DOI] [PubMed] [Google Scholar]
- Pistollato F, Abbadi S, Rampazzo E, Persano L, Della Puppa A, Frasson C, Sarto E, Scienza R, D’avella D, Basso G. Intratumoral hypoxic gradient drives stem cells distribution and MGMT expression in glioblastoma. Stem Cells Dayt Ohio. 2010;28:851–862. doi: 10.1002/stem.415. [DOI] [PubMed] [Google Scholar]
- Platet N, Liu SY, Atifi ME, Oliver L, Vallette FM, Berger F, Wion D. Influence of oxygen tension on CD133 phenotype in human glioma cell cultures. Cancer Lett. 2007;258:286–290. doi: 10.1016/j.canlet.2007.09.012. [DOI] [PubMed] [Google Scholar]
- Presente A, Andres A, Nye JS. Requirement of Notch in adulthood for neurological function and longevity. Neuroreport. 2001;12:3321–3325. doi: 10.1097/00001756-200110290-00035. [DOI] [PubMed] [Google Scholar]
- Pulvirenti T, Van Der Heijden M, Droms LA, Huse JT, Tabar V, Hall A. Dishevelled 2 signaling promotes self-renewal and tumorigenicity in human gliomas. Cancer Res. 2011;71:7280–7290. doi: 10.1158/0008-5472.CAN-11-1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puram SV, Yeung CM, Jahani-Asl A, Lin C, de la Iglesia N, Konopka G, Jackson-Grusby L, Bonni A. STAT3-iNOS Signaling Mediates EGFRvIII-Induced Glial Proliferation and Transformation. J Neurosci Off J Soc Neurosci. 2012;32:7806–7818. doi: 10.1523/JNEUROSCI.3243-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purow BW, Haque RM, Noel MW, Su Q, Burdick MJ, Lee J, Sundaresan T, Pastorino S, Park JK, Mikolaenko I, Maric D, Eberhart CG, Fine HA. Expression of Notch-1 and its ligands, Delta-like-1 and Jagged-1, is critical for glioma cell survival and proliferation. Cancer Res. 2005;65:2353–2363. doi: 10.1158/0008-5472.CAN-04-1890. [DOI] [PubMed] [Google Scholar]
- Radke J, Bortolussi G, Pagenstecher A. Akt and c-Myc induce stem-cell markers in mature primary p53−/− astrocytes and render these cells gliomagenic in the brain of immunocompetent mice. Plos One. 2013;8:e56691. doi: 10.1371/journal.pone.0056691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman M, Deleyrolle L, Vedam-Mai V, Azari H, Abd-El-Barr M, Reynolds BA. The cancer stem cell hypothesis: failures and pitfalls. Neurosurgery. 2011;68:531–545. doi: 10.1227/NEU.0b013e3181ff9eb5. discussion 545. [DOI] [PubMed] [Google Scholar]
- Read TA, Fogarty MP, Markant SL, McLendon RE, Wei Z, Ellison DW, Febbo PG, Wechsler-Reya RJ. Identification of CD15 as a marker for tumor-propagating cells in a mouse model of medulloblastoma. Cancer Cell. 2009;15:135–147. doi: 10.1016/j.ccr.2008.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- Ricci-Vitiani L, Pallini R, Biffoni M, Todaro M, Invernici G, Cenci T, Maira G, Parati EA, Stassi G, Larocca LM, De Maria R. Tumour vascularization via endothelial differentiation of glioblastoma stem-like cells. Nature. 2010;468:824–828. doi: 10.1038/nature09557. [DOI] [PubMed] [Google Scholar]
- Rich JN. The Implications of the Cancer Stem Cell Hypothesis for Neuro-Oncology and Neurology. Future Neurol. 2008;3:265–273. doi: 10.2217/14796708.3.3.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich JN, Eyler CE. Cancer stem cells in brain tumor biology. Cold Spring Harb Symp Quant Biol. 2008;73:411–420. doi: 10.1101/sqb.2008.73.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez FJ, Orr BA, Ligon KL, Eberhart CG. Neoplastic cells are a rare component in human glioblastoma microvasculature. Oncotarget. 2012;3:98–106. doi: 10.18632/oncotarget.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers HA, Sousa S, Salto C, Arenas E, Coyle B, Grundy RG. WNT/β-catenin pathway activation in Myc immortalised cerebellar progenitor cells inhibits neuronal differentiation and generates tumours resembling medulloblastoma. Br J Cancer. 2012;107:1144–1152. doi: 10.1038/bjc.2012.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ropolo M, Daga A, Griffero F, Foresta M, Casartelli G, Zunino A, Poggi A, Cappelli E, Zona G, Spaziante R, Corte G, Frosina G. Comparative analysis of DNA repair in stem and nonstem glioma cell cultures. Mol Cancer Res Mcr. 2009;7:383–392. doi: 10.1158/1541-7786.MCR-08-0409. [DOI] [PubMed] [Google Scholar]
- Rosen JM, Jordan CT. The increasing complexity of the cancer stem cell paradigm. Science. 2009;324:1670–1673. doi: 10.1126/science.1171837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi M, Magnoni L, Miracco C, Mori E, Tosi P, Pirtoli L, Tini P, Oliveri G, Cosci E, Bakker A. β-catenin and Gli1 are prognostic markers in glioblastoma. Cancer Biol Ther. 2011;11:753–761. doi: 10.4161/cbt.11.8.14894. [DOI] [PubMed] [Google Scholar]
- Rudin CM, Hann CL, Laterra J, Yauch RL, Callahan CA, Fu L, Holcomb T, Stinson J, Gould SE, Coleman B, LoRusso PM, Von Hoff DD, de Sauvage FJ, Low JA. Treatment of medulloblastoma with hedgehog pathway inhibitor GDC-0449. N Engl J Med. 2009;361:1173–1178. doi: 10.1056/NEJMoa0902903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz i Altaba A, Mas C, Stecca B. The Gli code: an information nexus regulating cell fate, stemness and cancer. Trends Cell Biol. 2007;17:438–447. doi: 10.1016/j.tcb.2007.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sai K, Wang S, Balasubramaniyan V, Conrad C, Lang FF, Aldape K, Szymanski S, Fokt I, Dasgupta A, Madden T, Guan S, Chen Z, Alfred Yung WK, Priebe W, Colman H. Induction of cell-cycle arrest and apoptosis in glioblastoma stem-like cells by WP1193, a novel small molecule inhibitor of the JAK2/STAT3 pathway. J Neurooncol. 2012;107:487–501. doi: 10.1007/s11060-011-0786-z. [DOI] [PubMed] [Google Scholar]
- Sanai N, Alvarez-Buylla A, Berger MS. Neural stem cells and the origin of gliomas. N Engl J Med. 2005;353:811–822. doi: 10.1056/NEJMra043666. [DOI] [PubMed] [Google Scholar]
- Sathornsumetee S, Cao Y, Marcello JE, Herndon JE, 2nd, McLendon RE, Desjardins A, Friedman HS, Dewhirst MW, Vredenburgh JJ, Rich JN. Tumor angiogenic and hypoxic profiles predict radiographic response and survival in malignant astrocytoma patients treated with bevacizumab and irinotecan. J Clin Oncol Off J Am Soc Clin Oncol. 2008;26:271–278. doi: 10.1200/JCO.2007.13.3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato A, Okada M, Shibuya K, Watanabe E, Seino S, Suzuki K, Narita Y, Shibui S, Kayama T, Kitanaka C. Resveratrol promotes proteasome-dependent degradation of Nanog via p53 activation and induces differentiation of glioma stem cells. Stem Cell Res. 2013;11:601–610. doi: 10.1016/j.scr.2013.04.004. [DOI] [PubMed] [Google Scholar]
- Schott AF, Landis MD, Dontu G, Griffith KA, Layman RM, Krop I, Paskett LA, Wong H, Dobrolecki LE, Lewis MT, Froehlich AM, Paranilam J, Hayes DF, Wicha MS, Chang JC. Preclinical and clinical studies of gamma secretase inhibitors with docetaxel on human breast tumors. Clin Cancer Res Off J Am Assoc Cancer Res. 2013;19:1512–1524. doi: 10.1158/1078-0432.CCR-11-3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüle R, Dictus C, Campos B, Wan F, Felsberg J, Ahmadi R, Centner FS, Grabe N, Reifenberger G, Bermejo JL, Unterberg A, Herold-Mende C. Potential canonical wnt pathway activation in high-grade astrocytomas. Scientific World Journal. 2012;2012:697313. doi: 10.1100/2012/697313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel S, Garvalov BK, Wirta V, von Stechow L, Schänzer A, Meletis K, Wolter M, Sommerlad D, Henze A-T, Nistér M, Reifenberger G, Lundeberg J, Frisén J, Acker T. A hypoxic niche regulates glioblastoma stem cells through hypoxia inducible factor 2 alpha. Brain J Neurol. 2010;133:983–995. doi: 10.1093/brain/awq042. [DOI] [PubMed] [Google Scholar]
- Shackleton M, Quintana E, Fearon ER, Morrison SJ. Heterogeneity in cancer: cancer stem cells versus clonal evolution. Cell. 2009;138:822–829. doi: 10.1016/j.cell.2009.08.017. [DOI] [PubMed] [Google Scholar]
- Shahi MH, Lorente A, Castresana JS. Hedgehog signalling in medulloblastoma, glioblastoma and neuroblastoma. Oncol Rep. 2008;19:681–688. [PubMed] [Google Scholar]
- Sheiness D, Fanshier L, Bishop JM. Identification of nucleotide sequences which may encode the oncogenic capacity of avian retrovirus MC29. J Virol. 1978;28:600–610. doi: 10.1128/jvi.28.2.600-610.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shervington A, Lu C. Expression of multidrug resistance genes in normal and cancer stem cells. Cancer Invest. 2008;26:535–542. doi: 10.1080/07357900801904140. [DOI] [PubMed] [Google Scholar]
- Shi L, Zhang S, Feng K, Wu F, Wan Y, Wang Z, Zhang J, Wang Y, Yan W, Fu Z, You Y. MicroRNA-125b-2 confers human glioblastoma stem cells resistance to temozolomide through the mitochondrial pathway of apoptosis. Int J Oncol. 2012;40:119–129. doi: 10.3892/ijo.2011.1179. [DOI] [PubMed] [Google Scholar]
- Shih AH, Holland EC. Notch signaling enhances nestin expression in gliomas. Neoplasia New York N. 2006;8:1072–1082. doi: 10.1593/neo.06526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiras A, Chettiar ST, Shepal V, Rajendran G, Prasad GR, Shastry P. Spontaneous transformation of human adult nontumorigenic stem cells to cancer stem cells is driven by genomic instability in a human model of glioblastoma. Stem Cells Dayt Ohio. 2007;25:1478–1489. doi: 10.1634/stemcells.2006-0585. [DOI] [PubMed] [Google Scholar]
- Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J, Dirks PB. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63:5821–5828. [PubMed] [Google Scholar]
- Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- Smith ZD, Meissner A. DNA methylation: roles in mammalian development. Nat Rev Genet. 2013;14:204–220. doi: 10.1038/nrg3354. [DOI] [PubMed] [Google Scholar]
- Soda Y, Marumoto T, Friedmann-Morvinski D, Soda M, Liu F, Michiue H, Pastorino S, Yang M, Hoffman RM, Kesari S, Verma IM. Transdifferentiation of glioblastoma cells into vascular endothelial cells. Proc Natl Acad Sci U S A. 2011;108:4274–4280. doi: 10.1073/pnas.1016030108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soeda A, Inagaki A, Oka N, Ikegame Y, Aoki H, Yoshimura S-I, Nakashima S, Kunisada T, Iwama T. Epidermal growth factor plays a crucial role in mitogenic regulation of human brain tumor stem cells. J Biol Chem. 2008;283:10958–10966. doi: 10.1074/jbc.M704205200. [DOI] [PubMed] [Google Scholar]
- Soeda A, Park M, Lee D, Mintz A, Androutsellis-Theotokis A, McKay RD, Engh J, Iwama T, Kunisada T, Kassam AB, Pollack IF, Park DM. Hypoxia promotes expansion of the CD133-positive glioma stem cells through activation of HIF-1alpha. Oncogene. 2009;28:3949–3959. doi: 10.1038/onc.2009.252. [DOI] [PubMed] [Google Scholar]
- Son MJ, Woolard K, Nam DH, Lee J, Fine HA. SSEA-1 is an enrichment marker for tumor-initiating cells in human glioblastoma. Cell Stem Cell. 2009;4:440–452. doi: 10.1016/j.stem.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stecca B, Ruiz i Altaba A. Brain as a paradigm of organ growth: Hedgehog-Gli signaling in neural stem cells and brain tumors. J Neurobiol. 2005;64:476–490. doi: 10.1002/neu.20160. [DOI] [PubMed] [Google Scholar]
- Stricker SH, Feber A, Engström PG, Carén H, Kurian KM, Takashima Y, Watts C, Way M, Dirks P, Bertone P, Smith A, Beck S, Pollard SM. Widespread resetting of DNA methylation in glioblastoma-initiating cells suppresses malignant cellular behavior in a lineage-dependent manner. Genes Dev. 2013;27:654–669. doi: 10.1101/gad.212662.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupp R, Hegi ME, Mason WP, van den Bent MJ, Taphoorn MJB, Janzer RC, Ludwin SK, Allgeier A, Fisher B, Belanger K, Hau P, Brandes AA, Gijtenbeek J, Marosi C, Vecht CJ, Mokhtari K, Wesseling P, Villa S, Eisenhauer E, Gorlia T, Weller M, Lacombe D, Cairncross JG, Mirimanoff R-O. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10:459–466. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- Sun P, Xia S, Lal B, Eberhart CG, Quinones-Hinojosa A, Maciaczyk J, Matsui W, Dimeco F, Piccirillo SM, Vescovi AL, Laterra J. DNER, an epigenetically modulated gene, regulates glioblastoma-derived neurosphere cell differentiation and tumor propagation. Stem Cells Dayt Ohio. 2009;27:1473–1486. doi: 10.1002/stem.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suvà ML, Riggi N, Bernstein BE. Epigenetic reprogramming in cancer. Science. 2013;339:1567–1570. doi: 10.1126/science.1230184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suvà M-L, Riggi N, Janiszewska M, Radovanovic I, Provero P, Stehle J-C, Baumer K, Le Bitoux M-A, Marino D, Cironi L, Marquez VE, Clément V, Stamenkovic I. EZH2 is essential for glioblastoma cancer stem cell maintenance. Cancer Res. 2009;69:9211–9218. doi: 10.1158/0008-5472.CAN-09-1622. [DOI] [PubMed] [Google Scholar]
- Syed N, Langer J, Janczar K, Singh P, Lo Nigro C, Lattanzio L, Coley HM, Hatzimichael E, Bomalaski J, Szlosarek P, Awad M, O’Neil K, Roncaroli F, Crook T. Epigenetic status of argininosuccinate synthetase and argininosuccinate lyase modulates autophagy and cell death in glioblastoma. Cell Death Dis. 2013;4:e458. doi: 10.1038/cddis.2012.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Takebayashi H, Yoshida S, Sugimori M, Kosako H, Kominami R, Nakafuku M, Nabeshima Y. Dynamic expression of basic helix-loop-helix Olig family members: implication of Olig2 in neuron and oligodendrocyte differentiation and identification of a new member, Olig3. Mech Dev. 2000;99:143–148. doi: 10.1016/s0925-4773(00)00466-4. [DOI] [PubMed] [Google Scholar]
- Tanaka S, Louis DN, Curry WT, Batchelor TT, Dietrich J. Diagnostic and therapeutic avenues for glioblastoma: no longer a dead end? Nat Rev Clin Oncol. 2013;10:14–26. doi: 10.1038/nrclinonc.2012.204. [DOI] [PubMed] [Google Scholar]
- Taylor MD, Poppleton H, Fuller C, Su X, Liu Y, Jensen P, Magdaleno S, Dalton J, Calabrese C, Board J, Macdonald T, Rutka J, Guha A, Gajjar A, Curran T, Gilbertson RJ. Radial glia cells are candidate stem cells of ependymoma. Cancer Cell. 2005;8:323–335. doi: 10.1016/j.ccr.2005.09.001. [DOI] [PubMed] [Google Scholar]
- Tchoghandjian A, Baeza N, Colin C, Cayre M, Metellus P, Beclin C, Ouafik L, Figarella- Branger D. A2B5 cells from human glioblastoma have cancer stem cell properties. Brain Pathol Zurich Switz. 2010;20:211–221. doi: 10.1111/j.1750-3639.2009.00269.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchorz JS, Tome M, Cloëtta D, Sivasankaran B, Grzmil M, Huber RM, Rutz-Schatzmann F, Kirchhoff F, Schaeren-Wiemers N, Gassmann M, Hemmings BA, Merlo A, Bettler B. Constitutive Notch2 signaling in neural stem cells promotes tumorigenic features and astroglial lineage entry. Cell Death Dis. 2012;3:e325. doi: 10.1038/cddis.2012.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Televantou D, Karkavelas G, Hytiroglou P, Lampaki S, Iliadis G, Selviaridis P, Polyzoidis KS, Fountzilas G, Kotoula V. DARPP32, STAT5 and STAT3 mRNA Expression Ratios in Glioblastomas are Associated with Patient Outcome. Pathol Oncol Res Por. 2013;19:329–343. doi: 10.1007/s12253-012-9588-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson MC, Fuller C, Hogg TL, Dalton J, Finkelstein D, Lau CC, Chintagumpala M, Adesina A, Ashley DM, Kellie SJ, Taylor MD, Curran T, Gajjar A, Gilbertson RJ. Genomics identifies medulloblastoma subgroups that are enriched for specific genetic alterations. J Clin Oncol Off J Am Soc Clin Oncol. 2006;24:1924–1931. doi: 10.1200/JCO.2005.04.4974. [DOI] [PubMed] [Google Scholar]
- Thon N, Damianoff K, Hegermann J, Grau S, Krebs B, Schnell O, Tonn J-C, Goldbrunner R. Presence of pluripotent CD133+ cells correlates with malignancy of gliomas. Mol Cell Neurosci. 2010;43:51–59. doi: 10.1016/j.mcn.2008.07.022. [DOI] [PubMed] [Google Scholar]
- Tobias A, Ahmed A, Moon KS, Lesniak MS. The art of gene therapy for glioma: a review of the challenging road to the bedside. J Neurol Neurosurg Psychiatry. 2013;84:213–222. doi: 10.1136/jnnp-2012-302946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todaro M, Alea MP, Di Stefano AB, Cammareri P, Vermeulen L, Iovino F, Tripodo C, Russo A, Gulotta G, Medema JP, Stassi G. Colon cancer stem cells dictate tumor growth and resist cell death by production of interleukin-4. Cell Stem Cell. 2007;1:389–402. doi: 10.1016/j.stem.2007.08.001. [DOI] [PubMed] [Google Scholar]
- Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, Campos C, Fabius AWM, Lu C, Ward PS, Thompson CB, Kaufman A, Guryanova O, Levine R, Heguy A, Viale A, Morris LGT, Huse JT, Mellinghoff IK, Chan TA. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483:479–483. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turchi L, Debruyne DN, Almairac F, Virolle V, Fareh M, Neirijnck Y, Burel-Vandenbos F, Paquis P, Junier M-P, Van Obberghen-Schilling E, Chneiweiss H, Virolle T. Tumorigenic Potential of miR-18A* in Glioma Initiating Cells Requires NOTCH-1 Signaling. Stem Cells Dayt Ohio. 2013 doi: 10.1002/stem.1373. [DOI] [PubMed] [Google Scholar]
- Uchida H, Arita K, Yunoue S, Yonezawa H, Shinsato Y, Kawano H, Hirano H, Hanaya R, Tokimura H. Role of sonic hedgehog signaling in migration of cell lines established from CD133-positive malignant glioma cells. J Neurooncol. 2011;104:697–704. doi: 10.1007/s11060-011-0552-2. [DOI] [PubMed] [Google Scholar]
- Ulasov IV, Nandi S, Dey M, Sonabend AM, Lesniak MS. Inhibition of Sonic hedgehog and Notch pathways enhances sensitivity of CD133(+) glioma stem cells to temozolomide therapy. Mol Med Camb Mass. 2011;17:103–112. doi: 10.2119/molmed.2010.00062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhaak RGW, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, Alexe G, Lawrence M, O’Kelly M, Tamayo P, Weir BA, Gabriel S, Winckler W, Gupta S, Jakkula L, Feiler HS, Hodgson JG, James CD, Sarkaria JN, Brennan C, Kahn A, Spellman PT, Wilson RK, Speed TP, Gray JW, Meyerson M, Getz G, Perou CM, Hayes DN Cancer Genome Atlas Research Network. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vescovi AL, Galli R, Reynolds BA. Brain tumour stem cells. Nat Rev Cancer. 2006;6:425–436. doi: 10.1038/nrc1889. [DOI] [PubMed] [Google Scholar]
- Vredenburgh JJ, Desjardins A, Herndon JE, 2nd, Dowell JM, Reardon DA, Quinn JA, Rich JN, Sathornsumetee S, Gururangan S, Wagner M, Bigner DD, Friedman AH, Friedman HS. Phase II trial of bevacizumab and irinotecan in recurrent malignant glioma. Clin Cancer Res Off J Am Assoc Cancer Res. 2007;13:1253–1259. doi: 10.1158/1078-0432.CCR-06-2309. [DOI] [PubMed] [Google Scholar]
- Wakimoto H, Kesari S, Farrell CJ, Curry WT, Zaupa C, Jr, Aghi M, Kuroda T, Stemmer-Rachamimov A, Shah K, Liu T-C, Jeyaretna DS, Debasitis J, Pruszak J, Martuza RL, Rabkin SD. Human glioblastoma-derived cancer stem cells: establishment of invasive glioma models and treatment with oncolytic herpes simplex virus vectors. Cancer Res. 2009;69:3472–3481. doi: 10.1158/0008-5472.CAN-08-3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Lathia JD, Wu Q, Wang J, Li Z, Heddleston JM, Eyler CE, Elderbroom J, Gallagher J, Schuschu J, MacSwords J, Cao Y, McLendon RE, Wang XF, Hjelmeland AB, Rich JN. Targeting interleukin 6 signaling suppresses glioma stem cell survival and tumor growth. Stem Cells Dayt Ohio. 2009;27:2393–2404. doi: 10.1002/stem.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Wakeman TP, Lathia JD, Hjelmeland AB, Wang XF, White RR, Rich JN, Sullenger BA. Notch promotes radioresistance of glioma stem cells. Stem Cells Dayt Ohio. 2010;28:17–28. doi: 10.1002/stem.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Wang H, Li Z, Wu Q, Lathia JD, McLendon RE, Hjelmeland AB, Rich JN. c-Myc is required for maintenance of glioma cancer stem cells. Plos One. 2008;3:e3769. doi: 10.1371/journal.pone.0003769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Chadalavada K, Wilshire J, Kowalik U, Hovinga KE, Geber A, Fligelman B, Leversha M, Brennan C, Tabar V. Glioblastoma stem-like cells give rise to tumour endothelium. Nature. 2010;468:829–833. doi: 10.1038/nature09624. [DOI] [PubMed] [Google Scholar]
- Wang W-J, Long L-M, Yang N, Zhang Q-Q, Ji W-J, Zhao J-H, Qin Z-H, Wang Z, Chen G, Liang Z-Q. NVP-BEZ235, a novel dual PI3K/mTOR inhibitor, enhances the radiosensitivity of human glioma stem cells in vitro. Acta Pharmacol Sin. 2013;34:681–690. doi: 10.1038/aps.2013.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Venugopal C, Manoranjan B, McFarlane N, O’Farrell E, Nolte S, Gunnarsson T, Hollenberg R, Kwiecien J, Northcott P, Taylor MD, Hawkins C, Singh SK. Sonic hedgehog regulates Bmi1 in human medulloblastoma brain tumor-initiating cells. Oncogene. 2012;31:187–199. doi: 10.1038/onc.2011.232. [DOI] [PubMed] [Google Scholar]
- Ward RJ, Lee L, Graham K, Satkunendran T, Yoshikawa K, Ling E, Harper L, Austin R, Nieuwenhuis E, Clarke ID, Hui C-C, Dirks PB. Multipotent CD15+ cancer stem cells in patched-1-deficient mouse medulloblastoma. Cancer Res. 2009;69:4682–4690. doi: 10.1158/0008-5472.CAN-09-0342. [DOI] [PubMed] [Google Scholar]
- Wei J, Barr J, Kong L-Y, Wang Y, Wu A, Sharma AK, Gumin J, Henry V, Colman H, Priebe W, Sawaya R, Lang FF, Heimberger AB. Glioblastoma cancer-initiating cells inhibit T-cell proliferation and effector responses by the signal transducers and activators of transcription 3 pathway. Mol Cancer Ther. 2010;9:67–78. doi: 10.1158/1535-7163.MCT-09-0734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J, Wu A, Kong LY, Wang Y, Fuller G, Fokt I, Melillo G, Priebe W, Heimberger AB. Hypoxia potentiates glioma-mediated immunosuppression. Plos One. 2011;6:e16195. doi: 10.1371/journal.pone.0016195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Jiang Y, Zou F, Liu Yingchao, Wang S, Xu N, Xu W, Cui C, Xing Y, Liu Ying, Cao B, Liu C, Wu G, Ao H, Zhang X, Jiang J. Activation of PI3K/Akt pathway by CD133-p85 interaction promotes tumorigenic capacity of glioma stem cells. Proc Natl Acad Sci U S A. 2013;110:6829–6834. doi: 10.1073/pnas.1217002110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams CK, Segarra M, Sierra MDLL, Sainson RCA, Tosato G, Harris AL. Regulation of CXCR4 by the Notch ligand delta-like 4 in endothelial cells. Cancer Res. 2008;68:1889–1895. doi: 10.1158/0008-5472.CAN-07-2181. [DOI] [PubMed] [Google Scholar]
- Woodward WA, Chen MS, Behbod F, Alfaro MP, Buchholz TA, Rosen JM. WNT/beta-catenin mediates radiation resistance of mouse mammary progenitor cells. Proc Natl Acad Sci U S A. 2007;104:618–623. doi: 10.1073/pnas.0606599104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu A, Wei J, Kong L-Y, Wang Y, Priebe W, Qiao W, Sawaya R, Heimberger AB. Glioma cancer stem cells induce immunosuppressive macrophages/microglia. Neuro-Oncol. 2010;12:1113–1125. doi: 10.1093/neuonc/noq082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulf GG, Wang RY, Kuehnle I, Weidner D, Marini F, Brenner MK, Andreeff M, Goodell MA. A leukemic stem cell with intrinsic drug efflux capacity in acute myeloid leukemia. Blood. 2001;98:1166–1173. doi: 10.1182/blood.v98.4.1166. [DOI] [PubMed] [Google Scholar]
- Xia H, Cheung WKC, Ng SS, Jiang X, Jiang S, Sze J, Leung GKK, Lu G, Chan DTM, Bian X-W, Kung H, Poon WS, Lin MC. Loss of brain-enriched miR-124 microRNA enhances stem-like traits and invasiveness of glioma cells. J Biol Chem. 2012;287:9962–9971. doi: 10.1074/jbc.M111.332627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Yuan X, Liu G, Black KL, Yu JS. Hedgehog signaling regulates brain tumor-initiating cell proliferation and portends shorter survival for patients with PTEN-coexpressing glioblastomas. Stem Cells Dayt Ohio. 2008;26:3018–3026. doi: 10.1634/stemcells.2008-0459. [DOI] [PubMed] [Google Scholar]
- Yang YP, Chien Y, Chiou GY, Cherng JY, Wang ML, Lo WL, Chang YL, Huang PI, Chen YW, Shih YH, Chen MT, Chiou SH. Inhibition of cancer stem cell-like properties and reduced chemoradioresistance of glioblastoma using microRNA145 with cationic polyurethane-short branch PEI. Biomaterials. 2012;33:1462–1476. doi: 10.1016/j.biomaterials.2011.10.071. [DOI] [PubMed] [Google Scholar]
- Yoon CH, Kim MJ, Kim RK, Lim EJ, Choi KS, An S, Hwang SG, Kang SG, Suh Y, Park MJ, Lee SJ. c-Jun N-terminal kinase has a pivotal role in the maintenance of self-renewal and tumorigenicity in glioma stem-like cells. Oncogene. 2012;31:4655–4666. doi: 10.1038/onc.2011.634. [DOI] [PubMed] [Google Scholar]
- Yoshida Y, Takahashi K, Okita K, Ichisaka T, Yamanaka S. Hypoxia enhances the generation of induced pluripotent stem cells. Cell Stem Cell. 2009;5:237–241. doi: 10.1016/j.stem.2009.08.001. [DOI] [PubMed] [Google Scholar]
- Yuan X, Curtin J, Xiong Y, Liu G, Waschsmann-Hogiu S, Farkas DL, Black KL, Yu JS. Isolation of cancer stem cells from adult glioblastoma multiforme. Oncogene. 2004;23:9392–9400. doi: 10.1038/sj.onc.1208311. [DOI] [PubMed] [Google Scholar]
- Yuki K, Natsume A, Yokoyama H, Kondo Y, Ohno M, Kato T, Chansakul P, Ito M, Kim S-U, Wakabayashi T. Induction of oligodendrogenesis in glioblastoma-initiating cells by IFN-mediated activation of STAT3 signaling. Cancer Lett. 2009;284:71–79. doi: 10.1016/j.canlet.2009.04.020. [DOI] [PubMed] [Google Scholar]
- Zbinden M, Duquet A, Lorente-Trigos A, Ngwabyt S-N, Borges I, Ruiz i Altaba A. NANOG regulates glioma stem cells and is essential in vivo acting in a cross-functional network with GLI1 and p53. Embo J. 2010;29:2659–2674. doi: 10.1038/emboj.2010.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Song T, Yang L, Chen R, Wu L, Yang Z, Fang J. Nestin and CD133: valuable stem cell-specific markers for determining clinical outcome of glioma patients. J Exp Clin Cancer Res Cr. 2008;27:85. doi: 10.1186/1756-9966-27-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Wei P, Gong A, Chiu WT, Lee HT, Colman H, Huang H, Xue J, Liu M, Wang Y, Sawaya R, Xie K, Yung WKA, Medema RH, He X, Huang S. FoxM1 promotes β-catenin nuclear localization and controls Wnt target-gene expression and glioma tumorigenesis. Cancer Cell. 2011;20:427–442. doi: 10.1016/j.ccr.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Dutta A, Abounader R. The role of microRNAs in glioma initiation and progression. Front Biosci J Virtual Libr. 2012;17:700–712. doi: 10.2741/3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Ying H, Yan H, Kimmelman AC, Hiller DJ, Chen A-J, Perry SR, Tonon G, Chu GC, Ding Z, Stommel JM, Dunn KL, Wiedemeyer R, You MJ, Brennan C, Wang YA, Ligon KL, Wong WH, Chin L, de Pinho RA. Pten and p53 converge on c-Myc to control differentiation, self-renewal, and transformation of normal and neoplastic stem cells in glioblastoma. Cold Spring Harb Symp Quant Biol. 2008;73:427–437. doi: 10.1101/sqb.2008.73.047. [DOI] [PubMed] [Google Scholar]
- Zheng H, Ying H, Yan H, Kimmelman AC, Hiller DJ, Chen AJ, Perry SR, Tonon G, Chu GC, Ding Z, Stommel JM, Dunn KL, Wiedemeyer R, You MJ, Brennan C, Wang YA, Ligon KL, Wong WH, Chin L, DePinho RA. p53 and Pten control neural and glioma stem/progenitor cell renewal and differentiation. Nature. 2008;455:1129–1133. doi: 10.1038/nature07443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B-BS, Zhang H, Damelin M, Geles KG, Grindley JC, Dirks PB. Tumour-initiating cells: challenges and opportunities for anticancer drug discovery. Nat Rev Drug Discov. 2009;8:806–823. doi: 10.1038/nrd2137. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Anderson DJ. The bHLH transcription factors OLIG2 and OLIG1 couple neuronal and glial subtype specification. Cell. 2002;109:61–73. doi: 10.1016/s0092-8674(02)00677-3. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Sun L, Wang Y, Wu Z, Geng J, Miu W, Pu Y, You Y, Yang Z, Liu N. Bone morphogenetic protein 4 inhibits cell proliferation and induces apoptosis in glioma stem cells. Cancer Biother Radiopharm. 2011;26:77–83. doi: 10.1089/cbr.2010.0857. [DOI] [PubMed] [Google Scholar]
- Zhuang W, Li B, Long L, Chen L, Huang Q, Liang Z. Induction of autophagy promotes differentiation of glioma-initiating cells and their radiosensitivity. Int J Cancer J Int Cancer. 2011;129:2720–2731. doi: 10.1002/ijc.25975. [DOI] [PubMed] [Google Scholar]