Abstract
Epigenetic control mechanisms determine active and silenced regions of the genome. It is known that the Polycomb Repressive Complex 2 (PRC2) and the Trithorax group/Mixed lineage leukemia (TrxG/Mll) complex are able to set repressive and active histone marks, respectively. Long non-coding RNAs (lncRNAs) can interact with either of these complexes and guide them to regulatory elements, thereby modifying the expression levels of target genes. The lncRNA Fendrr is transiently expressed in lateral mesoderm of mid-gestational mouse embryos and was shown to interact with both PRC2 and TrxG/Mll complexes in vivo. Gene targeting revealed that loss of Fendrr results in impaired differentiation of tissues derived from lateral mesoderm, the heart and the body wall, ultimately leading to embryonic death. Molecular data suggests that Fendrr acts via dsDNA/RNA triplex formation at target regulatory elements, and directly increases PRC2 occupancy at these sites. This, in turn, modifies the ratio of repressive to active marks, adjusting the expression levels of Fendrr target genes in lateral mesoderm. We propose that Fendrr also mediates long-term epigenetic marks to define expression levels of its target genes within the descendants of lateral mesoderm cells. Here we discuss approaches for lncRNA gene knockouts in the mouse, and suggest a model how Fendrr and possibly other lncRNAs act during embryogenesis.
Keywords: long non-coding RNA, lncRNA, Fendrr, histone modification, epigenetic control, Polycomb Repressive Complex 2, embryogenesis, mouse
Introduction
The process of gastrulation generates the three germ layers: (neuro-) ectoderm, mesoderm, and (gut-) endoderm, which give rise to all organs of the body. In the mouse, this process takes place over six days and consecutively forms the anlage of the head, trunk, and tail. The lineage choice made by cells during gastrulation is the most crucial one for determining their fate and further differentiation of all of their descendants. Following the first lineage choice, cells can be recruited to various sub-lineages. For instance, the lateral mesoderm subdivides into the somatic and splanchnic mesoderm. In this manner, with each consecutive lineage decision, cellular plasticity becomes more and more constrained. Thus, it is evident that the molecular activities controlling the first lineage decision during gastrulation have the most impact on cell fate.
An example of an essential regulator involved in early lineage decisions is the transcription factor Foxf1. The targeted disruption of the Foxf1 gene prevents the partition of lateral mesoderm into splanchnic and somatic mesoderm lineages, resulting in embryonic death.1 Moreover, the Foxf1 mutant is haploinsufficient, i.e., embryos expressing Foxf1 from a single active allele exhibit perinatal lethality with variable abnormalities, including lung defects. This underpins the crucial role of this transcription factor during embryonic development.2
Many other protein-coding genes, which play essential and specific roles during embryonic development, have been described over the past two decades. The discovery of microRNAs (miRNAs) as essential regulators of developmental programs has shifted the focus toward genes transcribed into non-coding RNA molecules.3–6 MiRNAs are a class of non-coding RNAs, which are processed into single-stranded RNAs of ~22 nucleotides in length, and form functional units with the RNA-induced silencing complex (RISC). They are known to be involved in post-transcriptional control of gene expression. LncRNAs constitute a distinct class of RNA molecules, which are typically over 200 nucleotides in length, highly heterogeneous, and associated with a plethora of different functions, but in particular epigenetic gene control (for review, see refs. 7 and 8).
Recent data from the human ENCODE project identified 13 333 genes encoding 22 631 transcripts in the human genome belonging to the lncRNA class (www.gencodegenes.org).9 They preferentially localize to the nucleus and, therefore, are thought to be involved in genome regulation.10 Although many of these lncRNAs contain short open reading frames and can be found associated with ribosomes, the majority of them are assumed to be non-coding.11,12
The identification and functional analysis of lncRNA transcripts encoded by the murine Xist (Xi-specific transcript) locus within the X-inactivation center (XIC) first revealed the importance of nuclear lncRNAs in genome regulation and embryonic development.13,14 The lncRNAs Xist and RepA, a PRC2 binding lncRNA transcribed from the Xist locus, were shown to play a role in X-chromosome inactivation.15 Moreover, the PRC2 component Enhancer of Zeste Homolog 2 (EZH2) was found to bind thousands of lncRNAs, further supporting a functional link between lncRNAs and histone-modifying protein complexes.16 Further importance of lncRNAs as factors regulating genome activity through histone-modifying complexes came from the discovery of HOTAIR, which is expressed from the HOXC locus and enhances PRC2 activity at the HOXD locus.17 HOTAIR binds to and might serve as a molecular scaffold for the histone-modifying complexes PRC2 and LSD1/CoREST/REST.18 In addition, the lncRNAs HOTTIP, Mistral, and NeST have been shown to associate with the TrxG/Mll component WDR5.19–21
The ability to survey the transcriptional landscape through the technique of RNA-seq allows screening for lncRNAs, which are differentially expressed during embryogenesis. We analyzed six tissues from mid-gestational mouse embryos (embryonic day [E] 8.25) for the presence of lncRNAs, and identified several hundred which are expressed in a tissue-restricted manner (unpublished). This data set provided the basis for determining whether lncRNAs are able to play an essential role in development.
Functional Analysis of lncRNAs in Embryonic Development
The functional analysis of numerous embryonic control genes has shown that the expression pattern of essential regulators in the embryo is often highly restricted. Thus, we presumed that restricted expression may also serve as a useful criterion for selecting lncRNA genes for functional analyses. We analyzed a number of lncRNAs showing differential expression in the RNA-seq data in more detail by whole-mount in situ hybridization. One of these, Fendrr, showed a remarkably restricted and specific expression in the nascent lateral mesoderm. It is divergently transcribed from the Foxf1 promoter and is co-expressed with Foxf1.22 Since divergently transcribed long non-coding/mRNA gene pairs are thought to be functionally linked and represent an intriguing gene class,23,24 we decided to proceed to a functional analysis of Fendrr in the mouse.
Most current studies on the function of lncRNAs involve RNAi or similar mechanisms to reduce transcript levels.6,25–29 It is, however, important to note that transcripts that localize to the nucleus are less susceptible to knockdown by RNAi. In addition, many lncRNAs exhibit a very low expression level.10,30 Hence, it cannot be excluded that even a few residual transcripts per cell that escape knockdown might be functionally sufficient. Initially, we attempted the functional analysis of Fendrr by using an in vivo knockdown system, which we had previously applied to the inactivation of several protein-coding genes.31 However, an shRNAmir construct expressing two different hairpins against Fendrr (best of six individually tested hairpins) achieved a maximal knockdown of only 60%. We observed no apparent phenotype in knockdown embryos up to embryonic day E18.0. Thus, 40% of the total Fendrr transcript was sufficient for embryonic development. This finding also holds true for many, if not most, transcription factors.
The gold standard for analyzing gene function in the mouse is gene targeting, resulting in a loss-of-function mutation. A number of approaches have been developed for targeting protein-coding genes, however, non-coding genes provide fewer options since an uncharacterized lncRNA sequence does not reveal the nucleotide stretch that is crucial for its function. Notably, it has been shown that nucleotides within the first 300 bases of the HOTAIR lncRNA transcript interact with EZH2.18 Thus, for a clear functional analysis of lncRNAs the complete transcript must be prevented from being expressed. One previously used option involves deletion of the complete gene.32,33 This approach is not ideal, since the deletion might also remove cis-acting regulatory sequences for neighboring genes.23,24,34 Another option is to integrate a strong transcriptional stop signal right at the start of the transcription unit. We consider this latter option superior, but it should be noted that possible alternative transcriptional start sites (TSS) downstream of the TSS that has been chosen for stop-cassette integration must not be overlooked. Also, since the insertion of genetic elements such as transcriptional stop signals can alter the chromatin landscape and, thereby, influence gene regulation at the site of insertion, it is crucial to rule out that the phenotype observed is due to a secondary effect. This can be done by a rescue experiment wherein the lncRNA transcript is expressed from a construct integrated randomly into the genome. If the phenotype is rescued it proves that the knockout phenotype was indeed caused by loss of the lncRNA transcript and, moreover, that the lncRNA acts in trans.
Two strong transcriptional stop signals have been previously used for gene knockout, a triple SV40 polyA signal and the chicken β-globin polyA signal.35,36 Both elements harbor an intron upstream of the polyadenylation signal that is presumed to increase their efficacy. Insertion of the triple SV40 polyA cassette into the first exon of Fendrr proved to be very efficient, but not absolutely transcriptionally tight. A residual transcript level of 0.1% was still detectable in Fendrr mutants following 3× SV40 pA insertion (data not shown). However, this residual activity was found to be insufficient for Fendrr function. Fendrr mutants presented with omphalocele and showed malfunctioning of the heart, which resulted in embryonic death by E13.75.22
To rescue the Fendrr-knockout phenotype, we integrated a mini-gene construct expressing the Fendrr transcript under control of the human ACTB promoter randomly into the genome of Fendrr-null ES cells. Ubiquitous expression of Fendrr at approximately 20% of its wild-type level was observed in embryos derived from the transgenic ES cells. This level of Fendrr transcript was not sufficient for a complete rescue of the knockout phenotype. For instance, though surviving embryos showed no omphalocele at E14.5 (Fendrr null: 100% omphalocele), the survival rate only increased moderately (from 21% [n = 9] to 60% [n = 5]). This partial rescue is most likely due to low Fendrr expression in the lateral mesoderm, but we cannot exclude the possibility that ubiquitous ectopic expression of Fendrr in the embryo may have contributed to the observed lethality.
We reasoned that expressing Fendrr under control of its endogenous promoter, providing correct spatial and temporal control as well as correct expression levels, would be more adequate for achieving rescue of the knockout phenotype. Therefore, we randomly integrated a BAC (bacterial artificial chromosome) containing an intact Fendrr gene in the middle of a 172194 bp region of the C57Bl/6J mouse genome, into Fendrr-null ES cells. The BAC clone was genetically modified by insertion of a Neomycin selection cassette into the translational start site of Foxf1. This inactivated Foxf1, thus avoiding an increase of Foxf1 activity, which might trigger unwanted effects in BAC transgenic embryos, while it left the Fendrr transcription unit on the BAC unchanged, and also allowed for antibiotic selection of ES clones, which had integrated the BAC. This construct faithfully reproduced the wild-type expression pattern of Fendrr, and produced approximately 50% of the Fendrr transcript level observed in wild-type embryos, corresponding to expression of one allele.22 This construct rescued the Fendrr-null phenotype until E17.5 illustrating that the Fendrr lncRNA acts in trans, and that loss of the Fendrr transcript is responsible for the lethal phenotype.
The rescue experiment was also necessary as a test of the genetic integrity of the mutant ES cells that were used for embryo production via tetraploid complementation.37 One advantage of tetraploid complementation over blastocyst injection for producing knockout mouse lines is the vast difference in the time required for obtaining homozygous embryos. While the standard approach takes 8–12 mo, tetraploid complementation allows the production of virtually pure ES cell-derived homozygous embryos directly from the genetically altered cells (consecutively mutated on both alleles, which takes around 2–3 mo). In a typical experiment, 8 to 12 genetically identical homozygous embryos can be obtained per mouse. Additional genetic manipulations, such as the introduction of a reporter, constructs for lineage tracing, rescue BAC, etc. can be performed in a reasonably short time frame. Importantly, the genetic background of the embryos produced for knockout and consecutive analyses can be kept virtually identical, which is highly desirable to avoid errors due to background effects. Therefore, the starting ES cell line does not need to be derived from an inbred strain.
Mode of Fendrr Action in Embryonic Development
Fendrr mutant embryos appear normal until E12.5, but by E14.5 most are dead. Both sub-lineages of the lateral plate mesoderm are affected: the heart, a derivative of the splanchnic mesoderm, and the ventral body wall, a product of the somatic mesoderm. The heart progenitors express Fendrr transiently at E6.5–7.0, and the progenitors of the ventral body wall at around E9.5 of development. We showed that proliferation of cardiomyocytes is impaired in Fendrr mutants, but not before E12.5.18 Thus, there is a delay of 6 d between the time of Fendrr expression in wild-type embryos and malfunctioning of the tissue derived from Fendrr-null cells in the mutant. How can this delay be explained? The molecular analysis provided some important hints toward answering this question.
Analogous to the human FENDRR (previously FOXF1-AS1) transcript, mouse Fendrr binds to the PRC2 complex.22,38 We showed that the occupancy of PRC2 was severely reduced at three putative target promoters in the mutant, which led to decreased trimethylation of lysine 27 on histone 3 (H3K27me3), the footprint of PRC2.22 For two of these targets, Foxf1 and Pitx2, we provided evidence that Fendrr interacts directly with the promoter DNA. Using an in silico approach, we identified a 40-nucleotide stretch of the Fendrr RNA, which is able to precipitate the Foxf1 and Pitx2 promoters in vitro.22 This interaction is not destroyed by RNaseH treatment, excluding heteroduplex formation as a binding mechanism between Fendrr and the promoter DNA. The 40-base binding sequence also closely resembles a purine-pyrimidine motif that has been previously shown to favor triplex formation.39,40
We confirmed by ChIRP (Chromatin isolation by RNA purification) analysis41 that interaction of Fendrr with the Foxf1 and Pitx2 promoters exists in a cellular context. For this we differentiated mouse ES cells to mesoderm in vitro for 4 d to reach peak expression of endogenous Fendrr. We then precipitated Fendrr RNA from cellular extracts with two pools of tiling oligonucleotides complementary to Fendrr. We found that Foxf1 and Pitx2 promoter DNA co-precipitated with at least one of the pools (Fig. 1).
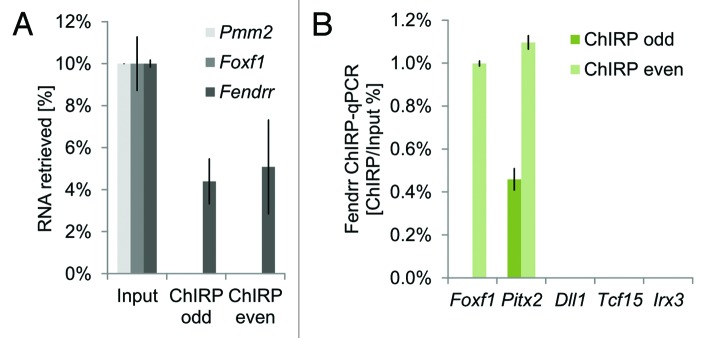
Figure 1. ChIRP (Chromatin Isolation by RNA purification) analysis shows that Fendrr interacts with target promoters in embryonic stem cells differentiated in vitro. Fourteen biotinylated oligonucleotides tiling the Fendrr transcript were combined in even and odd numbered pools. (A) Each oligo-pool specifically precipitated the Fendrr transcript. (B) At least one pool of tiling oligos co-precipitated the target promoters of Foxf1 and Pitx2, but not the control promoters of Dll1, Tcf15, or Irx3.
Taken together, these data suggest that Fendrr binds to target dsDNA sequences via formation of a dsDNA:RNA triplex structure. Fendrr binds to specific sites at target promoters as well as to PRC2. This increases PRC2 occupancy at the promoters, allowing for enhanced H3K27 trimethylation. Therefore, at least in the context of PRC2 binding, Fendrr acts as an anchor to promote repressive H3K27 trimethylation at target promoters (Fig. 2).
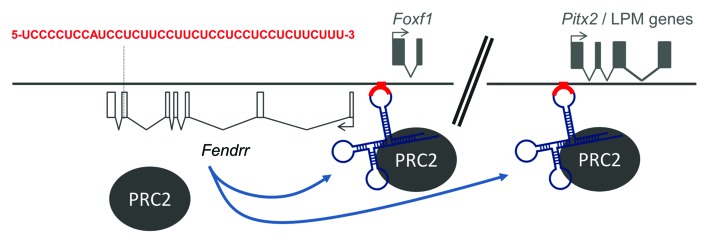
Figure 2.Fendrr anchors PRC2 to specific target sites. Schematic drawing illustrating binding of the Fendrr/PRC2 complex through a 40-nucleotide stretch (red) to specific recognition sequences in target promoters, e.g., that of Foxf1 and Pitx2, thus increasing PRC2 occupancy and H3K27 trimethylation at these sites.
We were able to show that Fendrr also interacts with the TrxG/Mll component WDR5. TrxG/Mll sets active marks by methylating lysine 4 on histone 3 (H3K4me3). We found that all genes showing higher mRNA expression in the Fendrr mutant in our analysis also displayed increased H3K4me3 levels at their promoters. However, WDR5 occupancy was not enhanced in parallel, suggesting an indirect effect of Fendrr loss on H3K4 trimethylation at these promoters. Indeed, the H3K4me3 active mark can be set by numerous methyl-transferase-containing complexes. Besides the MLL family of methyl-transferases from the TrxG/Mll complexes, SETD1A, SETD1B, SETD7, SMYD1/2/3, and ASH1L have also been shown to methylate H3K4.42–47 Moreover, WDR5 has also been found as a component of the histone acetylating complexes NSL/MSL1v and ATAC.48,49 Hence, at present it cannot be absolutely excluded that Fendrr might interact with any of these latter complexes involving WDR5.
The increase in target gene expression observed in Fendrr mutants is rather mild—on the order of 30–40% over the wild-type level. Is it conceivable that the altered expression of numerous (presumed) Fendrr target genes together causes sufficient cellular imbalance to lead to a phenotype? Some transcriptional regulators exhibit haploinsufficiency phenotypes, demonstrating that expression of a single allele can be insufficient for proper embryonic development, and the Fendrr targets Foxf1 and Pitx2 both belong to this group of genes.2,50 However, can a mild overexpression also cause impaired cellular function, or is the Fendrr phenotype due to a different mechanism?
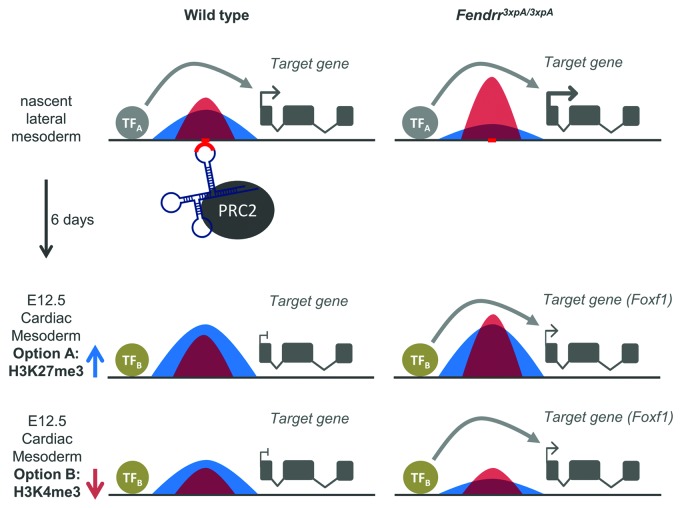
Foxf1 is completely silenced in the cardiac tissue of E12.5 wild-type embryos, but is ectopically expressed (at a low level) in the cardiac tissue of E12.5 Fendrr mutants.22 This finding, combined with the delay between Fendrr expression in the lateral plate mesoderm and the onset of heart dysfunction in Fendrr mutants, suggests a long-term effect of Fendrr action, a characteristic of epigenetic control mechanisms. In the mutants, failure to increase the level of repressive H3K27me3 marks, in combination with an increase in the active H3K4me3 mark at the Foxf1 promoter in the lateral mesoderm apparently affects the ability of descendants of these cells in the cardiac mesoderm to silence Foxf1. It is conceivable that, in wild-type cells, an increase of the H3K27me3/H3K4me3 ratio mediated by Fendrr at target promoters in nascent lateral mesoderm at a later stage of cardiac mesoderm differentiation is followed by a reduction of the active or an enhancement of the repressive mark, leading to a further increase of the H3K27me3/H3K4me3 ratio followed by gene silencing (Fig. 3). Since the H3K27me3/H3K4me3 ratio in lateral mesoderm of Fendrr mutants is reduced in favor of the active mark, a reduction of the resulting high H3K4me3 level or increase of the repressive marks in cardiac mesoderm might not be sufficient anymore to permit silencing of the Foxf1 promoter (Fig. 3). In this manner, setting and removal of active and repressive marks mediated by promoter-specific factors at consecutive stages of cell differentiation might control the activities of many genes, including important regulatory proteins, in a differential manner. Consecutive epigenetic modifications at particular promoters might be separated by hours or days and, thus, changes set early in a cell lineage may become effective much later in descendants of that lineage. This mode-of-action is quite different from the way transcription factors work, where a loss usually becomes effective right away in the very cells they are expressed in.
Figure 3. Model of immediate and long-term effects of Fendrr acting via PRC2 on epigenetic modification and gene expression. The ratio of repressive H3K27me3 (blue) to active H3K4me3 (red) marks determines the level of gene expression. Histone marks set in the lateral mesoderm are maintained in the descendants of these cells until they are modified by other site-specific regulators resulting, for instance, in an increase in the repressive or a reduction in the active marks within cardiac mesoderm (indicated by arrows). In the case of Foxf1, this leads to a dominating repressive mark and silencing of the gene in wild-type cardiac mesoderm. In Fendrr mutant cells, anchoring of PRC2 to the Foxf1 promoter fails, thus skewing the H3K27me3/H3K4me3 ratio and leading to an increased Foxf1 expression level. The subsequent histone modifications which normally take place in cardiac mesoderm are not able to correct the false prior epigenetic marks set in the lateral mesoderm cells and thus, Foxf1 cannot be silenced.
In summary, we propose that Fendrr functions in the lateral mesoderm via epigenetic modification of regulatory elements, thereby adjusting the expression level of target genes in this tissue and setting long-term marks, which allow for the proper control of Fendrr target genes in the descendants of lateral mesoderm cells.
In addition to its role in embryogenesis, the human FENDRR gene has been found to be expressed in the lung, and to be associated with a lethal lung developmental disorder called ACD/MPV (alveolar capillary dysplasia with misalignment of pulmonary veins).51 In one ACD/MPV case (#41.1) an 11 kb deletion removed the distal portion of the FOXF1 promoter and a portion of FENDRR (FOXF1-AS1).34 Notably, knockdown of FENDRR by siRNA in Human Pulmonary Microvascular Endothelial Cells (HPMEC) caused a weak increase of FOXF1 transcription, supporting the finding that FENDRR acts as transcriptional repressor.34
Perspectives
Fendrr is one of the first lncRNAs that has been shown to play an essential role in mammalian embryogenesis. Our preliminary data obtained by deep sequencing of the transcriptomes of six tissues dissected from E8.25 mouse embryos revealed several hundred differentially expressed lncRNAs, and many more are expected to arise from high-resolution RNA-seq analyses of different developmental stages. Current available information suggests that lncRNAs play a pivotal role in epigenetic control mechanisms acting in cells and on cell lineages. However, detailed functional analyses of many such genes are needed in order to derive a clearer picture of the roles of lncRNAs in epigenetic regulatory networks.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Tracie Pennimpede and Arica Beisaw for comments on the manuscript.
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/26165
References
- 1.Mahlapuu M, Ormestad M, Enerbäck S, Carlsson P. The forkhead transcription factor Foxf1 is required for differentiation of extra-embryonic and lateral plate mesoderm. Development. 2001;128:155–66. doi: 10.1242/dev.128.2.155. [DOI] [PubMed] [Google Scholar]
- 2.Mahlapuu M, Enerbäck S, Carlsson P. Haploinsufficiency of the forkhead gene Foxf1, a target for sonic hedgehog signaling, causes lung and foregut malformations. Development. 2001;128:2397–406. doi: 10.1242/dev.128.12.2397. [DOI] [PubMed] [Google Scholar]
- 3.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–54. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- 4.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–6. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 5.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–4. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 6.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–6. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 7.Lee JT. Epigenetic regulation by long noncoding RNAs. Science. 2012;338:1435–9. doi: 10.1126/science.1231776. [DOI] [PubMed] [Google Scholar]
- 8.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–66. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–89. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al. Landscape of transcription in human cells. Nature. 2012;489:101–8. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guttman M, Russell P, Ingolia NT, Weissman JS, Lander ES. Ribosome Profiling Provides Evidence that Large Noncoding RNAs Do Not Encode Proteins. Cell. 2013;154:240–51. doi: 10.1016/j.cell.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brown CJ, Hendrich BD, Rupert JL, Lafrenière RG, Xing Y, Lawrence J, Willard HF. The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992;71:527–42. doi: 10.1016/0092-8674(92)90520-M. [DOI] [PubMed] [Google Scholar]
- 14.Brockdorff N, Ashworth A, Kay GF, McCabe VM, Norris DP, Cooper PJ, Swift S, Rastan S. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell. 1992;71:515–26. doi: 10.1016/0092-8674(92)90519-I. [DOI] [PubMed] [Google Scholar]
- 15.Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–6. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao J, Ohsumi TK, Kung JT, Ogawa Y, Grau DJ, Sarma K, Song JJ, Kingston RE, Borowsky M, Lee JT. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol Cell. 2010;40:939–53. doi: 10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–23. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–93. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–4. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bertani S, Sauer S, Bolotin E, Sauer F. The noncoding RNA Mistral activates Hoxa6 and Hoxa7 expression and stem cell differentiation by recruiting MLL1 to chromatin. Mol Cell. 2011;43:1040–6. doi: 10.1016/j.molcel.2011.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Gomez JA, Wapinski OL, Yang YW, Bureau JF, Gopinath S, Monack DM, Chang HY, Brahic M, Kirkegaard K. The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the interferon-γ locus. Cell. 2013;152:743–54. doi: 10.1016/j.cell.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grote P, Wittler L, Hendrix D, Koch F, Währisch S, Beisaw A, Macura K, Bläss G, Kellis M, Werber M, et al. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev Cell. 2013;24:206–14. doi: 10.1016/j.devcel.2012.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–27. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sigova AA, Mullen AC, Molinie B, Gupta S, Orlando DA, Guenther MG, Almada AE, Lin C, Sharp PA, Giallourakis CC, et al. Divergent transcription of long noncoding RNA/mRNA gene pairs in embryonic stem cells. Proc Natl Acad Sci U S A. 2013;110:2876–81. doi: 10.1073/pnas.1221904110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun L, Goff LA, Trapnell C, Alexander R, Lo KA, Hacisuleyman E, Sauvageau M, Tazon-Vega B, Kelley DR, Hendrickson DG, et al. Long noncoding RNAs regulate adipogenesis. Proc Natl Acad Sci U S A. 2013;110:3387–92. doi: 10.1073/pnas.1222643110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klattenhoff CA, Scheuermann JC, Surface LE, Bradley RK, Fields PA, Steinhauser ML, Ding H, Butty VL, Torrey L, Haas S, et al. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell. 2013;152:570–83. doi: 10.1016/j.cell.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kretz M, Siprashvili Z, Chu C, Webster DE, Zehnder A, Qu K, Lee CS, Flockhart RJ, Groff AF, Chow J, et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature. 2013;493:231–5. doi: 10.1038/nature11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell. 2011;147:1537–50. doi: 10.1016/j.cell.2011.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ravasi T, Suzuki H, Pang KC, Katayama S, Furuno M, Okunishi R, Fukuda S, Ru K, Frith MC, Gongora MM, et al. Experimental validation of the regulated expression of large numbers of non-coding RNAs from the mouse genome. Genome Res. 2006;16:11–9. doi: 10.1101/gr.4200206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vidigal JA, Morkel M, Wittler L, Brouwer-Lehmitz A, Grote P, Macura K, Herrmann BG. An inducible RNA interference system for the functional dissection of mouse embryogenesis. Nucleic Acids Res. 2010;38:e122. doi: 10.1093/nar/gkq199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eißmann M, Gutschner T, Hämmerle M, Günther S, Caudron-Herger M, Groß M, Schirmacher P, Rippe K, Braun T, Zörnig M, et al. Loss of the abundant nuclear non-coding RNA MALAT1 is compatible with life and development. RNA Biol. 2012;9:1076–87. doi: 10.4161/rna.21089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones BK, Levorse JM, Tilghman SM. Igf2 imprinting does not require its own DNA methylation or H19 RNA. Genes Dev. 1998;12:2200–7. doi: 10.1101/gad.12.14.2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Szafranski P, Dharmadhikari AV, Brosens E, Gurha P, Kolodziejska KE, Zhishuo O, Dittwald P, Majewski T, Mohan KN, Chen B, et al. Small noncoding differentially methylated copy-number variants, including lncRNA genes, cause a lethal lung developmental disorder. Genome Res. 2013;23:23–33. doi: 10.1101/gr.141887.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lobe CG, Koop KE, Kreppner W, Lomeli H, Gertsenstein M, Nagy A. Z/AP, a double reporter for cre-mediated recombination. Dev Biol. 1999;208:281–92. doi: 10.1006/dbio.1999.9209. [DOI] [PubMed] [Google Scholar]
- 36.Sleutels F, Barlow DP. Investigation of elements sufficient to imprint the mouse Air promoter. Mol Cell Biol. 2001;21:5008–17. doi: 10.1128/MCB.21.15.5008-5017.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gertsenstein M. Tetraploid Complementation Assay. 1st Edition. (Pease S, Saunders TL, eds.). Berlin, Heidelberg Springer Berlin Heidelberg; 2011357–375. 10.1007/978-3-642-20792-1 [DOI] [Google Scholar]
- 38.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106:11667–72. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Buske FA, Bauer DC, Mattick JS, Bailey TL. Triplexator: detecting nucleic acid triple helices in genomic and transcriptomic data. Genome Res. 2012;22:1372–81. doi: 10.1101/gr.130237.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morgan AR, Wells RD. Specificity of the three-stranded complex formation between double-stranded DNA and single-stranded RNA containing repeating nucleotide sequences. J Mol Biol. 1968;37:63–80. doi: 10.1016/0022-2836(68)90073-9. [DOI] [PubMed] [Google Scholar]
- 41.Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell. 2011;44:667–78. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee J-S, Smith E, Shilatifard A. The language of histone crosstalk. Cell. 2010;142:682–5. doi: 10.1016/j.cell.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abu-Farha M, Lambert JP, Al-Madhoun AS, Elisma F, Skerjanc IS, Figeys D. The tale of two domains: proteomics and genomics analysis of SMYD2, a new histone methyltransferase. Mol Cell Proteomics. 2008;7:560–72. doi: 10.1074/mcp.M700271-MCP200. [DOI] [PubMed] [Google Scholar]
- 44.Gregory GD, Vakoc CR, Rozovskaia T, Zheng X, Patel S, Nakamura T, Canaani E, Blobel GA. Mammalian ASH1L is a histone methyltransferase that occupies the transcribed region of active genes. Mol Cell Biol. 2007;27:8466–79. doi: 10.1128/MCB.00993-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hamamoto R, Furukawa Y, Morita M, Iimura Y, Silva FP, Li M, Yagyu R, Nakamura Y. SMYD3 encodes a histone methyltransferase involved in the proliferation of cancer cells. Nat Cell Biol. 2004;6:731–40. doi: 10.1038/ncb1151. [DOI] [PubMed] [Google Scholar]
- 46.Tan X, Rotllant J, Li H, De Deyne P, Du SJ. SmyD1, a histone methyltransferase, is required for myofibril organization and muscle contraction in zebrafish embryos. Proc Natl Acad Sci U S A. 2006;103:2713–8. doi: 10.1073/pnas.0509503103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang H, Cao R, Xia L, Erdjument-Bromage H, Borchers C, Tempst P, Zhang Y. Purification and functional characterization of a histone H3-lysine 4-specific methyltransferase. Mol Cell. 2001;8:1207–17. doi: 10.1016/S1097-2765(01)00405-1. [DOI] [PubMed] [Google Scholar]
- 48.Li X, Wu L, Corsa CAS, Kunkel S, Dou Y. Two mammalian MOF complexes regulate transcription activation by distinct mechanisms. Mol Cell. 2009;36:290–301. doi: 10.1016/j.molcel.2009.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Y-L, Faiola F, Xu M, Pan S, Martinez E. Human ATAC Is a GCN5/PCAF-containing acetylase complex with a novel NC2-like histone fold module that interacts with the TATA-binding protein. J Biol Chem. 2008;283:33808–15. doi: 10.1074/jbc.M806936200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu W, Selever J, Lu M-F, Martin JF. Genetic dissection of Pitx2 in craniofacial development uncovers new functions in branchial arch morphogenesis, late aspects of tooth morphogenesis and cell migration. Development. 2003;130:6375–85. doi: 10.1242/dev.00849. [DOI] [PubMed] [Google Scholar]
- 51.Stankiewicz P, Sen P, Bhatt SS, Storer M, Xia Z, Bejjani BA, Ou Z, Wiszniewska J, Driscoll DJ, Maisenbacher MK, et al. Genomic and genic deletions of the FOX gene cluster on 16q24.1 and inactivating mutations of FOXF1 cause alveolar capillary dysplasia and other malformations. Am J Hum Genet. 2009;84:780–91. doi: 10.1016/j.ajhg.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]