Abstract
The human genome encodes several thousand long non-protein coding transcripts > 200 nucleotides in length, a subset of which were shown to play important roles in regulation of gene expression. We recently identified TINCR, a lncRNA required for induction of key differentiation genes in epidermal tissue, including genes mutated in human skin diseases characterized by disrupted epidermal barrier formation. High-throughput analyses of TINCR RNA- and protein-interactomes revealed TINCR interaction with differentiation mRNAs as well as the Staufen1 protein. TINCR, together with Staufen1, seems to stabilize a subset of mRNAs required for epidermal differentiation. Here, we discuss the emerging roles of Staufen1 and TINCR in the regulation of mammalian cell differentiation mediated by interaction with target mRNAs. We consider a role for TINCR as an epithelial-specific guide for targeting the Staufen1 protein to specific mRNAs, reflecting the increasing complexity of gene regulatory processes in mammalian cells and tissue.
Keywords: lncRNA, TINCR, STAU1, epidermis, skin, non-coding RNA, differentiation
Diverse Roles of Long Non-Coding RNAs in Gene Regulation
While most of the human genome is actively transcribed,1 only 1.9% encodes for proteins.2 A significant portion of the resulting non-coding transcripts appears to embody long non-coding RNAs (lncRNAs), a class of RNAs with no predicted protein coding potential, comprising up to 10000 transcripts in mammals.1,3–5 Although this large newly discovered portion of the human transcriptome is still poorly characterized to date, initial studies strongly indicate that lncRNAs are highly functionally relevant and not mere transcriptional waste. Accordingly, a rapidly growing number of studies have revealed important roles for lncRNAs in gene regulatory processes, such as recruitment of chromatin modifiers to target genomic sites6–11 or direct, promoter-specific modulation of target gene transcription.12–14 Additional roles of lncRNAs include control of nuclear shuttling of proteins,15 regulation of allelic expression ratios, such as genomic imprinting16–20 and mammalian X-chromosome inactivation.21 A number of recent studies report involvement of lncRNAs in cancer development, such as MALAT1,22 HOTAIR,9 GAS5,23 BANCR,24 H19,25,26 and others,27 strongly indicating that lncRNAs not only control gene regulatory pathways in normal cells and tissue but also play a role in tumor development.
Clearly, the classes of lncRNA transcripts known to date show a wide functional as well as mechanistic variety. Correspondingly, a number of lncRNAs can act as guides to direct proteins or protein complexes to their target sites, while others function as scaffolds to bring these proteins together in space and time. Some lncRNAs act as structural decoys to titrate away transcription factors or other proteins from their sites of action, thus altering transcriptional activity of nearby gene loci.28,29
Roles of lncRNAs in Cellular Differentiation
Transition from progenitor cells into highly differentiated cells involves tightly controlled gene regulatory changes. A growing number of lncRNAs has been implicated in such processes and, thus, help regulating mammalian differentiation in multiple tissues.30 While several lncRNAs appear to be required for maintaining the pluripotent state of embryonic stem cells,31–34 others were shown to be functionally important for differentiation of diverse cells and tissues.11,35–41 Interestingly, the majority of lncRNAs in mammalian embryonic stem cells is derived from divergently transcribed protein-coding genes, and protein-coding as well as non-coding transcripts coordinately change transcription patterns upon differentiation.33 In most cases, the mechanisms of lncRNA-mediated regulation of cellular differentiation are not well understood to date. Nevertheless, several recent studies shed some light on recurrent modes of action. A unique feature of a subset of lncRNAs is their ability to act as molecular decoys for microRNAs. Correspondingly, the lncRNA linc-MD1 harbors consensus sites for miR-135 as well as miR-133, two microRNAs important for regulation of muscle differentiation. Linc-MD1 thus acts as a sponge by titrating away these microRNAs from their target transcription factors, thus fine-tuning gene regulatory networks controlling muscle differentiation.40 In a similar fashion, the lncRNA linc-RoR shares microRNA response elements with transcription factors maintaining the progenitor cell status in embryonic stem cells and maintains a non-differentiated state by acting as a competing RNA for target microRNAs.34 A number of lncRNAs were shown to control tissue differentiation by recruiting chromatin-modifying protein complexes to their DNA target sites. In this way, the lncRNA Braveheart (Bvht) controls cardiomyocyte differentiation and maintenance of the cardiac cell fate, possibly by acting as a guide for Polycomb Repressive Complex 2 (PRC2) to mediate epigenetic regulation of the cardiac gene expression program.37 Similarly, the lncRNA Fendrr appears to bind to both activating and repressive chromatin complexes to control heart and body wall development in mice.11 In human epidermis, at least two lncRNAs control the balance between the progenitor and differentiated compartment of the tissue. The lncRNA ANCR represses differentiation in the progenitor containing basal layer.42 While its mode of action in keratinocytes is yet unclear, recent work using osteoblast differentiation as a model indicates a role for ANCR as guide for PRC2, mediating epigenetic gene regulation.43 In contrast to ANCR, the lncRNA TINCR regulates epidermal differentiation.36
TINCR-Mediated Control of Epidermal Differentiation
In an attempt to identify lncRNAs necessary for human epidermal homeostasis, we performed whole transcriptome RNA sequencing with progenitor and differentiated human keratinocytes and identified TINCR (terminal differentiation-induced non-coding RNA), a lncRNA highly induced during epidermal differentiation.36 TINCR is 3.7 kb in size, located in a gene desert on human chromosome 19, and highly enriched in the cytoplasmic compartment of differentiated keratinocytes. Its functional necessity for epidermal differentiation was verified by loss of function studies in epidermal tissue using RNA interference. TINCR-depletion resulted in lack of induction of key differentiation genes, including genes mutated in human skin diseases characterized by disrupted epidermal barrier formation. Consistent with this, TINCR-deficient epidermis showed abnormal late differentiation morphology, as indicated by lack of keratohyalin granules and lamellar bodies; both structures are essential for formation of a functional epidermal permeability barrier, which protects against the external environment as well as water loss.
In contrast to proteins, extensive data about lncRNA functional domains or interacting molecules is lacking, limiting our understanding of the modes of action for lncRNAs of interest. However, lncRNAs were shown to interact with other molecules, including protein, DNA, and RNA to mediate or modulate their functions.29 In this regard, we performed transcriptome-scale RNA interactome analysis and revealed that TINCR interacts with a suite of differentiation mRNAs. This direct interaction occurs through a 25-nucleotide motif (TINCR box) strongly enriched in interacting mRNAs as well as TINCR itself, and appears to be required for TINCR binding.
In an attempt to investigate TINCR binding capacity to ~9400 human recombinant proteins, we performed a high-throughput protein–RNA interaction analysis44 and revealed direct interaction of TINCR RNA with the Staufen1 protein (STAU1), with Staufen1-deficient tissue recapitulating the impaired differentiation seen with TINCR loss. Thus, the lncRNA TINCR, together with the Staufen1 protein, seem to mediate differentiation of human epidermal tissue. This protein–lncRNA complex, at least in part, acts through stabilization of differentiation mRNAs36 (Fig. 1A).
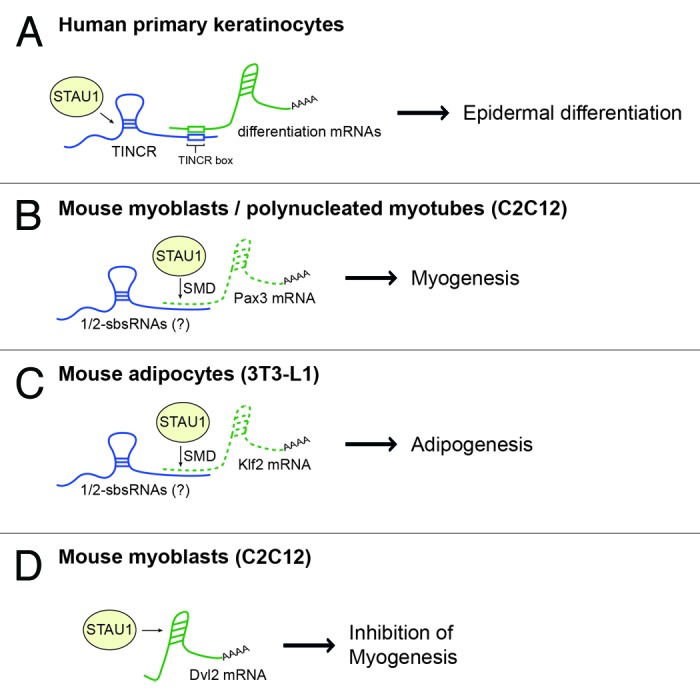
Figure 1. Stau1 is a regulator of terminal differentiation. (A) In differentiated keratinocytes, the lncRNA TINCR interacts with STAU1 as well as differentiation mRNAs. Together, STAUs1 and TINCR stabilize a subset of mRNAs required for epidermal differentiation. (B) In polynucleated myotubes, STAU1 interacts with and catalyzes degradation of Pax3 mRNA encoding an inhibitor of myogenesis. (C) STAU1-mediated decay of Klf2 mRNA, coding for an anti-adipogenic protein, promotes differentiation of pre-adipocytes to adipocytes. (D) Dvl2, an inhibitor of myogenesis, is expressed in non-differentiated myoblasts. STAU1 interaction with the Dvl2 mRNA in myoblasts leads to stabilization of the transcript and helps to negatively regulate myogenesis in these cells. SMD = STAU1-mediated decay.
The Two Faces of STAU1: RNA Stabilization Vs. Decay
STAU1 is a double-stranded RNA-binding protein first described as a mediator of maternal RNA localization in the Drosophila egg.45 Besides its role in localization of RNA to different subcellular compartments, it was shown to be implicated in RNA stability and promotion of mRNA translation in mammalian cells and tissues.46–49 Its ubiquitous expression in mammals50 suggests that STAU1 not only controls differentiation of human epidermis but might also be involved in regulating other terminal differentiating tissues. Indeed, STAU1 participates in controlling differentiation of pre-adipocytes into mature adipocyte cells51 as well as the process of myogenesis52–55 (Fig. 1), and appears to also play a role in early stages of mouse embryonic stem cell differentiation.56
STAU1 involvement in mammalian myogenesis has been extensively studied, revealing a dual role for STAU1 in this system: In non-differentiated myoblasts, it acts as an mRNA stabilizer,55 while in multi-nucleated myotubes, STAU1 controls mRNA decay.53,54 STAU1-dependent RNA degradation targets a subset of mRNAs acting as inhibitors of myogenesis, such as PAX3, through a process called Stau1-mediated mRNA decay (SMD)54 (Fig. 1B). Similarly, STAU1 promotes adipogenesis by SMD-mediated degradation of Klf2 mRNA encoding an anti-adipogenic factor51 (Fig. 1C). SMD can either occur through direct binding of STAU1 to double-stranded binding sites located in the 3′-UTR of target mRNAs or through a process involving a lncRNA.57 The latter is mediated by imperfect base pairing between an ALU element of an mRNA target of SMD and another ALU sequence in a half-STAU1-binding site lncRNA (1/2-sbsRNA).58 Binding of STAU leads to degradation of the mRNA in a UPF1-dependent manner. Similarly, SMD controls myogenesis in rodents. Instead of ALU elements, which are unique to primates, rodents create STAU1-binding sites through interaction of lncRNAs with target mRNAs, both containing short interspersed elements (SINE) of the B1, B2, B4, and ID families.53 In human epidermis, SMD seems not to be involved in regulation of differentiation since depletion of the helicase UPF1, which is required for SMD, does not recapitulate the effects seen in TINCR or STAU1 deficient tissue.36 Conversely, STAU1 and TINCR appear to increase stability of several differentiation mRNAs in differentiated keratinocytes, rather than destabilizing them. A similar, SMD-independent, mRNA stabilizing function of STAU1 seems to occur in myoblasts. STAU1 was shown to positively regulate expression of the Dvl2 gene, encoding for an inhibitor of myoblast differentiation into polynucleated myotubes. This regulation is mediated by binding of STAU1 to the 3′-UTR of Dvl2 mRNA and leads to stabilization of the transcript and consequentially to repression of differentiation55 (Fig. 1D).
Future Perspective
The mRNA stabilizing function of TINCR in keratinocytes adds to the growing number of lncRNAs with cytoplasmic functions,59,60 but the exact roles STAU1 and TINCR play in this process, as well as details about the formation of the complex consisting of TINCR, STAU1, and differentiation mRNAs are not completely clear and require further investigation.
While STAU1 stabilizes target mRNAs in myoblasts as well as differentiated keratinocytes, this might occur through different mechanisms in each of the two cell types. At least in vitro, STAU1 directly interacts with Dvl2 mRNA in myoblasts, while in keratinocytes, the lncRNA TINCR seems to link STAU1 and mRNA interaction. Whether STAU1 is able to bind differentiation mRNAs directly in this context still needs to be determined, but our studies with TINCR- or STAU1-deficient epidermal tissue showed that both components of the complex are required for normal differentiation to occur.
While the impact of TINCR on differentiation has been studied, not much is known about the factors controlling its expression in human epidermis. Further insight into the regulation of the TINCR gene locus would help placing TINCR into the context of known regulators of epidermal differentiation.
STAU1 is able to self-associate61 and also to associate with STAU2,62 raising the question whether homo- or heterodimerization of STAU1 is required for TINCR-mediated stabilization of mRNAs in human epidermis. While dimerization of STAU1 appears to be required for efficient wound healing of human keratinocytes,61 its impact on keratinocyte differentiation is not known to date. Also, a role of STAU2 in regulating differentiation in a TINCR-dependent manner has not been tested yet. Further studies are required to analyze potential roles of Staufen1/2 homo- or heterodimerization in epidermal differentiation.
The fact that STAU1 is ubiquitously expressed50 while most lncRNAs show a tissue-specific expression pattern,30 allows the hypothesis that lncRNAs might fine-tune target mRNA recruitment to STAU1 in different cellular environments, and therefore could provide tissue specificity for STAU1 function. TINCR expression appears to be mostly limited to differentiated keratinocytes and might therefore act as a tissue specific guide for pre-selection and shuttling of mRNAs to STAU1 in differentiated epidermal strata. In this light, the hypothesis would suggest the existence of tissue specific lncRNAs with similar function in other terminally differentiating tissues. Correspondingly, it might be of interest to investigate whether direct interaction of STAU1 with Dvl2 mRNA in myoblasts is sufficient to mediate stabilization of the transcript, or if a lncRNA is involved in this process.
Although the functional relevance of TINCR and STAU1 for epidermal differentiation has been shown, the exact mechanism by which this lncRNA–protein complex mediates stabilization of a subset of differentiation mRNAs remains unclear. STAU1 was previously shown to facilitate translation initiation of mRNAs through tethering of transcripts to polysomes.49 Lack of proper localization of differentiation mRNAs to ribosomes in STAU1- or TINCR-deficient keratinocytes might therefore lead to decreased translational efficiency and subsequent premature decay of these transcripts, probably resulting in the epidermal differentiation defect seen in TINCR- or STAU1-deficient tissue. While its exact role has yet to be defined, it appears that Stau1 plays multiple roles in terminal differentiation processes in human epidermis and other tissues. Thus, further study of the complex consisting of STAU1 protein, lncRNA and associated mRNAs will broaden our understanding of long non-coding RNA and STAU1 function in mammalian tissue.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/26249
References
- 1.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al. Landscape of transcription in human cells. Nature. 2012;489:101–8. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Consortium IHGS, International Human Genome Sequencing Consortium Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–45. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 3.Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–27. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jia H, Osak M, Bogu GK, Stanton LW, Johnson R, Lipovich L. Genome-wide computational identification and manual annotation of human long noncoding RNA genes. RNA. 2010;16:1478–87. doi: 10.1261/rna.1951310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–89. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–23. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–4. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heo JB, Sung S. Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science. 2011;331:76–9. doi: 10.1126/science.1197349. [DOI] [PubMed] [Google Scholar]
- 9.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai M-C, Hung T, Argani P, Rinn JL, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–6. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–93. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grote P, Wittler L, Hendrix D, Koch F, Währisch S, Beisaw A, Macura K, Bläss G, Kellis M, Werber M, et al. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev Cell. 2013;24:206–14. doi: 10.1016/j.devcel.2012.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martianov I, Ramadass A, Serra Barros A, Chow N, Akoulitchev A. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature. 2007;445:666–70. doi: 10.1038/nature05519. [DOI] [PubMed] [Google Scholar]
- 13.Feng J, Bi C, Clark BS, Mady R, Shah P, Kohtz JD. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev. 2006;20:1470–84. doi: 10.1101/gad.1416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bond AM, Vangompel MJW, Sametsky EA, Clark MF, Savage JC, Disterhoft JF, Kohtz JD. Balanced gene regulation by an embryonic brain ncRNA is critical for adult hippocampal GABA circuitry. Nat Neurosci. 2009;12:1020–7. doi: 10.1038/nn.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Willingham AT, Orth AP, Batalov S, Peters EC, Wen BG, Aza-Blanc P, Hogenesch JB, Schultz PG. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science. 2005;309:1570–3. doi: 10.1126/science.1115901. [DOI] [PubMed] [Google Scholar]
- 16.Bartolomei MS, Zemel S, Tilghman SM. Parental imprinting of the mouse H19 gene. Nature. 1991;351:153–5. doi: 10.1038/351153a0. [DOI] [PubMed] [Google Scholar]
- 17.Lyle R, Watanabe D, te Vruchte D, Lerchner W, Smrzka OW, Wutz A, Schageman J, Hahner L, Davies C, Barlow DP. The imprinted antisense RNA at the Igf2r locus overlaps but does not imprint Mas1. Nat Genet. 2000;25:19–21. doi: 10.1038/75546. [DOI] [PubMed] [Google Scholar]
- 18.Sleutels F, Zwart R, Barlow DP. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature. 2002;415:810–3. doi: 10.1038/415810a. [DOI] [PubMed] [Google Scholar]
- 19.Mancini-Dinardo D, Steele SJ, Levorse JM, Ingram RS, Tilghman SM. Elongation of the Kcnq1ot1 transcript is required for genomic imprinting of neighboring genes. Genes Dev. 2006;20:1268–82. doi: 10.1101/gad.1416906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Williamson CM, Ball ST, Dawson C, Mehta S, Beechey CV, Fray M, Teboul L, Dear TN, Kelsey G, Peters J. Uncoupling antisense-mediated silencing and DNA methylation in the imprinted Gnas cluster. PLoS Genet. 2011;7:e1001347. doi: 10.1371/journal.pgen.1001347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee JT, Bartolomei MS. X-inactivation, imprinting, and long noncoding RNAs in health and disease. Cell. 2013;152:1308–23. doi: 10.1016/j.cell.2013.02.016. [DOI] [PubMed] [Google Scholar]
- 22.Gutschner T, Hämmerle M, Eissmann M, Hsu J, Kim Y, Hung G, Revenko A, Arun G, Stentrup M, Gross M, et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013;73:1180–9. doi: 10.1158/0008-5472.CAN-12-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mourtada-Maarabouni M, Pickard MR, Hedge VL, Farzaneh F, Williams GT. GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene. 2009;28:195–208. doi: 10.1038/onc.2008.373. [DOI] [PubMed] [Google Scholar]
- 24.Flockhart RJ, Webster DE, Qu K, Mascarenhas N, Kovalski J, Kretz M, Khavari PA. BRAFV600E remodels the melanocyte transcriptome and induces BANCR to regulate melanoma cell migration. Genome Res. 2012;22:1006–14. doi: 10.1101/gr.140061.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoshimizu T, Miroglio A, Ripoche M-A, Gabory A, Vernucci M, Riccio A, Colnot S, Godard C, Terris B, Jammes H, et al. The H19 locus acts in vivo as a tumor suppressor. Proc Natl Acad Sci U S A. 2008;105:12417–22. doi: 10.1073/pnas.0801540105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shoshani O, Massalha H, Shani N, Kagan S, Ravid O, Madar S, Trakhtenbrot L, Leshkowitz D, Rechavi G, Zipori D. Polyploidization of murine mesenchymal cells is associated with suppression of the long noncoding RNA H19 and reduced tumorigenicity. Cancer Res. 2012;72:6403–13. doi: 10.1158/0008-5472.CAN-12-1155. [DOI] [PubMed] [Google Scholar]
- 27.Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 2012;9:703–19. doi: 10.4161/rna.20481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–14. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–66. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu W, Alvarez-Dominguez JR, Lodish HF. Regulation of mammalian cell differentiation by long non-coding RNAs. EMBO Rep. 2012;13:971–83. doi: 10.1038/embor.2012.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sheik Mohamed J, Gaughwin PM, Lim B, Robson P, Lipovich L. Conserved long noncoding RNAs transcriptionally regulated by Oct4 and Nanog modulate pluripotency in mouse embryonic stem cells. RNA. 2010;16:324–37. doi: 10.1261/rna.1441510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sigova AA, Mullen AC, Molinie B, Gupta S, Orlando DA, Guenther MG, Almada AE, Lin C, Sharp PA, Giallourakis CC, et al. Divergent transcription of long noncoding RNA/mRNA gene pairs in embryonic stem cells. Proc Natl Acad Sci U S A. 2013;110:2876–81. doi: 10.1073/pnas.1221904110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Y, Xu Z, Jiang J, Xu C, Kang J, Xiao L, Wu M, Xiong J, Guo X, Liu H. Endogenous miRNA sponge lincRNA-RoR regulates Oct4, Nanog, and Sox2 in human embryonic stem cell self-renewal. Dev Cell. 2013;25:69–80. doi: 10.1016/j.devcel.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 35.Hu W, Yuan B, Flygare J, Lodish HF. Long noncoding RNA-mediated anti-apoptotic activity in murine erythroid terminal differentiation. Genes Dev. 2011;25:2573–8. doi: 10.1101/gad.178780.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kretz M, Siprashvili Z, Chu C, Webster DE, Zehnder A, Qu K, Lee CS, Flockhart RJ, Groff AF, Chow J, et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature. 2013;493:231–5. doi: 10.1038/nature11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klattenhoff CA, Scheuermann JC, Surface LE, Bradley RK, Fields PA, Steinhauser ML, Ding H, Butty VL, Torrey L, Haas S, et al. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell. 2013;152:570–83. doi: 10.1016/j.cell.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun L, Goff LA, Trapnell C, Alexander R, Lo KA, Hacisuleyman E, Sauvageau M, Tazon-Vega B, Kelley DR, Hendrickson DG, et al. Long noncoding RNAs regulate adipogenesis. Proc Natl Acad Sci U S A. 2013;110:3387–92. doi: 10.1073/pnas.1222643110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bertani S, Sauer S, Bolotin E, Sauer F. The noncoding RNA Mistral activates Hoxa6 and Hoxa7 expression and stem cell differentiation by recruiting MLL1 to chromatin. Mol Cell. 2011;43:1040–6. doi: 10.1016/j.molcel.2011.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 40.Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tramontano A, Bozzoni I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–69. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Skreka K, Schafferer S, Nat I-R, Zywicki M, Salti A, Apostolova G, Griehl M, Rederstorff M, Dechant G, Hüttenhofer A. Identification of differentially expressed non-coding RNAs in embryonic stem cell neural differentiation. Nucleic Acids Res. 2012;40:6001–15. doi: 10.1093/nar/gks311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kretz M, Webster DE, Flockhart RJ, Lee CS, Zehnder A, Lopez-Pajares V, Qu K, Zheng GXY, Chow J, Kim GE, et al. Suppression of progenitor differentiation requires the long noncoding RNA ANCR. Genes Dev. 2012;26:338–43. doi: 10.1101/gad.182121.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu L, Xu P-C. Downregulated LncRNA-ANCR promotes osteoblast differentiation by targeting EZH2 and regulating Runx2 expression. Biochem Biophys Res Commun. 2013;432:612–7. doi: 10.1016/j.bbrc.2013.02.036. [DOI] [PubMed] [Google Scholar]
- 44.Siprashvili Z, Webster DE, Kretz M, Johnston D, Rinn JL, Chang HY, Khavari PA. Identification of proteins binding coding and non-coding human RNAs using protein microarrays. BMC Genomics. 2012;13:633. doi: 10.1186/1471-2164-13-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.St Johnston D, Beuchle D, Nüsslein-Volhard C. Staufen, a gene required to localize maternal RNAs in the Drosophila egg. Cell. 1991;66:51–63. doi: 10.1016/0092-8674(91)90138-O. [DOI] [PubMed] [Google Scholar]
- 46.Kim YK, Furic L, Desgroseillers L, Maquat LE. Mammalian Staufen1 recruits Upf1 to specific mRNA 3’UTRs so as to elicit mRNA decay. Cell. 2005;120:195–208. doi: 10.1016/j.cell.2004.11.050. [DOI] [PubMed] [Google Scholar]
- 47.Kiebler MA, Hemraj I, Verkade P, Köhrmann M, Fortes P, Marión RM, Ortín J, Dotti CG. The mammalian staufen protein localizes to the somatodendritic domain of cultured hippocampal neurons: implications for its involvement in mRNA transport. J Neurosci. 1999;19:288–97. doi: 10.1523/JNEUROSCI.19-01-00288.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thomas MG, Martinez Tosar LJ, Loschi M, Pasquini JM, Correale J, Kindler S, Boccaccio GL. Staufen recruitment into stress granules does not affect early mRNA transport in oligodendrocytes. Mol Biol Cell. 2005;16:405–20. doi: 10.1091/mbc.E04-06-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dugré-Brisson S, Elvira G, Boulay K, Chatel-Chaix L, Mouland AJ, DesGroseillers L. Interaction of Staufen1 with the 5′ end of mRNA facilitates translation of these RNAs. Nucleic Acids Res. 2005;33:4797–812. doi: 10.1093/nar/gki794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lebeau G, Maher-Laporte M, Topolnik L, Laurent CE, Sossin W, Desgroseillers L, Lacaille J-C. Staufen1 regulation of protein synthesis-dependent long-term potentiation and synaptic function in hippocampal pyramidal cells. Mol Cell Biol. 2008;28:2896–907. doi: 10.1128/MCB.01844-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cho H, Kim KM, Han S, Choe J, Park SG, Choi SS, Kim YK. Staufen1-mediated mRNA decay functions in adipogenesis. Mol Cell. 2012;46:495–506. doi: 10.1016/j.molcel.2012.03.009. [DOI] [PubMed] [Google Scholar]
- 52.Yamaguchi Y, Oohinata R, Naiki T, Irie K. Stau1 negatively regulates myogenic differentiation in C2C12 cells. Genes Cells. 2008;13:583–92. doi: 10.1111/j.1365-2443.2008.01189.x. [DOI] [PubMed] [Google Scholar]
- 53.Wang J, Gong C, Maquat LE. Control of myogenesis by rodent SINE-containing lncRNAs. Genes Dev. 2013;27:793–804. doi: 10.1101/gad.212639.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gong C, Kim YK, Woeller CF, Tang Y, Maquat LE. SMD and NMD are competitive pathways that contribute to myogenesis: effects on PAX3 and myogenin mRNAs. Genes Dev. 2009;23:54–66. doi: 10.1101/gad.1717309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yamaguchi Y, Naiki T, Irie K. Stau1 regulates Dvl2 expression during myoblast differentiation. Biochem Biophys Res Commun. 2012;417:427–32. doi: 10.1016/j.bbrc.2011.11.133. [DOI] [PubMed] [Google Scholar]
- 56.Gautrey H, McConnell J, Lako M, Hall J, Hesketh J. Staufen1 is expressed in preimplantation mouse embryos and is required for embryonic stem cell differentiation. Biochim Biophys Acta. 2008;1783:1935–42. doi: 10.1016/j.bbamcr.2008.05.017. [DOI] [PubMed] [Google Scholar]
- 57.Park E, Maquat LE. Staufen-mediated mRNA decay. Wiley Interdiscip Rev RNA. 2013;4:423–35. doi: 10.1002/wrna.1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gong C, Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature. 2011;470:284–8. doi: 10.1038/nature09701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Carrieri C, Cimatti L, Biagioli M, Beugnet A, Zucchelli S, Fedele S, Pesce E, Ferrer I, Collavin L, Santoro C, et al. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature. 2012;491:454–7. doi: 10.1038/nature11508. [DOI] [PubMed] [Google Scholar]
- 60.Yoon J-H, Abdelmohsen K, Srikantan S, Yang X, Martindale JL, De S, Huarte M, Zhan M, Becker KG, Gorospe M. LincRNA-p21 suppresses target mRNA translation. Mol Cell. 2012;47:648–55. doi: 10.1016/j.molcel.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gleghorn ML, Gong C, Kielkopf CL, Maquat LE. Staufen1 dimerizes through a conserved motif and a degenerate dsRNA-binding domain to promote mRNA decay. Nat Struct Mol Biol. 2013;20:515–24. doi: 10.1038/nsmb.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Park E, Gleghorn ML, Maquat LE. Staufen2 functions in Staufen1-mediated mRNA decay by binding to itself and its paralog and promoting UPF1 helicase but not ATPase activity. Proc Natl Acad Sci U S A. 2013;110:405–12. doi: 10.1073/pnas.1213508110. [DOI] [PMC free article] [PubMed] [Google Scholar]


