Abstract
Non-coding RNAs (ncRNAs) called Y RNAs are abundant components of both animal cells and a variety of bacteria. In all species examined, these ~100 nt RNAs are bound to the Ro 60 kDa (Ro60) autoantigen, a ring-shaped protein that also binds misfolded ncRNAs in some vertebrate nuclei. Although the function of Ro60 RNPs has been mysterious, we recently reported that a bacterial Y RNA tethers Ro60 to the 3′ to 5′ exoribonuclease polynucleotide phosphorylase (PNPase) to form RYPER (Ro60/Y RNA/PNPase Exoribonuclease RNP), a new RNA degradation machine. PNPase is a homotrimeric ring that degrades single-stranded RNA, and Y RNA-mediated tethering of Ro60 increases the effectiveness of PNPase in degrading structured RNAs. Single particle electron microscopy of RYPER suggests that RNA threads through the Ro60 ring into the PNPase cavity. Further studies indicate that Y RNAs may also act as gates to regulate entry of RNA substrates into the Ro60 channel. These findings reveal novel functions for Y RNAs and raise questions about how the bacterial findings relate to the roles of these ncRNAs in animal cells. Here we review the literature on Y RNAs, highlighting their close relationship with Ro60 proteins and the hypothesis that these ncRNAs function generally to tether Ro60 rings to diverse RNA-binding proteins.
Keywords: RNA degradation, RYPER, Y RNAs, exoribonucleases, non-coding RNAs
What Are Y RNAs?
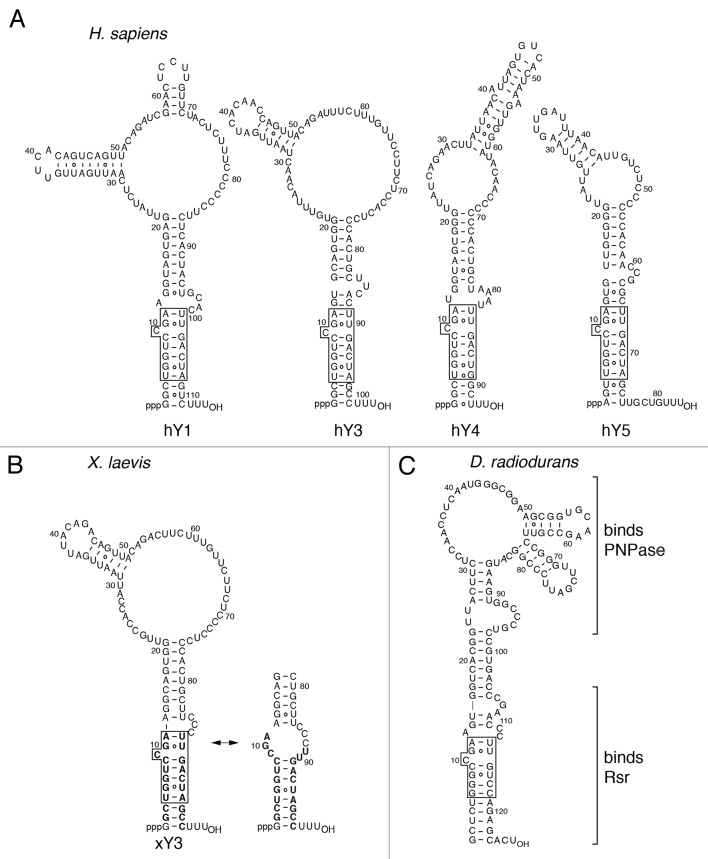
Y RNAs were discovered because these ncRNAs are complexed with the Ro60 protein, a frequent target of the immune system in patients suffering from two common rheumatic diseases, systemic lupus erythematosus, and Sjogren’s syndrome.1–4 Characterization of the four distinct Y RNAs in human cells revealed that these ncRNAs (called hY1, hY3, hY4, and hY5; h stands for human) are 83–112 nt long and transcribed by RNA polymerase III.3,5–10 The number of distinct Y RNAs varies between species. For example, mouse cells contain only two Y RNAs, mY1 and mY3, which are orthologs of hY1 and hY3.5 A defining feature of animal cell Y RNAs is that these RNAs fold into structures consisting of a large internal loop and a long stem formed by basepairing the 5′ and 3′ ends of the RNA.6–8,11 Near the base of the stem is a conserved sequence that is the high affinity binding site for Ro60 (refs. 12–15 and Fig. 1A).
Figure 1. Potential secondary structures for Y RNAs. (A) The four human Y RNAs. A conserved helix present in all animal cell Y RNAs is boxed. Nucleotides within this helix are important for Ro60 binding.13–15 The proposed structures are consistent with phylogenetic analyses52 and enzymatic probing experiments.11,53 (B) X. laevis Y3 RNA. Structural and enzymatic probing studies show that nucleotides in the conserved helix can form two alternate conformers.14,15,53 Sequences present in the X. laevis Ro60/Y3 RNA crystal structure are in bold type. (C) D. radiodurans Y RNA. Regions involved in Rsr and PNPase binding are shown.30
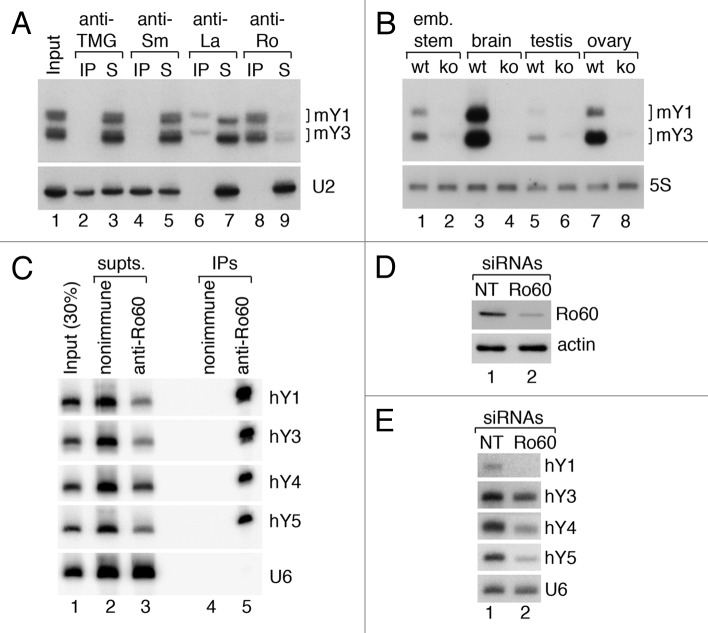
Most Y RNAs in cells are bound to Ro60, a doughnut-shaped RNA-binding protein that also binds misfolded ncRNA precursors in some animal cell nuclei.15–18 Ro60 orthologs are present in most animal cells and also in ~5% of sequenced bacterial genomes.19 Immunoprecipitation experiments from mouse and human cells have demonstrated that most Y RNAs are present as Ro60 RNPs (refs. 7 and 20 and Fig. 2A and C). As worms, mouse cells, and bacteria lacking Ro60 all have drastically reduced Y RNAs, Ro60 is required for stable accumulation of these RNAs (refs. 18 and 21–23 and Fig. 2B). Ro60 also stabilizes human Y RNAs, as experiments in which we used siRNAs to deplete Ro60 by 83% from human keratinocytes revealed that these RNAs were reduced between 30 and 90%, depending on the RNA (Fig. 2D and E).
Figure 2. Ro60 is a stable component of Y RNPs and is important for Y RNA integrity. (A) Mouse embryonic stem cell lysates were subjected to immunoprecipitation with antibodies against the trimethylguanosine (TMG) cap that is the 5′ end of many snRNAs (lanes 2–3), anti-Sm antibodies, which recognize the Sm proteins of the spliceosomal U snRNPs (lanes 4–5), anti-La (lanes 6–7) and anti-Ro60 antibodies (lanes 8–9). RNAs in immunoprecipitates (lanes 2,4,6,8), supernatants (lanes 3,5,7,9), and an equivalent amount of lysate (lane 1) were subjected to northern blotting to detect mY1 and mY3. As a control, the blot was reprobed to detect the spliceosomal U2 snRNA. Note that Y RNAs in the anti-La immunoprecipitate are slightly larger (lane 6) than the Y RNAs remaining in the supernatant (lane 7). (B) RNA extracted from wild-type and Ro60−/− embryonic stem cells (lanes 1–2), brain (lanes 3–4), testis (lanes 5–6), and ovary (lanes 7–8) were subjected to northern blotting to detect mY1 and mY3. As a loading control, the blot was reprobed to detect 5S rRNA. (C) HEK293 cell lysates were subjected to immunoprecipitation with anti-Ro60 antibodies or nonimmune isotype control IgG. RNAs extracted from immunoprecipitates (lanes 4 and 5), supernatants (lane 2 and 3), and the starting lysate (lane 1) were subjected to northern blotting to detect hY RNAs. As a negative control, the blot was probed to detect the spliceosomal U6 snRNA. (DandE) To determine if Ro60 stabilizes hY RNAs, siRNAs against Ro60, or control non-target (NT) siRNAs were transfected into human keratinocytes. After 72 h, lysates were prepared and subjected to western blotting (D) to detect Ro60. Actin was used as a loading control. RNA extracted from the lysates was subjected to northern blotting (E) to detect hY RNAs. U6 snRNA was used as a loading control. Quantitation revealed that Ro60 was reduced by 83%, while hY1, hY3, hY4, and hY5 were reduced by 90%, 30%, 59%, and 60%, respectively.
In addition to Ro60, some Y RNAs in animal cells are bound by the La autoantigen, a nuclear phosphoprotein that binds all newly synthesized RNA polymerase III transcripts.5,24 Like other La-bound RNAs, Y RNAs initially end in uridines, since RNA polymerase III terminates in a run of Ts and La recognizes the sequence UUUOH.24 Because trimming of the terminal uridines by exoribonuclease(s) removes the La binding site, the Y RNAs bound by La are slightly longer at the 3′ end than the bulk of the population (Fig. 2A). Ro60 and La can bind simultaneously to the same RNA.5 However, since the La binding site can be eliminated by end trimming, the fraction of Y RNAs bound by La varies between 0–100%, depending on the cell type.7,8,20Assigning Functions to Mammalian Y RNAs: A Work in Progress
One role of mammalian Y RNAs is to influence the subcellular location of Ro60. Ro60 is both nuclear and cytoplasmic, and its distribution between these compartments is at least partly Y RNA-mediated. Ro60 exits mouse cell nuclei as a Ro60/mY3 complex, and binding of the zipcode binding protein ZBP1 (also known as IMP1 and IGF2BP1) to mY3 RNA is important for export of this RNP.25 Additionally, binding of mY3 to Ro60 masks a nuclear accumulation signal on the Ro60 surface, thus retaining the RNP in the cytoplasm.26 Moreover, since both Ro60 and mY3 RNA become strongly nuclear following UV irradiation,18 the position of this RNA on Ro60 may be altered during environmental stress to allow the nuclear accumulation signal to become accessible.
Since Y RNAs are intimately associated with Ro60, our laboratory has used Ro60 as an entry point to identify additional roles of these RNAs. Because Ro60 binds misfolded 5S rRNA precursors and variant U2 small nuclear RNAs (snRNAs) in some animal cell nuclei, we proposed that Ro60 functions in ncRNA surveillance.16–18 Biochemical and crystallographic studies demonstrated that Ro60 binds misfolded ncRNAs that contain both single-strand 3′ ends and adjacent protein-free helices. Structural analyses revealed that the single-stranded 3′ end of a misfolded RNA fragment inserts through the Ro60 cavity, while a helix contacts the ring outer surface (ref. 27 and Fig. 3B). Because binding of Ro60 to misfolded ncRNAs is not strongly sequence-specific, Ro60 may scavenge RNAs that fail to assemble with their correct RNA-binding proteins. Further, the relative lack of sequence specificity suggests that Ro60 could potentially bind a wide range of RNAs.27
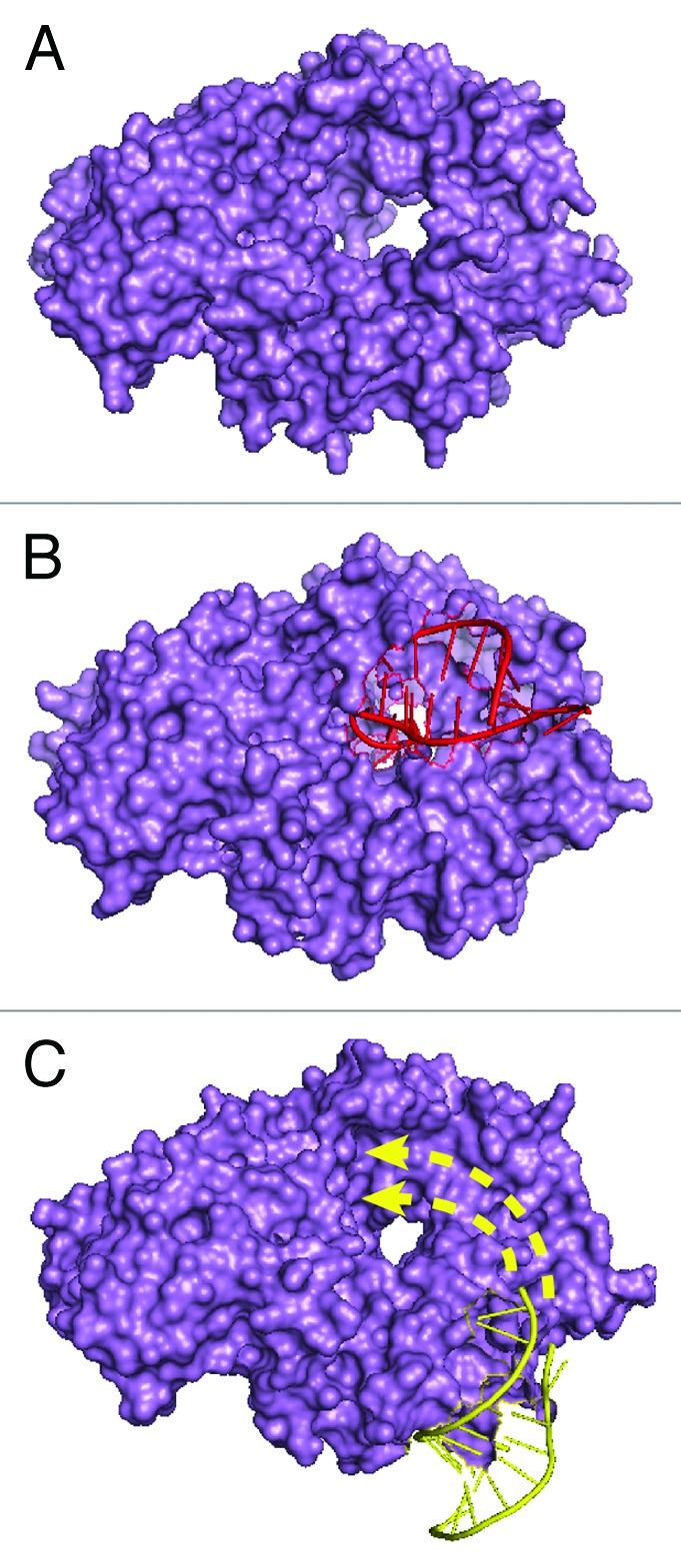
Figure 3. Structures of Ro60. (A) Molecular surface representation of X. laevis Ro60. The hole is 10–15 Å in diameter and binds single-stranded RNA.15 (B) X. laevis Ro60 bound to a misfolded pre-5S rRNA fragment consisting of a short duplex and a single-stranded 3′ extension. The duplex binds on the Ro outer surface, while the single-stranded end inserts through the cavity.27 (C) X. laevis Ro60 bound to a Y RNA fragment.15 The sequence used for crystallization is shown in bold in Figure 1B. Studies of Y RNA binding to mutant Ro60 proteins15,27 suggest that the remainder of the RNA interacts with portions of Ro60 that overlap the misfolded RNA binding site (arrows).
Structural and biochemical studies suggest that Y RNA binding could regulate access of misfolded RNAs to Ro60. A crystal structure of Ro60 complexed with a Y RNA fragment encompassing the Ro60 binding site revealed that this part of Y RNA binds on the outer edge of the ring (ref. 15 and Fig. 3C). However, both Y RNAs and misfolded RNAs are larger than the fragments present in the crystal structures, and biochemical experiments indicate that the two RNAs bind overlapping portions of Ro.15,27 Since Y RNAs bind Ro60 with higher affinity than misfolded RNAs, a bound Y RNA could sterically prevent misfolded RNA binding.14,15,27 It has also been proposed that Y RNAs could potentially contribute to recognition of misfolded ncRNAs and/or to recruiting helicases or nucleases that refold or degrade these RNAs.28 Excitingly, studies in bacteria29,30 demonstrate that Y RNAs both regulate access of Ro60 to some RNA substrates and recruit exoribonucleases involved in their degradation (described below).
Y RNAs may also function independently of Ro60. Specifically, it has been reported that vertebrate Y1 and Y3 RNAs are required for initiation of DNA replication.31,32 In these experiments, Y RNAs were found to stimulate DNA replication when added to isolated G1 phase nuclei.31 Moreover, when added to cell extracts, all four Y RNAs bound proteins involved in initiation of DNA replication.33 Although most of these experiments were performed in cell extracts, siRNA-mediated depletion of either hY1 or hY3 reduced the number of replicating HeLa cells31,34 and injection of antisense oligonucleotides into X. laevis and zebrafish embryos was found to block DNA replication.35 However, a role for Y RNAs in DNA replication must be reconciled with findings that Y RNA levels are reduced ~30-fold in mouse cells lacking Ro60, yet these cells grow indistinguishably from wild-type cells,18,36 and that mice lacking Ro60 are viable.23 One possibility is that the remaining Y RNAs, or fragments thereof, are sufficient to support DNA replication.37
A Bacterial Y RNA Tethers Ro60 to a Nuclease to Form an RNA Degradation Machine
Although Ro60 has not been detected in budding or fission yeast, likely orthologs are encoded in ~5% of sequenced bacterial genomes.19 To study Ro60 RNPs in a genetically tractable single-celled organism, we chose the first bacterium with a recognizable ortholog, Deinococcus radiodurans. D. radiodurans is best known for its remarkable resistance to severe oxidative stress and its ability to repair massive DNA damage.38 Our studies revealed that D. radiodurans lacking the Ro60 ortholog Rsr (Ro60-related) exhibit decreased survival after ultraviolet (UV), but not γ-irradiation, and are at a competitive disadvantage during growth in stationary phase.22,39 Remarkably, as in animal cells, Rsr associates with a ncRNA resembling Y RNA (Fig. 1C). Both Rsr and the Y RNA are upregulated after UV or γ-irradiation, and also during dessication, heat stress, and stationary phase.22,29,39,40 The role of Ro60 and Y RNA in cell stress responses is conserved, as mouse cells lacking Ro60 are sensitive to UV irradiation and both Ro60 and mY3 RNA accumulate in nuclei after UV irradiation.18 Additionally, Caenorhabditis elegans lacking Ro60 have abnormalities in dauer formation, a developmental stage adopted by larvae during unfavorable growth conditions.41
Molecular analyses revealed that Rsr and Y RNA function with 3′ to 5′ exoribonucleases to modulate RNA metabolism in response to environmental stress. During heat stress, Rsr, Y RNA, and the exoribonucleases RNase II and RNase PH function in 23S rRNA maturation.29 In stationary phase, both Rsr and the exoribonuclease polynucleotide phosphorylase (PNPase) contribute to rRNA degradation. Although Y RNA was not investigated in these initial studies, Rsr and PNPase co-purify, and the association of PNPase with ribosomal subunits requires Rsr.39 Moreover, PNPase exhibits genetic interactions with both Y RNA and Rsr during normal growth, growth at low temperature, and during oxidative stress.29
Characterization of the Rsr/PNPase complex revealed that Y RNA tethers Rsr to PNPase to form RYPER, an RNA degradation machine specialized for degrading structured RNA.30 PNPase forms a trimeric ring with a degradation cavity that is capped by single-stranded RNA-binding S1 and KH domains. These S1/KH domains bind RNA substrates and also channel single-stranded RNA into the PNPase cavity.42,43 In RYPER, the portion of Y RNA containing the high affinity Ro binding site interacts with Rsr, while the other end of Y RNA interacts with the PNPase S1/KH domains (ref. 30 and Fig. 1C). The Y RNA-mediated tethering of Rsr to PNPase results in a double-ringed complex that based on single particle electron microscopy, is oriented such that single-stranded RNA could pass from the Rsr ring into the PNPase central channel for degradation (ref. 30 and Fig. 4). Biochemical analyses revealed that RYPER degrades structured RNAs such as rRNAs more efficiently than PNPase, most likely because threading of RNA through the Rsr ring contributes to ATP-independent unwinding.30

Figure 4. Model for RYPER based on single particle electron microscopy reconstruction.30 Portions of the reconstruction corresponding to Rsr, Y RNA, and PNPase are depicted in magenta, yellow, and blue, respectively. A possible path for a duplex-containing RNA substrate is drawn in red.
Notably, although RYPER is more effective than PNPase in degrading structured RNAs, it is less effective on single-stranded substrates.30 One explanation for the decreased activity of RYPER on single-stranded RNA is that Y RNA-mediated tethering of Rsr to the PNPase KH/S1 motifs sterically blocks these RNA-binding domains, replacing the PNPase RNA-binding surface with that of Rsr.30 Although the RNA-binding specificity of Rsr has not been characterized, X. laevis Ro binds RNAs that contain both helices and single-stranded 3′ ends.27 If Rsr has similar RNA-binding requirements, RYPER would preferentially bind structured RNAs.
A key question raised by the discovery of RYPER is whether similar RNA degradation machines form in other bacteria with Ro60 orthologs. Preliminary studies in the human pathogen Salmonella enterica serovar Typhimurium revealed that the Ro60 ortholog and a ncRNA also co-purify with PNPase.30 Notably, both S. Typhimurium Ro60 and two associated ncRNAs are encoded within a σ54-regulated “RNA repair operon” that is transcribed in response to an unknown signal.30 Thus, as in D. radiodurans, the expression of S. Typhimurium Ro60 and its associated ncRNAs may be regulated in response to environmental stress.30
A Bacterial Y RNA May Also Function as a Gate
In addition to their role as tethers, Y RNAs can potentially regulate access of other RNAs to the Ro central cavity. As described above, Y RNA and misfolded RNAs bind overlapping sites on Ro60 and a bound Y RNA could sterically inhibit access of misfolded RNAs to the Ro cavity.15,27 Consistent with this hypothesis, Y RNAs and misfolded RNAs compete for binding to Ro60.14 However, since both these RNA binding experiments and the crystal structures employed only purified Ro60, the possibility that interactions with other proteins affect Y RNA positioning was not addressed. Importantly, the fact that RYPER degrades RNA substrates in the presence of Y RNA implies that substrates can enter the Ro60 cavity when PNPase is also complexed.
Single particle electron microscopy of RYPER suggested a model for how Y RNA binding can be modulated to allow RNA substrates to access the Ro60 surface. Specifically, in the three-dimensional reconstruction, the Rsr and PNPase rings are bridged by a rod shaped density (ref. 30 and Fig. 4). If as predicted from the biochemical experiments, this density corresponds to the Y RNA, binding of PNPase to the distal loops may remove this portion of the Y RNA from the Rsr surface, rendering the cavity accessible to RNA substrates (Fig. 5). In this model, a bound Y RNA would prevent RNA substrates from entering the Rsr cavity unless PNPase was also present.
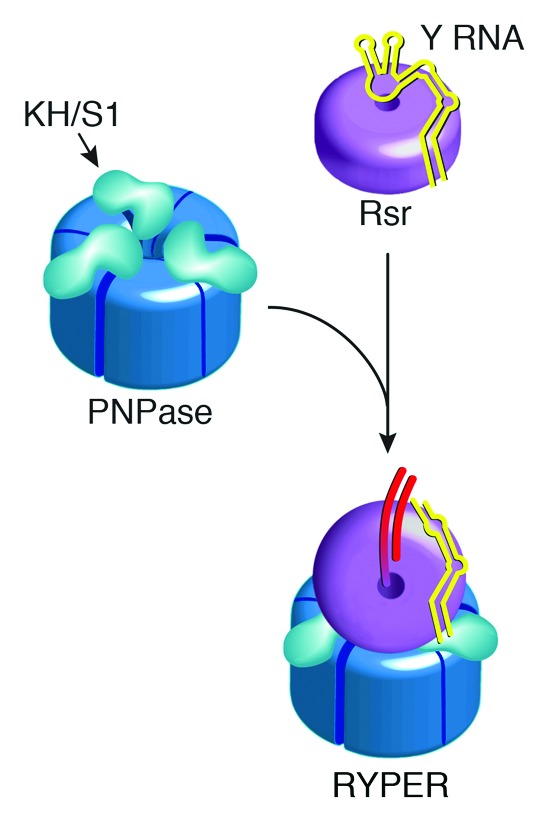
Figure 5. Model for RYPER formation. In the absence of interacting proteins, the Y RNA acts as a gate to prevent other RNAs from accessing the Ro60 cavity. In the presence of PNPase, the Y RNA loops interact with the KH and S1 domains, removing this part of the Y RNA from Ro60, and allowing the single-stranded ends of RNA substrates to enter the cavity.
Studies of the roles of Rsr and Y RNA during heat stress support the hypothesis that Y RNAs also function as gates.29 During normal growth, maturation of D. radiodurans 23S rRNA is very inefficient, as ~40% of these rRNAs contain 5′ and 3′ extensions. During heat stress, maturation becomes highly efficient and requires Rsr, RNase II, and RNase PH.29 As expected if Y RNAs block entry of pre-rRNAs into the Rsr cavity during normal growth, 23S rRNA is fully matured at all growth temperatures when either the Y RNA is deleted or a mutant Rsr that cannot bind Y RNA is overexpressed.29 These results also imply that Rsr is capable of assisting exoribonucleolytic maturation of at least some RNAs without a Y RNA tether.Evidence that Y RNAs Function as Tethers in Mammalian Cells
In mammalian cells, Y RNA-mediated tethering may be important for correct subcellular localization of Ro60. Specifically, binding of the zipcode-binding protein ZBP1 to mY3 RNA is important for nuclear export of the Ro60/mY3 RNP.25 ZBP1, which has well-characterized functions in mRNA post-transcriptional regulation,44 uses two of its four KH domains to bind mRNAs containing a short “zipcode” sequence.45 Since mY3 competes with a zipcode-containing RNA fragment for ZBP1 binding,46 formation of the Ro60/Y RNA/ZBP1 RNP may involve binding of one or both of these KH domains to the mY3 RNA large internal loop.
Because Ro60 RNPs are largely cytosolic and mammalian PNPase localizes to the mitochondrial intermembrane space,47 RYPER may not form in mammalian cells. However, Y RNAs could potentially tether Ro60 to other RNA remodeling proteins, such as exoribonucleases, helicases, or RNA chaperones to assist unwinding of structured RNAs. In this scenario, Ro60 and their associated Y RNAs would function as modules that attach in trans to diverse proteins involved in RNA metabolism. Moreover, the multiple distinct Y RNAs found in mammalian cells could allow Ro60 to be tethered to a greater range of RNA-binding proteins. Consistent with this proposal, several proteins have been shown to associate with Ro60 RNPs through binding distinct subsets of Y RNAs. These include two splicing factors, PUF60 and the polypyrimidine-tract binding protein PTB1, the multifunctional protein nucleolin, the putative helicase MOV10, and the cytidine deaminase APOBEC3G.25,28,48–50 Major goals of future studies will be to define the protein composition and RNA substrates of these complexes and to elucidate how Ro60 and Y RNAs contribute to their functions.Materials and Methods
RNA isolation and northern blotting
Total brain, testis, and ovary tissue was removed from wild-type and Ro60−/− mice,23 lysed in TRIzol (Invitrogen), and RNA isolated as described by the manufacturer. Wild-type and Ro60−/− embryonic stem (ES) cells were cultured as described18 and RNA isolated using TRIzol as above. For northern blotting, RNAs were fractionated in 5% polyacrylamide/8 M urea gels, transferred to Hybond-N membranes (GE Healthcare), and hybridized with [γ32-P]ATP-labeled oligonucleotides as described.51 Oligonucleotide probes were
my1: 5′-AAGGGGGGAA AGTGTAGAAC AGGA-3′,
my3: 5′-GAGCGGAGAA GGAACAAAGA AATCTG-3′,
mouse 5s: 5′-TCAGACGAGA TCGGGCGCGT TCAG-3′,
mouse u2: 5′-CAGATACTAC ACTTGATCTT AGCC-3′,
hy1: 5′-ATCTGTAACT GACTGTGAAC AATCAATTGA GATAA-3′,
hy3: 5′-GGAGAAGGAA CAAAGAAATC TGTAACTGGT TGTGAT-3′,
hy4: 5′-GGGTTGTATA CCAACTTTAG TGACAC-3′,
hy5: 5′-GGGAGACAAT GTTAAATCAA CTTAACAATA A-3′,
human u6: 5′-CACGAATTTG CGTGTCATCC TT-3′.
Immunoprecipitations
Mouse ES cells were maintained as described above. HEK293 cells were maintained in Dulbecco’s modified Eagle medium (Invitrogen) supplemented with 10% fetal bovine serum (FBS) and 2 mM L-glutamine. Cells were sonicated in NET-2 (40 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.1% NP-40) containing 1 mM phenylmethylsulfonyl fluoride and 1x protease inhibitor cocktail (Roche Applied Science). After clearing by centrifugation at 100 000 × g in a Beckman TLA100.3 rotor for 20 min at 4 °C, ES cell lysates were incubated as described12 with antibodies bound to Protein A Sepharose (GE Healthcare). Antibodies used were rabbit anti-Ro60,18 human anti-La (gift of J. Harley, Cincinnati Children's Hospital), anti-TMG (Oncogene Science), and anti-Sm (Y12; gift of Mei-Di Shu and Joan Steitz, Yale University). HEK293 lysates were incubated with mouse anti-human Ro60 (1F2, Novus Biologicals) and isotype control IgG (M2AK), bound to Dynabeads Protein G (Invitrogen).
siRNA transfections
Adult human epidermal keratinocytes (Invitrogen) were maintained in EpiLife medium with 60 μM calcium and Human Keratinocyte Growth Supplement (Invitrogen). Cells were transfected with Lipofectamine RNAiMAX (Invitrogen) using a modified version of the manufacturer’s protocol for reverse transfection. Anti-Ro60 siRNA (siGENOME SMARTpool, Dharmacon) or non-targeting control siRNA (Ambion) was diluted in 125 μl of Opti-MEM (Invitrogen) in 10 cm dishes for a final concentration of 40 nM after addition of cells. Five μl of Lipofectamine RNAiMAX was mixed with 125 μl Opti-MEM, added to the dish and incubated for 15 min at room temperature. Two hundred and fifty thousand trypsinized keratinocytes were then added to each well in 1.75 ml of growth media. Fresh media was added the following day, and the cells incubated for 72 h before harvesting. Cells were sonicated in NET-2 containing 1 mM phenylmethylsulfonyl fluoride, 200 u/ml RNaseOUT (Invitrogen), and 1x protease inhibitor cocktail and cleared by sedimenting for 10 min at 4 °C in a microcentrifuge. Ten percent of the lysate was removed for western blotting using a monoclonal anti-mouse Ro60 antibody as described.23 RNA was extracted from the remaining lysate using phenol:chloroform:isoamyl alchohol, precipitated with ethanol, and subjected to northern blotting as described above.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Hong Shi for technical assistance. This work was supported by NIH grant R01 GM073863 (to SLW) and National Basic Research Program of China grant 2010CB912401 (to H-WW). DWT is an NSF Graduate Research Fellow.
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/26166
References
- 1.Clark G, Reichlin M, Tomasi TB., Jr. Characterization of a soluble cytoplasmic antigen reactive with sera from patients with systemic lupus erythmatosus. J Immunol. 1969;102:117–22. [PubMed] [Google Scholar]
- 2.Alspaugh MA, Tan EM. Antibodies to cellular antigens in Sjögren’s syndrome. J Clin Invest. 1975;55:1067–73. doi: 10.1172/JCI108007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lerner MR, Boyle JA, Hardin JA, Steitz JA. Two novel classes of small ribonucleoproteins detected by antibodies associated with lupus erythematosus. Science. 1981;211:400–2. doi: 10.1126/science.6164096. [DOI] [PubMed] [Google Scholar]
- 4.Lindop R, Arentz G, Thurgood LA, Reed JH, Jackson MW, Gordon TP. Pathogenicity and proteomic signatures of autoantibodies to Ro and La. Immunol Cell Biol. 2012;90:304–9. doi: 10.1038/icb.2011.108. [DOI] [PubMed] [Google Scholar]
- 5.Hendrick JP, Wolin SL, Rinke J, Lerner MR, Steitz JA. Ro small cytoplasmic ribonucleoproteins are a subclass of La ribonucleoproteins: further characterization of the Ro and La small ribonucleoproteins from uninfected mammalian cells. Mol Cell Biol. 1981;1:1138–49. doi: 10.1128/mcb.1.12.1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kato N, Hoshino H, Harada F. Nucleotide sequence of 4.5S RNA (C8 or hY5) from HeLa cells. Biochem Biophys Res Commun. 1982;108:363–70. doi: 10.1016/0006-291X(82)91875-7. [DOI] [PubMed] [Google Scholar]
- 7.Wolin SL, Steitz JA. Genes for two small cytoplasmic Ro RNAs are adjacent and appear to be single-copy in the human genome. Cell. 1983;32:735–44. doi: 10.1016/0092-8674(83)90059-4. [DOI] [PubMed] [Google Scholar]
- 8.O’Brien CA, Harley JB. A subset of hY RNAs is associated with erythrocyte Ro ribonucleoproteins. EMBO J. 1990;9:3683–9. doi: 10.1002/j.1460-2075.1990.tb07580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maraia RJ, Sasaki-Tozawa N, Driscoll CT, Green ED, Darlington GJ. The human Y4 small cytoplasmic RNA gene is controlled by upstream elements and resides on chromosome 7 with all other hY scRNA genes. Nucleic Acids Res. 1994;22:3045–52. doi: 10.1093/nar/22.15.3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maraia R, Sakulich AL, Brinkmann E, Green ED. Gene encoding human Ro-associated autoantigen Y5 RNA. Nucleic Acids Res. 1996;24:3552–9. doi: 10.1093/nar/24.18.3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Teunissen SW, Kruithof MJ, Farris AD, Harley JB, Venrooij WJ, Pruijn GJ. Conserved features of Y RNAs: a comparison of experimentally derived secondary structures. Nucleic Acids Res. 2000;28:610–9. doi: 10.1093/nar/28.2.610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wolin SL, Steitz JA. The Ro small cytoplasmic ribonucleoproteins: identification of the antigenic protein and its binding site on the Ro RNAs. Proc Natl Acad Sci U S A. 1984;81:1996–2000. doi: 10.1073/pnas.81.7.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pruijn GJM, Slobbe RL, van Venrooij WJ. Analysis of protein--RNA interactions within Ro ribonucleoprotein complexes. Nucleic Acids Res. 1991;19:5173–80. doi: 10.1093/nar/19.19.5173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Green CD, Long KS, Shi H, Wolin SL. Binding of the 60-kDa Ro autoantigen to Y RNAs: evidence for recognition in the major groove of a conserved helix. RNA. 1998;4:750–65. doi: 10.1017/S1355838298971667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stein AJ, Fuchs G, Fu C, Wolin SL, Reinisch KM. Structural insights into RNA quality control: the Ro autoantigen binds misfolded RNAs via its central cavity. Cell. 2005;121:529–39. doi: 10.1016/j.cell.2005.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O’Brien CA, Wolin SL. A possible role for the 60-kD Ro autoantigen in a discard pathway for defective 5S rRNA precursors. Genes Dev. 1994;8:2891–903. doi: 10.1101/gad.8.23.2891. [DOI] [PubMed] [Google Scholar]
- 17.Shi H, O’Brien CA, Van Horn DJ, Wolin SL. A misfolded form of 5S rRNA is complexed with the Ro and La autoantigens. RNA. 1996;2:769–84. [PMC free article] [PubMed] [Google Scholar]
- 18.Chen X, Smith JD, Shi H, Yang DD, Flavell RA, Wolin SL. The Ro autoantigen binds misfolded U2 small nuclear RNAs and assists mammalian cell survival after UV irradiation. Curr Biol. 2003;13:2206–11. doi: 10.1016/j.cub.2003.11.028. [DOI] [PubMed] [Google Scholar]
- 19.Sim S, Wolin SL. Emerging roles for the Ro 60-kDa autoantigen in noncoding RNA metabolism. Wiley Interdiscip Rev RNA. 2011;2:686–99. doi: 10.1002/wrna.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peek R, Pruijn GJM, van der Kemp AJ, van Venrooij WJ. Subcellular distribution of Ro ribonucleoprotein complexes and their constituents. J Cell Sci. 1993;106:929–35. doi: 10.1242/jcs.106.3.929. [DOI] [PubMed] [Google Scholar]
- 21.Labbé JC, Hekimi S, Rokeach LA. The levels of the RoRNP-associated Y RNA are dependent upon the presence of ROP-1, the Caenorhabditis elegans Ro60 protein. Genetics. 1999;151:143–50. doi: 10.1093/genetics/151.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen X, Quinn AM, Wolin SL. Ro ribonucleoproteins contribute to the resistance of Deinococcus radiodurans to ultraviolet irradiation. Genes Dev. 2000;14:777–82. [PMC free article] [PubMed] [Google Scholar]
- 23.Xue D, Shi H, Smith JD, Chen X, Noe DA, Cedervall T, Yang DD, Eynon E, Brash DE, Kashgarian M, et al. A lupus-like syndrome develops in mice lacking the Ro 60-kDa protein, a major lupus autoantigen. Proc Natl Acad Sci U S A. 2003;100:7503–8. doi: 10.1073/pnas.0832411100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bayfield MA, Yang R, Maraia RJ. Conserved and divergent features of the structure and function of La and La-related proteins (LARPs) Biochim Biophys Acta. 2010;1799:365–78. doi: 10.1016/j.bbagrm.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sim S, Yao J, Weinberg DE, Niessen S, Yates JR, 3rd, Wolin SL. The zipcode-binding protein ZBP1 influences the subcellular location of the Ro 60-kDa autoantigen and the noncoding Y3 RNA. RNA. 2012;18:100–10. doi: 10.1261/rna.029207.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sim S, Weinberg DE, Fuchs G, Choi K, Chung J, Wolin SL. The subcellular distribution of an RNA quality control protein, the Ro autoantigen, is regulated by noncoding Y RNA binding. Mol Biol Cell. 2009;20:1555–64. doi: 10.1091/mbc.E08-11-1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fuchs G, Stein AJ, Fu C, Reinisch KM, Wolin SL. Structural and biochemical basis for misfolded RNA recognition by the Ro autoantigen. Nat Struct Mol Biol. 2006;13:1002–9. doi: 10.1038/nsmb1156. [DOI] [PubMed] [Google Scholar]
- 28.Hogg JR, Collins K. Human Y5 RNA specializes a Ro ribonucleoprotein for 5S ribosomal RNA quality control. Genes Dev. 2007;21:3067–72. doi: 10.1101/gad.1603907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen X, Wurtmann EJ, Van Batavia J, Zybailov B, Washburn MP, Wolin SL. An ortholog of the Ro autoantigen functions in 23S rRNA maturation in D. radiodurans. Genes Dev. 2007;21:1328–39. doi: 10.1101/gad.1548207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen X, Taylor DW, Fowler CC, Galan JE, Wang HW, Wolin SL. An RNA degradation machine sculpted by Ro autoantigen and noncoding RNA. Cell. 2013;153:166–77. doi: 10.1016/j.cell.2013.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Christov CP, Gardiner TJ, Szüts D, Krude T. Functional requirement of noncoding Y RNAs for human chromosomal DNA replication. Mol Cell Biol. 2006;26:6993–7004. doi: 10.1128/MCB.01060-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krude T, Christov CP, Hyrien O, Marheineke KY. Y RNA functions at the initiation step of mammalian chromosomal DNA replication. J Cell Sci. 2009;122:2836–45. doi: 10.1242/jcs.047563. [DOI] [PubMed] [Google Scholar]
- 33.Zhang AT, Langley AR, Christov CP, Kheir E, Shafee T, Gardiner TJ, Krude T. Dynamic interaction of Y RNAs with chromatin and initiation proteins during human DNA replication. J Cell Sci. 2011;124:2058–69. doi: 10.1242/jcs.086561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Christov CP, Trivier E, Krude T. Noncoding human Y RNAs are overexpressed in tumours and required for cell proliferation. Br J Cancer. 2008;98:981–8. doi: 10.1038/sj.bjc.6604254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Collart C, Christov CP, Smith JC, Krude T. The midblastula transition defines the onset of Y RNA-dependent DNA replication in Xenopus laevis. Mol Cell Biol. 2011;31:3857–70. doi: 10.1128/MCB.05411-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garcia EL, Onafuwa-Nuga A, Sim S, King SR, Wolin SL, Telesnitsky A. Packaging of host mY RNAs by murine leukemia virus may occur early in Y RNA biogenesis. J Virol. 2009;83:12526–34. doi: 10.1128/JVI.01219-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gardiner TJ, Christov CP, Langley AR, Krude T. A conserved motif of vertebrate Y RNAs essential for chromosomal DNA replication. RNA. 2009;15:1375–85. doi: 10.1261/rna.1472009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Slade D, Radman M. Oxidative stress resistance in Deinococcus radiodurans. Microbiol Mol Biol Rev. 2011;75:133–91. doi: 10.1128/MMBR.00015-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wurtmann EJ, Wolin SL. A role for a bacterial ortholog of the Ro autoantigen in starvation-induced rRNA degradation. Proc Natl Acad Sci U S A. 2010;107:4022–7. doi: 10.1073/pnas.1000307107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tanaka M, Earl AM, Howell HA, Park MJ, Eisen JA, Peterson SN, Battista JR. Analysis of Deinococcus radiodurans’s transcriptional response to ionizing radiation and desiccation reveals novel proteins that contribute to extreme radioresistance. Genetics. 2004;168:21–33. doi: 10.1534/genetics.104.029249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Labbé JC, Burgess J, Rokeach LA, Hekimi S. ROP-1, an RNA quality-control pathway component, affects Caenorhabditis elegans dauer formation. Proc Natl Acad Sci U S A. 2000;97:13233–8. doi: 10.1073/pnas.230284297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hardwick SW, Gubbey T, Hug I, Jenal U, Luisi BF. Crystal structure of Caulobacter crescentus polynucleotide phosphorylase reveals a mechanism of RNA substrate channelling and RNA degradosome assembly. Open Biol. 2012;2:120028. doi: 10.1098/rsob.120028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Symmons MF, Williams MG, Luisi BF, Jones GH, Carpousis AJ. Running rings around RNA: a superfamily of phosphate-dependent RNases. Trends Biochem Sci. 2002;27:11–8. doi: 10.1016/S0968-0004(01)01999-5. [DOI] [PubMed] [Google Scholar]
- 44.Eliscovich C, Buxbaum AR, Katz ZB, Singer RH. mRNA on the move: the road to its biological destiny. J Biol Chem. 2013;288:20361–8. doi: 10.1074/jbc.R113.452094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chao JA, Patskovsky Y, Patel V, Levy M, Almo SC, Singer RH. ZBP1 recognition of beta-actin zipcode induces RNA looping. Genes Dev. 2010;24:148–58. doi: 10.1101/gad.1862910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Köhn M, Lederer M, Wächter K, Hüttelmaier S. Near-infrared (NIR) dye-labeled RNAs identify binding of ZBP1 to the noncoding Y3-RNA. RNA. 2010;16:1420–8. doi: 10.1261/rna.2152710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen HW, Rainey RN, Balatoni CE, Dawson DW, Troke JJ, Wasiak S, Hong JS, McBride HM, Koehler CM, Teitell MA, et al. Mammalian polynucleotide phosphorylase is an intermembrane space RNase that maintains mitochondrial homeostasis. Mol Cell Biol. 2006;26:8475–87. doi: 10.1128/MCB.01002-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bouffard P, Barbar E, Brière F, Boire G. Interaction cloning and characterization of RoBPI, a novel protein binding to human Ro ribonucleoproteins. RNA. 2000;6:66–78. doi: 10.1017/S1355838200990277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fabini G, Raijmakers R, Hayer S, Fouraux MA, Pruijn GJ, Steiner G. The heterogeneous nuclear ribonucleoproteins I and K interact with a subset of the ro ribonucleoprotein-associated Y RNAs in vitro and in vivo. J Biol Chem. 2001;276:20711–8. doi: 10.1074/jbc.M101360200. [DOI] [PubMed] [Google Scholar]
- 50.Fouraux MA, Bouvet P, Verkaart S, van Venrooij WJ, Pruijn GJ. Nucleolin associates with a subset of the human Ro ribonucleoprotein complexes. J Mol Biol. 2002;320:475–88. doi: 10.1016/S0022-2836(02)00518-1. [DOI] [PubMed] [Google Scholar]
- 51.Tarn W-Y, Yario TA, Steitz JA. U12 snRNA in vertebrates: evolutionary conservation of 5′ sequences implicated in splicing of pre-mRNAs containing a minor class of introns. RNA. 1995;1:644–56. [PMC free article] [PubMed] [Google Scholar]
- 52.Perreault J, Perreault J-P, Boire G. Ro-associated Y RNAs in metazoans: evolution and diversification. Mol Biol Evol. 2007;24:1678–89. doi: 10.1093/molbev/msm084. [DOI] [PubMed] [Google Scholar]
- 53.van Gelder CWG, Thijssen JPHM, Klaassen ECJ, Sturchler C, Krol A, van Venrooij WJ, Pruijn GJ. Common structural features of the Ro RNP associated hY1 and hY5 RNAs. Nucleic Acids Res. 1994;22:2498–506. doi: 10.1093/nar/22.13.2498. [DOI] [PMC free article] [PubMed] [Google Scholar]