Abstract
RNA editing by ADARs can change the coding potential of protein-coding mRNAs. So far, this type of RNA editing has mainly been shown to affect RNAs expressed in the nervous system with much lower editing levels being observed in other tissues.
The actin crosslinking proteins filamin α and filamin β are widely expressed in most tissues. The mRNAs encoding either protein are edited at the same position leading to a conserved Q to R exchange in both proteins. Using bar-coded next generation sequencing, we show that editing of filamin α is most abundant in the gastrointestinal tract and only to a lesser extent in the nervous system. Using knockout mice, we show that ADARB1 (ADAR2) is responsible for the majority of FLNA editing, while ADAR1 can edit filamin α mRNA in some tissues quite efficiently. Interestingly, editing levels of filamin α and β do not follow the same trend across tissues, suggesting a substrate-specific regulation of editing.
Keywords: RNA-editing, ADARs, adenosine deamination, filamin A, mouse, development
Introduction
Adenosine to inosine deamination of RNAs is a conserved RNA-editing mechanism that is widespread in metazoa. The enzymatic reaction is performed by adenosine deaminases that act on RNA (ADARs) that recognize structured and double-stranded (ds) RNA.1 Inosines are recognized as guanosines by many cellular processes and, therefore, editing can alter the amino acid sequence of encoded proteins, change splice patterns, alter RNA-folding, or modify binding sites for associated proteins.2 In all metazoa tested so far, ADARs have been shown to be essential for development and normal life.2,3
Mice have two active ADAR enzymes, ADAR1 and ADARB1 (generally referred to as ADAR2), which exhibit overlapping, yet distinct substrate specificity.4-6 So far, editing events that lead to an amino acid exchange in the encoded protein were primarily observed in RNAs transcribed in neuronal tissues that encode neuronal receptors and ion channels.7 In most cases, these editing events have significant impact on protein structure and function.8-13 The majority of editing events observed in neuronally expressed protein coding regions are mediated by ADAR2. Editing events in protein coding regions are less abundant outside the nervous system and may also be attributed to ADAR1 activity.14,15 Moreover, abundant editing is found in non-coding regions of mRNAs such as introns and UTRs where repetitive elements lead to the formation of extensive double-stranded regions.16-18 Editing in UTRs can be performed by both ADAR1 and ADAR2.
Consistently, lack of ADAR1 or ADAR2 leads to different phenotypes. ADAR1−/− mice die at embryonic stage (E12.5) due to liver disintegration and defects in hematopoiesis, whereas ADAR2−/− mice are prone to seizures and die shortly after birth.12,19-21 Altered editing patterns are also found in patients suffering from psychiatric disorders, neurodegenerative diseases, or malignant gliomas.22-24 Following editing patterns throughout development, it was also shown that editing frequencies in the nervous system increase, despite a constant expression of ADARs.25
Comprehensive computational and sequence analysis has revealed novel editing events in the coding region of mRNAs outside neuronal tissues.26,27 One editing event that is found highly conserved among vertebrates affects the widely expressed cytoskeletal protein Filamin A (FLNA).26,28
Filamin A is a dimeric protein that binds and crosslinks F-actin.29 Besides its function as an actin crosslinker, novel roles for Filamin A have been described that can be attributed to many interactions at its C-terminal part. More than 90 binding partners including channels, receptors, intracellular signaling molecules, and even transcription factors have been reported to interact with Filamin A.30 Thus, Filamin A may anchor membrane receptors to the actin cytoskeleton and regulate their precise location and transport.31-33 Moreover, Filamin A also acts as a co-localization factor for signaling pathways and a mechanical element for caveolae and membrane ruffle formation.34-39
The Filamin A transcript is edited at a single position in exon 42, which leads to a Q/R substitution in Ig-repeat 22 of the encoded protein.26 This site is located in the very interactive rod 2 domain, implying that it could affect the FLNA interactome. The editing site and sequence context have been found conserved from birds to mammals suggesting that FLNA editing must have functional consequences, which are currently not known.26 Moreover, the very same position is also edited in the highly conserved filamin B pre-mRNA, also giving rise to a Q/R substitution in repeat 22 of the encoded protein.27 To get some clues on the functional relevance of FLNA editing, to determine the extent of editing in different tissues and developmental stages, and to determine the enzyme responsible for editing we applied Illumina deep sequencing to FLNA amplicons of spliced mRNAs isolated from different tissues throughout development. Based on these data we generated the spatio-temporal profile of Filamin A editing in mature mRNAs.
Results and Discussion
Previous studies have shown that A to I editing levels for protein-coding targets expressed in the mammalian brain increase gradually during development, from embryo to adulthood.25,40 To test whether this trend would also apply to Filamin A and whether editing can also be found in other organs of non-neuronal origin we determined FLNA editing levels using Illumina deep sequencing. Using bar-coded amplicons of FLNA cDNA we compared editing levels in various tissues from developmental stages E15, P0, P20, and adult (> 4 mo) mice. Depending on the sample, read depths varied from a minimum of 450 reads to 961000 reads maximum with an average of 161500 reads per sample. As the overall mistakes in base calling were around 0.3%, A to G transitions that were lower than 0.5% were not considered for further analyses.
Due to the small size of E15 embryos we separated head, brain, whole thorax and abdomen, and abdomen only in these early stages. In contrast to the majority of mammalian protein coding targets of A to I editing, filamin A mRNA showed highest editing in the abdomen, reaching up to 12%, while editing levels in the brain were around 2%, similar to the editing levels previously observed for the R/G site in Gria225 (Fig. 1A; Table 1).
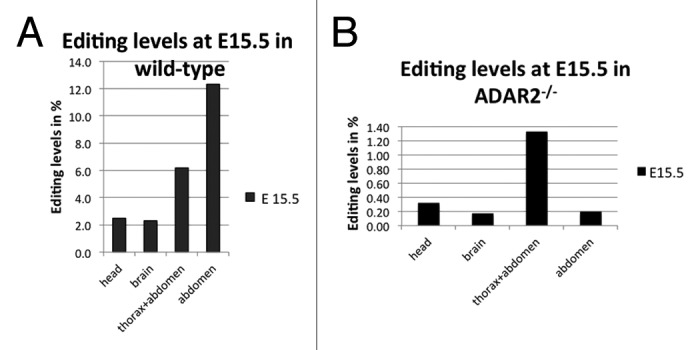
Figure 1. Editing levels in filamin α in wild-type (A) and ADAR2-knockout (B) mice at E15.5. (A) In the embryo, editing levels are highest in the abdomen and thorax, reaching up to 12% while editing in the brain only reaches 2%. (B) In ADAR2−/− animals, editing levels are dramatically reduced, only reaching 1.3% in thorax and abdomen while editing in all other parts is around the base calling error of 0.3%.
Table 1. Editing levels at E 15.5 in wild-type mice.
| Region | Editing in % |
|---|---|
|
head |
2.5 (145k) |
|
brain |
2.3 (385k) |
|
thorax+abdomen |
6.2 (73k) |
| abdomen | 12.3 (9k) |
E 15.5 embryos were dissected into the regions indicated and cDNAs of the FLNA mRNA were amplified, bar coded, and sequenced on an Illumina GA II platform. Editing levels are indicated in %. Read depth is indicated in 1000 (k) in brackets.
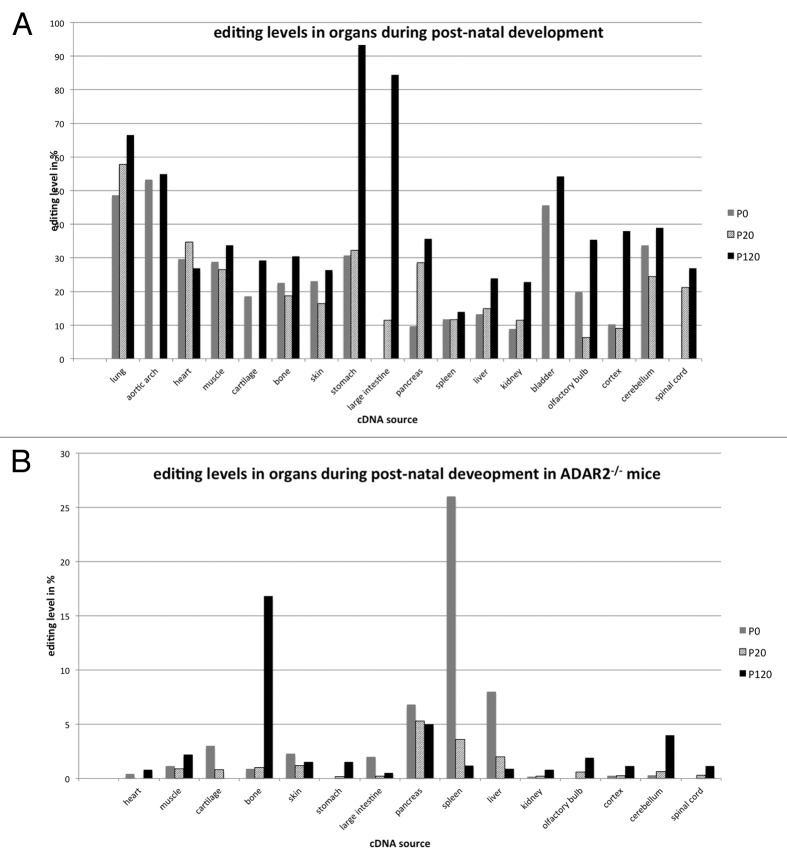
Filamin A editing levels increased with age and, hence, higher editing levels were observed in young and adult mice. Most interestingly, the overall frequency distribution of editing events varied dramatically from previous studies on other substrates.41 Our analysis revealed that editing levels of Filamin A mRNA are highest in 4-mo-old mice with stomach showing 95% A–G transitions, large intestine (86% A–G transitions), and lung (68% A–G transitions). Nevertheless, editing levels were also high in different parts of the brain (30–40% of A–G transitions), as well as in heart (56% A–G transitions), and bladder (55% A-G transitions) (Fig. 2A; Table 2). To verify the high editing levels observed in non-neuronal tissues and to compensate for any inter-individual differences, we have tested editing levels in the gastro-intestinal tract by Sanger sequencing on three additional mice. These data independently confirmed the very high editing levels in stomach (~85–90%), large intestine (~80%), and heart (~60%) (data not shown). Thus, while direct Sanger sequencing generally detected slightly less editing events the general observation of very high editing levels in the gastrointestinal tract could be verified. This also demonstrates that inter-individual differences are low, as already discussed elsewhere.42
Figure 2. Post-natal editing levels in filamin in wild-type (A) and ADAR2-knockout animals (B). (A) In the wild-type, post-natal editing levels in filamin A increase from P0 to P120. Editing levels are highest in the gastrointestinal tract, reaching more than 90% in the stomach. Editing in neuronal tissue is comparatively modest, reaching almost 40% in the cerebellum. (B) In ADAR2−/− animals, editing rates are strongly reduced and decrease from P0 to P120 in spleen pancreas and liver while editing increases in the nervous system. Highest editing levels are reached in spleen, pancreas, liver, and bone, reaching at least 25% in the spleen.
Table 2. Editing levels in filamin A pre-mRNA during post-natal development.
| Organ | P0 | P20 | P120 |
|---|---|---|---|
|
lung |
48.6 (961k) |
57.8 (331k) |
66.5 (176k) |
|
aortic arch |
53.3 (550k) |
nd |
54.9 (Sanger) |
|
heart |
29.6 (595k) |
34.7 (16k) |
26.9 (134k) |
|
muscle |
28.9 (213k) |
26.6 (13k) |
33.7 (1k) |
|
cartilage |
18.5 (420k) |
nd |
29.0 (27k) |
|
bone |
22.6 (102k) |
18.7 (354k) |
30.5 (95k) |
|
skin |
23.1 (142k) |
16.3 (140k) |
26.3 (9k) |
|
stomach |
30.7 (163k) |
32.2 (135k) |
93.4 (146k) |
|
small intestine |
nd |
2.3 (127k) |
15.9 (74k) |
|
large intestine |
nd |
11.5 (24k) |
84.5 (138k) |
|
pancreas |
9.7 (414k) |
28.6 (558k) |
35.6 (166k) |
|
spleen |
11.7 (488k) |
11.6 (48k) |
13.9 (19k) |
|
Liver |
13.3 (55k) |
14.9 (2k) |
24.0 (0.5k) |
|
kidney |
8.8 (76k) |
11.5 (300k) |
22.8 (6k) |
|
bladder |
45.6 (392k) |
nd |
54.2 (114k) |
|
brown fat |
7.1 (228k) |
nd |
nd |
|
eyes |
11.1 (431k) |
nd |
51.3 (355k) |
|
olfactory bulb |
19.8 (288k) |
6.3 (29k) |
35.3 (28k) |
|
cortex |
10.3 (143k) |
9.0 (10k) |
38.0 (127k) |
|
hippocampus |
nd |
32.6 (230k) |
38.8 (41k) |
|
thalamus/hypot. |
nd |
33.4 (119k) |
32.0 (12k) |
|
cerebellum |
33.7 (5k) |
24.5 (28k) |
38.0 (111k) |
| spinal cord | nd | 21.2 (6k) | 27.0 (5k) |
cDNAs prepared from the organs indicated were amplified, bar-coded, and sequenced on an Illumina GAII platform. Editing levels were calculated for each tissue and are listed in %. Nd, not determined. Read depth is indicated in 1000 (k) in brackets.
FLNA editing levels had also been determined for a few human embryonic and adult tissues.43 The limited number of human tissues investigated makes a direct comparison of human and mouse editing levels difficult. However, in humans—like in the mouse—embryonic editing levels were very low. In adult human brain, liver, kidney, and spleen editing reached about 25%. In the mouse, in contrast, editing levels of up to 40% were observed in the brain, while editing levels in kidney, liver and spleen were in a similar range between 15–25%. Whether humans also show high editing levels in the gastrointestinal tract remains to be determined.
ADAR1 and ADAR2 have different, yet overlapping target specificities.6,44 Moreover, ADAR1 and ADAR2 show different but partially overlapping expression patterns.45-47 We therefore wanted to determine which enzyme would be largely responsible for the observed editing events. In a previous study, we had already shown that editing of FLNA in post-natal brain can be achieved by both ADAR1 and ADAR2.4
We therefore performed deep sequencing of organ and development-specific cDNA samples derived from ADAR2−/− mice and embryos. In the absence of ADAR2, editing levels were strongly reduced, indicating that ADAR2 is the main enzyme responsible for editing of FLNA RNA. In embryos lacking ADAR2, the rate of A to G transitions dropped below the background level in head and brain (Fig. 1B; Table 3). The only significant editing levels of 1.2% were detectable in samples derived from thorax and abdomen. In post-natal tissues only few organs showed editing levels well above 1% (Fig. 2B; Table 4). The highest editing levels were detected in bone, pancreas, spleen, and liver. In the spleen of P0 animals editing reached 26%, followed by 17% editing detected in adult bone. Editing in liver varied from 8% to 1% while editing in pancreas varied from 7 to 5% between P0 and P120, respectively (Fig. 2B; Table 4). Again, editing in neuronal tissues was lower than in other organs but reached at least 4% in adult cerebellum.
Table 3. Editing levels at E 15.5 in ADAR2−/− mice.
| Region | Editing in % |
|---|---|
|
head |
0.3 (614k) |
|
brain |
0.2 (305k) |
|
thorax+abdomen |
1.3 (243k) |
| abdomen | 0.2 (675k) |
E 15.5 embryos were dissected into the regions indicated and cDNAs of the FLNA mRNA were amplified, bar coded, and sequenced on an Illumina GA II platform. Editing levels are indicated in %. As background basecall errors lay around 0.3%, significant editing levels can only be identified in thorax+abdomen. Read depth is indicated in 1000 (k) in brackets.
Table 4. Editing levels in filamin A pre-mRNA during post-natal development in ADAR2−/− mice.
| Organ | P0 | P20 | P120 |
|---|---|---|---|
|
lung |
0.2 (930k) |
0.2 (76k) |
0.2 (26k) |
|
heart |
0.4 (77k) |
nd |
0.8 (8k) |
|
muscle |
1.2 (401k) |
0.9 (13k) |
2.2 (6k) |
|
cartilage |
3.0 (61k) |
0.8 (104k) |
nd |
|
bone |
0.9 (36k) |
1.0 (3k) |
16.8 (28k) |
|
Skin |
2.3 (57k) |
1.2 (137k) |
1.5 (84k) |
|
stomach |
nd |
0.2 (56k) |
1.5 (8k) |
|
small intestine |
nd |
3.0 (0.4k) |
3.3 (23k) |
|
large intestine |
2.0 (60k) |
0.2 (66k) |
0.5 (25k) |
|
pancreas |
6.8 (1k) |
5.3 (2k) |
5.0 (3k) |
|
spleen |
26.0 (31k) |
3.6 (0.4k) |
1.2 (32k) |
|
liver |
8.0 (86k) |
2.0 (0.5k) |
0.9 (9k) |
|
kidney |
0.2 (297k) |
0.2 (51k) |
0.8 (34k) |
|
bladder |
0.1 (688k) |
0.1 (159k) |
0.4 (24k) |
|
brown fat |
nd |
nd |
1.4 (8k) |
|
eyes |
0.2 (55k) |
0.4 (30k) |
nd |
|
olfactory bulb |
nd |
0.6 (181k) |
1.9 (66k) |
|
cortex |
0.2 (212k) |
0.2 (208k) |
1.1 (50k) |
|
hippocampus |
nd |
0.8 (83k) |
1.2 (117k) |
|
thalamus/hypot. |
nd |
0.8 (3k) |
6.1 (458k) |
|
cerebellum |
0.3 (351k) |
0.6 (71k) |
4.0 (45k) |
| spinal cord | nd | 0.3 (39k) | 1.1 (145k) |
cDNAs prepared from the organs indicated were amplified, bar-coded, and sequenced on an Illumina GAII platform. Editing levels were calculated for each tissue and are listed in %. Nd, not determined. Read depth is indicated in 1000 (k) in brackets.
Taken together, our data indicates that ADAR2 is the primary enzyme responsible for editing of FLNA mRNA. Nonetheless, in some tissues and some developmental stages editing by ADAR1 still reaches significant levels, even in the absence of ADAR2. In spleen, for instance, editing levels peak at 14% at p120 in the wild type, but can increase to 26% at P0 in ADAR2−/− knockout animals. Also, in bone, editing levels reach 30% in the wild type vs. 16% in the ADAR2−/− knockout.
We have obviously considered the possibility of barcoding or basecalling problems. We have therefore confirmed the most important samples derived from ADAR2−/− samples by conventional Sanger sequencing (data not shown). Increased editing in the absence of one ADAR enzyme suggests competitive binding of different ADARs to one and the same site. Lack of one ADAR would then facilitate access to an editing site for another ADAR enzyme that might act more efficiently at this site. Similar phenomena had been described before.4
Both, in the presence and absence of ADAR2, editing levels in most tissues increase with age. However, in some non-neuronal tissues like large intestine, cartilage, skin, liver, or spleen, editing levels decreased from P0 to P120; sometimes even below the levels of basecalling errors, which we defined around 0.3% (compare Fig. 2A and B).
Recently, it was noted that the mRNA encoding the paralogous protein filamin B is edited at exactly the same position as filamin A, also leading to a Q to R exchange in Ig repeat 22.27 We therefore wanted to test whether editing levels in FLNB mRNA would follow the same trend as editing levels in FLNA. We therefore amplified and sequenced the FLNB mRNA from the same cDNAs used for the analysis of FLNA editing. cDNAs from stomach, large intestine, heart, bladder, and lung were analyzed using Sanger sequencing. Interestingly, while FLNA editing levels were highest in stomach, followed by large intestine, lung, bladder, and heart, the situation was different in filamin B. FLNB editing was highest in heart, followed by stomach, lung, large intestine, and bladder (Fig. 3A; Table 5). As the very same cDNA samples to determine editing in filamin A and filamin B mRNAs were used, the observed differences in editing levels cannot reflect differences in expression of ADARs in these samples. We therefore compared the ratios of FLNA and ADAR2 expression in a few isolated tissues (Fig. 3B). However, no clear trend between editing levels and FLNA/ADAR2 ratios could be observed. Stomach, showing very high editing levels had a very high FLNA to ADAR2 ratio while cerebellum with also high editing levels had a low FLNA to ADAR2 ratio. It therefore appears more likely that other factors that can enhance ore repress editing in a tissue- and substrate-specific manner will show different expression levels in those tissues that have a different trend for FLNA and FLNB editing. In fact, we have recently shown that some RNA-binding proteins can selectively modulate editing of some substrates, most likely by competitive binding to editing sites.48,49 Therefore, if some RNA-binding proteins can specifically interact with the editing sites in either FLNA or FLNB pre-mRNAs, editing in either one of these substrates might be specifically repressed.
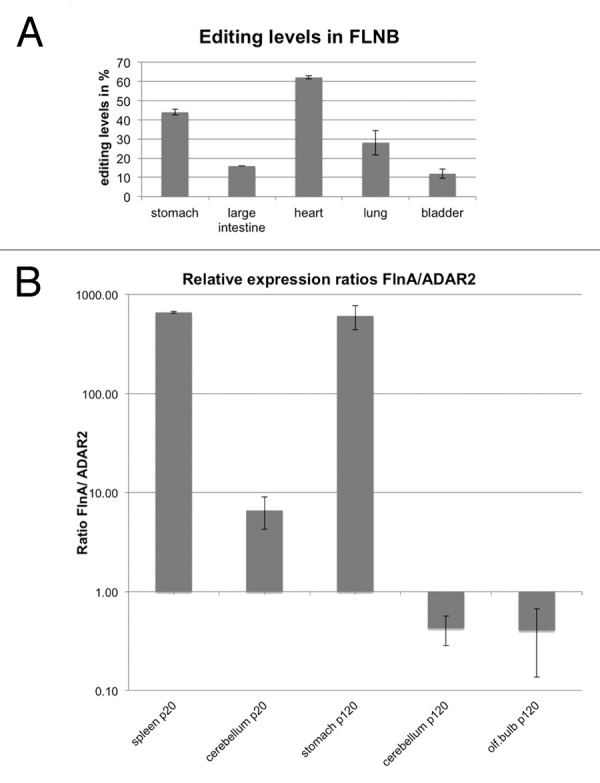
Figure 3. Different levels of filamin A and filamin B editing. (A) Filamin B cDNAs from selected tissues used in the analysis shown in Figure 2 were amplified, subjected to Sanger sequencing, and editing levels were determined by calculating A to G peak ratios. Filamin B editing was highest in heart, followed (in decreasing order) by stomach, lung, large intestine, and bladder. Therefore, the extent of filamin A and filamin B editing does not correlate in these tissues. (B) Expression ratios of filamin A and ADAR2 do not correlate with editing levels. To determine whether the ratios of substrate to enzyme might govern editing levels, expression levels of filamin A and ADAR2 were determined relative to actin mRNA using qPCR. Highest substrate (FLNA) to ADAR2 ratio was observed in spleen of P20 animals and in stomach of P120 animals. These two tissues show 10 and 90% editing levels, respectively. Adult neuronal tissues, in contrast, showed a rather low substrate to enzyme ratio but only exhibited moderate editing levels of 30–40%. Hence, differences in editing levels cannot be explained by different ratios of substrate RNA to editing enzyme.
Table 5. Editing levels in FLNB at P120 in wild-type mice.
| Organ | Editing level in % |
|---|---|
|
stomach |
44 |
|
large intestine |
16 |
|
heart |
62 |
|
lung |
28 |
| bladder | 11.50 |
cDNAs prepared from the organs indicated were amplified and sequenced by Sanger sequencing. Peak heights were measured at the editing site and the percentage of editing was calculated by dividing the G peak height by the sum of the A and G peak heights.
In summary, our comprehensive analysis of editing in the filamin A encoding mRNA shows very high editing levels outside the nervous system. Our study shows further that FLNA editing is mainly achieved by ADAR2 but that in some cases ADAR1 can efficiently compensate for ADAR2. High editing levels in some tissues in the absence of ADAR2 might possibly result from facilitating access of ADAR1.4 However, clarification of this point will require further experimentation. Most interestingly, our finding that editing levels in FLNA are highest outside neuronal tissues suggests further novel and unexplored roles for RNA editing. Given the many functions of filamin A, ranging from actin crosslinking to receptor interaction it may be too premature to speculate about the function of FLNA editing. However, editing may well alter the interaction repertoire of FLNA. With editing levels as high as 90% in the gastrointestinal tract of adult mice dropping to a mere 4–5% of editing in ADAR2−/− mice and editing levels of more than 12% in the abdomen of wild-type embryos it is unlikely that the high editing levels in the gastrointestinal tract represent an ADAR1-mediated anti-inflammatory response. Instead, a constitutive function for the edited version of FLNA seems most likely.
Materials and Methods
Tissue isolation
Adarb1−/− (ADAR2)-knockout mice were a kind gift of Peter Seeburg (MPI Heidelberg).12 Tissues expressing both active versions of ADAR were obtained from c57bl/6 wt mouse. Mice were sacrificed and organs were dissected and kept in liquid nitrogen. The tissues were macerated using 10% SDS and 0.5M EDTA before RNA isolation. For deep sequencing, tissues from single mice were analyzed per time point. For some tissues (stomach, intestine, heart), sequences from three additional mice were determined by Sanger sequencing.
RNA isolation, reverse transcription (RT), and Filamin amplification
Total RNA was isolated from homogenized organs by using PeqGOLD TRIzol reagent (PEQLAB Biotechnologie GmbH) using manufacturers protocols. Aliquots of isolated RNAs were treated with DNase I (Fermentas 1 U/μL, 10U) at 37 °C for 1 h and kept in 96% ethanol prior to reverse transcription. cDNAs were synthesized using M-MLV Reverse Transcriptase kit (Invitrogen) and random hexamer primers in a total volume of 20 μl. Filamin A fragments were amplified in a 15 cycle PCR reactions using Phusion High Fidelity Polymerase (Finnzymes). Forward primers were designed to bind Filamin A exon 42 and contained the sequencing primer sequence and a 4-nucleotide barcode. The forward primer sequence was ACACGACGCT CTTCCGATCT NNNNCCGCCTT ACTGTTTCTA GTCT. The sequencing primer part is in italics while NNNN is the position of the barcode. The FLNA hybridizing part is underlined. The reverse primer was designed to bind Filamin A exon 43 and had the sequence GCTGGTTGAC CTTTAACCCT G. PCR products (50 nt long, spanning spliced exons 42–43) were purified from polyacrylamide gels by elution with 2.5M NH4OAc o/n at +4 °C, followed by ethanol precipitation. The eluted samples were used for second round of PCR with Phusion High Fidelity Polymerase (Finnzymes) and Illumina sequencing adaptor primers (15 cycles). Forward primer sequence AATGATACGG CGACCACCGA GATCTACACT CTTTCCCTAC ACGACGCTCT TCCGATCT; reverse primer sequence: CAAGCAGAAG ACGGCATACG AGCTCTTCCG ATCTGCTGGT TGACCTTTAA CCCTG. PCR fragments were again purified by PAGE and resuspended in water.
The same cDNAs were used to amplify filamin B with the forward primer in exon 41: CAGCCCTTAC CTGGTGCCCG T and the reverse primer in exon 42: CTTTGCATCA ATCTTCCCTT TCG
Deep sequencing
Cluster generation and DNA sequencing was performed using the Illumina Genome analyzer (GA) IIx system. After sequencing, the adaptor and barcode sequences were removed using Cutadapt.50 Sequences were aligned to the predicted spliced product. Sequences not fitting the predicted spliced product except for the edited nucleotide were discarded.
Basecalling tables were used to define the level of noise at 0.3% for all possible transitions over the entire sequence length. A to G transitions above 0.5% at position 3 of the actual sequence, representing the editing site, were considered as true editing events.
Real-time PCR
To quantify the expression of filamin A, ADAR2, and actin in selected tissues, RNAs prepared for the deep sequencing was DNase I digested (Fermentas 1 U/μl, 10 U) and reverse transcribed using M-MLV Reverse Transcriptase kit (Invitrogen) and random hexamer primers in a total volume of 20 μl. Real-time PCR was performed on an Eppendorf realplex 2 mastercycler, whereas using a GoTaq qPCR master mix (Promega) and following primers and reaction conditions:
Filamin A sense 5′-GGTGACGCCC GCCGCCTTAC- 3′,
anti- sense 5′-AAGATGCTGG CTGGTTGACC-3′,
ADARb1 sense 5′-TCCTGCAGTG ACAAGATAGC A-3′,
anti-sense 5′- GGTTCCACGA AAATGCTGAG 3′
Actin β sense 5′-CTTTGCAGCT CCTTCGTTGC-3′,
antisense 5′ ACGATGGAGG GGAATACAGC 3′
40 cycles of 95 °C/15 s, 57.5 °C/30 s, 72 °C/1 min, preceded by denaturation at 95 °C/3 min.
Relative difference in expression of FilaminA and ADARb1 was calculated by the ΔCt method using actin β as a reference gene.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors would like to thank the Vienna Campus Support Facility (CSF) for performing the Illumina deep sequencing, de-barcoding, and aligning of sequences. The authors would also like to thank Prof Peter Seeburg, MPI Heidelberg for the kind gift of ADAR2-knockout mice. This work was supported by the Austrian Science Foundation grant Nr. SFB 4313 to MFJ. MS was supported by the Austrian Science Foundation Doctoral Program W1207.
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/26216
References
- 1.Bass BL. RNA editing and hypermutation by adenosine deamination. Trends Biochem Sci. 1997;22:157–62. doi: 10.1016/S0968-0004(97)01035-9. [published erratum appears in Trends Biochem Sci 1997 Jul;22(7):278] [DOI] [PubMed] [Google Scholar]
- 2.Nishikura K. Functions and regulation of RNA editing by ADAR deaminases. Annu Rev Biochem. 2010;79:321–49. doi: 10.1146/annurev-biochem-060208-105251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bass BL. RNA editing by adenosine deaminases that act on RNA. Annu Rev Biochem. 2002;71:817–46. doi: 10.1146/annurev.biochem.71.110601.135501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Riedmann EM, Schopoff S, Hartner JC, Jantsch MF. Specificity of ADAR-mediated RNA editing in newly identified targets. RNA. 2008;14:1110–8. doi: 10.1261/rna.923308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wong SK, Sato S, Lazinski DW. Substrate recognition by ADAR1 and ADAR2. RNA. 2001;7:846–58. doi: 10.1017/S135583820101007X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eggington JM, Greene T, Bass BL. Predicting sites of ADAR editing in double-stranded RNA. Nat Commun. 2011;2:319. doi: 10.1038/ncomms1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tariq A, Jantsch MF. Transcript diversification in the nervous system: a to I RNA editing in CNS function and disease development. Front Neurosci. 2012;6:99. doi: 10.3389/fnins.2012.00099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morabito MV, Abbas AI, Hood JL, Kesterson RA, Jacobs MM, Kump DS, Hachey DL, Roth BL, Emeson RB. Mice with altered serotonin 2C receptor RNA editing display characteristics of Prader-Willi syndrome. Neurobiol Dis. 2010;39:169–80. doi: 10.1016/j.nbd.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burns CM, Chu H, Rueter SM, Hutchinson LK, Canton H, Sanders-Bush E, Emeson RB. Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature. 1997;387:303–8. doi: 10.1038/387303a0. [DOI] [PubMed] [Google Scholar]
- 10.Kawahara Y, Grimberg A, Teegarden S, Mombereau C, Liu S, Bale TL, Blendy JA, Nishikura K. Dysregulated editing of serotonin 2C receptor mRNAs results in energy dissipation and loss of fat mass. J Neurosci. 2008;28:12834–44. doi: 10.1523/JNEUROSCI.3896-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Olaghere da Silva UB, Morabito MV, Canal CE, Airey DC, Emeson RB, Sanders-Bush E. Impact of RNA editing on functions of the serotonin 2C receptor in vivo. Front Neurosci. 2010;4:26. doi: 10.3389/neuro.23.001.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brusa R, Zimmermann F, Koh DS, Feldmeyer D, Gass P, Seeburg PH, Sprengel R. Early-onset epilepsy and postnatal lethality associated with an editing-deficient GluR-B allele in mice. Science. 1995;270:1677–80. doi: 10.1126/science.270.5242.1677. [DOI] [PubMed] [Google Scholar]
- 13.Maas S, Rich A, Nishikura K. A-to-I RNA editing: recent news and residual mysteries. J Biol Chem. 2003;278:1391–4. doi: 10.1074/jbc.R200025200. [DOI] [PubMed] [Google Scholar]
- 14.Yeo J, Goodman RA, Schirle NT, David SS, Beal PA. RNA editing changes the lesion specificity for the DNA repair enzyme NEIL1. Proc Natl Acad Sci U S A. 2010;107:20715–9. doi: 10.1073/pnas.1009231107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jayan GC, Casey JL. Inhibition of hepatitis delta virus RNA editing by short inhibitory RNA-mediated knockdown of ADAR1 but not ADAR2 expression. J Virol. 2002;76:12399–404. doi: 10.1128/JVI.76.23.12399-12404.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levanon EY, Eisenberg E, Yelin R, Nemzer S, Hallegger M, Shemesh R, Fligelman ZY, Shoshan A, Pollock SR, Sztybel D, et al. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat Biotechnol. 2004;22:1001–5. doi: 10.1038/nbt996. [DOI] [PubMed] [Google Scholar]
- 17.Athanasiadis A, Rich A, Maas S. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol. 2004;2:e391. doi: 10.1371/journal.pbio.0020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nishikura K. Editing the message from A to I. Nat Biotechnol. 2004;22:962–3. doi: 10.1038/nbt0804-962. [DOI] [PubMed] [Google Scholar]
- 19.Horsch M, Seeburg PH, Adler T, Aguilar-Pimentel JA, Becker L, Calzada-Wack J, Garrett L, Götz A, Hans W, Higuchi M, et al. Requirement of the RNA-editing enzyme ADAR2 for normal physiology in mice. J Biol Chem. 2011;286:18614–22. doi: 10.1074/jbc.M110.200881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Q, Miyakoda M, Yang W, Khillan J, Stachura DL, Weiss MJ, Nishikura K. Stress-induced apoptosis associated with null mutation of ADAR1 RNA editing deaminase gene. J Biol Chem. 2004;279:4952–61. doi: 10.1074/jbc.M310162200. [DOI] [PubMed] [Google Scholar]
- 21.Higuchi M, Maas S, Single FN, Hartner J, Rozov A, Burnashev N, Feldmeyer D, Sprengel R, Seeburg PH. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature. 2000;406:78–81. doi: 10.1038/35017558. [DOI] [PubMed] [Google Scholar]
- 22.Hideyama T, Yamashita T, Suzuki T, Tsuji S, Higuchi M, Seeburg PH, Takahashi R, Misawa H, Kwak S. Induced loss of ADAR2 engenders slow death of motor neurons from Q/R site-unedited GluR2. J Neurosci. 2010;30:11917–25. doi: 10.1523/JNEUROSCI.2021-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Silberberg G, Lundin D, Navon R, Öhman M. Deregulation of the A-to-I RNA editing mechanism in psychiatric disorders. Hum Mol Genet. 2012;21:311–21. doi: 10.1093/hmg/ddr461. [DOI] [PubMed] [Google Scholar]
- 24.Maas S, Patt S, Schrey M, Rich A. Underediting of glutamate receptor GluR-B mRNA in malignant gliomas. Proc Natl Acad Sci U S A. 2001;98:14687–92. doi: 10.1073/pnas.251531398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wahlstedt H, Daniel C, Ensterö M, Ohman M. Large-scale mRNA sequencing determines global regulation of RNA editing during brain development. Genome Res. 2009;19:978–86. doi: 10.1101/gr.089409.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levanon EY, Hallegger M, Kinar Y, Shemesh R, Djinovic-Carugo K, Rechavi G, Jantsch MF, Eisenberg E. Evolutionarily conserved human targets of adenosine to inosine RNA editing. Nucleic Acids Res. 2005;33:1162–8. doi: 10.1093/nar/gki239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li JB, Levanon EY, Yoon JK, Aach J, Xie B, Leproust E, Zhang K, Gao Y, Church GM. Genome-wide identification of human RNA editing sites by parallel DNA capturing and sequencing. Science. 2009;324:1210–3. doi: 10.1126/science.1170995. [DOI] [PubMed] [Google Scholar]
- 28.Feng Y, Walsh CA. The many faces of filamin: a versatile molecular scaffold for cell motility and signalling. Nat Cell Biol. 2004;6:1034–8. doi: 10.1038/ncb1104-1034. [DOI] [PubMed] [Google Scholar]
- 29.Hartwig JH, Stossel TP. Isolation and properties of actin, myosin, and a new actinbinding protein in rabbit alveolar macrophages. J Biol Chem. 1975;250:5696–705. [PubMed] [Google Scholar]
- 30.Nakamura F, Stossel TP, Hartwig JH. The filamins: organizers of cell structure and function. Cell Adh Migr. 2011;5:160–9. doi: 10.4161/cam.5.2.14401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith L, Page RC, Xu Z, Kohli E, Litman P, Nix JC, Ithychanda SS, Liu J, Qin J, Misra S, et al. Biochemical basis of the interaction between cystic fibrosis transmembrane conductance regulator and immunoglobulin-like repeats of filamin. J Biol Chem. 2010;285:17166–76. doi: 10.1074/jbc.M109.080911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang HY, Burns LH. Naloxone’s pentapeptide binding site on filamin A blocks Mu opioid receptor-Gs coupling and CREB activation of acute morphine. PLoS One. 2009;4:e4282. doi: 10.1371/journal.pone.0004282. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33.Jiménez-Baranda S, Gómez-Moutón C, Rojas A, Martínez-Prats L, Mira E, Ana Lacalle R, Valencia A, Dimitrov DS, Viola A, Delgado R, et al. Filamin-A regulates actin-dependent clustering of HIV receptors. Nat Cell Biol. 2007;9:838–46. doi: 10.1038/ncb1610. [DOI] [PubMed] [Google Scholar]
- 34.Muriel O, Echarri A, Hellriegel C, Pavón DM, Beccari L, Del Pozo MA. Phosphorylated filamin A regulates actin-linked caveolae dynamics. J Cell Sci. 2011;124:2763–76. doi: 10.1242/jcs.080804. [DOI] [PubMed] [Google Scholar]
- 35.Sverdlov M, Shinin V, Place AT, Castellon M, Minshall RD. Filamin A regulates caveolae internalization and trafficking in endothelial cells. Mol Biol Cell. 2009;20:4531–40. doi: 10.1091/mbc.E08-10-0997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ravid D, Chuderland D, Landsman L, Lavie Y, Reich R, Liscovitch M. Filamin A is a novel caveolin-1-dependent target in IGF-I-stimulated cancer cell migration. Exp Cell Res. 2008;314:2762–73. doi: 10.1016/j.yexcr.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 37.Head BP, Patel HH, Roth DM, Murray F, Swaney JS, Niesman IR, Farquhar MG, Insel PA. Microtubules and actin microfilaments regulate lipid raft/caveolae localization of adenylyl cyclase signaling components. J Biol Chem. 2006;281:26391–9. doi: 10.1074/jbc.M602577200. [DOI] [PubMed] [Google Scholar]
- 38.Hjälm G, MacLeod RJ, Kifor O, Chattopadhyay N, Brown EM. Filamin-A binds to the carboxyl-terminal tail of the calcium-sensing receptor, an interaction that participates in CaR-mediated activation of mitogen-activated protein kinase. J Biol Chem. 2001;276:34880–7. doi: 10.1074/jbc.M100784200. [DOI] [PubMed] [Google Scholar]
- 39.Fiori JL, Zhu TN, O’Connell MP, Hoek KS, Indig FE, Frank BP, Morris C, Kole S, Hasskamp J, Elias G, et al. Filamin A modulates kinase activation and intracellular trafficking of epidermal growth factor receptors in human melanoma cells. Endocrinology. 2009;150:2551–60. doi: 10.1210/en.2008-1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Venø MT, Bramsen JB, Bendixen C, Panitz F, Holm IE, Öhman M, Kjems J. Spatio-temporal regulation of ADAR editing during development in porcine neural tissues. RNA Biol. 2012;9:1054–65. doi: 10.4161/rna.21082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Danecek P, Nellåker C, McIntyre RE, Buendia-Buendia JE, Bumpstead S, Ponting CP, Flint J, Durbin R, Keane TM, Adams DJ. High levels of RNA-editing site conservation amongst 15 laboratory mouse strains. Genome Biol. 2012;13:26. doi: 10.1186/gb-2012-13-4-r26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Greenberger S, Levanon EY, Paz-Yaacov N, Barzilai A, Safran M, Osenberg S, Amariglio N, Rechavi G, Eisenberg E. Consistent levels of A-to-I RNA editing across individuals in coding sequences and non-conserved Alu repeats. BMC Genomics. 2010;11:608. doi: 10.1186/1471-2164-11-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shtrichman R, Germanguz I, Mandel R, Ziskind A, Nahor I, Safran M, Osenberg S, Sherf O, Rechavi G, Itskovitz-Eldor J. Altered A-to-I RNA editing in human embryogenesis. PLoS One. 2012;7:e41576. doi: 10.1371/journal.pone.0041576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lehmann KA, Bass BL. Double-stranded RNA adenosine deaminases ADAR1 and ADAR2 have overlapping specificities. Biochemistry. 2000;39:12875–84. doi: 10.1021/bi001383g. [DOI] [PubMed] [Google Scholar]
- 45.Melcher T, Maas S, Herb A, Sprengel R, Seeburg PH, Higuchi M. A mammalian RNA editing enzyme. Nature. 1996;379:460–4. doi: 10.1038/379460a0. [DOI] [PubMed] [Google Scholar]
- 46.Jacobs MM, Fogg RL, Emeson RB, Stanwood GD. ADAR1 and ADAR2 expression and editing activity during forebrain development. Dev Neurosci. 2009;31:223–37. doi: 10.1159/000210185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hartner JC, Schmittwolf C, Kispert A, Müller AM, Higuchi M, Seeburg PH. Liver disintegration in the mouse embryo caused by deficiency in the RNA-editing enzyme ADAR1. J Biol Chem. 2004;279:4894–902. doi: 10.1074/jbc.M311347200. [DOI] [PubMed] [Google Scholar]
- 48.Garncarz W, Tariq A, Handl C, Pusch O, Jantsch MF. A high-throughput screen to identify enhancers of ADAR-mediated RNA-editing. RNA Biol. 2013;10:192–204. doi: 10.4161/rna.23208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tariq A, Garncarz W, Handl C, Balik A, Pusch O, Jantsch MF. RNA-interacting proteins act as site-specific repressors of ADAR2-mediated RNA editing and fluctuate upon neuronal stimulation. Nucleic Acids Res. 2013;41:2581–93. doi: 10.1093/nar/gks1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing. EMBnetjournal. 2011;17:10–2. [Google Scholar]