Abstract
CRISPR/Cas systems are bacterial RNA-guided endonuclease machineries that target foreign nucleic acids. Recently, we demonstrated that the Cas protein Cas9 controls gene expression and virulence in Francisella novicida by altering the stability of the mRNA for an immunostimulatory bacterial lipoprotein (BLP). Genomic analyses, however, revealed that Francisella species with increased virulence harbor degenerated CRISPR/Cas systems. We hypothesize that CRISPR/Cas degeneration removed a barrier against genome alterations, which resulted in enhanced virulence. Importantly, the BLP locus was also lost; likely a necessary adaptation in the absence of Cas9-mediated repression. CRISPR/Cas systems likely play regulatory roles in numerous bacteria, and these data suggest additional genomic changes may be required to maintain fitness after CRISPR/Cas loss in such bacteria, having important evolutionary implications.
Keywords: CRISPR/Cas, regulatory RNA, gene expression, bacterial evolution, bacterial pathogenesis
CRISPR/Cas (clustered, regularly interspaced, short palindromic repeats/CRISPR- associated) systems are well-described RNA-guided endonuclease complexes that act to target and degrade foreign nucleic acids, such as those derived from bacteriophages.1 They consist of genomic or plasmid-encoded arrays of repetitive sequences that are interspaced by unique “spacer” sequences. These arrays are encoded adjacent to groups of conserved Cas genes, which distinguish three primary CRISPR/Cas subtypes.1 Following transcription of the CRISPR array, the transcript is processed into individual CRISPR RNAs (crRNAs) each containing partial repeat sequences and one unique spacer.2 These crRNAs form complexes with Cas proteins, hybridize to complementary nucleic acid targets, and the associated Cas genes catalyze the degradation of the target. Additionally, CRISPR arrays are adaptive. The Cas proteins Cas1 and Cas2 act to integrate new spacer sequences derived from invading foreign nucleic acids into the CRISPR array, allowing CRISPR systems to adapt and target these sequences in the future.3,4 Due to their specificity and adaptivity, CRISPR/Cas systems are well established to play an important role in mediating defense against invading bacteriophages. These systems can also prevent transformation by plasmids as well as chromosomal DNA, clearly demonstrating that they represent broad barriers to horizontal gene transfer (HGT).5,6
The Gram-negative intracellular pathogen, Francisella novicida, encodes a Type-II CRISPR/Cas system, which is characterized by the presence of the Cas9 endonuclease.1,7 We recently established the importance of this system in the pathogenesis of F. novicida.8 Like other Francisella species, F. novicida is capable of infecting and replicating within the cytosol of a variety of host cells, including phagocytic cells of the innate immune system.9 Upon phagocytosis by macrophages, Francisella spp evade or block numerous phagosomal host defenses, before rapidly escaping this compartment to reach the host cell cytosol where they replicate to high titers (reviewed by Jones, et al.10). During this process, the bacteria can be detected by the host innate immune protein Toll-like Receptor 2 (TLR2), which recognizes bacterial lipoproteins (BLP) and is present at both the plasma membrane and in the phagosome. TLR2 plays a critical role in recognizing Francisella and mounting a proinflammatory response (reviewed by Jones, et al.10). Therefore, in order to reach its replicative niche in the cytosol without inducing a significant inflammatory response, Francisella dampens recognition by, and activation of, TLR2. We have demonstrated that components of the F. novicida Type II CRISPR/Cas system are capable of targeting and repressing the expression of an endogenous transcript (FTN_1103) encoding a TLR2-activating BLP.8 Specifically, Cas9 forms a complex with the tracrRNA and a novel small RNA, termed small, CRISPR/Cas-associated RNA (scaRNA). Together, these components allow tracrRNA to interact with and target the FTN_1103 transcript and alter its stability.8 Using this system, F. novicida is able to rapidly decrease the abundance of the FTN_1103 transcript specifically when the bacteria are in the phagosome and in the presence of TLR2. Repression of this BLP via Cas9-dependent regulation allows F. novicida to dampen activation of TLR2.8 Since mutants lacking components of the Cas9 regulatory complex are severely attenuated, the innate immune evasion mediated by this system is absolutely critical for F. novicida pathogenesis.8
In addition to the role of components of the Type II CRISPR/Cas system in F. novicida pathogenesis, this system is predicted to be functional in the canonical role of targeting foreign nucleic acid.7 The Type II CRISPR/Cas locus in F. novicida genomes encodes full-length forms of all the necessary components for the adaptation (cas1, cas2, cas4) and effector phases (cas9, crRNA, tracrRNA—also required for crRNA processing and interaction of the crRNA with Cas9) of targeting foreign DNA, as compared with functional Type II systems in Streptococcus spp and other bacteria. Further suggesting that the Type II system is active in the targeting of foreign nucleic acid, most F. novicida genomes encode spacers identical to sequences in a predicted prophage present in the genome of a single known isolate of F. novicida.7 Interestingly, this isolate does not encode such spacers, potentially explaining why it harbors this prophage.7 In addition, F. novicida genomes encode a second CRISPR/Cas locus that most closely resembles a Type II locus in its architecture, but rather than Cas9, it encodes a novel Cas protein with no homology to known proteins.7 In contrast, we and others observe that the CRISPR/Cas systems present in the more virulent F. holarctica, F. mediasiatica, and F. tularensis species, have degenerated and lack critical components for CRISPR/Cas functionality (Fig. 1A).7
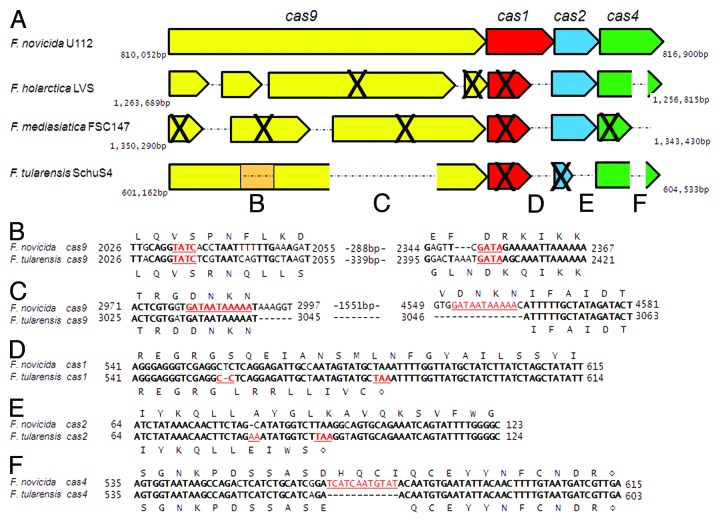
Figure 1. Analysis of the Type II CRISPR/Cas locus of Francisella species. (A) Operon alignment of the complete set of cas genes shared between four species, with the regions of significant difference between F. novicida and F. tularensis (insertions, deletions) indicated by letters. Bold X’s indicate predicted pseudogenes or gene fragments. The brown box depicted within cas9 of F. tularensis SchuS4 represents a region of significant dissimilarity with cas9 of F. novicida U112. Operons are adapted from NCBI, F. novicida U112 (Accession #: NC_008601), F. holarctica LVS (NC_007880), F. mediasiatica FSC147 (NC_010677), and F. tularensis SchuS4 (NC_006570). (B) Highly dissimilar nucleotide sequence between cas9 genes of F. novicida and F. tularensis. Four bp inverted repeats flanking the region highlighted in red (TATC/GATA). (C) Region of predicted intramolecular recombination, leading to excision within cas9 of F. tularensis. GATAATAAAAA direct repeats highlighted by red bars. (D) cas1 nucleotide alignment demonstrating the -1 frameshift within F. tularensis (C-C, red) leading to an early stop codon (TAA, red, diamond). (E) cas2 nucleotide alignment, demonstrating the +1 frameshift within F. tularensis (highlighted in red) leading to an early stop codon (TAA, red, diamond). (F) cas4 nucleotide alignment showing in-frame loss of 12 nucleotides of the F. novicida sequence (red) in F. tularensis. For all alignments, bold text indicates identical nucleotides within the alignment. Amino acid sequences are above and below for F. novicida and F. tularensis, respectively, and diamonds indicate stop codons.
Analysis of the genome of highly virulent F. tularensis (strain SchuS4) provides strong evidence for the degeneration of its CRISPR/Cas systems compared with F. novicida (strain U112). Specifically, there are disruptions within all four cas genes. While F. tularensis encodes the full-length DNA sequence for cas1, it contains a single base deletion (thymine 556 [815 478]) resulting in a -1 frame-shift mutation, leading to truncation of the protein by 125 amino acids (Fig. 1D). Similarly, this truncation of the cas1 gene is also present in F. holarctica (strain LVS) and F. mediasiatica (strain FSC147). The cas2 gene of F. tularensis contains a single base insertion (adenine 83 [816 119–816 120]) resulting in a +1 frame-shift mutation and a Cas2 protein only 31 amino acids in length, compared with 98 in F. novicida (Fig. 1E), whereas the cas2 open reading frames in F. holarctica and F. mediasiatica appear to be full-length in comparison to F. novicida. Since cas1 is likely non-functional in F. tularensis and other virulent Francisella species, these species would lack the ability to integrate new spacer sequences and therefore to adapt to new target sequences.3,4 F. tularensis cas4 has an internal deletion of 12 bases (567–578 [816 853–816 864]) resulting in a loss of four amino acids (Fig. 1F). However, the predicted protein is in-frame, and it is therefore unclear if it retains function. A similar in-frame mutation is present in F. holarctica, while F. mediasiatica contains an early stop codon, resulting in truncation of this protein. The cas9 open reading frame is the most divergent between these species. While F. novicida and F. tularensis have a single open reading frame corresponding to a cas9 protein predicted to be produced, F. holarctica and F. mediasiatica contain a cas9 sequence that has been degenerated into four or three truncated open reading frames, respectively, with the majority of these predicted to be pseudogenes (Fig. 1A). On the other hand, detailed analysis of F. tularensis cas9 has revealed some striking differences. F. tularensis cas9 has a large internal deletion of 1572 bases (corresponding to bases 2992 through 4563 of F. novicida cas9 [813 044–814 617], and 524 amino acids)(Fig. 1C). This deletion includes the predicted RuvC-IV endonuclease domain,8,11 as well as a portion of the predicted HNH endonuclease domain, necessary for Cas9 cleavage of DNA targets.12 However, the deletion does not disrupt the conserved HNH catalytic residues. It is striking that this cas9 sequence excised from F. tularensis is flanked by the sequence GATAATAAAAA as a direct repeat in F. novicida (Fig. 1C). In F. tularensis, there is only a single copy of this flanking sequence, highly suggestive of an intramolecular recombination event, which would have led to the excision of the 1572 nucleotides present in F. novicida.
Furthermore, there is a large span of amino acids (681 aa through 784 aa in F. novicida Cas9) that are highly dissimilar between the two species (Fig. 1B). Flanking this region of dissimilarity is a small, 4 bp, inverted repeat (TATC–GATA) that may be an indication of the occurrence of an illegitimate recombination event or the product of double strand break repair. Small, inverted repeats may also be scars of transposition events; however, we find no evidence of an inserted transposon within this sequence.
Not only are Cas proteins disrupted in the F. tularensis SchuS4 genome, but the content of CRISPR/Cas system RNAs is also altered. F. tularensis SchuS4 contains a transposable element (Is-Ftu2) inserted at the site within the F. novicida genome that encodes the crRNA array and the scaRNA, resulting in the deletion of these CRISPR/Cas components. The tracrRNA is still present in the F. tularensis genome, but in the absence of the crRNA array, it is unclear if, or how, this RNA would function to target foreign nucleic acid. Similarly, in the absence of scaRNA, which is critical for the ability of F. novicida to repress production of FTN_1103,8 it is unlikely that the remaining components in the F. tularensis system could function equally in its regulation. Since this regulatory pathway is essential for evasion of TLR2 and virulence in F. novicida, its inactivation in F. tularensis would potentially be highly detrimental to the pathogen’s ability to survive in mammalian hosts.8 This raises two important questions: what evolutionary pressures would select against a functional CRISPR/Cas locus in F. tularensis, and were there coincident changes that occurred in order to prevent the induction of the host TLR2 response?
Recent work has very clearly demonstrated that CRISPR/Cas systems represent a strong barrier to HGT. This restriction is extremely broad, as these systems prevent not only infection by bacteriophages (as well as their integration and the subsequent potential for lysogenic conversion), but also acquisition of plasmids, and both conjugative and free linear DNA.5,6,13,14 Therefore, acquisition of new genetic information from many sources is significantly inhibited by CRISPR/Cas systems. Additionally, since plasmids and bacteriophages can be carriers of transposons,15 CRISPR/Cas systems also present a blockade to prevent uptake of these and other mobile elements. Furthermore, CRISPR/Cas systems have been shown to prevent the induction of prophages.13 Therefore, it is interesting to speculate that this type of action may also inhibit the excision of other mobile elements, and may therefore prevent other mechanistically similar recombination events within the chromosome, such as gene duplication or phase variation through inversion or gene conversion.14 Thus, in the absence of a functional CRISPR/Cas system, as observed in the highly virulent Francisella spp, genomes would be more likely to undergo these numerous types of genetic alterations.
Sequence analysis of highly virulent Francisella species reveals that they underwent a number of genomic changes compared with F. novicida, during their hypothesized patho-adaptation to mammalian hosts.16-18 F. holarctica and F. tularensis species contain 41 genes not present in F. novicida, that are predicted to play important roles during infection of mammals.18 While the function of the majority of these unique genes has not been determined, six are predicted to play roles in the biosynthesis of O-antigen, a critical surface structure necessary for pathogenesis.18 Additionally, F. tularensis strains have nine genes unique to their genomes.18 Eight of these are located in a predicted remnant of a prophage or other mobile element flanked by transposons, and while its function is unknown, it has been postulated to be an F. tularensis-specific pathogenicity island.18 Furthermore, the F. tularensis genome has no less than 20 genetic duplication events.16-18 Notably, this includes duplication of the Francisella Pathogenicity Island (FPI), an event observed in all highly virulent species of Francisella.16-18 The FPI encodes a Type VI secretion system that is absolutely essential for intracellular replication and virulence of Francisella spp in mammals.19 The FPI is flanked by transposable elements that likely facilitated its duplication by non-reciprocal recombination.17 Duplication of the region may result in an increased gene dosage and/or altered pattern of expression, enhancing the virulence of highly pathogenic strains. Additionally, F. tularensis genomes as a whole have gained a number of transposable elements (79 in F. tularensis SchuS4, compared with 26 in F. novicida U112), which may have facilitated the aforementioned genetic duplications and acquisitions, as well as large-scale transposon-mediated inversions.16-18 Together, these global genetic changes are generally thought to have been essential for the increased virulence of F. tularensis.
Because CRISPR/Cas systems are capable of inhibiting the acquisition of new genetic information, we hypothesize that the loss of functional CRISPR/Cas systems facilitated those widespread genomic changes that occurred in highly virulent Francisella species. However, since F. novicida absolutely requires the Cas9 regulatory system to repress an immunostimulatory BLP (FTN_1103),8 the loss of CRISPR/Cas systems in highly virulent species would have likely come at the cost of a decreased ability to dampen BLP levels and, thus, recognition by the host innate immune receptor TLR2. This is paradoxical in light of the many studies that have clearly demonstrated that F. tularensis is much less inflammatory, and in some cases even anti-inflammatory, compared with the less virulent F. novicida.10 Therefore, additional changes likely occurred in highly virulent Francisella species to prevent the activation of TLR2 and host innate immune defenses, even in the absence of the Cas9-encoding CRISPR/Cas locus.
DNA sequence analysis demonstrates that significant degeneration of the FTN_1103 region occurred in F. tularensis (Fig. 2). The region encompassing FTN_1103 degenerated completely in F. tularensis (as well as in F. holarctica and F. mediasiatica), lacking any nucleotide sequence directly corresponding to the gene. Furthermore, there is no FTN_1103 ortholog elsewhere within the F. tularensis genome. FTN_1102 is also absent, and the FTN_1104 ortholog (FTT1122c) is truncated by 78 bases (Fig. 2). There is evidence of a transposon insertion occurring within this region of the more virulent strains (Fig. 2), as each contains an ISFtu6 sequence (now predicted to be a non-functional pseudogene). This insertion may have facilitated the loss of the ~2 kbp region that contains FTN_1101, FTN_1102, and FTN_1103 in the F. novicida genome. The FTN_1103 region within F. novicida is flanked by ygiH, a gene predicted to be involved in glycerolipid metabolism and tgt, a predicted queuine-tRNA ribosyltransferase. Both of these genes remain highly conserved between F. novicida and the more virulent Francisella genomes, providing boundaries to the genetic changes which occurred. These data clearly delineate the widespread loss of FTN_1103 among virulent Francisella species, as well as the degeneration of the surrounding genomic region.
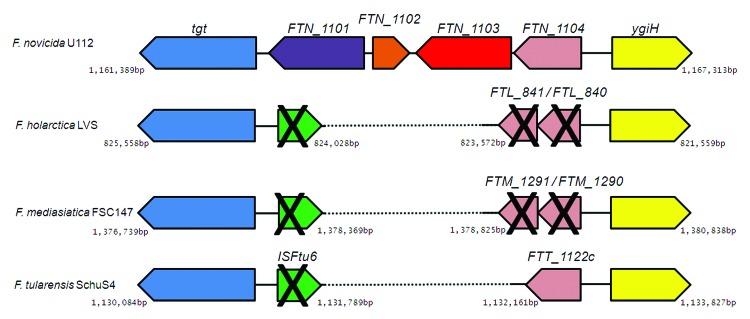
Figure 2. Operon alignment of the FTN_1103 region between F. novicida and more virulent species. Operon alignments, with each color corresponding to orthologous genes between species, black X’s representing predicted pseudogenes or gene fragments, and dashed lines indicating nucleotide deletions. ISFtu6 (green) represents a transposon insertion.
While its physiological function is unknown, FTN_1103 is dispensable for F. novicida virulence since mutants lacking this gene are not significantly attenuated in a mouse model of infection.8,20 Thus, loss of this gene does not have a significant adverse effect on the fitness of the organism. Taken together, we hypothesize that virulent Francisella species lost the FTN_1103 coding sequence (as well as some of the surrounding genetic region) previously to, or concurrently with, the degeneration of the CRISPR/Cas locus. This coincident change would have allowed F. tularensis to undergo significant genome alterations (in the absence of CRISPR/Cas–mediated HGT restriction), facilitating its increased virulence, while also preventing increased activation of the host innate immune system. This provides a parsimonious explanation for the apparent paradox between the striking importance of the CRISPR/Cas system as a critical virulence factor in the pathogenic lifestyle of F. novicida, and the non-functionality of the system in the most virulent Francisella species.
Here, we correlate CRISPR/Cas system degradation and the subsequent increase in HGT, with the coincident loss of a CRISPR/Cas-regulated locus. Loss of CRISPR/Cas systems may provide a fitness advantage to organisms that undergo frequent and beneficial genetic exchange, particularly during patho-adaptation. For example, Streptococcus pneumoniae is unable to acquire critical virulence factors when a functional CRISPR/Cas system is present (and the system is engineered to contain spacers targeting those genes), demonstrating that CRISPR/Cas systems can directly restrict DNA acquisition and the emergence of virulence during in vivo infection.6 Further, many S. pyogenes crRNA arrays contain targets against lysogenic bacteriophages, suggesting that they may prevent acquisition of phage-encoded virulence factors or act as regulators of virulence traits.2 It has also been suggested that a functional CRISPR/Cas system prevents HGT in Staphylococcus epidermidis, but that the more virulent S. aureus is able to acquire genes horizontally due to a lack of a functional CRISPR/Cas system.21 Similarly, antibiotic sensitive strains of Enterococcus faecalis often encode CRISPR/Cas systems whereas highly antibiotic resistant strains are less likely to encode these loci, suggesting that CRISPR/Cas systems prevent the acquisition of antibiotic resistance.22
In the event that CRISPR/Cas systems play additional roles in bacterial physiology beyond their action in defense against foreign DNA, as we have demonstrated for the Cas9 system in F. novicida,8 their loss or degeneration might have more complex effects on bacterial physiology. For example, loss of cas9 in Neisseria meningitidis or Campylobacter jejuni results in a decreased ability to attach, invade, and replicate within host cells.8,23 Further, in C. jejuni, increased degeneration of the CRISPR/Cas system correlates with loss of a specific gene encoding a sialyltransferase, suggesting that the Cas9 system may be a regulator of this specific gene, and potentially providing another example of coincident evolution between a CRISPR/Cas system and a regulatory target.23 Furthermore, loss of cas2 in Legionella pneumophila results in an inability to replicate within amoeba.24 Loss of the Type-I cas genes in Pseudomonas aeruginosa results in dysfunctional biofilm formation (an important virulence trait), suggestive of broader CRISPR/Cas functionality in regulation beyond Cas9 and the Type II systems alone.25 The data presented here suggest that in those bacteria in which Cas9 or other CRISPR/Cas components play a role in gene regulation or other alternative functions, loss of CRISPR/Cas functionality would not only facilitate HGT, but would have a disruptive effect on gene regulation. Since these regulatory changes might negatively impact bacterial fitness, compensatory changes may be required to prevent this loss of fitness (as we describe here for FTN_1103 deletion in highly virulent Francisella species). Thus, we propose that coincident loss of regulatory targets or other compensatory genomic changes may be a common and necessary occurrence in the face of CRISPR/Cas loss in diverse bacteria.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We would like to thank V Band, CY Chin, W Shafer, A Sjöstedt, and D Stephens for helpful discussions and critical reading of this manuscript. This work was supported by National Institutes of Health (NIH) grants U54-AI057157 from the Southeastern Regional Center of Excellence for Emerging Infections and Biodefense and R56-AI87673 to DSW, who is also supported by a Burroughs Wellcome Fund Investigator in the Pathogenesis of Infectious Disease award. TRS was supported by the NSF Graduate Research Fellowship, as well as the ARCS Foundation.
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/26423
References
- 1.Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, Moineau S, Mojica FJ, Wolf YI, Yakunin AF, et al. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol. 2011;9:467–77. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–7. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yosef I, Goren MG, Qimron U. Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res. 2012;40:5569–76. doi: 10.1093/nar/gks216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Datsenko KA, Pougach K, Tikhonov A, Wanner BL, Severinov K, Semenova E. Molecular memory of prior infections activates the CRISPR/Cas adaptive bacterial immunity system. Nat Commun. 2012;3:945. doi: 10.1038/ncomms1937. [DOI] [PubMed] [Google Scholar]
- 5.Marraffini LA, Sontheimer EJ. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322:1843–5. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bikard D, Hatoum-Aslan A, Mucida D, Marraffini LA. CRISPR interference can prevent natural transformation and virulence acquisition during in vivo bacterial infection. Cell Host Microbe. 2012;12:177–86. doi: 10.1016/j.chom.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 7.Schunder E, Rydzewski K, Grunow R, Heuner K. First indication for a functional CRISPR/Cas system in Francisella tularensis. Int J Med Microbiol. 2013;303:51–60. doi: 10.1016/j.ijmm.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 8.Sampson TR, Saroj SD, Llewellyn AC, Tzeng YL, Weiss DSA. A CRISPR/Cas system mediates bacterial innate immune evasion and virulence. Nature. 2013;497:254–7. doi: 10.1038/nature12048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hall JD, Woolard MD, Gunn BM, Craven RR, Taft-Benz S, Frelinger JA, Kawula TH. Infected-host-cell repertoire and cellular response in the lung following inhalation of Francisella tularensis Schu S4, LVS, or U112. Infect Immun. 2008;76:5843–52. doi: 10.1128/IAI.01176-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones CL, Napier BA, Sampson TR, Llewellyn AC, Schroeder MR, Weiss DS. Subversion of host recognition and defense systems by Francisella spp. Microbiol Mol Biol Rev. 2012;76:383–404. doi: 10.1128/MMBR.05027-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Makarova KS, Aravind L, Wolf YI, Koonin EV. Unification of Cas protein families and a simple scenario for the origin and evolution of CRISPR-Cas systems. Biol Direct. 2011;6:38. doi: 10.1186/1745-6150-6-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–21. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edgar R, Qimron U. The Escherichia coli CRISPR system protects from λ lysogenization, lysogens, and prophage induction. J Bacteriol. 2010;192:6291–4. doi: 10.1128/JB.00644-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nozawa T, Furukawa N, Aikawa C, Watanabe T, Haobam B, Kurokawa K, Maruyama F, Nakagawa I. CRISPR inhibition of prophage acquisition in Streptococcus pyogenes. PLoS One. 2011;6:e19543. doi: 10.1371/journal.pone.0019543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sampson T, Broussard GW, Marinelli LJ, Jacobs-Sera D, Ray M, Ko CC, Russell D, Hendrix RW, Hatfull GF. Mycobacteriophages BPs, Angel and Halo: comparative genomics reveals a novel class of ultra-small mobile genetic elements. Microbiology. 2009;155:2962–77. doi: 10.1099/mic.0.030486-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Champion MD, Zeng Q, Nix EB, Nano FE, Keim P, Kodira CD, Borowsky M, Young S, Koehrsen M, Engels R, et al. Comparative genomic characterization of Francisella tularensis strains belonging to low and high virulence subspecies. PLoS Pathog. 2009;5:e1000459. doi: 10.1371/journal.ppat.1000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Larsson P, Elfsmark D, Svensson K, Wikström P, Forsman M, Brettin T, Keim P, Johansson A. Molecular evolutionary consequences of niche restriction in Francisella tularensis, a facultative intracellular pathogen. PLoS Pathog. 2009;5:e1000472. doi: 10.1371/journal.ppat.1000472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rohmer L, Fong C, Abmayr S, Wasnick M, Larson Freeman TJ, Radey M, Guina T, Svensson K, Hayden HS, Jacobs M, et al. Comparison of Francisella tularensis genomes reveals evolutionary events associated with the emergence of human pathogenic strains. Genome Biol. 2007;8:R102. doi: 10.1186/gb-2007-8-6-r102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barker JR, Chong A, Wehrly TD, Yu JJ, Rodriguez SA, Liu J, Celli J, Arulanandam BP, Klose KE. The Francisella tularensis pathogenicity island encodes a secretion system that is required for phagosome escape and virulence. Mol Microbiol. 2009;74:1459–70. doi: 10.1111/j.1365-2958.2009.06947.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones CL, Sampson TR, Nakaya HI, Pulendran B, Weiss DS. Repression of bacterial lipoprotein production by Francisella novicida facilitates evasion of innate immune recognition. Cell Microbiol. 2012;14:1531–43. doi: 10.1111/j.1462-5822.2012.01816.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Otto M. Coagulase-negative staphylococci as reservoirs of genes facilitating MRSA infection: Staphylococcal commensal species such as Staphylococcus epidermidis are being recognized as important sources of genes promoting MRSA colonization and virulence. Bioessays. 2013;35:4–11. doi: 10.1002/bies.201200112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palmer KL, Gilmore MS. Multidrug-resistant enterococci lack CRISPR-cas. MBio. 2010;1:e00227–10. doi: 10.1128/mBio.00227-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Louwen R, Horst-Kreft D, de Boer AG, van der Graaf L, de Knegt G, Hamersma M, et al. A novel link between Campylobacter jejuni bacteriophage defence, virulence and Guillain-Barre syndrome. Eur J Clin Microbiol Infect Dis. 2012;32:207–26. doi: 10.1007/s10096-012-1733-4. [DOI] [PubMed] [Google Scholar]
- 24.Gunderson FF, Cianciotto NP. The CRISPR-associated gene cas2 of Legionella pneumophila is required for intracellular infection of amoebae. MBio. 2013;4:e00074–13. doi: 10.1128/mBio.00074-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zegans ME, Wagner JC, Cady KC, Murphy DM, Hammond JH, O’Toole GA. Interaction between bacteriophage DMS3 and host CRISPR region inhibits group behaviors of Pseudomonas aeruginosa. J Bacteriol. 2009;191:210–9. doi: 10.1128/JB.00797-08. [DOI] [PMC free article] [PubMed] [Google Scholar]