Abstract
High concentrations (> 100 µM) of the ribonucleoside analog 4-thiouridine (4sU) is widely used in methods for RNA analysis like photoactivatable-ribonucleoside-enhanced crosslinking and immunoprecipitation (PAR-CLIP) and nascent messenger (m)RNA labeling (4sU-tagging). Here, we show that 4sU-tagging at low concentrations ≤ 10 µM can be used to measure production and processing of ribosomal (r)RNA. However, elevated concentrations of 4sU (> 50 µM), which are usually used for mRNA labeling experiments, inhibit production and processing of 47S rRNA. The inhibition of rRNA synthesis is accompanied by nucleoplasmic translocation of nucleolar nucleophosmin (NPM1), induction of the tumor suppressor p53, and inhibition of proliferation. We conclude that metabolic labeling of RNA by 4sU triggers a nucleolar stress response, which might influence the interpretation of results. Therefore, functional ribosome biogenesis, nucleolar integrity, and cell cycle should be addressed in 4sU labeling experiments.
Keywords: 4-thiouridine, ribosomal RNA, rRNA processing, p53, nucleolar stress, RNA labeling, nucleophosmin
Introduction
Metabolic pulse labeling of nascent RNA is a powerful approach to assess the kinetics of RNA metabolism. There are several approaches to measure production, processing, and turnover of nascent RNA. First, labeling experiments of nascent RNA with radioactive uracil (3H or 14C), methyl-methionine (3H), or phosphoric acid (32P) allow direct autoradiographic analysis of highly abundant transcripts such as ribosomal (r)RNAs or transfer (t)RNAs.1,2 Second, nascent RNA synthesis can alternatively be analyzed by 5-bromouridine (5-BrU) labeling. 5-BrU allows non-radioactive detection of less abundant transcripts like pre-messenger (m)RNAs. However, issues with cell permeability may restrain 5-BrU labeling to certain cell types and tissues. Detection of 5-BrU-labeled transcripts is based on the recognition of antibodies. More recently, activated uridine analogs like 4-thiouridine (4sU) or 5-ethynyluridine (5-eU) have been used to measure nascent RNA synthesis.3,4 Labeling with activated uridine analogs enables analysis of nascent RNA synthesis and metabolism at single nucleotide level with high sensitivity, and is applicable to a broad range of RNA species. For example, combination of 4sU labeling with crosslinking and precipitation (CLIP) can identify binding sites of cellular RNA-binding proteins.5
We could recently show that the incorporation of the uridine analog 5-fluorouridine (5-FU) into nascent rRNA inhibits rRNA processing.6 A complete block of rRNA processing was achieved at concentrations > 100 µM. Here we show that 4sU treatment of cells inhibits rRNA synthesis and nucleolar function in a concentration-dependent manner. While concentrations of ≤ 10 µM 4sU are sufficient for labeling and purification of nascent rRNA without significantly affecting rRNA synthesis, concentrations ≥ 50 µM 4sU strongly inhibit production and processing of rRNA. With regard to the high concentrations of 4sU required for current RNA labeling protocols and the high sensitivity of rRNA synthesis to 4sU, we conclude that 4sU itself has a strong impact on rRNA metabolism of cells and can influence the outcome and interpretation of experiments
Results
Measurement of rRNA production and processing by 4sU-tagging
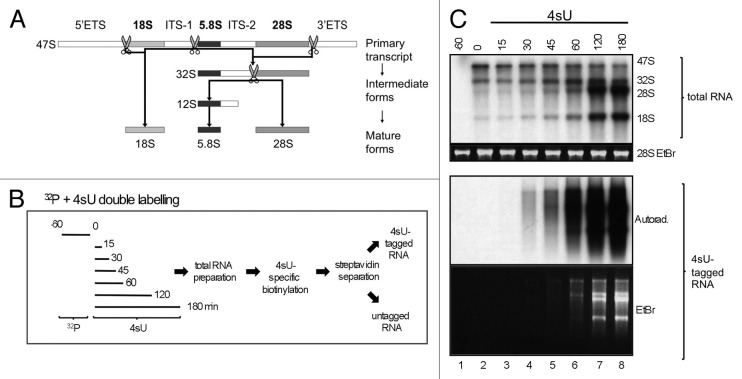
Nascent RNA labeling with uridine analogs like 4-thiouridine (4sU) allows global quantification of gene expression and kinetic analysis of RNA production, processing, and degradation at nucleotide resolution.7,8 Most 4sU labeling approaches focus on the metabolism of mRNAs and neglect rRNA synthesis, which comprises up to 70% of total RNA metabolism.9 In rRNA synthesis, a 47S rRNA primary transcript is produced by RNA polymerase (RNAP)I and subsequently processed to various intermediate (32S, 12S) and three mature (18S, 5.8S, 28S) rRNA forms (Fig. 1A). Processing of rRNA involves about 200 factors such as nucleases, helicases, and small nucleolar (sno)RNAs.10,11 To investigate the possible impact of 4sU-tagging on human rRNA processing, cells were pre-labeled with [32P]-ortho-phosphate as tracer, followed by incubation with 10 µM 4sU and purification of 4sU-tagged RNA as indicated (Fig. 1B). Double labeling with 32P and 4sU allows direct comparison of both labeling approaches (Fig. 1C). Total RNA samples were purified and used to measure production and processing of 32P-labeled nascent rRNA. 47S rRNA signals are visible after 60 min 32P labeling (Fig. 1C lane 2, upper panel), and decrease with prolonged 4sU incubation (lanes 3–8). Signals for intermediate and mature rRNAs are weak initially, but concomitantly increase over time. Analysis of 4sU-tagged RNA shows a time-dependent increase of all rRNA signals, both, in the autoradiograph and ethidium bromide gel (Fig. 1C, lower panel). Note that labeling with 10 µM 4sU for 120–180 min is required to detect mature rRNA forms, but does not impair production and processing of 47S rRNA. Thus, labeling with low concentrations of 4sU is a well-suited alternative to [32P]-ortho-phosphate to analyze rRNA synthesis.
Figure 1. Analysis of rRNA synthesis by 4sU-tagging. (A) Transcription and processing of rRNA. The polycistronic transcription unit encodes a 47S precursor rRNA containing 5′ and 3′ external transcribed spacers (5′ETS, 3′ETS), internal transcribed spacers 1 and 2 (ITS-1, ITS-2), and 18S, 5.8S, and 28S rRNAs. The 47S rRNA undergoes a cascade of endonucleolytic cleavages (scissors) and exonucleolytic degradation steps. (B) Metabolic in vivo double-labeling workflow. 2fTGH cells were pre-labeled with [32P]-ortho-phosphate for one hour, 4sU (10 µM) was added as indicated, total RNA was prepared, 4sU-tagged RNA was biotinylated and separated from untagged RNA by streptavidin-coated magnetic beads. (C) Comparison of 32P labeling and 4sU-tagging approaches for the analysis of rRNA synthesis. 2fTGH cells were seeded and cultured overnight. Nascent RNA was prepared as indicated in (B). Total or 4sU tagged RNA was separated by agarose gel electrophoresis and analyzed by autoradiography and ethidium bromide (EtBr) staining. 28S rRNA EtBr signals serve as loading control in this and subsequent experiments. A representative of two experiments is shown.
High concentrations of 4sU inhibit the production and processing of rRNA
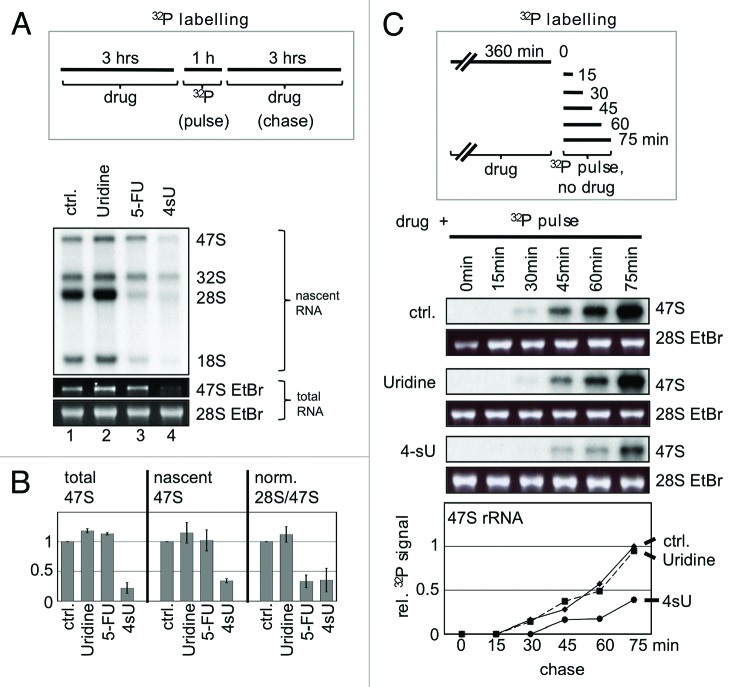
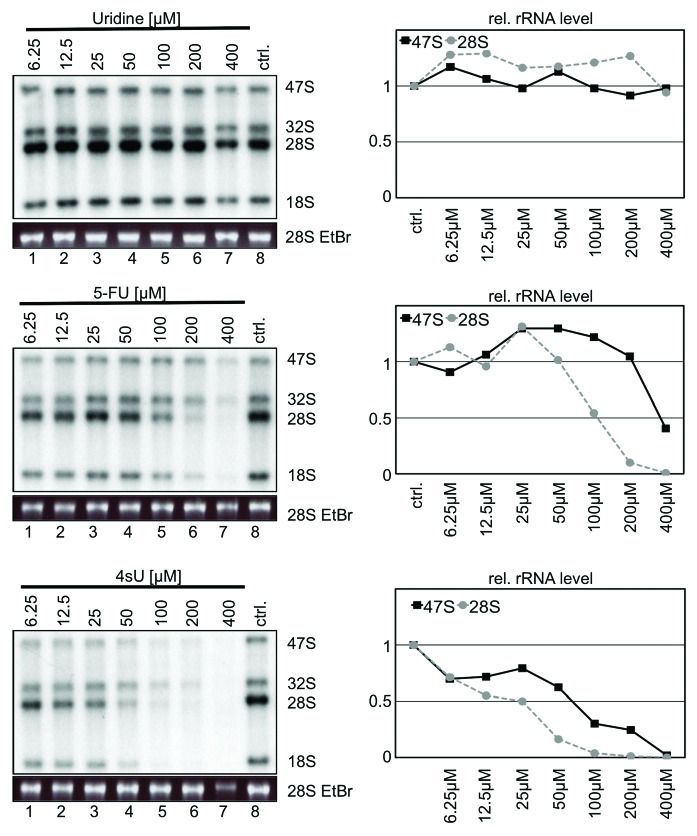
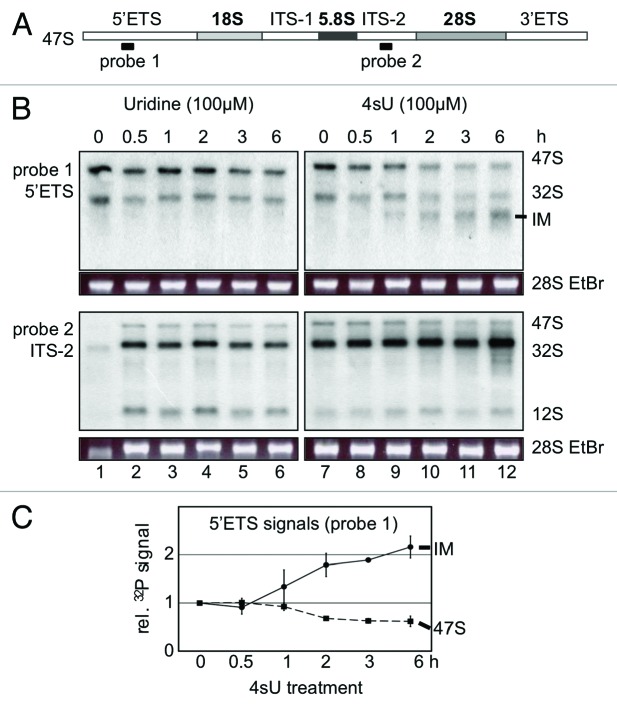
Previously, we could show that the uridine analog 5-fluorouridine (5-FU) predominantly inhibits the maturation of 28S and 18S rRNA, but not the production of 47S rRNA.6 We performed [32P]-ortho-phosphate pulse chase labeling in human U2OS cells to analyze rRNA synthesis in presence of high levels of 4sU (100 µM), a concentration typically used in 4sU-tagging and photoactivatable-ribonucleoside-enhanced crosslinking and immunoprecipitation (PAR-CLIP).12 While uridine has no impact on rRNA synthesis and 5-FU predominantly impairs rRNA processing, incubation of cells with 4sU inhibits both production and processing of 47S rRNA (Fig. 2A). 4sU reduces 47S rRNA levels by about 75% and the processing of the remaining primary transcript by about 60% (Fig. 2A and B). Next, the 4sU-mediated inhibition of 47S rRNA production was analyzed in more detail. While the levels of 47S rRNA showed a comparable time-dependent increase in control cells and cells treated with uridine, this increase was significantly reduced in the presence of 4sU after 75 min chase (Fig. 2C). Delayed production and impaired processing of 47S rRNA was also obtained after prolonged chase times (Fig. S1). The inhibition of rRNA metabolism was also measured in concentration kinetics. Fifty µM 4sU for 6 h are sufficient to impair 28S rRNA maturation comparable to 200 µM 5-FU treatment (Fig. 3). Similar results were obtained for HeLa cells and H1299 cells (Fig. S2). When cells were treated with 50 µM 4sU, 47S rRNA production is reduced by 50%. Defective rRNA processing could be associated with the stabilization of unprocessed rRNA precursors. We found that 4sU induces the accumulation of an aberrant 28S sized rRNA in U2OS cells (Fig. 4). The aberrant 28S rRNA is only detectable with a 5′ETS probe, but not with an ITS-2-specific probe in Northern analysis. Whether 4sU stabilizes the aberrant 28S rRNA due to perturbations in rRNA processing steps and/or defects in the removal of unprocessed precursors by exosomal degradation is currently unclear. Taken together, the results indicate that high amounts of 4sU strongly and rapidly inhibit rRNA synthesis at the levels of production and processing of rRNA. Notably, 4sU-mediated rRNA synthesis inhibition differs from 5-FU treatment, which predominantly blocks only late rRNA processing into mature 28S and 18S rRNA forms, but has little implication for 47S rRNA production.
Figure 2. Inhibition of rRNA production and processing by 4sU treatment. (A) U2OS cells were treated with uridine (100 μM), 5-FU (100 µM), or 4sU (100 µM) and nascent RNA was labeled by [32P]-ortho-phosphate as indicated. Total RNA was purified and analyzed as described in Materials and Methods. Nascent RNA was visualized by autoradiography, total RNA was visualized by ethidium bromide (EtBr) staining under UV-light. A representative of four experiments is shown. (B) Quantitation of nascent and total RNA levels from A. rRNA signals and ratios were measured by PhosphorImager or AIDA and plotted relative to signals from control cells (0.1% DMSO). Error bars: Standard deviation (n = 4). (C) U2OS cells were treated with drugs (100 µM), and nascent RNA was pulse-labeled by [32P]-ortho-phosphate as indicated. Newly synthesized 47S rRNA was analyzed and quantified as in (B). 47S signals from uridine-treated cells after 75 min pulse were set as one. 28S rRNA EtBr staining under UV-light was used to monitor equal loading, a representative gel is shown. Drug: uridine, 5-FU, or 4sU.
Figure 3. Analysis of rRNA synthesis in presence of uridine, 5-fluorouridine (5-FU), and 4-thiouridine (4sU). U2OS cells were incubated with increasing concentrations of drugs as indicated. Nascent RNA was labeled by [32P]-ortho-phosphate, total RNA was purified and analyzed as described in Materials and Methods. Nascent RNA was visualized by autoradiography, total RNA was visualized by ethidium bromide (EtBr) staining under UV-light. Signals were quantified by a PhosphorImager and plotted as relative rRNA levels. Signals from control cells treated with 0.1% DMSO was set as one. A representative experiment is shown.
Figure 4. Accumulation of an aberrant 28S rRNA intermediate (IM) upon 4sU treatment. (A) Scheme of the 47S rRNA primary transcript sequence composition and hybridization sites of Northern probes. (B) Analysis of rRNA synthesis by northern blot hybridization. U2OS cells were treated with uridine (100 µM) or 4sU (100 µM) for 6 h and total RNA was purified. Northern blot analysis was performed with probes recognizing 5′ external transcribed spacer (5′ETS, probe 1) or internal transcribed spacer-2 (ITS-2, probe 2) sequences. (C) Quantitation of RNA signals gained by hybridization with probe 1 upon 4sU treatment. Signals corresponding to 47S rRNA and an aberrant intermediate rRNA form (IM) were quantified by PhosphorImager analysis and plotted relative to signals from control cells (0.1% DMSO). Error bars: standard deviation (n = 3).
4sU triggers translocation of NPM1, stabilization of p53, and inhibition of proliferation
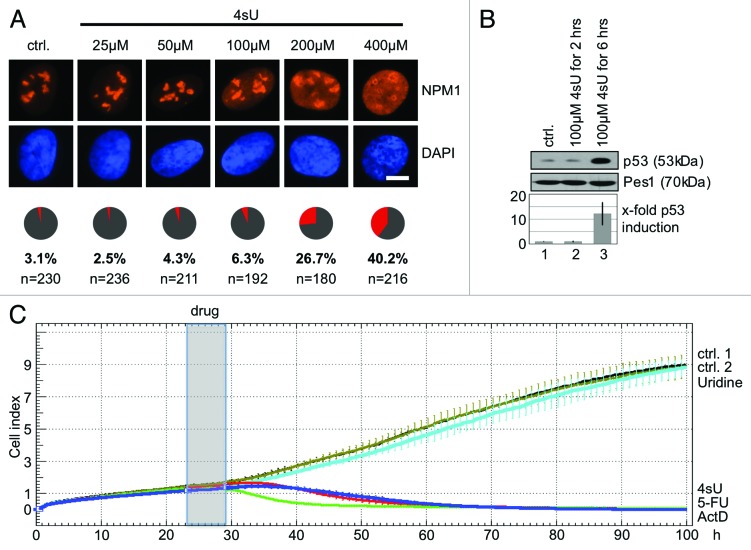
Having established that elevated concentrations of 4sU inhibit rRNA synthesis, we next asked whether 4sU could interfere with nucleolar integrity and trigger a nucleolar stress response. We assessed the sub-cellular localization of nucleophosmin (NPM1), a nucleolar phosphoprotein, which is involved in the processing of 32S rRNA13 and has been shown to translocate to the nucleoplasm upon stress.14 We found that treatment of U2OS cells with 4sU causes a dose-dependent translocation of nucleolar NPM1 to the nucleoplasm in a subset of cells (Fig. 5A). In contrast, incubation with uridine or 5-FU did not change NPM1 localization significantly (Fig. S3). Translocation of NPM1 (and other nucleolar proteins) to the nucleoplasm has been shown to stabilize p53 and block cell cycle progression.15,16 In line with that, we measured a strong induction of p53 (Fig. 5B) and severe reduction of cell proliferation (Fig. 5C; Fig. S4) upon treatment of cells with 4sU. In summary, the data suggest that high concentrations of 4sU are sensed by a nucleolar stress response, which involves nucleolar disintegration, p53 stabilization, and inhibition of proliferation.
Figure 5. Induction of nucleolar stress upon 4sU treatment. (A) Analysis of nucleophosmin (NPM1) localization in presence of 4sU. U2OS cells were treated with 4sU for 6 h as indicated. NPM1 localization was analyzed by immunofluorescence analysis using a specific antibody. Nuclei were stained with DAPI. Cake diagrams indicate the number of cells with nucleolar (gray) and nuclear (red) NPM1 staining. Scale bar: 1 µm. (B) Analysis of p53 levels upon 4sU treatment. U2OS cells were treated with 4sU as indicated, whole cell lysates were separated by SDS-PAGE and analyzed by Western Blot. P53 levels were quantified by AIDA software and plotted relative to signals from control cells (0.1% DMSO). (C) Analysis of proliferation upon 4sU treatment. U2OS cells were seeded and cultured overnight. Drugs (uridine, 100 µM; 4sU 100 µM; 5-FU 100 µM; ActD 1 µM) were added for 6 h (gray box), medium was replaced and cells were cultured for 100 h. Cell number was measured in real time and compared with control cells (DMEM, ctrl. 1; 0.1% DMSO, ctrl. 2). The cell number correlates to changes in impedance, which is termed “cell index” (see details in Material and Methods). Error bars: Standard deviation (n = 3).
Discussion
Inhibition of ribosome biogenesis is a drawback of 4sU labeling
Data from this work show that 4sU treatment strongly impacts on the metabolism of rRNAs. 4sU inhibits the production and processing of rRNA. Importantly, 4sU-mediated inhibition of rRNA synthesis strongly depends on the concentrations of 4sU and incubation times. Our data demonstrate that labeling of nascent RNA using low concentrations of 4sU (e.g., 10 µM) in principle is a suitable approach to analyze nascent rRNA synthesis. Interestingly, exposure of murine fibroblasts to 200 µM 4sU for 1 and 2 h did not result in any significant effects on cellular mRNA levels detectable by microarray analysis of total cellular RNA.4,17 While the sensitivity of gene expression profiling on total cellular RNA in these rather short time-scales is low4 substantial alterations in RNA synthesis rates of short-lived transcripts would have likely been picked up. Nevertheless, the impact of short-term 4sU exposure (≤ 2 h) on RNA metabolism may not only be restricted to rRNAs in other cell types or conditions. Recently, we applied ultra-short and progressive 4sU labeling to study the kinetics of RNA processing. Of note, we observed both substantially delayed splicing of numerous known retained introns as well as very rapid co-transcriptional splicing of other transcripts indicating that 4sU exposure does not impact on RNA processing in general.8 It is, however, important to note that long-term (> 12 h) 100 µM 4sU exposure, as commonly employed in PAR-CLIP experiments, does result in reduced cell proliferation (data not shown) and may thus result in experimental bias. We conclude that the impact of 4sU on RNA metabolism may not only be restricted to rRNAs, but might cause issues in the analysis of mRNAs when applying 4sU-tagging or PAR-CLIP.
How can the inhibition of rRNA synthesis by 4sU be explained on the molecular level? Correct processing of rRNA depends on the correct formation of stem loops and long-range secondary structures. For example, processing of the 3′ external transcribed spacer (3′ETS) requires snoRNA U8 in mammals.18 Hybridization of U8 with unprocessed rRNA allows formation of secondary structures and recruitment of downstream processing factors.19 Depletion of Ddx51, a helicase regulating U8 hybridization, blocks 3′ETS processing.20 It might well be that incorporation of 4sU into rRNA (and/or snoRNAs) prevents U8 hybridization to 3′ETS and other hybridization steps, thereby inhibiting processing of rRNA. In a recent study, we could show that inhibition of 3′ end processing of 47S rRNA negatively feeds back on RNAPI transcription upon reduction of U8 level.21 Thus, defective rRNA processing upon 4sU treatment could also be sensed by the RNAPI transcription machinery and reduce 47S rRNA synthesis rate.
We show that 4sU treatment causes the time-dependent decrease of 47S rRNA and concomitant accumulation of an aberrant 28S rRNA intermediate in northern blot hybridizations with a 5′ETS probe. No aberrant rRNA forms could be detected when hybridizing with a probe against the internal transcribed spacer 2 (ITS-2), indicating that 4sU preferentially inhibits processing of the primary transcript in the 5′ETS region rather than ITS-2 processing. However, a more detailed analysis of individual cleavage forms is required to describe defective rRNA processing upon 4sU treatment in detail and shed light into the sequence composition of the 28S-sized aberrant rRNA intermediate.
4sU and 5-FU inhibit ribosome biogenesis differently
Interestingly, inhibition of rRNA synthesis by 4sU differed from 5-FU treatment. While treatment with 4sU inhibits production and processing of rRNA, 5-FU more specifically blocks processing of 32S rRNA, with limited influence on the production of 47S rRNA.6,22 How can this difference be explained? Both drugs are uridine analogs, but modified at different positions and resemble different structures. 4sU and 5-FU are substrates for RNA polymerases and can be incorporated at sites, which require modifications (e.g., pseudouridylation) for correct function. For example, incorporation of 5-FU into U2 small nuclear (sn)RNA prevents pseudouridylation and inhibits the formation of functional snRNPs for pre-mRNA splicing.23 In analogy, inhibition of 32S rRNA processing by 5-FU could be explained by its incorporation into rRNA and/or snoRNAs, to allow proper rRNA processing. In analogy, we speculate that incorporation of 4sU into nascent rRNA impairs correct rRNA pseudouridylation, changes the secondary structure of unprocessed rRNAs, and correct rRNA processing. It is tempting to speculate, if 4sU treatment also reduces translational fidelity of ribosomes, as reported for ribosomes lacking pseudouridine synthase dyskerin.24 It also remains to be established, whether other uridine analogs, such as 5-BrU or 5-eU impact on rRNA metabolism.
Stabilization of p53 is a key feature of 4sU-mediated nucleolar stress
The structural and functional integrity of the nucleolus is directly involved in the control and turnover of the p53 tumor suppressor by the E3 ubiquitin ligase Hdm2.25-28 In brief, nucleolar disintegration facilitates p53 induction by the inactivation of Hdm2 by translocation of various nucleolar proteins, including L5, L11, and L23, to the nucleoplasm.29-33 We find that 4sU treatment causes a nucleolar disruption phenotype, which is accompanied by induction of p53 and reduction of cellular proliferation. Inhibition of rRNA synthesis by 4sU directly triggers the onset of a nucleolar stress response.
Taken together, 4sU-tagging of nascent RNA is a sophisticated technique to measure transcription, processing, and turnover of newly transcribed RNA.4,9,34,35 However, 4sU labeling time and concentration should be kept as short as possible, with regard to the severe nucleolar stress 4sU can cause.
Materials and Methods
Tissue culture and treatments
Human U2OS and 2fTGH cells were cultured in Dulbecco's modified Eagle's medium with 10% fetal bovine serum at 8% CO2. Cells were treated with uridine (Sigma), 5-fluorouracil (Sigma), 4-thiouridine (Sigma), or actinomycin D (Sigma) as indicated.
[32P]-ortho-phosphate metabolic labeling and RNA extraction
2.5 × 105 U2OS cells were incubated in phosphate-free Dulbecco's modified Eagle's medium/10% fetal bovine serum for 1 h and then incubated for 1 h in presence 15 μCi/ml [32P]-ortho-phosphate (pulse). Labeling medium was removed and cells were further cultivated for 3 h in Dulbecco's modified Eagle's medium/10% fetal bovine serum (chase). RNA was extracted using the PeqGOLD total RNA kit (PeqLab). One μg total RNA was separated on a 1% agarose-formaldehyde gel. The gel was dried on a Whatman paper using a regular gel drier (Bio-Rad) connected to a vacuum pump for 4 h at 80 °C. Metabolically labeled RNA was visualized by autoradiography and quantified by a PhosphorImager and AIDA software.
4-thiouridine-(4sU)-tagging
1.5 × 106 2fTGH cells were cultured in presence of 4sU (10 µM), lysed, and total RNA was extracted as described for metabolic in vivo labeling. 50 μg total RNA was incubated with biotin-HPDP (Pierce, 1 mg/ml; 2 μl/μg RNA) in biotinylation buffer (100 mM Tris, 10 mM EDTA, pH 7.4, 1 μl/μg RNA) for 1.5 h at RT. An equal volume of chloroform was added, mixed, and incubated with biotinylated RNA for 3 min. The mixture was separated in pre-spun Phase Trap Gel Heavy Tubes (5 min, 16000 rpm). For RNA precipitation and removal of unincorporated biotin-HPDP, a 1/10 volume 5 M NaCl and an equal volume of absolute isopropanol were added to the aqueous phase and centrifuged (20 min, 16000 rpm). The pellet was washed in an equal volume of 75% ethanol and centrifuged (10 min, 16000 rpm). RNA was resuspended in 100 μl RNase-free H2O. For separation, untagged and 4sU-tagged RNA was first heated to 65 °C for 10 min and cooled on ice for 5 min. RNA was incubated with 75 μl streptavidin-coated magnetic beads (Miltenyi) for 15 min with rotation. The reaction volume was applied to μMACs columns (Miltenyi), placed in an OctoMACS Seperator magnetic stand (Miltenyi), and equilibrated with 900 μl μMACS washing buffer (100 mM Tris, 10 mM EDTA, 1 M NaCl, 0.1% Tween-20, pH 7.5). The columns were washed with μMACS washing buffer. 4sU-biotin-streptavidin-tagged RNA was eluted in 700 μl RLT lysis buffer (PeqLab) with dithioerythritol (100 mM). 4sU-tagged RNA was recovered with the PeqGOLD total RNA kit as described above. 4sU-tagged RNA was separated on a 1.5% agarose-gel, containing ethidium bromide (37.5 μg/100 ml). Signals of RNA under UV-light were quantified by AIDA software.
Northern blot hybridization
Two μg of U2OS total RNA was separated on a 1% agarose-formaldehyde gel and blotted on Hybond N+ membranes (Amersham Biosciences). Probes were as following: 5′ETS, CGGTACCCCC AAGGCACGCC TCTCAGATCG CTAGAGAAGG CTTTTCTC; ITS-2, CTCTCTTTCC CTCTCCGTCT TCCGGCGGCG GCGCCGCCCT CCCCGTCT.
Immunoblotting and immunofluorescence
For immunoblotting, 2.5 × 105 U2OS cells were washed with phosphate-buffered saline and directly lysed in 2 × SDS loading buffer (100 mM Tris/HCl, 200 mM dithioerythritol, 4% SDS, 10 mM EDTA, 0.2% bromophenol blue, 20% glycerol). Whole cell lysates were separated by SDS-PAGE and blotted on nitrocellulose membranes (Amersham Biosciences). Immunodetection was performed with antibodies as following: human anti-p53 (Santa Cruz, sc-126, DO-1) and human anti-Pes1.36 Signals were quantified by AIDA software. For immunofluorescence, 8 × 104 U2OS cells were grown on coverslips and fixed in 2% formaldehyde for 2 min and permeabilised with PBS/Tween 0.04% for 7 min at RT. Unspecific binding was blocked with PBS/10% FBS for 2 h. Human anti-NPM1 (Sigma, B0556) primary antibody was incubated over night at 4 °C in a humidified chamber. Cy3-labeled secondary antibody (Dianova) was incubated for 30 min at RT. Nuclei were counterstained with DAPI (Sigma-Aldrich). Images were acquired using the Openlab software (Improvision) and a microscope (model Axiovert 200M; Carl Zeiss MicroImaging, Inc.)
Proliferation assay
Cell proliferation was measured by electric impedance detection using the xCELLigence device (Roche). Three × 103 U2OS cells were plated on an electronic plate (E-plate), which is capable of measuring electric impedance in real time by electrodes in direct contact with adherent cells. Impedance increases with the area on electrodes, which is covered by proliferating cells and termed “cell index.” Cells were cultured overnight, drugs were added for 6 h, and cells were measured for 100 h in 15 min intervals.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft (SFB684 and SFB-TR5), and Deutsche José Carreras Leukämie-Stiftung e.V. (DJCLS, project F09/03) to DE.
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/rnabiology/article/26214
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/26214
References
- 1.Warner JR, Soeiro R. Nascent ribosomes from HeLa cells. Proc Natl Acad Sci USA. 1967;58:1984–90. doi: 10.1073/pnas.58.5.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pestov DG, Lapik YR, Lau LF. Assays for ribosomal RNA processing and ribosome assembly. Current protocols in cell biology / editorial board, Juan S Bonifacino [et al] 2008; Chapter 22:Unit 22 11 [DOI] [PubMed] [Google Scholar]
- 3.Cleary MD, Meiering CD, Jan E, Guymon R, Boothroyd JC. Biosynthetic labeling of RNA with uracil phosphoribosyltransferase allows cell-specific microarray analysis of mRNA synthesis and decay. Nat Biotechnol. 2005;23:232–7. doi: 10.1038/nbt1061. [DOI] [PubMed] [Google Scholar]
- 4.Dölken L, Ruzsics Z, Rädle B, Friedel CC, Zimmer R, Mages J, Hoffmann R, Dickinson P, Forster T, Ghazal P, et al. High-resolution gene expression profiling for simultaneous kinetic parameter analysis of RNA synthesis and decay. RNA. 2008;14:1959–72. doi: 10.1261/rna.1136108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jr., Jungkamp AC, Munschauer M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–41. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burger K, Mühl B, Harasim T, Rohrmoser M, Malamoussi A, Orban M, Kellner M, Gruber-Eber A, Kremmer E, Hölzel M, et al. Chemotherapeutic drugs inhibit ribosome biogenesis at various levels. J Biol Chem. 2010;285:12416–25. doi: 10.1074/jbc.M109.074211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–42. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 8.Windhager L, Bonfert T, Burger K, Ruzsics Z, Krebs S, Kaufmann S, Malterer G, L’Hernault A, Schilhabel M, Schreiber S, et al. Ultrashort and progressive 4sU-tagging reveals key characteristics of RNA processing at nucleotide resolution. Genome Res. 2012;22:2031–42. doi: 10.1101/gr.131847.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rabani M, Levin JZ, Fan L, Adiconis X, Raychowdhury R, Garber M, Gnirke A, Nusbaum C, Hacohen N, Friedman N, et al. Metabolic labeling of RNA uncovers principles of RNA production and degradation dynamics in mammalian cells. Nat Biotechnol. 2011;29:436–42. doi: 10.1038/nbt.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fatica A, Tollervey D. Making ribosomes. Curr Opin Cell Biol. 2002;14:313–8. doi: 10.1016/S0955-0674(02)00336-8. [DOI] [PubMed] [Google Scholar]
- 11.Henras AK, Soudet J, Gérus M, Lebaron S, Caizergues-Ferrer M, Mougin A, Henry Y. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell Mol Life Sci. 2008;65:2334–59. doi: 10.1007/s00018-008-8027-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.König J, Zarnack K, Luscombe NM, Ule J. Protein-RNA interactions: new genomic technologies and perspectives. Nat Rev Genet. 2012;13:77–83. doi: 10.1038/nrg3141. [DOI] [PubMed] [Google Scholar]
- 13.Itahana K, Bhat KP, Jin A, Itahana Y, Hawke D, Kobayashi R, Zhang Y. Tumor suppressor ARF degrades B23, a nucleolar protein involved in ribosome biogenesis and cell proliferation. Mol Cell. 2003;12:1151–64. doi: 10.1016/S1097-2765(03)00431-3. [DOI] [PubMed] [Google Scholar]
- 14.Chan PK, Qi Y, Amley J, Koller CA. Quantitation of the nucleophosmin/B23-translocation using imaging analysis. Cancer Lett. 1996;100:191–7. doi: 10.1016/0304-3835(95)04100-1. [DOI] [PubMed] [Google Scholar]
- 15.Kurki S, Peltonen K, Latonen L, Kiviharju TM, Ojala PM, Meek D, Laiho M. Nucleolar protein NPM interacts with HDM2 and protects tumor suppressor protein p53 from HDM2-mediated degradation. Cancer Cell. 2004;5:465–75. doi: 10.1016/S1535-6108(04)00110-2. [DOI] [PubMed] [Google Scholar]
- 16.Boulon S, Westman BJ, Hutten S, Boisvert FM, Lamond AI. The nucleolus under stress. Mol Cell. 2010;40:216–27. doi: 10.1016/j.molcel.2010.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kenzelmann M, Maertens S, Hergenhahn M, Kueffer S, Hotz-Wagenblatt A, Li L, Wang S, Ittrich C, Lemberger T, Arribas R, et al. Microarray analysis of newly synthesized RNA in cells and animals. Proc Natl Acad Sci USA. 2007;104:6164–9. doi: 10.1073/pnas.0610439104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peculis BA, Steitz JA. Disruption of U8 nucleolar snRNA inhibits 5.8S and 28S rRNA processing in the Xenopus oocyte. Cell. 1993;73:1233–45. doi: 10.1016/0092-8674(93)90651-6. [DOI] [PubMed] [Google Scholar]
- 19.Peculis BA. The sequence of the 5′ end of the U8 small nucleolar RNA is critical for 5.8S and 28S rRNA maturation. Mol Cell Biol. 1997;17:3702–13. doi: 10.1128/mcb.17.7.3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Srivastava L, Lapik YR, Wang M, Pestov DG. Mammalian DEAD box protein Ddx51 acts in 3′ end maturation of 28S rRNA by promoting the release of U8 snoRNA. Mol Cell Biol. 2010;30:2947–56. doi: 10.1128/MCB.00226-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burger K, Mühl B, Rohrmoser M, Coordes B, Heidemann M, Kellner M, Gruber-Eber A, Heissmeyer V, Strässer K, Eick D. Cyclin-dependent kinase 9 links RNA Polymerase II transcription to processing of ribosomal RNA. J Biol Chem. 2013;288:21173–83. doi: 10.1074/jbc.M113.483719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghoshal K, Jacob ST. Specific inhibition of pre-ribosomal RNA processing in extracts from the lymphosarcoma cells treated with 5-fluorouracil. Cancer Res. 1994;54:632–6. [PubMed] [Google Scholar]
- 23.Zhao X, Yu YT. Incorporation of 5-fluorouracil into U2 snRNA blocks pseudouridylation and pre-mRNA splicing in vivo. Nucleic Acids Res. 2007;35:550–8. doi: 10.1093/nar/gkl1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jack K, Bellodi C, Landry DM, Niederer RO, Meskauskas A, Musalgaonkar S, Kopmar N, Krasnykh O, Dean AM, Thompson SR, et al. rRNA pseudouridylation defects affect ribosomal ligand binding and translational fidelity from yeast to human cells. Mol Cell. 2011;44:660–6. doi: 10.1016/j.molcel.2011.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stommel JM, Marchenko ND, Jimenez GS, Moll UM, Hope TJ, Wahl GM. A leucine-rich nuclear export signal in the p53 tetramerization domain: regulation of subcellular localization and p53 activity by NES masking. EMBO J. 1999;18:1660–72. doi: 10.1093/emboj/18.6.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ljungman M. Dial 9-1-1 for p53: mechanisms of p53 activation by cellular stress. Neoplasia. 2000;2:208–25. doi: 10.1038/sj.neo.7900073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xirodimas DP, Stephen CW, Lane DP. Cocompartmentalization of p53 and Mdm2 is a major determinant for Mdm2-mediated degradation of p53. Exp Cell Res. 2001;270:66–77. doi: 10.1006/excr.2001.5314. [DOI] [PubMed] [Google Scholar]
- 28.Boyd MT, Vlatkovic N, Rubbi CP. The nucleolus directly regulates p53 export and degradation. J Cell Biol. 2011;194:689–703. doi: 10.1083/jcb.201105143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lohrum MA, Ludwig RL, Kubbutat MH, Hanlon M, Vousden KH. Regulation of HDM2 activity by the ribosomal protein L11. Cancer Cell. 2003;3:577–87. doi: 10.1016/S1535-6108(03)00134-X. [DOI] [PubMed] [Google Scholar]
- 30.Dai MS, Lu H. Inhibition of MDM2-mediated p53 ubiquitination and degradation by ribosomal protein L5. J Biol Chem. 2004;279:44475–82. doi: 10.1074/jbc.M403722200. [DOI] [PubMed] [Google Scholar]
- 31.Bhat KP, Itahana K, Jin A, Zhang Y. Essential role of ribosomal protein L11 in mediating growth inhibition-induced p53 activation. EMBO J. 2004;23:2402–12. doi: 10.1038/sj.emboj.7600247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dai MS, Zeng SX, Jin Y, Sun XX, David L, Lu H. Ribosomal protein L23 activates p53 by inhibiting MDM2 function in response to ribosomal perturbation but not to translation inhibition. Mol Cell Biol. 2004;24:7654–68. doi: 10.1128/MCB.24.17.7654-7668.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hernandez-Verdun D, Roussel P, Thiry M, Sirri V, Lafontaine DL. The nucleolus: structure/function relationship in RNA metabolism. Wiley Interdiscip Rev RNA. 2010;1:415–31. doi: 10.1002/wrna.39. [DOI] [PubMed] [Google Scholar]
- 34.Friedel CC, Dölken L, Ruzsics Z, Koszinowski UH, Zimmer R. Conserved principles of mammalian transcriptional regulation revealed by RNA half-life. Nucleic Acids Res. 2009;37:e115. doi: 10.1093/nar/gkp542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Friedel CC, Dölken L. Metabolic tagging and purification of nascent RNA: implications for transcriptomics. Mol Biosyst. 2009;5:1271–8;. doi: 10.1039/b911233b. [DOI] [PubMed] [Google Scholar]
- 36.Hölzel M, Rohrmoser M, Schlee M, Grimm T, Harasim T, Malamoussi A, Gruber-Eber A, Kremmer E, Hiddemann W, Bornkamm GW, et al. Mammalian WDR12 is a novel member of the Pes1-Bop1 complex and is required for ribosome biogenesis and cell proliferation. J Cell Biol. 2005;170:367–78. doi: 10.1083/jcb.200501141. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.