Abstract
MicroRNA duplices are separated into a guide and a passenger strand. By convention, the guide represents the active microRNA while the passenger is supposedly degraded. However, passenger strands also emerge as active microRNAs. It is unknown whether the guide-to-passenger-strand ratio can be actively regulated and which factors influence strand incorporation into the RISC. Here, we identify a microRNA with a variable guide-to-passenger-strand ratio along with its regulatory factor: Human Argonaute-3 specifically enhances the passenger strand expression and activity of the tumor suppressor microRNA let-7a. This post-maturational effect is mediated by the Ago3 PAZ and MID domains yielding an elevated affinity for let-7a-3p. Notably, this is independent of the 5′-terminal basepair stability, challenging the universality of the respective rule for microRNA strand selection. Thus, this study uncovers the first protein regulator of the ratio between microRNA guide and passenger strand expression and activity.
Keywords: Argonaute-3, microRNA, let-7a, passenger strand, thermodynamic stability
Introduction
MicroRNAs (miRNAs) are essential post-transcriptional gene regulators inhibiting target mRNAs through translational repression, deadenylation, or mRNA cleavage.1-4 The primary miRNA transcript, the pri-miRNA, is cleaved by the microprocessor complex of Drosha and DGCR8, exported into the cytoplasm by Exportin-5 and subsequently processed by Dicer into a miRNA duplex. This duplex is unwound by a so far unknown mechanism into two single-stranded RNA fragments of 21 nucleotides (nt) in length, the guide and the passenger strand. In the generally accepted model, only the guide strand is incorporated into the RNA-Induced Silencing Complex (RISC) while the passenger strand is destined for degradation. However, highly conserved passenger strands5,6 can also be incorporated into RISCs and serve as active miRNAs bound to Argonaute (Ago) proteins. Additionally, the loop region can give rise to a loop-miR.7,8 For some miRNAs, both arms of the pre-miRNA hairpin are found in RISCs in significant amounts underlining the functional relevance of these less abundant products of miRNA biogenesis.9 Recent publications demonstrate tissue- and developmental stage-specific differences in the ratios of guide and passenger strands10-12 and their impact on the cellular phenotype, e.g., by targeting tumor suppressor genes.5,6,13-18 While miR-29c* is linked to the prognosis of malignant pleural mesothelioma,16 miR-378* mediates the metabolic shift in breast cancer cells19 and miR-199a* regulates the MET protooncogene.20 Thus far, two mechanisms are described to regulate the ratio between guide and passenger strand expression in the human system: the identity of the 5′ terminal nucleotide and the relative thermodynamic stability of the two ends of the duplex. The strand with the less stable basepair at the 5′-end is more frequently incorporated into the RISC.21,22 If this was the main or only mechanism of miRNA strand selection, this model would predict a rather constant ratio of guide-to-passenger strands in the cell dictated by the structure of the miRNA duplex.
Let-7a, one of the first miRNAs described, is highly conserved among species in sequence and function,23 exists in several isoforms,24 and dysregulation of let-7 leads to a less differentiated cellular state and the development of cancer: By targeting RAS25,26 and HMGA2,27-29 let-7 operates as a tumor suppressor in human cells. However, the infrequent upregulation of certain let-7 family members has also been observed. This indicates that individual let-7 family members can have different activities, e.g., let-7b* upregulation coincides with let-7e* downregulation in malignant mesothelioma,30 which demonstrates the functionality of passenger strands derived from let-7 family members. Due to its important physiological and pathological effects, let-7 expression and activity is tightly regulated at the transcriptional level,31,32 as well as by a wide range of processing factors. While hnRNP A1 inhibits Drosha-mediated pri-let-7 cleavage,33,34 its antagonist KSRP promotes Drosha- and Dicer-mediated processing of several miRNAs, including let-7a.35 Furthermore, lin-28 recognizes pre-let-7a via a motif in its terminal loop and recruits a TUTase to mark pre-let-7a for degradation.36
Argonaute (Ago) proteins are highly conserved and ubiquitously expressed in all higher eukaryotes.37,38 Next to their role as RISC effectors, Argonaute proteins regulate miRNA expression post-transcriptionally. Recent publications have shown that the stability39 and, consequently, the abundance of mature miRNAs is regulated by the levels of Ago protein expression.40-43 Moreover, Ago2 possesses the potential to increase the efficacy and specificity of target silencing41 and to cleave the 3′-arm of siRNAs (small interfering RNAs)44-46 or some highly complementary pre-miRNA hairpins to generate the ac-pre-miRNA (Ago2-cleaved precursor-microRNA).40 This cleavage facilitates strand separation by affecting the thermodynamic stability as for small interfering RNAs.44-47 Additionally, Ago2-mediated pre-miRNA cleavage can induce Dicer-independent miRNA processing: The ac-pre-miRNA of miR-451 can be exonucleolytically degraded into a mature miRNA.48-50 Lastly, human Ago2 or C. elegans ALG-1 can bind to the pri-miRNA of let-7 and promote its processing.51 The human Ago proteins Ago1 and Ago3 do not possess endonucleolytic slicer activity, but can be activated by synthetic mutations.52,53 Nevertheless, no specific functions have been identified for the individual Ago1, Ago3, and Ago4 proteins in mammalian systems.
Here, we provide evidence that protein factors possess the ability to shift the equilibrium between the let-7a guide and passenger strands and that Argonaute-3 can have specific effects among the other members of the human Argonaute family. Thus, this is the first study identifying a differential guide-to-passenger-strand microRNA ratio along with its regulatory factor.
Results
Argonaute-3 specifically regulates let-7a-3p expression
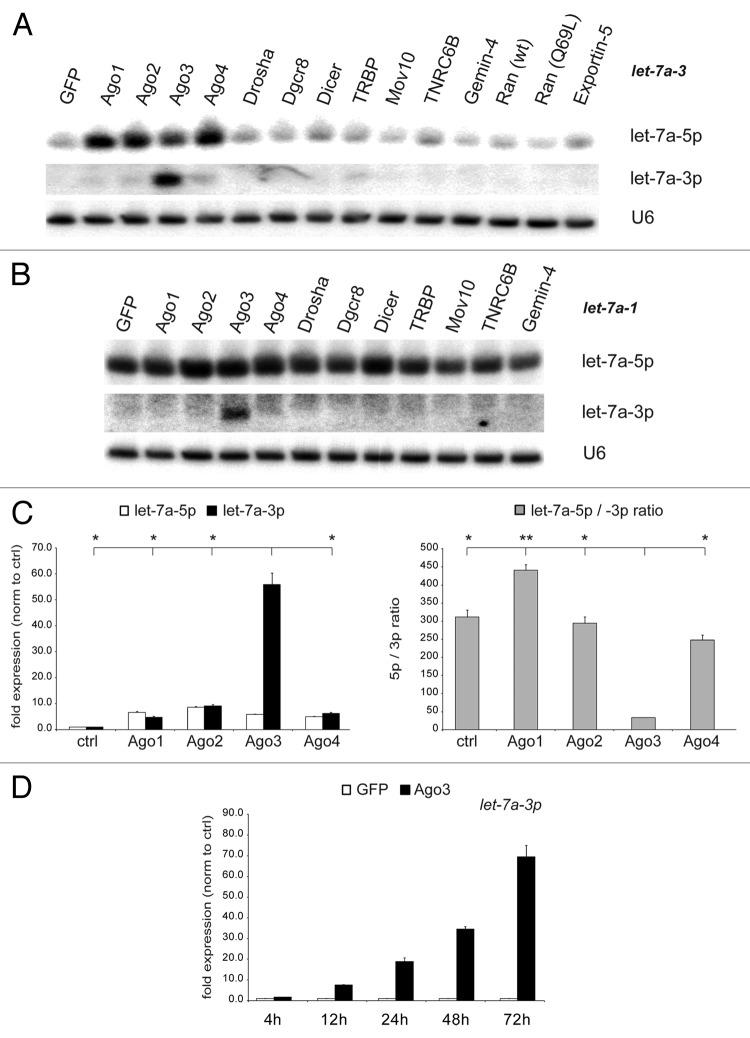
Previous studies reported that processing factors post-transcriptionally affect miRNA expression and activity.40-42,54 Hence, we aimed to determine whether human proteins that have either been linked to the miRNA biogenesis pathway or to RNA interference (RNAi) could have differential effects on guide and passenger strand expression and, thereby, shift their equilibrium. We transfected a series of miRNA processing factors into human HEK293 cells to assess their impact on microRNA maturation. Due to its important role in health and disease, we selected the miRNA let-7a and characterized the let-7a guide strand, let-7a-5p, as well as its passenger strand, let-7a-3p. Since cell lines often express low levels of miRNAs,55 we transiently co-transfected a CMV-driven let-7a primary transcript to saturate the miRNA biogenesis machinery along with the processing factors. Northern blot analysis revealed that the expression of each member of the Argonaute family gave rise to increased levels of mature let-7a-5p expression, confirming our previous report.40 Surprisingly, passenger strand (let-7a-3p) expression was markedly and specifically increased only by Ago3 but not by any other protein tested, not even by the closely related Ago protein family members (Fig. 1A). The enhanced mature let-7a-3p expression by Ago3, further referred to as the Ago3 effect, was consistently observed with different primary let-7a constructs generated from different loci in the genome, let-7a-3 and let-7a-1 (Fig. 1A and B). To independently confirm this Ago3 function, the results were validated by quantitative RT-PCR (qRT-PCR): While each Argonaute upregulated ectopic let-7a-5p guide strand expression 5-fold, exclusively Ago3 resulted in a 55-fold increase in let-7a-3p expression, leading to a significant change of the let-7a-5p/-3p ratio (TTest; all P values can be found in Table S2). Ago1, Ago2, or Ago4 did not significantly affect this equilibrium (Fig. 1C). To validate this result in an independent cell line to exclude a cell line-specific effect, the experiments were reproduced in HeLa cells with similar results (Fig. S1). Furthermore, let-7a expression was determined at different time points confirming its time-dependent increase after Ago3 overexpression (Fig. 1D).
Figure 1. Argonaute-3 overexpression increases exogenous let-7a-3p expression. (A and B) HEK293 cells were co-transfected with constructs encoding let-7a-3 pri-miRNA (A) and let-7a-1 pri-miRNA (B) and protein factors involved in miRNA biogenesis (Argonautes, Drosha, DGCR8, Dicer, TRBP, Mov10, TNRC6B, Gemin-4, Ran, Exportin-5). Mature miRNA expression was determined by northern blotting. Reprobing of the blot for U6 snRNA served as loading control. (C) Results were confirmed by qRT-PCR analysis for let-7a-5p (white bars) and let-7a-3p (black bars) and the let-7a-5p/-3p ratio (gray bars) was calculated. (D) Let-7a-3p expression upon Ago3 overexpression (black bars) in comparison to control transfections (white bars) was determined over time. Depicted is the mean expression (+SEM) of three independent experiments as compared with EGFP-transfected cells.
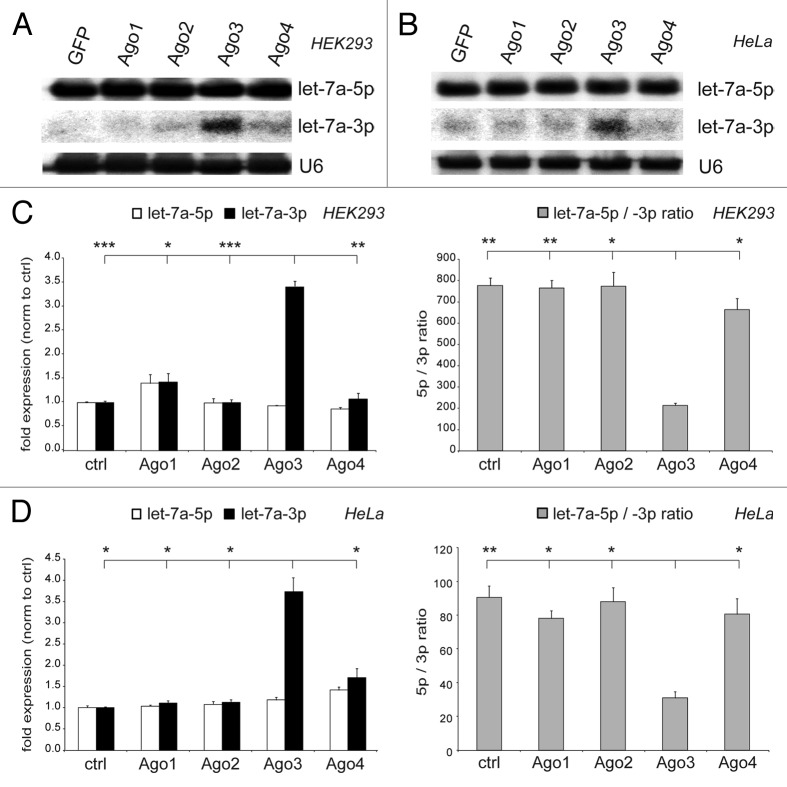
To analyze the physiological effect of Ago3 on the let-7a strand ratio, endogenous let-7a was tested for its regulation by Ago3. Ago3 overexpression specifically increased the expression of the endogenous passenger strand, thereby significantly decreasing the let-7a-5p/-3p ratio, confirming the data presented above. Again, the data were validated in two cell lines, HEK293 (Fig. 2A and C) and HeLa (Fig. 2B and D), and verified by two independent methods, northern blotting (Fig. 2A and B) and qRT-PCR analysis (Fig. 2C and D). As described previously, Argonaute overexpression did not alter the expression of the mature endogenous guide strands, probably due to sufficient Ago expression in the cell when the miRNA processing machinery was not saturated with exogenous pri-miRNAs.
Figure 2. Argonaute-3 overexpression increases endogenous let-7a-3p expression. (A and B) Endogenous mature miRNA expression in HEK293 (A) and HeLa (B) cells transfected with Argonaute proteins 1–4 was determined by northern blotting. Reprobing of the blot for U6 snRNA served as loading control. (C and D) Results were confirmed by qRT-PCR analysis for let-7a-5p (white bars) and let-7a-3p (black bars) in HEK293 (C) and HeLa (D) cells and the let-7a-5p/-3p ratio (gray bars) was calculated. Depicted is the mean expression (+SEM) of three independent experiments as compared with the EGFP-transfected control cells.
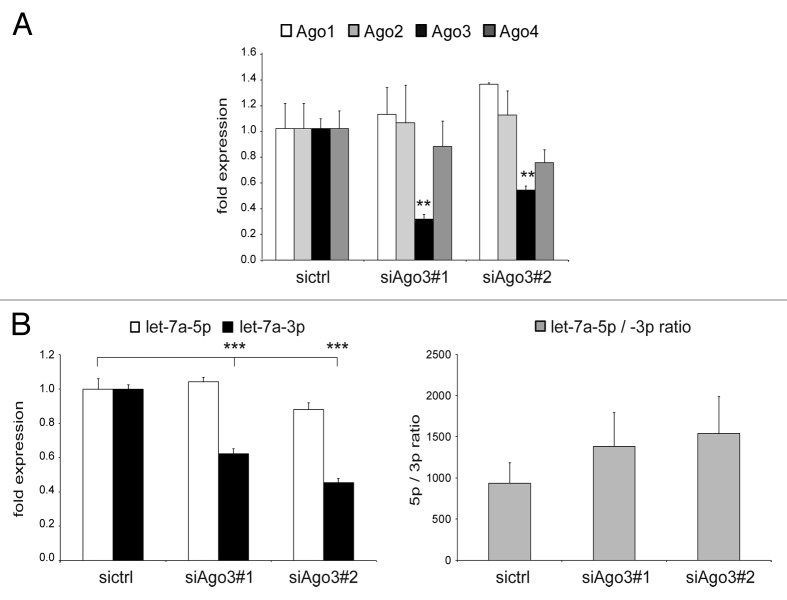
After uncovering the effect of exogenous Ago3 on ectopic as well as endogenous let-7a, we tested whether endogenous Ago3 also had an impact on the expression level of endogenous let-7a-3p. We used two different siRNAs that specifically reduced the expression level of Ago3 (70–80% reduction), but did not significantly affect Ago1, Ago2, or Ago4 expression (Fig. 3A). Both siRNAs gave rise to a strongly reduced expression of let-7a-3p coinciding with constant let-7a-5p expression. Thus, loss of endogenous Ago3 decreased endogenous let-7a-3p expression without affecting let-7a-5p (Fig. 3B).
Figure 3. Loss of endogenous Argonaute-3 decreases endogenous let-7a-3p expression. (A and B) HEK293 cells were transfected with two different siRNAs against Argonaute-3 and knockdown efficiency (A) and mature let-7a-5p (white bars) and let-7a-3p (black bars) expression (B) were determined by qRT-PCR analysis. Additionally, let-7a-5p/-3p ratios (gray bars) were calculated. Depicted is the mean expression (+SEM) of three independent experiments as compared with the control-siRNA-transfected cells.
The impact of Ago3 on endogenous let-7a-3p raised the question whether this was a specific effect or whether other miRNAs might be differentially affected by different Ago proteins. Hence, mature endogenous miRNA expression was also determined by qRT-PCR analysis for other miRNA guides as well as a number of additional miRNA/miRNA* pairs upon Ago overexpression (Figs. S2 and 3) or knockdown (Fig. S4). Several of these miRNAs were selected from deep sequencing studies after Argonaute immunoprecipitation for their putative strong binding to Ago3.56,57 None of these miRNAs displayed an effect comparable in strength or specificity to the effect of Ago3 on the let-7a passenger strand. None of the endogenous miRNAs was more than 1.6-fold induced by any of the Argonaute proteins. This indicates a highly specific interaction between Ago3 and the let-7a passenger strand.
Argonaute-3 specifically regulates let-7a-3p activity
After discovering the specific effect of Ago3 on let-7a-3p passenger strand expression, we addressed the question whether let-7a-3p was an active miRNA—serving in this sense as a “guide strand”—and whether its activity was affected by Ago3.
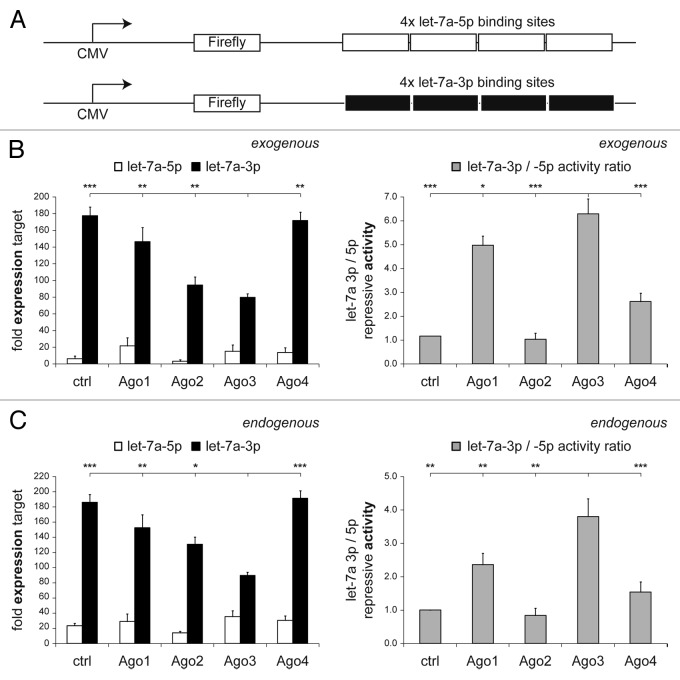
To analyze let-7a-3p activity, we created a reporter construct encoding luciferase fused to a 3′ untranslated region (3′-UTR) with four binding sites complementary to the guide (let-7a-5p) or the passenger strand (let-7a-3p) of let-7a, respectively (Fig. 4A).
Figure 4. Overexpression of Argonaute-3 increases let-7a-3p activity. HEK293 cells were transfected with firefly luciferase constructs containing four let-7a-5p or let-7a-3p-binding sites in their 3′-UTRs (A) as well as let-7a, and Ago1–4, as indicated. Renilla luciferase was co-transfected for normalization. (B and C) Depicted is the mean target expression (+SEM) of six experiments for let-7a-5p (white bars) and let-7a-3p (black bars) targets either regulated by exogenous (B) or endogenous let-7a (C) compared with an empty control plasmid. Additionally, the ratio of the repressive activity of the let-7a-3p divided by the let-7a-5p repressive activity is provided (gray bars).
HEK293 cells were co-transfected with the reporter constructs and Argonaute proteins. Forty-eight hours after transfection, exogenous (Fig. 4B) and endogenous (Fig. 4C) let-7a activities were quantified by luciferase assays. MiRNA target expression of both strands and the ratio of the miRNA strand activities were calculated. Compared with a control vector or with the other Ago proteins, Ago3 overexpression specifically increased exogenous let-7a-3p activity—as indicated by a decrease in luciferase target gene expression—activity while let-7a-5p activity was not affected (Fig. 4B). As described before,41 overexpression of non-slicing Ago1 protein competed with endogenous Ago2 and, hence, inhibited let-7a-5p activity. Luciferase assays were repeated and results were confirmed for endogenous let-7a activity (Fig. 4C).
While targets for various miRNA guide and passenger strands have already been identified and experimentally validated, let-7a-3p targets remained elusive. To analyze whether let-7a-3p could function as an active miRNA and whether Ago3 overexpression could increase its silencing activity, we predicted let-7a-3p targets and tested their regulation upon let-7a and Ago3 overexpression. Let-7a-3p binding sites were identified in silico in the 3′-UTR of the Ras-related GTP-binding protein RAB10. For target validation, HEK293 cells were co-transfected with let-7a, Ago3, and reporter constructs encoding luciferase fused to the 3′-UTR of RAB10. Luciferase assays confirmed a decrease of luciferase expression upon let-7a overexpression, validating this gene as a target. A small but significant additional target silencing was detected upon let-7a and Ago3 co-transfection corroborating the increase of let-7a-3p activity by Ago3 (Fig. S5A) and underlining the data obtained with synthetic let-7a* target constructs (Fig. 4). For control purposes, reporter constructs encoding luciferase fused to the 3′-UTR of RAB10 that were mutated in the let-7a-3p binding sites were not regulated by let-7a-3p or Ago3. To further validate this effect, endogenous RAB10 protein expression levels were detected upon Ago3 and let-7a overexpression by western blotting (Fig. S5B and C).
Taken together, overexpression of Ago3 did not only enhance let-7a-3p expression but also increased let-7a-3p activity, indicating that let-7a-3p could be incorporated into active and functional RISCs.
Argonaute-3 affects late phases of the miRNA lifecycle
MiRNA biogenesis can be regulated at each step of the processing pathway.4,58 Consequently, we investigated whether pri- or pre-let-7a levels were affected by Ago3. Quantitative RT-PCR analysis and northern blotting revealed that exogenous and endogenous pri- and pre-let-7a levels were slightly upregulated due to overexpression of Ago1–4. However, this effect was not specific for Ago3 (Fig. S6A–D) and could thus not explain the specific induction of let-7a-3p. Moreover, Ago3 could specifically increase the expression of the passenger strand from a co-transfected miRNA duplex, indicating that Ago3 affects late phases of the let-a7-3p miRNA lifecycle (Fig. S6E).
Furthermore, several groups recently reported that RNA-binding factors interacted with the terminal loop of miRNAs to regulate their processing.33-35,59 To uncover whether Ago3 or an Ago3-affected cofactor could act in a similar way, we swapped the loop of let-7a and a miRNA that did not show the Ago3 effect, miR-193a, generating hybrid pri-miRNA structures of a let-7a duplex + miR-193a loop and a miR-193a duplex + let-7a loop (Fig. S7A). For the wild-type let-7a as well as the wild-type miR-193, the results were reproduced as expected: exogenous Ago3 specifically increased let-7a-3p (Fig. S7B), but neither let-7a-5p nor any strand of miR-193 (Fig. S7C) were specifically affected by Ago3. The let-7a duplex fused to the miR-193 loop structure also gave rise to a significantly increased level of let-7a-3p upon Ago3 expression (Fig. S7D), indicating that all information for the Ago3 effect was included in the let-7a duplex and independent of the loop structure or sequence. Vice versa, the miR-193 duplex fused to the let-7a loop did not specifically respond to Ago3 (Fig. S7E), corroborating that the let-7a duplex rather than the loop of the miRNA was recognized by Ago3.
Taken together, we concluded that the Ago3 effect likely occurred either in the late phases of the processing pathway after Drosha-, Dicer-, or Ago-mediated cleavage, or after maturation at the level of RISC incorporation or miRNA stability. It did not rely on the terminal loop structure of let-7a since the miRNA duplex contained all required information.
The Argonaute-3 effect is not sensitive to the thermodynamic stability of the first basepair in the let-7a microRNA duplex
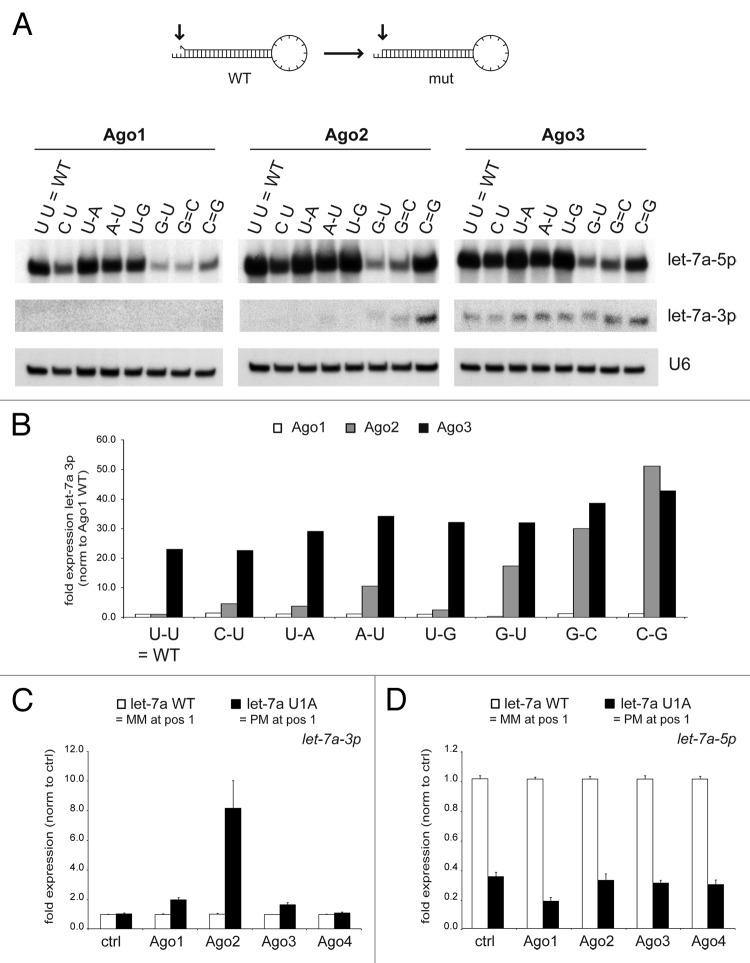
The dominant rule for human miRNA strand selection depends on the thermodynamic stability of the first basepair at either end of the miRNA duplex.21,22 Thus, the stability of the terminal basepair would be one possible signal within the let-7a duplex for the Ago3 effect. Therefore, let-7a mutants were generated that displayed differences in the complementarity at the 5′-terminal basepair of the guide strand. Northern blotting analysis revealed that increased complementarity at the 5′-end of the guide strand increased the expression of let-7a-3p in combination with Ago2—as predicted by the terminal basepair stability rule (Fig. 5A and B). However, the expression of let-7a-3p was surprisingly not significantly affected by the different mutations in the presence of Ago3. In contrast, the let-7a-3p passenger strand expression, together with Ago3, was in all cases similar to the expression reached for Ago2 only by the addition of a strong G:C basepair at the 5′-end. For Ago1 overexpression, the let-7a-3p passenger strand was not detected, also indicating that the terminal basepair stability rule in the case of let-7a might apply mainly to Ago2—for which it was also first described.
Figure 5. Thermodynamic stability of the terminal basepair mainly controls miRNA strand selection in the presence of Ago2. (A and B) HEK293 cells were co-transfected with Ago1–3 and constructs encoding let-7a-3 pri-miRNA that were mutated at the primary basepair of the 5′-arm to affect thermodynamic stability. Mature let-7a-5p and 3p expression was determined by northern blotting (A). Reprobing of the blot for U6 snRNA served as loading control. (B) Signal intensity was quantified by densitometry and normalized to wild-type miRNA expression with ectopic Ago1. Mature let-7a-3p (C) and -5p (D) expression in HEK293 cells co-transfected with wild-type let-7a (white bars) and a let-7a construct that was mutated at position 1 to induce perfect complementarity at the first basepair (let-7a U1A, black bars), was determined by qRT-PCR. Depicted is the mean expression (+SEM) of three independent experiments.
These results were validated and quantified by qRT-PCR analysis for let-7a WT (U U) and let-7a U1A (A-U) (Fig. 5C). Again, the stabilized terminal basepair increased let-7a-3p only in the presence of ectopic Ago2. In contrast, the terminal basepair stability only weakly affected the impact of other Ago proteins. In turn, let-7a-5p guide strand expression was always reduced by stabilizing its terminal basepair. In line with this data, mouse embryonic fibroblasts (MEFs) lacking the expression of Ago2 displayed reduced levels of let-7a-5p but constant expression of let-7a-3p (Fig. S8). However, this effect was not universal to all miRNAs since let-7c and miR-27b mutants that displayed differences in the complementarity at the 5′-terminal basepair of the guide strand did not display significant differences upon the overexpression of the four Argonaute proteins (Fig. S9). Furthermore, we tested whether regions other than the first basepair (Fig. 5A) of the miRNA duplex might affect the Ago3 effect. Therefore, let-7a mutants were generated altering the complementarity in the center of the duplex (position 10, 11, and the combination of both). qRT-PCR analysis upon co-transfection of these let-7a mutants together with Ago1–4 revealed that all three mutants still displayed increased let-7a-3p expression upon Ago3 overexpression, providing evidence that the Ago3 effect was neither dependent on the sequence nor the complementarity of the terminal or central basepairs of the let-7a duplex (Fig. S10).
This indicated that the let-7a guide strand selected through the terminal basepair stability was mainly affected by Ago2 loss, whereas the passenger strand expression was less dependent on the terminal basepair expression and more dependent on Ago3.
The Argonaute-3 effect is mediated by its PAZ and MID domain
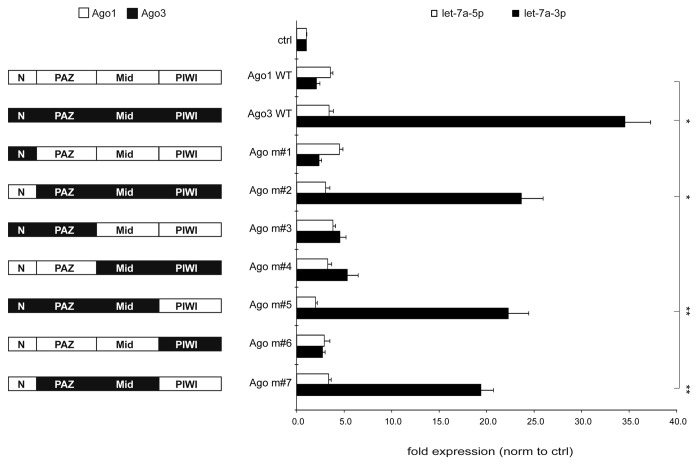
To uncover the domain responsible for the Ago3 effect on let-7a passenger strand expression, we swapped the N-terminal, PAZ, MID, or PIWI domains of Ago3 with its closest homolog Ago1 (Fig. 6). Expression of the four Ago proteins and the respective mutants was verified by western blotting (Fig. S11). These two Argonaute proteins were chosen since they are expressed at comparable levels in human cells and are both slicing-deficient. We transiently co-transfected CMV-driven let-7a primary transcripts and constructs encoding the indicated Ago WT or Ago domain swap proteins into HEK293 cells for 48 h and determined let-7a guide and passenger strand expression by qRT-PCR analysis. Consistent with the data presented above, overexpression of Ago3, but not Ago1, increased let-7a-3p expression while let-7a-5p expression remained constant. While Ago3 constructs lacking the Ago3 N terminus (Ago m#2) or PIWI domain (Ago m#5) or both (Ago m#7) still displayed increased let-7a-3p expression, replacing the PAZ or MID domain completely abolished the upregulation of the let-7a passenger strand (Fig. 6). Thus, the domains that are binding the 5′- and 3′-end of the mature miRNA, PAZ and MID (Ago m#7), were crucial to maintain the Ago3 effect on let-7a-3p. To confirm the specificity of the Ago3 effect, domain swap experiments were repeated with a miRNA that did not show specific upregulation upon Ago3 overexpression, miR-20a (Fig. S12).
Figure 6. The MID and PAZ domain mediate the increased let-7a-3p expression upon Argonaute-3 overexpression. Argonaute domain swap mutants were cloned exchanging individual domains of Argonaute-1 and Argonaute-3. These mutants were co-transfected together with a construct encoding the let-7a-3 pri-miRNA. Let-7a-5p (white bars) and 3p (black bars) expression was detected by qRT-PCR. Depicted is the mean expression (+SEM) of three independent experiments.
Argonaute-3 preferentially binds to let-7a-3p
Since the PAZ and the MID domain of Ago3 were the two domains directly involved in miRNA binding by anchoring the 3′-end and the 5′-end, respectively, we hypothesized that Ago3 might have an increased binding affinity toward let-7a-3p compared with other Ago proteins. Since multiple Ago proteins were available for RISC formation, single-stranded miRNAs could also be distributed into various RISCs. While miRNA sorting had been described for several species,60-62 a common sorting step in humans has not been identified to date.56,57 To investigate whether Ago3 preferentially bound to the passenger strand of let-7a, we transiently transfected HEK293 cells with epitope-tagged human Argonaute proteins and determined the binding affinity of the let-7a guide and passenger strands to Ago1, Ago2, and Ago3 (Fig. S13A). Ago4 was excluded from the analysis due to its low precipitation efficiency. Ago3 displayed the highest 3p/5p binding affinity for exogenous (Fig. S13B) and endogenous (Fig. S13C) let-7a, in comparison to Ago1 and Ago2. Thus, an increased affinity of Ago3 for let-7a-3p could be a part of the mechanism elevating the let-7a-3p expression and activity by Ago3. Furthermore, to test whether this effect was dependent on direct binding of the miRNA to the Ago3 PAZ domain, we transiently co-transfected the let-7a primary transcript together with an Ago3 construct that contained a deletion in the PAZ domain and was therefore binding-defective but still expressed (Fig. S13D and E). In comparison to wild-type Ago3, the PAZ mutant did not upregulate let-7a-3p expression (Fig. S13D).
Hemin treatment increases Ago3 and let-7a-3p expression
As mentioned above, the let-7a-3p passenger strand displayed lower expression values than the let-7a-5p guide strand. However, to verify that this miRNA could be of biological relevance, even though displaying a lower abundance, its expression was compared with other miRNA guide and passenger strands and mirtrons. These had previously been shown to execute cellular functions and mediate gene silencing despite their low level of expression.15,17-20,63-70 Let-7a-3p expression ranked well among these functional miRNAs (Fig. S14A).
Lastly, we identified a physiological example of Ago3 and let-7a-3p co-regulation. In an in-depth analysis of the GEO database, we found an induction of Ago3 upon a physiological stimulus: when K562 cells were treated with Hemin, an inducer of erythroid commitment, Ago3 expression was specifically induced while the other Ago proteins remained unchanged. qRT-PCR analysis verified that Hemin treatment upregulated the expression of Ago3 as it was shown previously.71 This Ago3 induction was associated with an increase in endogenous let-7a passenger strand expression while let-7a-5p expression was not significantly altered (Fig. S14B).
Discussion
Human Argonaute-3 activates the let-7a passenger strand
Our study uncovers a first example of a vertebrate protein factor, Argonaute-3, specifically affecting the guide-to-passenger-strand ratio of the miRNA let-7a. In summary, we propose a multi-layered mechanism for the observed impact of Ago3 on the let-7a-3p passenger strand expression and activity. It can be subdivided into at least two Ago3-specific effects: First, the thermodynamic stability at either end of the let-7a duplex has only a limited impact on the strand selection in the presence of Ago3 in stark contrast to Ago2. Second, Ago3 binds more strongly to the let-7a-3p passenger strand than other Argonaute proteins. Ago3 differentially regulates the expression of let-7a-3p in a post-maturational effect mediated via its PAZ and MID domain—the two domains anchoring both ends of the mature miRNA (Fig. 6). In contrast to previously published mechanisms regulating miRNA biogenesis,33-35 the Ago3 effect is independent of the sequence and structure of the loop in the pre-miRNA hairpin. All information for this specific effect is included in the let-7a miRNA duplex structure and sequence of the two mature miRNA strands. Thus, protein factors can shift the equilibrium between the let-7a guide and passenger strand expression and activity.
The Ago3-induced let-7a-3p is an active miRNA regulating synthetic as well as endogenous target gene sequences, which is in good concordance with recent studies corroborating the emerging role of miRNA passenger strands as important active players in health and disease.5,6,13-18 While let-7a-5p and the proto-oncogene p60/RAS regulate each other via a feedback loop, the let-7a-3p passenger strand might negatively regulate RAB10, another member of the Ras oncogene family of small GTPases that are involved in cellular signal transduction.72 Hence, since the expression and the activity of the let-7a passenger strand is increased upon Ago3 overexpression, this Ago3 effect is more likely to be explained by differences in Ago loading rather than a simple unwinding defect, which should not impact let-7a-3p activity.
MicroRNA sorting and selection: Beyond terminal basepair stability
MicroRNA biogenesis requires multiple sorting decisions during its final stages: First, the miRNA duplex can give rise to two different mature miRNAs derived from the former 5′- or 3′-arm of the pre-miRNA hairpin. Second, multiple Ago proteins are available in a cell so that miRNAs can be incorporated into different RISCs.
One mechanism regulating the ratio between guide and passenger strand expression in human cells is the relative thermodynamic stability of the terminal basepairs at either end of the miRNA duplex. This rule was developed for Ago2-interacting siRNA guide strands with perfect complementarity.21,22 As inferred from these studies, miRNA strand selection has also been proposed as a process dominated by the thermodynamic stability of the first basepair of the miRNA duplex giving rise to more or less stable ratios of the active miRNA guide strand and the often degraded passenger strand. However, this dogma had already been challenged by several observations that (1) the relative expression ratios between guide and passenger strands differ between different cell types12 as well as (2) several miRNA duplexes give rise to two active miRNAs that are sometimes even expressed at comparable levels.9,11
In A. thaliana61,73,74 and D. melanogaster,75-77 sorting and RISC incorporation depend on the 5′ terminal nucleotide. Complementarity of the miRNA duplex is critical for strand selection in D. melanogaster.60,62 However, a common sorting step in mammals has not been identified to date, even though several publications point toward the fact that it might be linked to the 5′ terminal nucleotide.56,78 Since the Ago3 effect has been unraveled for only one mature miRNA, any impact of the terminal nucleotide identity cannot be statistically analyzed. However, many miRNA strands with the same starting nucleotide as let-7a-3p and let-7a-5p, respectively, have been analyzed by qRT-PCR for the effect of different Ago proteins (Fig. S15). Since none of these was specifically regulated by Ago3 (Figs. S2–4), it appears unlikely that the terminal nucleotide identity controls the Ago3 effect.
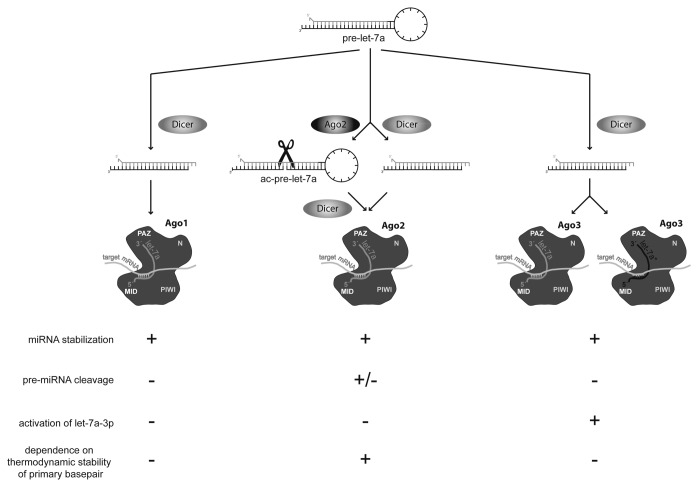
On the one hand, our experiments confirmed the 5′-terminal basepair stability rule for let-7a-5p in the presence of Ago2: the guide strand with the less stable basepair at its 5′-end was incorporated into the RISC more frequently and mutations altering the complementarity at the 5′-end affected the equilibrium between guide and passenger strand. In contrast, in the presence of Ago3, the stability of the terminal ends had little or no impact on the expression of let-7a-3p, further challenging the universal applicability of the terminal basepair rule for all Ago proteins and miRNAs (Fig. 7).
Figure 7. Argonaute proteins handle the let-7a microRNA precursor differently. Three human Ago proteins possess the potential to mediate target silencing by binding the single-stranded mature miRNA via their PAZ and MID domains and to increase miRNA stability. In contrast, due to its slicing-proficient PIWI domain, Ago2 is the only member of this family that is able to generate the ac-pre-let-7a, which is further processed into an active miRNA. Here, we provide evidence that Argonaute-3 specifically enhances the passenger strand expression and activity of let-7a and, thereby, identify a specific function for Ago3 and a protein regulator of a miRNA guide-to-passenger-strand ratio. This effect is mediated by the Ago3 PAZ and MID domains, yielding an elevated affinity for let-7a-3p. Moreover, the rule of strand selection based on terminal basepair stability mainly applies to let-7a incorporated into Argonaute-2 complexes.
A similar mechanism has recently been identified in plants: miR-393/393* represents an example of a miRNA where guide and passenger strands are both active players, displaying distinct binding affinities to Ago2 and Ago1, respectively, and, consequently, silencing different target genes, both involved in the plant’s antibacterial immunity.79 Even though animals and plants differ significantly in miRNA processing, the number of Argonaute proteins, and the process of strand selection,61,80,81 this study underlines the fact that both guide and passenger strands could act as active miRNAs in complex with different Ago proteins.
Dual functionality: MicroRNA processing factors as microRNA regulators
This study also recapitulates that factors linked to miRNA biogenesis regulate the abundance of mature let-7a. Elevated expression of the let-7a-5p guide strand induced by all four Argonaute proteins is consistent with previous findings40,42 and mechanistically due to the stabilization of the mature miRNAs.39
In contrast, exclusively Ago3, but none of the other Argonaute proteins or processing factors, affects the let-7a-3p passenger strand expression and activity. While several publications characterize the different roles of Ago2 in microRNA and siRNA processing due to its slicing-proficient PIWI domain,40,41,44-46 this is the first specific function for mammalian Ago3.
This effect appears to be specific for let-7a even though this seems to be surprising given that functional equivalence and similar binding affinities for all four human Ago proteins have been reported.82-84 However, we cannot formally exclude that other miRNAs could also be differentially regulated by Ago3. We tested 33 candidate miRNAs, including miRNAs that were found to be preferentially bound to Ago3,56,57 but none of them was affected to a similar extent as let-7a-3p. Thus, let-7a-3p appears as a specific partner for Ago3. This correlates well with previous publications identifying factors that regulate processing of either one single miRNA48-50,85 or a small group of miRNAs,33-35,59,86-92 providing mechanisms to precisely regulate miRNA biogenesis, abundance, and activity. Given the multitude of miRNAs and miRNA processing factors or proteins affecting miRNA activity, we expect that many more examples of protein factors differentially regulating miRNA guide and so-called passenger strands could be found in the future that allow the cell to tightly control these important regulators of gene expression in health and disease.
Materials and Methods
Cell lines, transfections, and knockdown
HEK293 and HeLa cells were cultured in DMEM + 10% FCS + Glutamine (Sigma) at 37 °C in 5% CO2. Plasmids have been described above or previously.40,41 For transfection, cells were seeded to reach 90% confluence on the day of transfection and 3 µg plasmid DNA were transfected with 10 µl Polyethylenimine in TBS buffer per well in 6-well plates. Co-transfections were performed with 20 pmoles duplex and 200 ng plasmid DNA in a 24-well format. For siRNA knockdown, cells were reverse transfected in 6-well plates with 100 pmoles siRNA and 5 µl RNAiMAX (Invitrogen) according to the manufacturer’s instruction and lysed 72 h after transfection. 2 × 105 K562-cells were plated in a 24-well plate format and cultivated in the absence or presence of Hemin (50 µM) for 96 h.
RNA extraction, qRT-PCR, and northern blotting
Cells were lysed in 1 ml TRIzol (Invitrogen) at the indicated time points and RNA was isolated as previously described.40,93 MicroRNA expression was detected by TaqMan assays (Life Technologies) according to the manufacturer’s instructions or by stem-loop TaqMan assays as previously described.94 MiRNA northern blotting was performed as previously described40 and quantifications were performed using ImageJ software. Probe sequences are listed in Table S1.
Expression plasmid cloning and mutagenesis
Cloning or sources of expression plasmids were described previously.40 Mutagenesis of let-7a and miR-193a loop exchange, single point mutations in the miRNA duplex, and Argonaute-3 deltaPAZ mutant was performed by QuikChange PCR using Pfu Ultra AD (Stratagene) followed by a DpnI digest (Fermentas) and transformation into MACH1 chemocompetent E. coli cells. Argonaute domain swap mutants were cloned using XhoI and XbaI restriction sites. Primer sequences are listed in Table S1.
Luciferase assays
HEK293 cells were seeded to reach 80% confluency and transfected with 1.2 μg plasmid DNA and 4.0 μl Polyethylenimine (1 mg / ml) in a 24-well plate. Each transfection contained 50 ng pRL-TK or pRL-SV40 Renilla expression construct for normalization purposes (Promega), 150 ng pcDNA3.1D-Firefly expression construct, including the 3′-UTR of interest, 500 ng expression plasmid for the miRNA of interest, and 500 ng expression plasmid for the protein factor to be analyzed. After 48 h, cells were washed with 500 μl PBS and lysed in 120 μl Passive Lysis Buffer (Promega). Twenty µl lysate was subsequently analyzed using 50 μl LAR II substrate or Stop&Glow (1:5 dilutions) to determine Firefly and Renilla luciferase activity, respectively (Dual Luciferase Reporter Assay System; Promega). Each sample was analyzed in quadruplicates, and each transfection was performed six times. Firefly activity was normalized to Renilla activity, and mean values plus standard error of mean are depicted. Sequences are listed in Table S1.
Co-immunoprecipitation
For co-immunoprecipitation (Co-IP) experiments, two 10 cm plates of HEK293 cells per sample were transfected with appropriate plasmids for 2 d. Cells were washed twice with PBS and lysed in 500 μl lysis buffer (150 mM KCl; 25 mM Tris-HCl [pH 7.5]; 0.5% NP-40; 1x Complete Protease Inhibitor Cocktail Tablets [Roche]; 1 µl/ml Ribolock [Fermentas]) per plate. Lysates were cleared by centrifugation at 12000 g for 15 min at 4 °C and incubated with 60 μl anti-FLAG M2 (Sigma-Aldrich) agarose beads overnight at 4 °C. All IP samples were washed three times with 500 µl IP wash buffer (300 mM NaCl; 50 mM Tris [pH 7.5]; 1 mM NaF; 0.01% NP-40; 5 mM MgCl2) and once with PBS. For the detection of proteins, beads were boiled in protein sample buffer. For the detection of associated RNAs, beads were lysed in 1 ml Trizol.
Western blotting
Cells were lysed in 200 µl RIPA buffer, lysates were denatured, separated on 10% SDS-PAGE gels, and electroblotted onto Nitrocellulose membranes (Amersham). α-HA (Covance), α-FLAG (Sigma-Aldrich), and α-RAB10 (Abcam) primary antibodies were used for detection, α-Tubulin (Cell Signaling) antibody as loading control.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
Our research is supported by the Helmholtz Society (VH-NG-504), the DKFZ-MOST-Cooperation Program (Ca-135), the Marie Curie Program of the European Union (239308) and the German Research Foundation (TRR77 TP B03). We thank Catherina Hildenbrand for excellent technical assistance and all members of the Diederichs lab for helpful discussions. We are grateful to Drs Thomas Tuschl, Ramin Shiekhattar, Gideon Dreyfuss, Mien-Chie Hung, and Ian G Macara for providing expression plasmids.
Author Contributions
Diederichs S conceived the idea; Winter J and Diederichs S performed experiments, analyzed the results and wrote the paper. The authors declare no conflict of interest. This manuscript is part of the PhD thesis of Winter J.
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/rnabiology/article/26424
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/26424
References
- 1.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–14. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 3.Winter J, Diederichs S. MicroRNA biogenesis and cancer. Methods Mol Biol. 2011;676:3–22. doi: 10.1007/978-1-60761-863-8_1. [DOI] [PubMed] [Google Scholar]
- 4.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11:228–34. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 5.Okamura K, Phillips MD, Tyler DM, Duan H, Chou YT, Lai EC. The regulatory activity of microRNA* species has substantial influence on microRNA and 3′ UTR evolution. Nat Struct Mol Biol. 2008;15:354–63. doi: 10.1038/nsmb.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang JS, Phillips MD, Betel D, Mu P, Ventura A, Siepel AC, Chen KC, Lai EC. Widespread regulatory activity of vertebrate microRNA* species. RNA. 2011;17:312–26. doi: 10.1261/rna.2537911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Okamura K, Ladewig E, Zhou L, Lai EC. Functional small RNAs are generated from select miRNA hairpin loops in flies and mammals. Genes Dev. 2013;27:778–92. doi: 10.1101/gad.211698.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Winter J, Link S, Witzigmann D, Hildenbrand C, Previti C, Diederichs S. Loop-miRs: active microRNAs generated from single-stranded loop regions. Nucleic Acids Res. 2013;41:5503–12. doi: 10.1093/nar/gkt251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–14. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuchenbauer F, Mah SM, Heuser M, McPherson A, Rüschmann J, Rouhi A, Berg T, Bullinger L, Argiropoulos B, Morin RD, et al. Comprehensive analysis of mammalian miRNA* species and their role in myeloid cells. Blood. 2011;118:3350–8. doi: 10.1182/blood-2010-10-312454. [DOI] [PubMed] [Google Scholar]
- 11.Kuchenbauer F, Morin RD, Argiropoulos B, Petriv OI, Griffith M, Heuser M, Yung E, Piper J, Delaney A, Prabhu AL, et al. In-depth characterization of the microRNA transcriptome in a leukemia progression model. Genome Res. 2008;18:1787–97. doi: 10.1101/gr.077578.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ro S, Park C, Young D, Sanders KM, Yan W. Tissue-dependent paired expression of miRNAs. Nucleic Acids Res. 2007;35:5944–53. doi: 10.1093/nar/gkm641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Y, Wang X, Jiang J, Cao Z, Yang B, Cheng X. Modulation of T cell cytokine production by miR-144* with elevated expression in patients with pulmonary tuberculosis. Mol Immunol. 2011;48:1084–90. doi: 10.1016/j.molimm.2011.02.001. [DOI] [PubMed] [Google Scholar]
- 14.Musiyenko A, Bitko V, Barik S. Ectopic expression of miR-126*, an intronic product of the vascular endothelial EGF-like 7 gene, regulates prostein translation and invasiveness of prostate cancer LNCaP cells. J Mol Med (Berl) 2008;86:313–22. doi: 10.1007/s00109-007-0296-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Packer AN, Xing Y, Harper SQ, Jones L, Davidson BL. The bifunctional microRNA miR-9/miR-9* regulates REST and CoREST and is downregulated in Huntington’s disease. J Neurosci. 2008;28:14341–6. doi: 10.1523/JNEUROSCI.2390-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pass HI, Goparaju C, Ivanov S, Donington J, Carbone M, Hoshen M, Cohen D, Chajut A, Rosenwald S, Dan H, et al. hsa-miR-29c* is linked to the prognosis of malignant pleural mesothelioma. Cancer Res. 2010;70:1916–24. doi: 10.1158/0008-5472.CAN-09-3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsang WP, Kwok TT. The miR-18a* microRNA functions as a potential tumor suppressor by targeting on K-Ras. Carcinogenesis. 2009;30:953–9. doi: 10.1093/carcin/bgp094. [DOI] [PubMed] [Google Scholar]
- 18.Xu Y, Fang F, Zhang J, Josson S, St Clair WH, St Clair DK. miR-17* suppresses tumorigenicity of prostate cancer by inhibiting mitochondrial antioxidant enzymes. PLoS One. 2010;5:e14356. doi: 10.1371/journal.pone.0014356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eichner LJ, Perry MC, Dufour CR, Bertos N, Park M, St-Pierre J, Giguère V. miR-378(∗) mediates metabolic shift in breast cancer cells via the PGC-1β/ERRγ transcriptional pathway. Cell Metab. 2010;12:352–61. doi: 10.1016/j.cmet.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 20.Kim S, Lee UJ, Kim MN, Lee EJ, Kim JY, Lee MY, Choung S, Kim YJ, Choi YC. MicroRNA miR-199a* regulates the MET proto-oncogene and the downstream extracellular signal-regulated kinase 2 (ERK2) J Biol Chem. 2008;283:18158–66. doi: 10.1074/jbc.M800186200. [DOI] [PubMed] [Google Scholar]
- 21.Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–16. doi: 10.1016/S0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 22.Schwarz DS, Hutvágner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/S0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 23.Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Müller P, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–9. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 24.Griffiths-Jones S. miRBase: the microRNA sequence database. Methods Mol Biol. 2006;342:129–38. doi: 10.1385/1-59745-123-1:129. [DOI] [PubMed] [Google Scholar]
- 25.Kumar MS, Erkeland SJ, Pester RE, Chen CY, Ebert MS, Sharp PA, Jacks T. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci U S A. 2008;105:3903–8. doi: 10.1073/pnas.0712321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Esquela-Kerscher A, Trang P, Wiggins JF, Patrawala L, Cheng A, Ford L, Weidhaas JB, Brown D, Bader AG, Slack FJ. The let-7 microRNA reduces tumor growth in mouse models of lung cancer. Cell Cycle. 2008;7:759–64. doi: 10.4161/cc.7.6.5834. [DOI] [PubMed] [Google Scholar]
- 27.Lee YS, Dutta A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 2007;21:1025–30. doi: 10.1101/gad.1540407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. 2007;315:1576–9. doi: 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shell S, Park SM, Radjabi AR, Schickel R, Kistner EO, Jewell DA, Feig C, Lengyel E, Peter ME. Let-7 expression defines two differentiation stages of cancer. Proc Natl Acad Sci U S A. 2007;104:11400–5. doi: 10.1073/pnas.0704372104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guled M, Lahti L, Lindholm PM, Salmenkivi K, Bagwan I, Nicholson AG, Knuutila S. CDKN2A, NF2, and JUN are dysregulated among other genes by miRNAs in malignant mesothelioma -A miRNA microarray analysis. Genes Chromosomes Cancer. 2009;48:615–23. doi: 10.1002/gcc.20669. [DOI] [PubMed] [Google Scholar]
- 31.Chang TC, Yu D, Lee YS, Wentzel EA, Arking DE, West KM, Dang CV, Thomas-Tikhonenko A, Mendell JT. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet. 2008;40:43–50. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sampson VB, Rong NH, Han J, Yang Q, Aris V, Soteropoulos P, Petrelli NJ, Dunn SP, Krueger LJ. MicroRNA let-7a down-regulates MYC and reverts MYC-induced growth in Burkitt lymphoma cells. Cancer Res. 2007;67:9762–70. doi: 10.1158/0008-5472.CAN-07-2462. [DOI] [PubMed] [Google Scholar]
- 33.Guil S, Cáceres JF. The multifunctional RNA-binding protein hnRNP A1 is required for processing of miR-18a. Nat Struct Mol Biol. 2007;14:591–6. doi: 10.1038/nsmb1250. [DOI] [PubMed] [Google Scholar]
- 34.Michlewski G, Guil S, Semple CA, Cáceres JF. Posttranscriptional regulation of miRNAs harboring conserved terminal loops. Mol Cell. 2008;32:383–93. doi: 10.1016/j.molcel.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trabucchi M, Briata P, Garcia-Mayoral M, Haase AD, Filipowicz W, Ramos A, Gherzi R, Rosenfeld MG. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature. 2009;459:1010–4. doi: 10.1038/nature08025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heo I, Joo C, Cho J, Ha M, Han J, Kim VN. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol Cell. 2008;32:276–84. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 37.Höck J, Meister G. The Argonaute protein family. Genome Biol. 2008;9:210. doi: 10.1186/gb-2008-9-2-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hutvagner G, Simard MJ. Argonaute proteins: key players in RNA silencing. Nat Rev Mol Cell Biol. 2008;9:22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- 39.Winter J, Diederichs S. Argonaute proteins regulate microRNA stability: Increased microRNA abundance by Argonaute proteins is due to microRNA stabilization. RNA Biol. 2011;8:1149–57. doi: 10.4161/rna.8.6.17665. [DOI] [PubMed] [Google Scholar]
- 40.Diederichs S, Haber DA. Dual role for argonautes in microRNA processing and posttranscriptional regulation of microRNA expression. Cell. 2007;131:1097–108. doi: 10.1016/j.cell.2007.10.032. [DOI] [PubMed] [Google Scholar]
- 41.Diederichs S, Jung S, Rothenberg SM, Smolen GA, Mlody BG, Haber DA. Coexpression of Argonaute-2 enhances RNA interference toward perfect match binding sites. Proc Natl Acad Sci U S A. 2008;105:9284–9. doi: 10.1073/pnas.0800803105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.O’Carroll D, Mecklenbrauker I, Das PP, Santana A, Koenig U, Enright AJ, Miska EA, Tarakhovsky A. A Slicer-independent role for Argonaute 2 in hematopoiesis and the microRNA pathway. Genes Dev. 2007;21:1999–2004. doi: 10.1101/gad.1565607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou Y, Chen L, Barlogie B, Stephens O, Wu X, Williams DR, Cartron MA, van Rhee F, Nair B, Waheed S, et al. High-risk myeloma is associated with global elevation of miRNAs and overexpression of EIF2C2/AGO2. Proc Natl Acad Sci U S A. 2010;107:7904–9. doi: 10.1073/pnas.0908441107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leuschner PJ, Ameres SL, Kueng S, Martinez J. Cleavage of the siRNA passenger strand during RISC assembly in human cells. EMBO Rep. 2006;7:314–20. doi: 10.1038/sj.embor.7400637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607–20. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- 46.Rand TA, Petersen S, Du F, Wang X. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell. 2005;123:621–9. doi: 10.1016/j.cell.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 47.Miyoshi K, Tsukumo H, Nagami T, Siomi H, Siomi MC. Slicer function of Drosophila Argonautes and its involvement in RISC formation. Genes Dev. 2005;19:2837–48. doi: 10.1101/gad.1370605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cheloufi S, Dos Santos CO, Chong MM, Hannon GJ. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010;465:584–9. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cifuentes D, Xue H, Taylor DW, Patnode H, Mishima Y, Cheloufi S, Ma E, Mane S, Hannon GJ, Lawson ND, et al. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science. 2010;328:1694–8. doi: 10.1126/science.1190809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang JS, Maurin T, Robine N, Rasmussen KD, Jeffrey KL, Chandwani R, Papapetrou EP, Sadelain M, O’Carroll D, Lai EC. Conserved vertebrate mir-451 provides a platform for Dicer-independent, Ago2-mediated microRNA biogenesis. Proc Natl Acad Sci U S A. 2010;107:15163–8. doi: 10.1073/pnas.1006432107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zisoulis DG, Kai ZS, Chang RK, Pasquinelli AE. Autoregulation of microRNA biogenesis by let-7 and Argonaute. Nature. 2012;486:541–4. doi: 10.1038/nature11134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hauptmann J, Dueck A, Harlander S, Pfaff J, Merkl R, Meister G. Turning catalytically inactive human Argonaute proteins into active slicer enzymes. Nat Struct Mol Biol. 2013;20:814–7. doi: 10.1038/nsmb.2577. [DOI] [PubMed] [Google Scholar]
- 53.Schürmann N, Trabuco LG, Bender C, Russell RB, Grimm D. Molecular dissection of human Argonaute proteins by DNA shuffling. Nat Struct Mol Biol. 2013;20:818–26. doi: 10.1038/nsmb.2607. [DOI] [PubMed] [Google Scholar]
- 54.Yi R, Doehle BP, Qin Y, Macara IG, Cullen BR. Overexpression of exportin 5 enhances RNA interference mediated by short hairpin RNAs and microRNAs. RNA. 2005;11:220–6. doi: 10.1261/rna.7233305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–8. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 56.Azuma-Mukai A, Oguri H, Mituyama T, Qian ZR, Asai K, Siomi H, Siomi MC. Characterization of endogenous human Argonautes and their miRNA partners in RNA silencing. Proc Natl Acad Sci U S A. 2008;105:7964–9. doi: 10.1073/pnas.0800334105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Landthaler M, Gaidatzis D, Rothballer A, Chen PY, Soll SJ, Dinic L, Ojo T, Hafner M, Zavolan M, Tuschl T. Molecular characterization of human Argonaute-containing ribonucleoprotein complexes and their bound target mRNAs. RNA. 2008;14:2580–96. doi: 10.1261/rna.1351608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Breving K, Esquela-Kerscher A. The complexities of microRNA regulation: mirandering around the rules. Int J Biochem Cell Biol. 2010;42:1316–29. doi: 10.1016/j.biocel.2009.09.016. [DOI] [PubMed] [Google Scholar]
- 59.Piskounova E, Viswanathan SR, Janas M, LaPierre RJ, Daley GQ, Sliz P, Gregory RI. Determinants of microRNA processing inhibition by the developmentally regulated RNA-binding protein Lin28. J Biol Chem. 2008;283:21310–4. doi: 10.1074/jbc.C800108200. [DOI] [PubMed] [Google Scholar]
- 60.Förstemann K, Horwich MD, Wee L, Tomari Y, Zamore PD. Drosophila microRNAs are sorted into functionally distinct argonaute complexes after production by dicer-1. Cell. 2007;130:287–97. doi: 10.1016/j.cell.2007.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mi S, Cai T, Hu Y, Chen Y, Hodges E, Ni F, Wu L, Li S, Zhou H, Long C, et al. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell. 2008;133:116–27. doi: 10.1016/j.cell.2008.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tomari Y, Matranga C, Haley B, Martinez N, Zamore PD. A protein sensor for siRNA asymmetry. Science. 2004;306:1377–80. doi: 10.1126/science.1102755. [DOI] [PubMed] [Google Scholar]
- 63.Zhou Q, Gallagher R, Ufret-Vincenty R, Li X, Olson EN, Wang S. Regulation of angiogenesis and choroidal neovascularization by members of microRNA-23~27~24 clusters. Proc Natl Acad Sci U S A. 2011;108:8287–92. doi: 10.1073/pnas.1105254108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Westholm JO, Lai EC. Mirtrons: microRNA biogenesis via splicing. Biochimie. 2011;93:1897–904. doi: 10.1016/j.biochi.2011.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Veronese A, Lupini L, Consiglio J, Visone R, Ferracin M, Fornari F, Zanesi N, Alder H, D’Elia G, Gramantieri L, et al. Oncogenic role of miR-483-3p at the IGF2/483 locus. Cancer Res. 2010;70:3140–9. doi: 10.1158/0008-5472.CAN-09-4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sangokoya C, Telen MJ, Chi JT. microRNA miR-144 modulates oxidative stress tolerance and associates with anemia severity in sickle cell disease. Blood. 2010;116:4338–48. doi: 10.1182/blood-2009-04-214817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Miko E, Margitai Z, Czimmerer Z, Várkonyi I, Dezso B, Lányi A, Bacsó Z, Scholtz B. miR-126 inhibits proliferation of small cell lung cancer cells by targeting SLC7A5. FEBS Lett. 2011;585:1191–6. doi: 10.1016/j.febslet.2011.03.039. [DOI] [PubMed] [Google Scholar]
- 68.Kondo N, Toyama T, Sugiura H, Fujii Y, Yamashita H. miR-206 Expression is down-regulated in estrogen receptor alpha-positive human breast cancer. Cancer Res. 2008;68:5004–8. doi: 10.1158/0008-5472.CAN-08-0180. [DOI] [PubMed] [Google Scholar]
- 69.Jones D, Li Y, He Y, Xu Z, Chen H, Min W. Mirtron microRNA-1236 inhibits VEGFR-3 signaling during inflammatory lymphangiogenesis. Arterioscler Thromb Vasc Biol. 2012;32:633–42. doi: 10.1161/ATVBAHA.111.243576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen X, Yan Q, Li S, Zhou L, Yang H, Yang Y, Liu X, Wan X. Expression of the tumor suppressor miR-206 is associated with cellular proliferative inhibition and impairs invasion in ERα-positive endometrioid adenocarcinoma. Cancer Lett. 2012;314:41–53. doi: 10.1016/j.canlet.2011.09.014. [DOI] [PubMed] [Google Scholar]
- 71.Addya S, Keller MA, Delgrosso K, Ponte CM, Vadigepalli R, Gonye GE, Surrey S. Erythroid-induced commitment of K562 cells results in clusters of differentially expressed genes enriched for specific transcription regulatory elements. Physiol Genomics. 2004;19:117–30. doi: 10.1152/physiolgenomics.00028.2004. [DOI] [PubMed] [Google Scholar]
- 72.Subramani D, Alahari SK. Integrin-mediated function of Rab GTPases in cancer progression. Mol Cancer. 2010;9:312. doi: 10.1186/1476-4598-9-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Montgomery TA, Howell MD, Cuperus JT, Li D, Hansen JE, Alexander AL, Chapman EJ, Fahlgren N, Allen E, Carrington JC. Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell. 2008;133:128–41. doi: 10.1016/j.cell.2008.02.033. [DOI] [PubMed] [Google Scholar]
- 74.Takeda A, Iwasaki S, Watanabe T, Utsumi M, Watanabe Y. The mechanism selecting the guide strand from small RNA duplexes is different among argonaute proteins. Plant Cell Physiol. 2008;49:493–500. doi: 10.1093/pcp/pcn043. [DOI] [PubMed] [Google Scholar]
- 75.Czech B, Zhou R, Erlich Y, Brennecke J, Binari R, Villalta C, Gordon A, Perrimon N, Hannon GJ. Hierarchical rules for Argonaute loading in Drosophila. Mol Cell. 2009;36:445–56. doi: 10.1016/j.molcel.2009.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ghildiyal M, Xu J, Seitz H, Weng Z, Zamore PD. Sorting of Drosophila small silencing RNAs partitions microRNA* strands into the RNA interference pathway. RNA. 2010;16:43–56. doi: 10.1261/rna.1972910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Okamura K, Liu N, Lai EC. Distinct mechanisms for microRNA strand selection by Drosophila Argonautes. Mol Cell. 2009;36:431–44. doi: 10.1016/j.molcel.2009.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Frank F, Sonenberg N, Nagar B. Structural basis for 5′-nucleotide base-specific recognition of guide RNA by human AGO2. Nature. 2010;465:818–22. doi: 10.1038/nature09039. [DOI] [PubMed] [Google Scholar]
- 79.Zhang X, Zhao H, Gao S, Wang WC, Katiyar-Agarwal S, Huang HD, Raikhel N, Jin H. Arabidopsis Argonaute 2 regulates innate immunity via miRNA393(∗)-mediated silencing of a Golgi-localized SNARE gene, MEMB12. Mol Cell. 2011;42:356–66. doi: 10.1016/j.molcel.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Czech B, Hannon GJ. Small RNA sorting: matchmaking for Argonautes. Nat Rev Genet. 2011;12:19–31. doi: 10.1038/nrg2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Axtell MJ, Westholm JO, Lai EC. Vive la différence: biogenesis and evolution of microRNAs in plants and animals. Genome Biol. 2011;12:221. doi: 10.1186/gb-2011-12-4-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gu S, Jin L, Zhang F, Huang Y, Grimm D, Rossi JJ, Kay MA. Thermodynamic stability of small hairpin RNAs highly influences the loading process of different mammalian Argonautes. Proc Natl Acad Sci U S A. 2011;108:9208–13. doi: 10.1073/pnas.1018023108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Su H, Trombly MI, Chen J, Wang X. Essential and overlapping functions for mammalian Argonautes in microRNA silencing. Genes Dev. 2009;23:304–17. doi: 10.1101/gad.1749809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yoda M, Kawamata T, Paroo Z, Ye X, Iwasaki S, Liu Q, Tomari Y. ATP-dependent human RISC assembly pathways. Nat Struct Mol Biol. 2010;17:17–23. doi: 10.1038/nsmb.1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hagan JP, Piskounova E, Gregory RI. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat Struct Mol Biol. 2009;16:1021–5. doi: 10.1038/nsmb.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Newman MA, Thomson JM, Hammond SM. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA. 2008;14:1539–49. doi: 10.1261/rna.1155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rybak A, Fuchs H, Smirnova L, Brandt C, Pohl EE, Nitsch R, Wulczyn FG. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat Cell Biol. 2008;10:987–93. doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- 89.Sakamoto S, Aoki K, Higuchi T, Todaka H, Morisawa K, Tamaki N, Hatano E, Fukushima A, Taniguchi T, Agata Y. The NF90-NF45 complex functions as a negative regulator in the microRNA processing pathway. Mol Cell Biol. 2009;29:3754–69. doi: 10.1128/MCB.01836-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Trabucchi M, Briata P, Filipowicz W, Rosenfeld MG, Ramos A, Gherzi R. How to control miRNA maturation? RNA Biol. 2009;6:536–40. doi: 10.4161/rna.6.5.10080. [DOI] [PubMed] [Google Scholar]
- 91.Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yamagata K, Fujiyama S, Ito S, Ueda T, Murata T, Naitou M, Takeyama K, Minami Y, O’Malley BW, Kato S. Maturation of microRNA is hormonally regulated by a nuclear receptor. Mol Cell. 2009;36:340–7. doi: 10.1016/j.molcel.2009.08.017. [DOI] [PubMed] [Google Scholar]
- 93.Müller-Tidow C, Diederichs S, Schrader MG, Vogt U, Miller K, Berdel WE, Serve H. Cyclin A1 is highly expressed in aggressive testicular germ cell tumors. Cancer Lett. 2003;190:89–95. doi: 10.1016/S0304-3835(02)00582-7. [DOI] [PubMed] [Google Scholar]
- 94.Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.