Correction
After the publication of this work [1], we became aware of the fact that from No. 10 onwards the numbering of the references in the reference list lost congruence with citations in the body of the text. Please see below the corrected list of references. The citations in the original manuscript refer to the corrected list below.
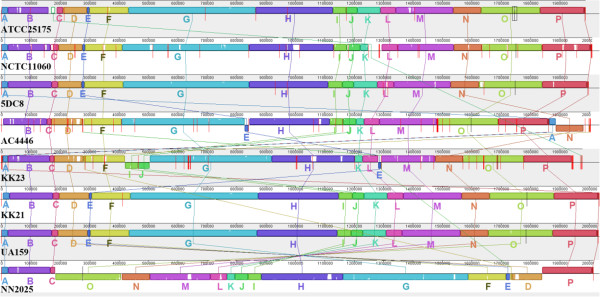
Furthermore we like to correct two additional errors: In Figure two (Figure 1 here) the strain number KK26 has to be corrected to KK21 and on page 13 of the manuscript, chapter “Antibiotic resistance-related proteins” the unit of the minimum inhibitory concentration of Streptococcus mutans against bacitracin has to be changed from μg/l to μg/ml, reading now:
Figure 1.
Comparison of local collinear blocks (LCBs) of chromosomal sequences of the eight S. mutans strains. In total 16 local LCBs, marked as A to P, were generated and compared by applying the MAUVE software [13,14] with default settings and using strain UA159 as reference. The red vertical bars indicate contig ends. The white areas inside each LCB show regions with low similarities.
The S. mutans species is known to be intrinsically resistant to bacitracin produced by Bacillus subtilis. We confirmed this by testing all the 10 strains with a bacitracin-E-test (data not shown). All strains including S. ratti DSM 20564 and S. sobrinus DSM 20742 had a minimum inhibitory concentration between 128 and >256 μg/ml.
We regret any inconvenience that these inaccuracies might have caused. We wish to thank Prof. Dr. Rudolf Lütticken for bringing this matter to our attention.
References (corrected)
1. Tapp J, Thollesson M, Herrmann B: Phylogenetic relationships and genotyping of the genus Streptococcus by sequence determination of the RNase P RNA gene, rnpB.Int J Syst Evol Microbiol 2003, 53:1861–1871.
2. Loesche WJ: Role of Streptococcus mutans in human dental decay.Microbiol Rev 1986, 50:353–380.
3. Lemos JA, Burne RA: A model of efficiency: stress tolerance by Streptococcus mutans.Microbiology 2008, 154:3247–3255.
4. Nakano K, Nomura R, Matsumoto M, Ooshima T: Roles of oral bacteria in cardiovascular diseases--from molecular mechanisms to clinical cases: Cell-surface structures of novel serotype k Streptococcus mutans strains and their correlation to virulence.J Pharmacol Sci 2010, 113:120–125.
5. Nomura R, Nakano K, Taniguchi N, Lapirattanakul J, Nemoto H, Gronroos L, Alaluusua S, Ooshima T: Molecular and clinical analyses of the gene encoding the collagen-binding adhesin of Streptococcus mutans.J Med Microbiol 2009, 58:469–475.
6. Redfield RJ, Findlay WA, Bosse J, Kroll JS, Cameron AD, Nash JH: Evolution of competence and DNA uptake specificity in the Pasteurellaceae.BMC Evol Biol 2006, 6:82.
7. Ehrlich GD, Hu FZ, Shen K, Stoodley P, Post JC: Bacterial plurality as a general mechanism driving persistence in chronic infections.Clin Orthop Relat Res 2005:20–24.
8. Ajdic D, McShan WM, McLaughlin RE, Savic G, Chang J, Carson MB, Primeaux C, Tian R, Kenton S, Jia H, et al.: Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen.Proc Natl Acad Sci U S A 2002, 99:14434–14439.
9. Maruyama F, Kobata M, Kurokawa K, Nishida K, Sakurai A, Nakano K, Nomura R, Kawabata S, Ooshima T, Nakai K, et al.: Comparative genomic analyses of Streptococcus mutans provide insights into chromosomal shuffling and species-specific content.BMC Genomics 2009, 10:358.
10. Cornejo OE, Lefebure T, Pavinski Bitar PD, Lang P, Richards VP, Eilertson K, Do T, Beighton D, Zeng L, Ahn SJ, et al.: Evolutionary and population genomics of the cavity causing bacteria Streptococcus mutans.Mol Biol Evol 2013, 30:881–893.
11. Zhao Y, Wu J, Yang J, Sun S, Xiao J, Yu J: PGAP: pan-genomes analysis pipeline.Bioinformatics 2012, 28:416–418.
12. Darling AE, Miklos I, Ragan MA: Dynamics of genome rearrangement in bacterial populations.PLoS Genet 2008, 4:e1000128.
13. Darling AC, Mau B, Blattner FR, Perna NT: Mauve: multiple alignment of conserved genomic sequence with rearrangements.Genome Res 2004, 14:1394–1403.
14. Darling AE, Mau B, Perna NT: progressive Mauve: multiple genome alignment with gene gain, loss and rearrangement.PLoS One 2010, 5:e11147.
15. Waterhouse JC, Swan DC, Russell RR: Comparative genome hybridization of Streptococcus mutans strains.Oral Microbiol Immunol 2007, 22:103–110.
16. Wu C, Cichewicz R, Li Y, Liu J, Roe B, Ferretti J, Merritt J, Qi F: Genomic island TnSmu2 of Streptococcus mutans harbors a nonribosomal peptide synthetase-polyketide synthase gene cluster responsible for the biosynthesis of pigments involved in oxygen and H 2 O 2 tolerance.Appl Environ Microbiol 2010, 76:5815–5826.
17. Waterhouse JC, Russell RR: Dispensable genes and foreign DNA in Streptococcus mutans.Microbiology 2006, 152:1777–1788.
18. Muzzi A, Donati C: Population genetics and evolution of the pan-genome of Streptococcus pneumoniae.Int J Med Microbiol 2011, 301:619–622.
19. Tettelin H, Masignani V, Cieslewicz MJ, Donati C, Medini D, Ward NL, Angiuoli SV, Crabtree J, Jones AL, Durkin AS, et al.: Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome”.Proc Natl Acad Sci U S A 2005, 102:13950–13955.
20. Tettelin H, Riley D, Cattuto C, Medini D: Comparative genomics: the bacterial pan-genome.Curr Opin Microbiol 2008, 11:472–477.
21. Mira A, Martin-Cuadrado AB, D’Auria G, Rodriguez-Valera F: The bacterial pan-genome: a new paradigm in microbiology.Int Microbiol 2010, 13:45–57.
22. Donati C, Hiller NL, Tettelin H, Muzzi A, Croucher NJ, Angiuoli SV, Oggioni M, Dunning Hotopp JC, Hu FZ, Riley DR, et al.: Structure and dynamics of the pan-genome of Streptococcus pneumoniae and closely related species.Genome Biol 2010, 11:R107.
23. Lefebure T, Stanhope MJ: Evolution of the core and pan-genome of Streptococcus: positive selection, recombination, and genome composition.Genome Biol 2007, 8:R71.
24. Hogg JS, Hu FZ, Janto B, Boissy R, Hayes J, Keefe R, Post JC, Ehrlich GD: Characterization and modeling of the Haemophilus influenzae core and supragenomes based on the complete genomic sequences of Rd and 12 clinical nontypeable strains.Genome Biol 2007, 8:R103.
25. Li L, Stoeckert CJ, Jr., Roos DS: OrthoMCL: identification of ortholog groups for eukaryotic genomes.Genome Res 2003, 13:2178–2189.
26. Song L, Sudhakar P, Wang W, Conrads G, Brock A, Sun J, Wagner-Dobler I, Zeng AP: A genome-wide study of two-component signal transduction systems in eight newly sequenced mutans streptococci strains.BMC Genomics 2012, 13:128.
27. Tanzer JM, Livingston J, Thompson AM: The microbiology of primary dental caries in humans.J Dent Educ 2001, 65:1028–1037.
28. Hale JD, Heng NC, Jack RW, Tagg JR: Identification of nlmTE, the locus encoding the ABC transport system required for export of nonlantibiotic mutacins in Streptococcus mutans.J Bacteriol 2005, 187:5036–5039.
29. Hossain MS, Biswas I: An extracelluar protease, SepM, generates functional competence-stimulating peptide in Streptococcus mutans UA159.J Bacteriol 2012, 194:5886–5896.
30. Li YH, Tang N, Aspiras MB, Lau PC, Lee JH, Ellen RP, Cvitkovitch DG: A quorum-sensing signaling system essential for genetic competence in Streptococcus mutans is involved in biofilm formation.J Bacteriol 2002, 184:2699–2708.
31. Petersen FC, Scheie AA: Genetic transformation in Streptococcus mutans requires a peptide secretion-like apparatus.Oral Microbiol Immunol 2000, 15:329–334.
32. Petersen FC, Fimland G, Scheie AA: Purification and functional studies of a potent modified quorum-sensing peptide and a two-peptide bacteriocin in Streptococcus mutans.Mol Microbiol 2006, 61:1322–1334.
33. Allan E, Hussain HA, Crawford KR, Miah S, Ascott ZK, Khwaja MH, Hosie AH: Genetic variation in comC, the gene encoding competence-stimulating peptide (CSP) in Streptococcus mutans.FEMS Microbiol Lett 2007, 268:47–51.
34. Ahn SJ, Wen ZT, Burne RA: Multilevel control of competence development and stress tolerance in Streptococcus mutans UA159.Infect Immun 2006, 74:1631–1642.
35. Kreth J, Hung DC, Merritt J, Perry J, Zhu L, Goodman SD, Cvitkovitch DG, Shi W, Qi F: The response regulator ComE in Streptococcus mutans functions both as a transcription activator of mutacin production and repressor of CSP biosynthesis.Microbiology 2007, 153:1799–1807.
36. Kreth J, Merritt J, Shi W, Qi F: Co-ordinated bacteriocin production and competence development: a possible mechanism for taking up DNA from neighbouring species.Mol Microbiol 2005, 57:392–404.
37. Kreth J, Merritt J, Zhu L, Shi W, Qi F: Cell density- and ComE-dependent expression of a group of mutacin and mutacin-like genes in Streptococcus mutans.FEMS Microbiol Lett 2006, 265:11–17.
38. van der Ploeg JR: Regulation of bacteriocin production in Streptococcus mutans by the quorum-sensing system required for development of genetic competence.J Bacteriol 2005, 187:3980–3989.
39. Mashburn-Warren L, Morrison DA, Federle MJ: A novel double-tryptophan peptide pheromone controls competence in Streptococcus spp. via an Rgg regulator.Mol Microbiol 2010, 78:589–606.
40. Okinaga T, Niu G, Xie Z, Qi F, Merritt J: The hdrRM operon of Streptococcus mutans encodes a novel regulatory system for coordinated competence development and bacteriocin production.J Bacteriol 2010, 192:1844–1852.
41. Okinaga T, Xie Z, Niu G, Qi F, Merritt J: Examination of the hdrRM regulon yields insight into the competence system of Streptococcus mutans.Mol Oral Microbiol 2010, 25:165–177.
42. Xie Z, Okinaga T, Niu G, Qi F, Merritt J: Identification of a novel bacteriocin regulatory system in Streptococcus mutans.Mol Microbiol 2010, 78:1431–1447.
43. Mair RW, Senadheera DB, Cvitkovitch DG: CinA is regulated via ComX to modulate genetic transformation and cell viability in Streptococcus mutans.FEMS Microbiol Lett 2012, 331:44–52.
44. Alaluusua S, Takei T, Ooshima T, Hamada S: Mutacin activity of strains isolated from children with varying levels of mutants streptococci and caries.Arch Oral Biol 1991, 36:251–255.
45. Baba T, Schneewind O: Instruments of microbial warfare: bacteriocin synthesis, toxicity and immunity.Trends Microbiol 1998, 6:66–71.
46. Hossain MS, Biswas I: Mutacins from Streptococcus mutans UA159 are active against multiple streptococcal species.Appl Environ Microbiol 2011, 77:2428–2434.
47. Yonezawa H, Kuramitsu HK: Genetic analysis of a unique bacteriocin, Smb, produced by Streptococcus mutans GS5.Antimicrob Agents Chemother 2005, 49:541–548.
48. Hyink O, Balakrishnan M, Tagg JR: Streptococcus rattus strain BHT produces both a class I two-component lantibiotic and a class II bacteriocin.FEMS Microbiol Lett 2005, 252:235–241.
49. Nguyen T, Zhang Z, Huang IH, Wu C, Merritt J, Shi W, Qi F: Genes involved in the repression of mutacin I production in Streptococcus mutans.Microbiology 2009, 155:551–556.
50. Qi F, Chen P, Caufield PW: Purification and biochemical characterization of mutacin I from the group I strain of Streptococcus mutans, CH43, and genetic analysis of mutacin I biosynthesis genes.Appl Environ Microbiol 2000, 66:3221–3229.
51. Chen P, Qi F, Novak J, Caufield PW: The specific genes for lantibiotic mutacin II biosynthesis in Streptococcus mutans T8 are clustered and can be transferred en bloc.Appl Environ Microbiol 1999, 65:1356–1360.
52. Qi F, Chen P, Caufield PW: Purification of mutacin III from group III Streptococcus mutans UA787 and genetic analyses of mutacin III biosynthesis genes.Appl Environ Microbiol 1999, 65:3880–3887.
53. Robson CL, Wescombe PA, Klesse NA, Tagg JR: Isolation and partial characterization of the Streptococcus mutans type AII lantibiotic mutacin K8.Microbiology 2007, 153:1631–1641.
54. Qi F, Chen P, Caufield PW: The group I strain of Streptococcus mutans, UA140, produces both the lantibiotic mutacin I and a nonlantibiotic bacteriocin, mutacin IV.Appl Environ Microbiol 2001, 67:15–21.
55. Hale JD, Ting YT, Jack RW, Tagg JR, Heng NC: Bacteriocin (mutacin) production by Streptococcus mutans genome sequence reference strain UA159: elucidation of the antimicrobial repertoire by genetic dissection.Appl Environ Microbiol 2005, 71:7613–7617.
56. Dufour D, Cordova M, Cvitkovitch DG, Levesque CM: Regulation of the competence pathway as a novel role associated with a streptococcal bacteriocin.J Bacteriol 2011, 193:6552–6559.
57. Nes IF, Diep DB, Holo H: Bacteriocin diversity in Streptococcus and Enterococcus.J Bacteriol 2007, 189:1189–1198.
58. Bekal-Si Ali S, Hurtubise Y, Lavoie MC, LaPointe G: Diversity of Streptococcus mutans bacteriocins as confirmed by DNA analysis using specific molecular probes.Gene 2002, 283:125–131.
59. Johnson DW, Tagg JR, Wannamaker LW: Production of a bacteriocine-like substance by group-A streptococci of M-type 4 and T-pattern 4.J Med Microbiol 1979, 12:413–427.
60. Perry JA, Jones MB, Peterson SN, Cvitkovitch DG, Levesque CM: Peptide alarmone signalling triggers an auto-active bacteriocin necessary for genetic competence.Mol Microbiol 2009, 72:905–917.
61. Chatterjee AK, Starr MP: Transfer among Erwinia spp. and other enterobacteria of antibiotic resistance carried on R factors.J Bacteriol 1972, 112:576–584.
62. Yano H, Kuga A, Okamoto R, Kitasato H, Kobayashi T, Inoue M: Plasmid-encoded metallo-beta-lactamase (IMP-6) conferring resistance to carbapenems, especially meropenem.Antimicrob Agents Chemother 2001, 45:1343–1348.
63. Ouyang J, Tian XL, Versey J, Wishart A, Li YH: The BceABRS four-component system regulates the bacitracin-induced cell envelope stress response in Streptococcus mutans.Antimicrob Agents Chemother 2010, 54:3895–3906.
64. Tsuda H, Yamashita Y, Shibata Y, Nakano Y, Koga T: Genes involved in bacitracin resistance in Streptococcus mutans.Antimicrob Agents Chemother 2002, 46:3756–3764.
65. El Ghachi M, Bouhss A, Blanot D, Mengin-Lecreulx D: The bacA gene of Escherichia coli encodes an undecaprenyl pyrophosphate phosphatase activity.J Biol Chem 2004, 279:30106–30113.
66. Bernard R, El Ghachi M, Mengin-Lecreulx D, Chippaux M, Denizot F: BcrC from Bacillus subtilis acts as an undecaprenyl pyrophosphate phosphatase in bacitracin resistance.J Biol Chem 2005, 280:28852–28857.
67. McCord JM, Fridovich I: Superoxide dismutase: the first twenty years (1968–1988).Free Radic Biol Med 1988, 5:363–369.
68. Yamamoto Y, Higuchi M, Poole LB, Kamio Y: Role of the dpr product in oxygen tolerance in Streptococcus mutans.J Bacteriol 2000, 182:3740–3747.
69. Higuchi M, Yamamoto Y, Kamio Y: Molecular biology of oxygen tolerance in lactic acid bacteria: Functions of NADH oxidases and Dpr in oxidative stress.J Biosci Bioeng 2000, 90:484–493.
70. Yamamoto Y, Poole LB, Hantgan RR, Kamio Y: An iron-binding protein, Dpr, from Streptococcus mutans prevents iron-dependent hydroxyl radical formation in vitro.J Bacteriol 2002, 184:2931–2939.
71. Mustacich D, Powis G: Thioredoxin reductase.Biochem J 2000, 346 Pt 1:1–8.
72. Arner ES, Holmgren A: Physiological functions of thioredoxin and thioredoxin reductase.Eur J Biochem 2000, 267:6102–6109.
73. Seo HJ, Lee YN: Characterization of Deinococcus radiophilus thioredoxin reductase active with both NADH and NADPH.J Microbiol 2010, 48:637–643.
74. Holmgren A: Thioredoxin and glutaredoxin systems.J Biol Chem 1989, 264:13963–13966.
75. Fernandes AP, Holmgren A: Glutaredoxins: glutathione-dependent redox enzymes with functions far beyond a simple thioredoxin backup system.Antioxid Redox Signal 2004, 6:63–74.
76. Fahey RC, Brown WC, Adams WB, Worsham MB: Occurrence of glutathione in bacteria.J Bacteriol 1978, 133:1126–1129.
77. Janowiak BE, Griffith OW: Glutathione synthesis in Streptococcus agalactiae. One protein accounts for gamma-glutamylcysteine synthetase and glutathione synthetase activities.J Biol Chem 2005, 280:11829–11839.
78. Zhang J, Biswas I: 3′-Phosphoadenosine-5′-phosphate phosphatase activity is required for superoxide stress tolerance in Streptococcus mutans.J Bacteriol 2009, 191:4330–4340.
79. Ma H, Zeng AP: Reconstruction of metabolic networks from genome data and analysis of their global structure for various organisms.Bioinformatics 2003, 19:270–277.
80. Kohl M, Wiese S, Warscheid B: Cytoscape: software for visualization and analysis of biological networks.Methods Mol Biol 2011, 696:291–303.
81. Taniai H, Iida K, Seki M, Saito M, Shiota S, Nakayama H, Yoshida S: Concerted action of lactate oxidase and pyruvate oxidase in aerobic growth of Streptococcus pneumoniae: role of lactate as an energy source.J Bacteriol 2008, 190:3572–3579.
82. Kreth J, Merritt J, Shi W, Qi F: Competition and coexistence between Streptococcus mutans and Streptococcus sanguinis in the dental biofilm.J Bacteriol 2005, 187:7193–7203.
83. Okahashi N, Nakata M, Sumitomo T, Terao Y, Kawabata S: Hydrogen peroxide produced by oral streptococci induces macrophage cell death.PLoS One 2013, 8:e62563.
84. Subramanian S, Sivaraman C: Bacterial citrate lyase.Journal of Biosciences 1984, 6:379–401.
85. Evans HJ, Wood HG: The mechanism of the pyruvate, phosphate dikinase reaction.Proc Natl Acad Sci U S A 1968, 61:1448–1453.
86. Benziman M, Eisen N, Palgi A: Properties and physiological role of the pep-synthase of A. xylinum.FEBS Lett 1969, 3:156–159.
87. Sauer U, Eikmanns BJ: The PEP-pyruvate-oxaloacetate node as the switch point for carbon flux distribution in bacteria.FEMS Microbiol Rev 2005, 29:765–794.
88. Cvitkovitch DG, Gutierrez JA, Bleiweis AS: Role of the citrate pathway in glutamate biosynthesis by Streptococcus mutans.J Bacteriol 1997, 179:650–655.
89. Chain PS, Grafham DV, Fulton RS, Fitzgerald MG, Hostetler J, Muzny D, Ali J, Birren B, Bruce DC, Buhay C, et al.: Genomics. Genome project standards in a new era of sequencing.Science 2009, 326:236–237.
90. Li R, Yu C, Li Y, Lam TW, Yiu SM, Kristiansen K, Wang J: SOAP2: an improved ultrafast tool for short read alignment.Bioinformatics 2009, 25:1966–1967.
91. Li H, Durbin R: Fast and accurate short read alignment with Burrows-Wheeler transform.Bioinformatics 2009, 25:1754–1760.
92. de la Bastide M, McCombie WR: Assembling genomic DNA sequences with PHRAP.Curr Protoc Bioinformatics 2007, Chapter 11:Unit11 14.
93. Rissman AI, Mau B, Biehl BS, Darling AE, Glasner JD, Perna NT: Reordering contigs of draft genomes using the Mauve aligner.Bioinformatics 2009, 25:2071–2073.
94. Delcher AL, Bratke KA, Powers EC, Salzberg SL: Identifying bacterial genes and endosymbiont DNA with Glimmer.Bioinformatics 2007, 23:673–679.
95. Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M: Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research.Bioinformatics 2005, 21:3674–3676.
96. Karp PD, Paley S, Romero P: The Pathway Tools software.Bioinformatics 2002, 18 Suppl 1:S225-232.
97. Stelzer M, Sun J, Kamphans T, Fekete SP, Zeng AP: An extended bioreaction database that significantly improves reconstruction and analysis of genome-scale metabolic networks.Integr Biol (Camb) 2011, 3:1071–1086.
98. Lau PC, Sung CK, Lee JH, Morrison DA, Cvitkovitch DG: PCR ligation mutagenesis in transformable streptococci: application and efficiency.J Microbiol Methods 2002, 49:193–205.
99. Reck M, Rutz K, Kunze B, Tomasch J, Surapaneni SK, Schulz S, Wagner-Dobler I: The biofilm inhibitor carolacton disturbs membrane integrity and cell division of Streptococcus mutans through the serine/threonine protein kinase PknB.J Bacteriol 2011, 193:5692–5706.
100. Lefrancois J, Samrakandi MM, Sicard AM: Electrotransformation and natural transformation of Streptococcus pneumoniae: requirement of DNA processing for recombination.Microbiology 1998, 144 (Pt 11):3061–3068.
101. Ween O, Teigen S, Gaustad P, Kilian M, Havarstein LS: Competence without a competence pheromone in a natural isolate of Streptococcus infantis.J Bacteriol 2002, 184:3426–3432.
102. Li YH, Lau PC, Lee JH, Ellen RP, Cvitkovitch DG: Natural genetic transformation of Streptococcus mutans growing in biofilms.J Bacteriol 2001, 183:897–908.
103. LeBlanc D, Chen Y-Y, Buckley N, Lee L: Genetic transfer methods for Streptococcus sobrinus and other oral streptococci.Methods in Cell Science 1998, 20:85–93.
104. Caparon MG, Scott JR: Genetic manipulation of pathogenic streptococci.Methods Enzymol 1991, 204:556–586.
105. McLaughlin RE, Ferretti JJ: Electrotransformation of Streptococci.Methods Mol Biol 1995, 47:185–193.
Contributor Information
Lifu Song, Email: lifu.song@tuhh.de.
Wei Wang, Email: wei.wang@tuhh.de.
Georg Conrads, Email: gconrads@ukaachen.de.
Anke Rheinberg, Email: Anke.Rheinberg@rwth-aachen.de.
Helena Sztajer, Email: helena.sztajer@helmholtz-hzi.de.
Michael Reck, Email: Michael.Reck@helmholtz-hzi.de.
Irene Wagner-Döbler, Email: Irene.Wagner-Doebler@helmholtz-hzi.de.
An-Ping Zeng, Email: aze@tuhh.de.
References
- Song L, Wang W, Conrads G, Rheinberg A, Sztajer H, Reck M, Wagner-Dobler I, Zeng AP. Genetic variability of mutans streptococci revealed by wide whole-genome sequencing. BMC Genomics. 2013;14:430. doi: 10.1186/1471-2164-14-430. [DOI] [PMC free article] [PubMed] [Google Scholar]