Abstract
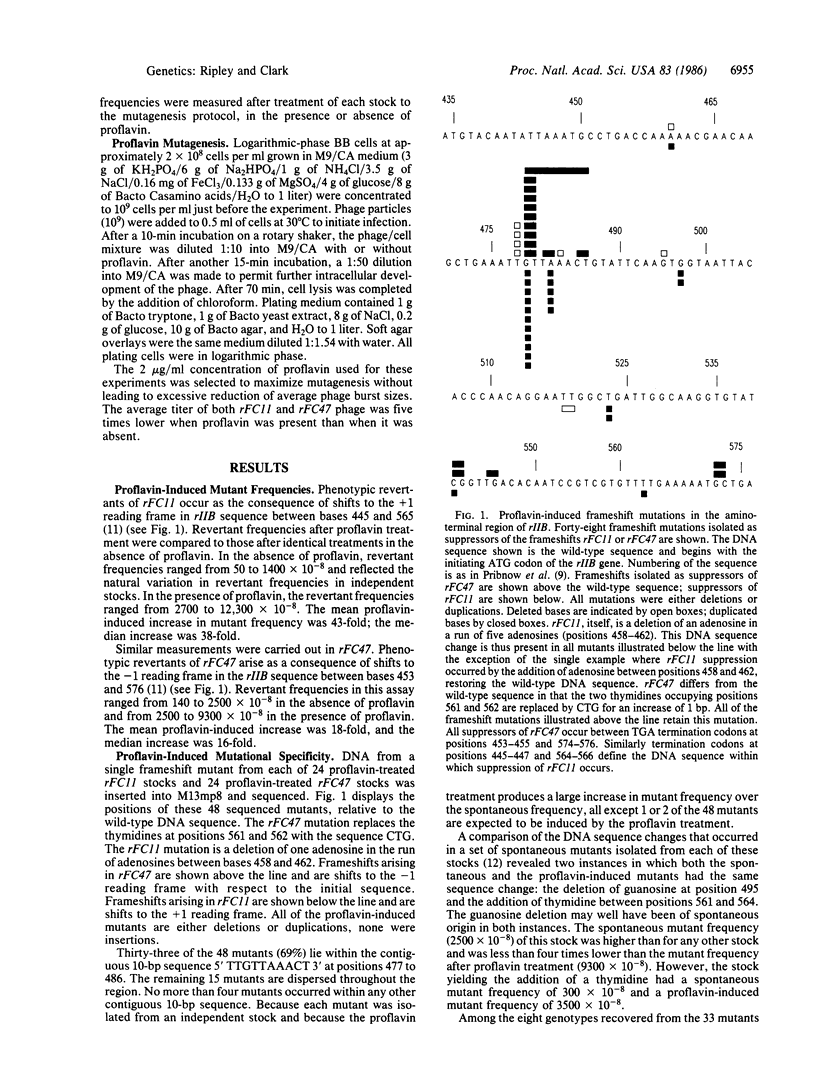
Frameshift mutations were induced by proflavin in the rIIB gene of bacteriophage T4. rIIB DNA from each of 48 independent frameshifts was inserted into M13mp8 and sequenced. Two-thirds of the frameshifts (33/48) lie contiguous to one another in 10 base pairs of the rIIB sequence. This hotspot differs markedly from previously characterized mutagen-induced frameshift hotspots. Distinctive features of the hotspot include the absence of locally repetitive sequences, particularly G X C runs, and the fact that many different sequence changes are induced within the hotspot sequence at appreciable frequencies. Among the 33 mutants at the hotspot, 8 distinguishable DNA sequence changes were seen. All of the mutations were deletions of a single base or duplications of one or more bases. Duplications were more frequent than deletions. The patterns of the base sequence changes suggest that two specific phosphodiester bonds within the hotspot sequence are sites at which proflavin-induced mutation is initiated.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altman S., Lerman L. S. Effects of 9-aminoacridine on bacteriophage T4 deoxyribonucleic acid synthesis. J Mol Biol. 1970 Jun 14;50(2):263–277. doi: 10.1016/0022-2836(70)90191-9. [DOI] [PubMed] [Google Scholar]
- BRENNER S., BENZER S., BARNETT L. Distribution of proflavin-induced mutations in the genetic fine structure. Nature. 1958 Oct 11;182(4641):983–985. doi: 10.1038/182983a0. [DOI] [PubMed] [Google Scholar]
- Benzer S. ON THE TOPOLOGY OF THE GENETIC FINE STRUCTURE. Proc Natl Acad Sci U S A. 1959 Nov;45(11):1607–1620. doi: 10.1073/pnas.45.11.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calos M. P., Miller J. H. Genetic and sequence analysis of frameshift mutations induced by ICR-191. J Mol Biol. 1981 Nov 25;153(1):39–64. doi: 10.1016/0022-2836(81)90525-8. [DOI] [PubMed] [Google Scholar]
- Ernst J. F., Hampsey D. M., Sherman F. DNA sequences of frameshift and other mutations induced by ICR-170 in yeast. Genetics. 1985 Oct;111(2):233–241. doi: 10.1093/genetics/111.2.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickman B. W., Ripley L. S. Structural intermediates of deletion mutagenesis: a role for palindromic DNA. Proc Natl Acad Sci U S A. 1984 Jan;81(2):512–516. doi: 10.1073/pnas.81.2.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koffel-Schwartz N., Verdier J. M., Bichara M., Freund A. M., Daune M. P., Fuchs R. P. Carcinogen-induced mutation spectrum in wild-type, uvrA and umuC strains of Escherichia coli. Strain specificity and mutation-prone sequences. J Mol Biol. 1984 Jul 25;177(1):33–51. doi: 10.1016/0022-2836(84)90056-1. [DOI] [PubMed] [Google Scholar]
- Kohno T., Roth J. R. Proflavin mutagenesis of bacteria. J Mol Biol. 1974 Oct 15;89(1):17–32. doi: 10.1016/0022-2836(74)90160-0. [DOI] [PubMed] [Google Scholar]
- Kreuzer K. N., Alberts B. M. Site-specific recognition of bacteriophage T4 DNA by T4 type II DNA topoisomerase and Escherichia coli DNA gyrase. J Biol Chem. 1984 Apr 25;259(8):5339–5346. [PubMed] [Google Scholar]
- Kreuzer K. N. Recognition of single-stranded DNA by the bacteriophage T4-induced type II topoisomerase. J Biol Chem. 1984 Apr 25;259(8):5347–5354. [PubMed] [Google Scholar]
- LERMAN L. S. Structural considerations in the interaction of DNA and acridines. J Mol Biol. 1961 Feb;3:18–30. doi: 10.1016/s0022-2836(61)80004-1. [DOI] [PubMed] [Google Scholar]
- Lindstrom D. M., Drake J. W. Mechanics of frameshift mutagenesis in bacteriophage T4: role of chromosome tips. Proc Natl Acad Sci U S A. 1970 Mar;65(3):617–624. doi: 10.1073/pnas.65.3.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ORGEL A., BRENNER S. Mutagenesis of bacteriophage T4 by acridines. J Mol Biol. 1961 Dec;3:762–768. doi: 10.1016/s0022-2836(61)80081-8. [DOI] [PubMed] [Google Scholar]
- Owen J. E., Schultz D. W., Taylor A., Smith G. R. Nucleotide sequence of the lysozyme gene of bacteriophage T4. Analysis of mutations involving repeated sequences. J Mol Biol. 1983 Apr 5;165(2):229–248. doi: 10.1016/s0022-2836(83)80255-1. [DOI] [PubMed] [Google Scholar]
- Pribnow D., Sigurdson D. C., Gold L., Singer B. S., Napoli C., Brosius J., Dull T. J., Noller H. F. rII cistrons of bacteriophage T4. DNA sequence around the intercistronic divide and positions of genetic landmarks. J Mol Biol. 1981 Jul 5;149(3):337–376. doi: 10.1016/0022-2836(81)90477-0. [DOI] [PubMed] [Google Scholar]
- Ripley L. S., Glickman B. W. Unique self-complementarity of palindromic sequences provides DNA structural intermediates for mutation. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):851–861. doi: 10.1101/sqb.1983.047.01.097. [DOI] [PubMed] [Google Scholar]
- Ripley L. S. Model for the participation of quasi-palindromic DNA sequences in frameshift mutation. Proc Natl Acad Sci U S A. 1982 Jul;79(13):4128–4132. doi: 10.1073/pnas.79.13.4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripley L. S., Shoemaker N. B. A major role for bacteriophage T4 DNA polymerase in frameshift mutagenesis. Genetics. 1983 Mar;103(3):353–366. doi: 10.1093/genetics/103.3.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skopek T. R., Hutchinson F. Frameshift mutagenesis of lambda prophage by 9-aminoacridine, proflavin and ICR-191. Mol Gen Genet. 1984;195(3):418–423. doi: 10.1007/BF00341442. [DOI] [PubMed] [Google Scholar]
- Streisinger G., Okada Y., Emrich J., Newton J., Tsugita A., Terzaghi E., Inouye M. Frameshift mutations and the genetic code. This paper is dedicated to Professor Theodosius Dobzhansky on the occasion of his 66th birthday. Cold Spring Harb Symp Quant Biol. 1966;31:77–84. doi: 10.1101/sqb.1966.031.01.014. [DOI] [PubMed] [Google Scholar]
- Streisinger G., Owen J. Mechanisms of spontaneous and induced frameshift mutation in bacteriophage T4. Genetics. 1985 Apr;109(4):633–659. doi: 10.1093/genetics/109.4.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topal M. D. Molecular mechanisms of chemical mutagenesis: 9-aminoacridine inhibits DNA replication in vitro by destabilizing the DNA growing point and interacting with the DNA polymerase. Biochemistry. 1984 May 22;23(11):2367–2372. doi: 10.1021/bi00306a007. [DOI] [PubMed] [Google Scholar]
- de Boer J. G., Ripley L. S. Demonstration of the production of frameshift and base-substitution mutations by quasipalindromic DNA sequences. Proc Natl Acad Sci U S A. 1984 Sep;81(17):5528–5531. doi: 10.1073/pnas.81.17.5528. [DOI] [PMC free article] [PubMed] [Google Scholar]



