Abstract
Alzheimer's disease (AD) is an important social and economic issue for our societies. The development of therapeutics against this severe dementia requires assessing the effects of new drugs in animal models thanks to dedicated biomarkers. According to the amyloid cascade hypothesis, β–amyloid deposits are at the origin of most of the lesions associated with AD. These extracellular deposits are therefore one of the main targets in therapeutical strategies. Aβ peptides can be revealed histologically with specific dyes or antibodies, or by magnetic resonance microscopy (µMRI) that uses their association with iron as a source of signal. The microscopic size of the lesions necessitates the development of specific imaging protocols. Most protocols use T2-weighted sequences that reveal the aggregates as hypointense spots. This chapter describes histological methods that reveal amyloid plaques with specific stains and MR imaging protocols for in vivo and ex vivo MR imaging of AD mice.
Keywords: Animal model, mouse, amyloid, APP, PS1, imaging, MRI
1. Introduction
Alzheimer's disease (AD) is a severe dementia with critical social and economic consequences. Senile plaques are one of the hallmarks of this disease. They are microscopic lesions that measure less than 20 µm in humans (1). These lesions are constituted of aggregated extracellular deposition of beta-amyloid (Aβ) peptides. Amyloid deposits are believed to occur in the brain a long time, maybe decades, before the occurrence of clinical AD (2) and according to the amyloid cascade hypothesis, amyloid is at the origin of most of the pathological processes associated with AD (3).
To date, there is no curative treatment against AD, but many disease-modifying treatments are under investigation (4). The development of these treatments relies on the use of animals such as transgenic mouse models of amyloidosis (5). Most of these models are based on the overexpression of mutated forms of APP alone or with an additional mutation of presenilin (PS1 or PS2) genes (6). The generation and use of these models require the ability to phenotype these animals and to evaluate AD-like pathologies.
MRI can play a critical role to follow-up these models. First, MRI can be used to follow-up cerebral atrophy in transgenic mouse models of AD (Fig. 1) (7). This biomarker is critical because in humans, cerebral atrophy appears progressively during the evolution of AD (8) and it is associated with disease progression in clinical trials (9). Imaging amyloid plaques would also be critical to follow-up the AD pathology in animals. Today, in humans, amyloid plaque imaging relies mainly on positron emission tomography and on specific ligands such as PIB (2). Such ligands are however difficult to use in small rodents (10).
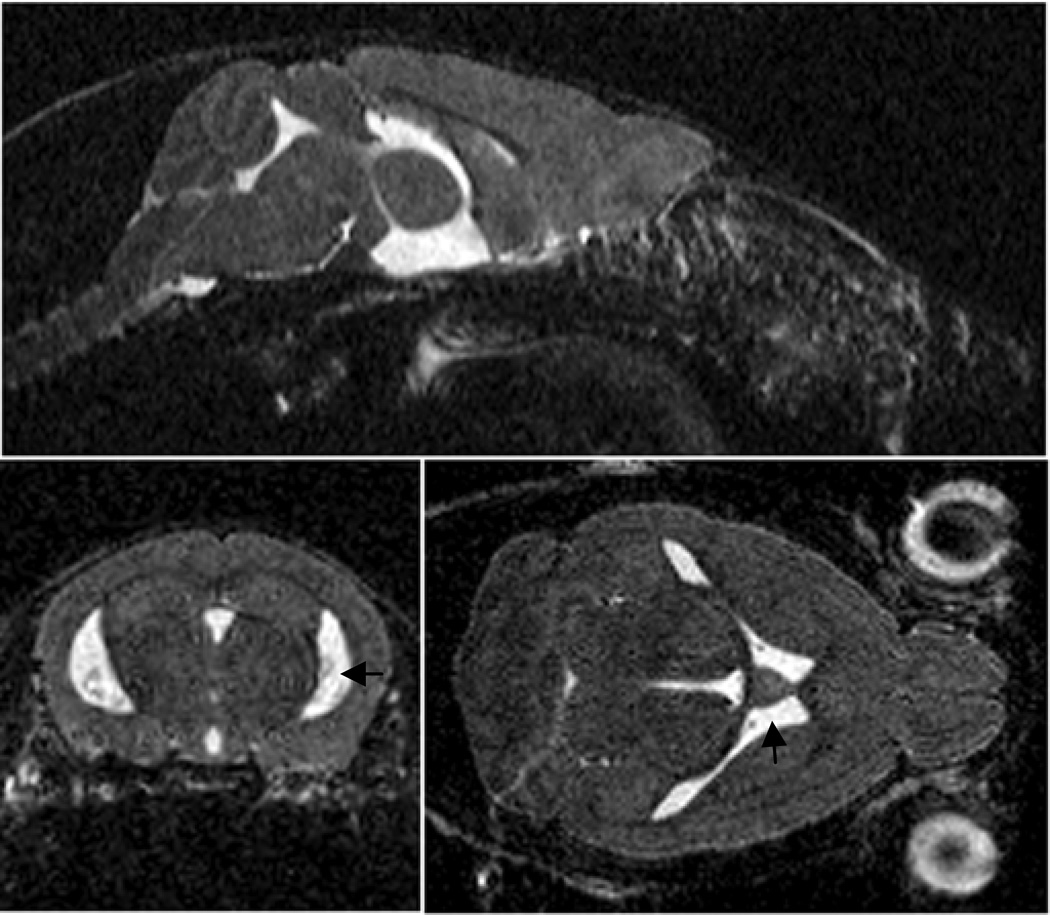
Fig. 1.
In vivo 3D acquisition of a mouse brain: sagittal (top), coronal (left) and axial (right) views. Cerebral ventricles filled with CSF appear hyperintense on these images (arrows). The measure of their volumes can be used as an index of atrophy.
MR methods using MRI are under development to either quantify some parameters that reflect amyloid load (11, 12) or to directly detect amyloid plaques (13, 14).
This chapter describes methods to detect amyloid plaques in vivo and ex vivo in the brains of transgenic mouse models of AD.
2. Material
2.1. Mouse models
APP/PS1dE9 mouse models of AD can be purchased from the Jackson Laboratory (http://www.jax.org/; 600 Main Street Bar Harbor, Maine 04609 USA) (see Note 1).
2.2. MR imaging systems for in vivo imaging
MRI spectrometer and MR probes: 7-Tesla spectrometer (Pharmascan, Bruker Biospin GmbH) equipped with a 9-cm inner diameter gradient system (760 mT/m strength and 6836 T/m/s slew rate) and interfaced to a console running Paravision 5.0. Birdcage coil (Bruker) of 38 mm diameter for power transmission–reception. Other high field MRI systems can also be used for in vivo imaging of mouse models of AD.
Mouse holder and monitoring (Fig. 2): the head of the animal is stabilized in a head holder using ear bars and a bite bar built in a dedicated cradle (available from RAPID Biomedical GmbH, Germany). The head holder is inserted into the radio frequency (RF) coils during the imaging session. This setup prevents movements of the animals during the long imaging acquisitions. Monitoring devices, such as the MR compatible small animal monitoring and gating system (respiration / IBP module) from SA Instrument Inc (Stony Brook, NY 11790, USA), are used to follow the animal’s physiological parameters. The animals are warmed with a water-filled heating blanket connected to a thermoregulated water bath (circulating thermostat system from Bruker).
Anesthetic devices: isoflurane vaporiser connected to several debitmeters for O2 / N2O / Air and one cage dedicated to anesthesia induction (Minerve, Estrernay, France).
Isoflurane gas (Belamont, Paris, France) is used for anesthesia.
Air, O2, N2O tanks.
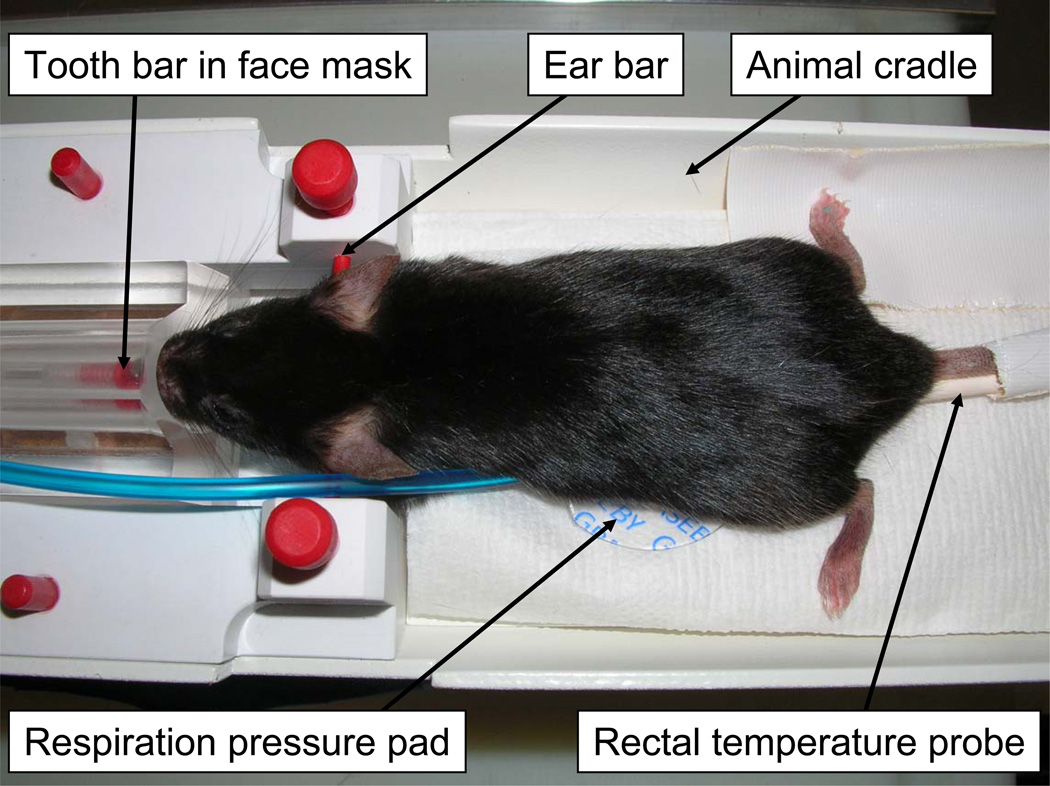
Fig. 2.
Mouse setup for in vivo MR imaging. The animal is held still with tooth and ear bars. The isoflurane anesthesia is delivered via a face mask. Respiration is recorded through a pressure pad and body temperature is recorded through a rectal probe.
2.3. Animal sacrifice (Fig. 3)
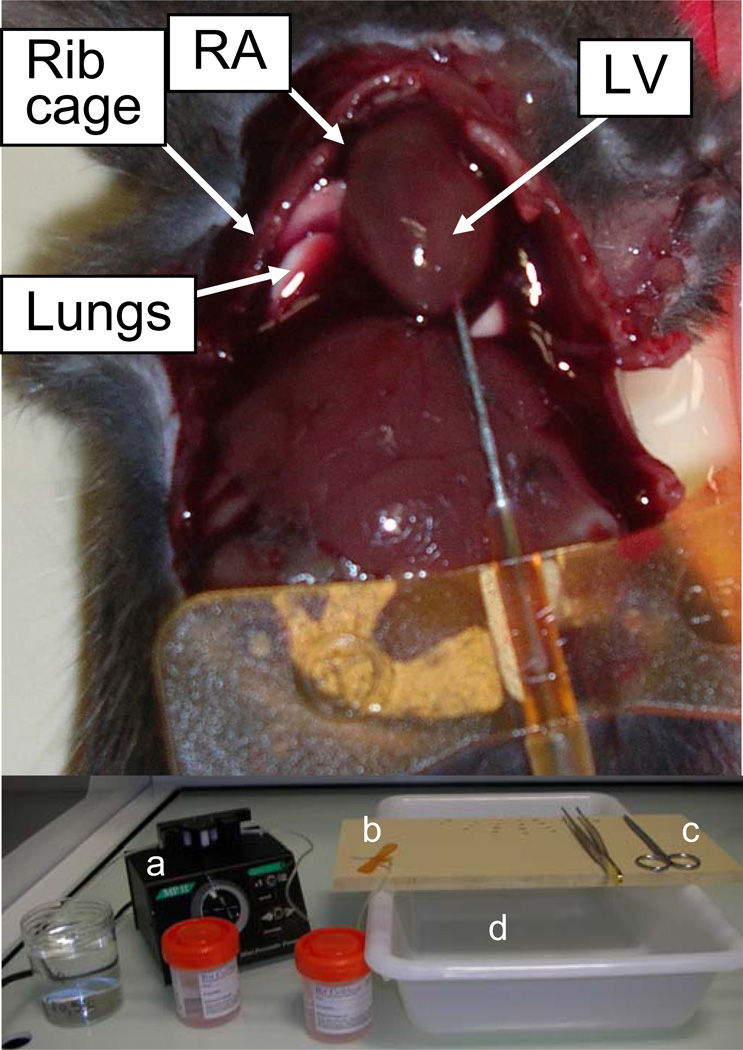
Fig. 3.
Mouse intracardiac perfusion. The top picture shows the rib cage cut off and the butterfly needle inserted into the left ventricle (LV). The bottom picture shows the material used: formalin beakers and PBS beaker, peristaltic pump (a), butterfly needle (b), forceps, scissors (c), surgical board and draining bucket (d). RA = right atrium.
Peristaltic pump for intracardiac perfusion: Miniperistaltic pump II by Harvard Apparatus.
Surgical supplies such as scissors (available from WPI Europe (http://www.wpi-europe.com), dressing forceps (10–cm long), micro Bulldog clamp (3–cm long), butterfly needle (25 gauge).
Perfusion fluids: phosphate buffered saline and 10% buffered formalin at room temperature.
Surgery board and draining bucket.
2.4. MR imaging systems for ex vivo imaging
Clinical 7-Tesla spectrometer (Syngo MR, VB15, Siemens), equipped with an AC84 head gradient set with 36-cm available bore (80 mT/m strength and slew rate of 333 mT/m/s). Birdcage coils can be used (inner diameter = 24 mm) for signal transmit–receive. Other MR systems can also be used for ex vivo MRI. For example, we performed previous studies on a 4.7-Tesla spectrometer (Bruker) (13) or on the 7 Tesla spectrometer used for in vivo imaging.
10-mL syringes can be used to make containers that allow keeping the brain still in place and soaked.
Fluorinert™ Electronic Liquid FC-4 (3M™, Cergy-Pontoise, France), a fully fluorinated liquid is used to embed the sample before MRI and to remove any background signal.
2.5. Histological analysis
2.5.1. General histology
Sliding freezing microtome (e.g. LEICA SM2400).
Homemade baskets to rinse tissue. Commercial systems (15–mm netwell insert with 74 µm mesh size polyester membrane from Corning Life Sciences) are available as an alternative.
Slow orbital agitator and routine small equipment for histology lab.
Phosphate buffer (PB).
Dimethyl sulfoxide.
Glycerol.
Dry ice.
Superfrost+ glass slides.
2.5.2. β-Amyloid staining with BAM10 antibody
Usual glassware and small equipment for histology lab.
Phosphate buffered saline (PBS).
Hydrogen peroxide 30% (Sigma H 0904).
Octylphenol ethylene oxide condensate 0.2% (Triton X-100™, Sigma).
Normal rabbit serum, can be aliquoted at –20°C (Vector® Labs S 5000).
Monoclonal BAM10 clone A3981 (Sigma).
Biotinylated IgG anti-mouse, BA–9200 (Vector®).
Sodium azide 8%, stored at room temperature (Sigma S 8032).
Avidin-biotin Complex (ABC Vectastain kit, Elite PK 6100).
Tyramin Biotin Reagent, stored at 4°C (Blast PC 2815-0897)
Peroxidase Substrate kit (VIP SK 4600 substrate kit for peroxidase, Vector®).
2.5.3. β-Amyloid staining with Congo red
Labeling of amyloid deposits is done by standard Congo red staining (adapted from ref. 15).
Usual glassware and small equipment for histology lab.
Solution S1: 80° ethanol saturated with NaCl.
-
Solution S2: Saturated solution of Congo red (Fluka, ref 60910) made in saline ethanol.
S1 and S2 solutions are stable for weeks / months but it is recommended to prepare them immediately before staining.
Sodium hydroxide.
2.5.4. Iron staining with Perls-DAB Method
Staining of iron deposits is performed by means of the Perls’ method with diaminobenzidine intensification (16).
Usual glassware and small equipment for histology lab.
Potassium ferrocyanide (Sigma ref. P 9387).
Tris buffer.
Diaminobenzidine (DAB) dissolved in distilled water (1g/1000ml). DAB solution can be prepared and aliquoted at –20°C before use.
Methanol.
Hydrogen peroxide 35%.
Hydrochloric acid 35%.
2.5.5. Analysis and quantification of histological stainings
Slide scanner with high optical resolution (e.g. Super CoolScan 8000 ED scanner, Nikon, Champigny sur Marne, France) (see Note 2). Use a scanner that has a 4000 dpi in-plane digitization resolution (pixel size 6.35 µm2) to allow quantification of large objects (e.g. plaques and big focal iron deposits). Work under calibrated and constant illumination conditions.
Optical microscope equipped with a digital camera (optional).
ImageJ freeware (Rasband, W.S., ImageJ, U. S. National Institutes of Health, Bethesda, Maryland, USA, http://rsb.info.nih.gov/ij/, 1997–2005).
Adobe Photoshop® software or any image manipulation program (e.g. The Gimp, http://www.gimp.org/, can be a good freeware alternative).
3. Methods
3.1. Mouse preparation for in vivo MRI
Place the mouse in the induction cage.
Turn the oxygen or air tank on.
Turn the flowmeter up to 1–1.5 L/min.
Induce anesthesia by turning the isoflurane level to 5% until the animal is in lateral decubitus for 2 minutes.
Maintain anesthesia at a concentration of 1.0–1.5% (see Note 3).
Place the mouse prone in the animal cradle of the MR scanner; insert the teeth into the tooth bar and the ear bars into the ear canal; put the respiration and temperature probes in place; cover the animal with the warming blanket.
Insert the head of the animal in the RF coil and slide the animal into the magnet for imaging.
3.2. In vivo MR brain imaging
The parameters for a typical T2-weighted spin echo sequence are presented in Table 1. The resolution with these parameters is about 117 µm isotropic, and the acquisition time is about 42 min.
Table 1.
Example of acquisition parameters for the fast spin echo sequence used to record T2-weighted MR images in vivo
| Parameter | Value | Unit | |
|---|---|---|---|
| Repetition time | TR | 2500 | ms |
| Echo time | TE | 92.3 | ms |
| Field-of-view | FOVx | 15 | mm |
| FOVy | 30 | ||
| FOVz | 15 | ||
| Matrix | MATx | 128 | |
| MATy | 256 | ||
| MATz | 128 | ||
| Rare factor | 16 | ||
3.3. Mouse sacrifice and preparation for ex vivo MRI
3.3.1. Mouse sacrifice and fixation
The entire perfusion fixation procedure should be performed under a hood, possibly equipped with a sink connected to adhoc disposal to evacuate perfused fluids.
Prepare the pump: pour about 500 mL of PBS in a beaker and 500 mL of formalin in another beaker. Connect a butterfly needle to the outflow end of the peristaltic pump and drop the inflow end of the pump into the PBS beaker. Run the pump until no more air bubbles are visible in the perfusion line.
Anesthetize the mouse with an intraperitoneal injection of pentobarbital sodium (at a dose of 100 mg / kg).
When the animal is deeply anesthetized and all reflexes are lost (toe- and tail-pinch checks), place the animal in supine position on a dissecting board placed over a draining bucket. Tape or pushpin the limbs away from the body to hold the animal still. Proceed fairly quickly to begin the perfusion before the heart stops beating.
Perform a bilateral thoracotomy: reach the sternum with the tooth forceps and, just under the ribs, make a bilateral incision into the skin and through the pleural cavity. Use scissors to cut the ribs towards the shoulders and remove them to expose the heart cavity. Cut through the diaphragm then through the pericardium to expose the heart. Be careful not to puncture any organ. Make sure you can clearly identify the left and right ventricles (that are colored with slightly contrasted red nuances) and the right atrium.
Hold the heart with smooth forceps or thumb-index soft pinch and insert the butterfly needle into the left ventricle from the apex up, without piercing into the right ventricle. When the needle is in place (Fig. 3), turn the pump on (flow rate ~ 2 mL / min), and immediately make an incision into the right atrium with a pair of fine scissors to let the blood flow out. Flush the blood out with PBS until the perfusate runs clear. If the perfusion is properly performed, all organs should turn white and the tail should briefly stiffen up.
When all the blood is flushed out, turn the pump off and switch the perfusate line to formalin. Turn the pump back on and perfuse the fixative for about 5 min or until the mouse limb are stiff.
Upon completion of the fixation, turn the pump off and remove the needle from the heart.
Release the mouse, cut its head off, dispose the carcass in a biological hazard bag, then proceed to brain dissection.
3.3.2. Brain dissection
After the mouse has been perfusion fixed and the head has been cut off, quickly proceed to brain dissection to limit tissue degradation.
Make a medial incision through the skin of the head from the base of the neck to the nose to expose the skull.
Localize the olfactory bulbs through the skull, between the two eyes. Insert the tip of sharp scissors through the skull on the medial line at the tip of the olfactory bulbs. Open the inserted scissors to crack open the skull and to separate it in two halves.
With tooth forceps, pull away the two skull halves. Then remove the skull from around the cerebellum and other pieces that might still be covering the brain. Be very careful not to pull apart, squeeze or slice the brain while removing the skull (see Note 4).
The brain should now be free from the skull. With a small spatula, reach under the brain and gently release it from its cup; the cranial nerves should easily break.
Drop the brain in a container with formalin for a 24-hour post-fixation at 4°C, and dispose of the rest of the head carcass in a biological hazard bag.
3.3.3. Passive staining
We adapted and optimized previously published "staining” protocols (13, 17) to Gd-stain the fixed brains.
After a post-fixation of at least 24 hours, soak the brain sample in a solution of phosphate buffered saline (PBS) and 0.5 M gadoterate meglumine at a dilution of 1:200 (2.5 M) (see Note 5).
Store at 4°C for at least 24 hours prior to imaging.
3.3.4. Imaging holder
Imaging holders can easily be home built with 10-mL syringes (Fig. 4).
Use two syringe pistons to close each end of the holder made with the syringe. You can use plastic pieces to hold the brain tight in place in the holder but be careful not to squeeze it (see Note 6).
Place the brain in the imaging holder and fill it half way (up to the beginning of the brain) with Fluorinert®.
Close the holder and remove all air bubbles. To do so, insert a 26-gauge needle filled with Fluorinert® between the cap and the wall of the holder. Push in some fluid; the air bubbles will exit from the small gap created by the needle. Slowly remove the needle while still pushing in some fluid. All air bubbles should be gone. If not, insert again the needle and repeat step 3.
Fig. 4.
Mouse brain holder for ex vivo imaging. The brain sample is held still in a 10-mL syringe filled with Fluorinert.
3.4. Ex-vivo MR brain imaging
A 3D gradient echo sequence can be used (FLASH) to acquire T2*-weighted images. Table 2 gives typical parameters to acquire 72 images in about 14 hrs at a resolution of about 23×23×90 µm3.
Table 2.
Example of acquisition parameters for the 3D gradient echo sequence used to record T2*-weighted MR images ex vivo
| Parameter | Value | Unit | |
|---|---|---|---|
| Repetition time | TR | 100 | ms |
| Echo time | TE | 21 | ms |
| Field-of-view | FOVx | 24 | mm |
| FOVy | 20.25 | ||
| Matrix | MATx | 1024 | |
| MATy | 864 | ||
| Slice thickness | 0.09 | mm | |
| Number of slices | 72 | ||
| Flip angle | FA | 25 | 0 |
3.5. Histological studies
3.5.1. General histology
Section whole brains or single hemispheres (frontal 40 µm-thick sections) on a freezing microtome after a one-week fixation in 10% formalin (see Note 7) and subsequent cryoprotection in 20% glycerin and 2% DMSO in 0.1 M PB.
Collect twelve batches of serial sections (ranging from the frontal pole to the end of the caudal part of the hippocampus). Immediately rinse in 0.1 M PB the series of free floating sections to be stained and mount them on Superfrost+ glass slides before drying them overnight at room temperature (or in an oven at 40°C). Remaining tissue can be stored at –20°C in cryoprotectant as backup material.
For each mouse it is suggested to perform a Nissl (thionin) stain to control for tissue quality and/or cytoarchitectonic anomalies before processing Congo red and Perls-DAB stains.
3.5.2. β–Amyloid staining with BAM10 antibody (Fig. 5)
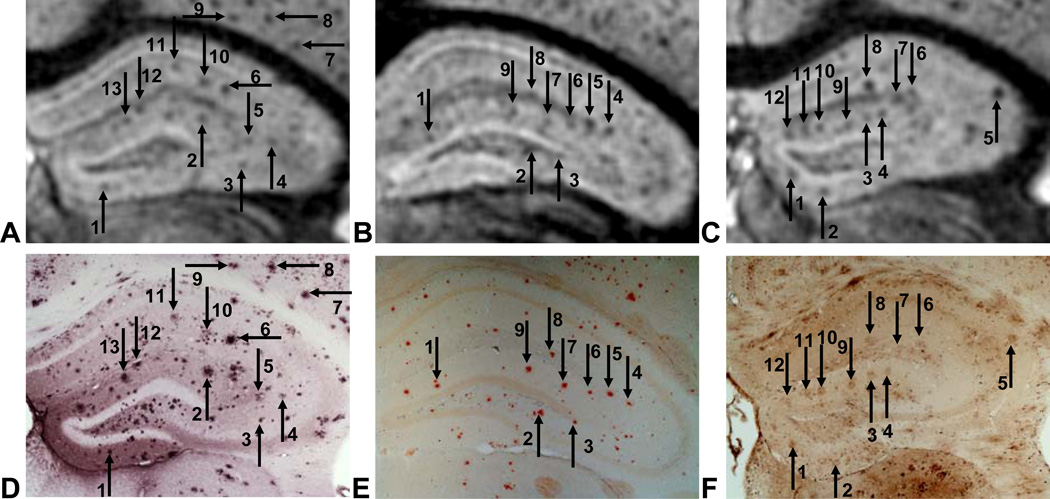
Fig. 5.
Hypointense spots detected at the level of the hippocampus with MRI (top, A-C) match anti-Aβ staining (D), Congo red staining (E) and Perls’ staining (F). This indicates that, in the mouse model that we used, these spots correspond to amyloid plaques and that they are loaded with iron.
Follow these steps for free-floating sections (total solution volume of 5 mL)
Day 1
Rinse the sections 6 times in PBS for 5 min.
Incubate in H202 (0.3%) for 20 min.
Rinse 3 times in PBS for 10 min.
To the (PBS + 0.2% Triton) solution, add 4.5% of normal rabbit serum. Incubate for 30 min.
To the (PBS + 0.2% Triton) solution, add 3% of normal rabbit serum and 0.1% of the primary antibody. Incubate for 48 hrs at room temperature (or 3 days at 4° C).
Day 3
Rinse 3 times in PBS for 10 min.
To the (PBS + 0.2% Triton) solution, add 3% of normal rabbit serum and 0.1% of the secondary antibody and incubate for 1 hr.
Rinse 3 times in PBS for 10 min.
To the (PBS + 0.2% Triton) solution, add kit reagent A (avidin DH) at dilution 1/250 and kit reagent B (biotinylated enzyme) at dilution 1/250. Incubate in this avidin-biotin complex for 1 hr.
Rinse 3 times in PBS for 10 min.
Revelation with VIP substrate: to 5 mL of PBS, add 3 drops of each of the kit reagents (reagents 1, 2, 3 and hydrogen peroxide) and mix well. Incubate the free-floating sections in this solution for about 2 min.
3.5.3. β–Amyloid staining with Congo red (Fig. 5)
Labeling of amyloid deposits is done by standard Congo red staining (adapted from ref. 15).
Prepare S1 & S2; filter S2 before use.
Put slides under running tap water for 20 min.
Add NaOH [10−4] to S1 and incubate the slides for 30 min.
Add NaOH [10−4] to S2 and incubate the slides for 30 min.
Rinse under tap water.
Dehydrate in alcohols, clear in xylen and coverslip with Eukitt.
3.5.4. Iron staining with Perls-DAB Method (Fig. 5)
Staining of iron deposits is performed by means of the Perls’ method with diaminobenzidine intensification (16).
Rehydrate the slides under running tap water for 20 min.
Inactivate endogenous peroxidase activity by immersing the tissue in a methanol (20%) / H202 (3%) solution made in distilled water for 10 min.
Rinse in distilled water twice for 5 min.
Incubate in acid potassium ferrocyanide solution (for 100 mL of distilled water, add 1 g of potassium ferrocyanide and 1 mL of 35% HCl) for 20 min.
Rinse in distilled water for 5 min and then twice in 0.1M Tris buffer for 5 min.
Dilute DAB 2X in 0.2 M Tris; add 30–40 µL of H202 per 100 mL of final volume just before reaction.
Incubate the slides in DAB until a good signal-to-noise ratio is obtained (reaction is monitored under the microscope).
Rinse in distilled water. Store DAB in waste container or detoxify it.
Dehydrate in alcohols, clear in xylen and coverslip with Eukitt.
3.5.5. Analysis and quantification of histological stainings
Amyloid deposits
If the purpose of the study is to register MR images with histologically assessed topography of plaques, no additional processing is required (13).
For quantitative analysis, amyloid loads are evaluated using computer-based thresholding methods. Scans are prepared using Photoshop software to outline selected regions of interest (ROI). Images are then processed with ImageJ freeware using a dedicated macrocommand that extracts amyloid deposits from background tissue (18, 19). Briefly, image processing to detect plaques relies on RGB color component adjustment, global automated threshold based on entropy criterion and morphometric filtering according to Feret's diameter. Macro is available on demand.
The step-by-step instructions are as follows:
Run ImageJ and open the macrocommand.
Select the folder containing the images to be analyzed (tiff format).
Automatic processing of all images is then initiated (this can take few minutes in case numerous files have to be processed).
After batch processing, a new subdirectory named "processed" is automatically created in the parent directory and contains (a) thresholded images allowing quick visual inspection of the results of plaque segmentation, (b) a text file that can be imported (tab format) in a spreadsheet program and that contains all morphological data (eg total surface, thresholded surface) required to calculate amyloid loads for each images.
Regional amyloid loads are expressed as percent of tissue surface stained by the Congo red dye that corresponds to the proportion of plaque volume according to Delesse’s principle (20). Evaluation of amyloid loads can be performed in multiple ROIs. Quantitative analyses are usually performed on several serial sections to sample the whole rostro-caudal extent of each ROI.
Iron deposits
If the purpose of the study is to register MR images with histologically-assessed topography of iron deposits, no additional processing is required (14).
Considering now more quantitative aspects (e.g. measuring iron loads), it is known that there is a relationship between “true” tissue iron content (as assessed for instance by atomic absorption spectroscopy) and intensity of Perls’ staining (21, 22). We and others perform an analysis of Perls-stained brain tissue by means of optical densitometry -OD (11, 23, 24).
OD determines levels of iron deposition on the basis of transmitted light in the stained tissue. OD can be automatically calculated in selected ROIs using ImageJ or Photoshop. The optical density of each pixel is derived from its gray level and mean iron load is defined as the mean OD from all pixels of the ROI. Standards to assess absolute iron quantities are not easily available for the analysis of Perls-stained material; therefore only relative quantities of iron deposition can be calculated, still allowing inter-group comparisons.
The step-by-step instructions are as follows:
Run image J and set measurements to include the mean gray level variable as output.
Open image to be analyzed.
Outline the region of interest using the polygon tool.
Run the measure command (shortcut: ctrl+M).
Uncalibrated optical density of Perls staining is calculated using the following formula: OD = log10 (255–MGL) with MGL as mean gray level of the region of interest.
ACKNOWLEDGEMENTS
We thank Fanny Petit from CEA for her assistance in histology processing for the figures presented. Our work was supported by the France-Alzheimer association, the National Foundation for Alzheimer’s Disease and Related Disorders, and the NIH (R01-AG020197).
Footnotes
Notes
The APP/PS1dE9 double transgenic mice express a chimeric mouse/human amyloid precursor protein (Mo/HuAPP695swe) and a mutant human presenilin 1 (PS1-dE9) both directed to CNS neurons. Both mutations are associated with early-onset of AD. The "humanized" Mo/HuAPP695swe transgene allows the mice to secrete a human A-beta peptide. The included Swedish mutations (K595N/M596L) elevate the amount of A-beta produced from the transgene by favoring processing through the beta-secretase pathway. The PS1 mutation elevates the amount of A-beta produced from the transgene by favoring processing through the gamma-secretase pathway. We used the strain referenced as "Stock Number: 004462". This strain is maintained as a hemizygote line by crossing transgenic mice to B6C3F1/J mice. The strain referenced as "Stock Number: 005864" can also be used. These mice are based on the 004462 strain but were backcrossed to C57BL/6J for at least 8 generations.
If the resolution of the scanner appears to be insufficient (e.g. tiny deposits are not detected or with very blurred edges), it is recommended to digitize material at higher resolution using a microscope coupled to a camera. For optimal results, images should be stored as tiff files (jpeg compression has detrimental effects when images are further processed).
If O2 is used as a pushing gas for isoflurane, the percentage of isoflurane will have to be increased to achieve the same level of anesthesia.
To avoid pressing against the brain and damaging the tissue during dissection, you can fold the skin around the inferior (ventral) part of the head and hold it between your fingers, this way you should not touch the brain itself at all.
By soaking the brain sample in a PBS solution (and 0.5 M gadoterate meglumine) for at least 24 hrs prior to MR imaging, you will regain some signal due to tissue rehydration.
The length of the holder should be at least 4 times as long as the brain so that the brain sits away from the ends to avoid imaging artifacts that might arise from the pistons. It is also recommended to wipe off the syringe graduation marks with ethanol as they might also cause imaging artifacts.
As a general rule, if the animal is fixed via a peri-mortem intracardiac perfusion, no further tissue fixation is necessary. For fresh brains or brains perfused with saline / PBS only, a one-week fixation should be done in 10% formalin. However, in the ex vivo protocol for MR imaging, the brains are soaked for days in the staining solution containing PBS instead of formalin. It is therefore recommended to soak the brains again in formalin before sectioning them for histology evaluation.
Perls-DAB staining can alternatively be performed on free-floating sections to maximize penetration of reagents.
Before dehydrating tissue, a nuclear counterstain can be applied using thionin, nuclear red, or Harris hematoxylin. Do not counterstain when quantification of iron deposition has to be performed by optical densitometry.
REFRENCES
- 1.Hyman BT, Marzloff K, Arriagada PV. The lack of accumulation of senile plaques or amyloid burden in Alzheimer's disease suggests a dynamic balance between amyloid deposition and resolution. J. Neuropath. Exp. Neur. 1993;52:594–600. doi: 10.1097/00005072-199311000-00006. [DOI] [PubMed] [Google Scholar]
- 2.Jack CR, Jr, Lowe VJ, Weigand SD, Wiste HJ, Senjem ML, Knopman DS, Shiung MM, Gunter JL, Boeve BF, Kemp BJ, Weiner M, Petersen RC. Serial PIB and MRI in normal, mild cognitive impairment and Alzheimer's disease: implications for sequence of pathological events in Alzheimer's disease. Brain. 2009;132:1355–1365. doi: 10.1093/brain/awp062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer's disease: progress and problems on the road to therapeutics. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 4.Blennow K, de Leon MJ, Zetterberg H. Alzheimer's disease. Lancet. 2006;368:387–403. doi: 10.1016/S0140-6736(06)69113-7. [DOI] [PubMed] [Google Scholar]
- 5.Duyckaerts C, Potier MC, Delatour B. Alzheimer disease models and human neuropathology: similarities and differences. Acta Neuropathol. 2008;115:5–38. doi: 10.1007/s00401-007-0312-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delatour B, Le Cudennec C, El Tannir-El Tayara N, Dhenain M. Transgenic models of Alzheimer's pathology: Success and caveat. In: Welsh EM, editor. Topics in Alzheimer's Disease. Nova Publishers; 2006. pp. 1–34. [Google Scholar]
- 7.Delatour B, Guegan M, Volk A, Dhenain M. In vivo MRI and histological evaluation of brain atrophy in APP/PS1 transgenic mice. Neurobiol. Aging. 2006;27:835–847. doi: 10.1016/j.neurobiolaging.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 8.Valk J, Barkhof F, Scheltens P. Magnetic resonance in dementia. Heidelberg Berlin New York: Springer-Verlag; 2002. [Google Scholar]
- 9.Albert M, DeCarli C, DeKosky S, de Leon M, Foster NL, Fox N, Frank R, Frackowiak R, Jack C, Jagust WJ, Knopman D, Morris JC, Petersen RC, Reiman E, Scheltens P, Small G, Soininen H, Thal L, Wahlund L-O, Thies W, Weiner MW, Khachaturian Z. Report of the neuroimaging work group of the Alzheimer's Association. 2005 Available from http://www.alz.org/national/documents/Imaging_consensus_report.pdf. [Google Scholar]
- 10.Klunk WE, Lopresti BJ, Ikonomovic MD, Lefterov IM, Koldamova RP, Abrahamson EE, Debnath ML, Holt DP, Huang GF, Shao L, DeKosky ST, Price JC, Mathis CA. Binding of the positron emission tomography tracer Pittsburgh compound-B reflects the amount of amyloid-beta in Alzheimer's disease brain but not in transgenic mouse brain. J. Neurosci. 2005;25:10598–10606. doi: 10.1523/JNEUROSCI.2990-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.El Tannir El Tayara N, Delatour B, Le Cudennec C, Guegan M, Volk A, Dhenain M. Age-related evolution of amyloid burden, iron load, and MR relaxation times in a transgenic mouse model of Alzheimer's disease. Neurobiol. Dis. 2006;22:199–208. doi: 10.1016/j.nbd.2005.10.013. [DOI] [PubMed] [Google Scholar]
- 12.El Tayara Nel T, Volk A, Dhenain M, Delatour B. Transverse relaxation time reflects brain amyloidosis in young APP/PS1 transgenic mice. Magn. Reson. Med. 2007;58:179–184. doi: 10.1002/mrm.21266. [DOI] [PubMed] [Google Scholar]
- 13.Dhenain M, Delatour B, Walczak C, Volk A. Passive staining: A novel ex vivo MRI protocol to detect amyloid deposits in mouse models of Alzheimer's disease. Magn. Reson. Med. 2006;55:687–693. doi: 10.1002/mrm.20810. [DOI] [PubMed] [Google Scholar]
- 14.Dhenain M, El Tannir El Tayara N, Wu T-D, Guegand M, Volk A, Quintana C, Delatour B. Characterization of in vivo MRI detectable thalamic amyloid plaques from APP/PS1 mice. Neurobiol. Aging. 2009;30:41–53. doi: 10.1016/j.neurobiolaging.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 15.Puchtler H, Sweat F, Levine M. On the binding of Congo red by amyloid. J. Histochem. Cytochem. 1962;10:355–364. [Google Scholar]
- 16.Nguyen-Legros J, Bizot J, Bolesse M, Pulicani J-P. "Noir de diaminobenzidine" : une nouvelle méthode histochimique de révélation du fer exogène ("Diaminobenzidine black": A new histochemical method for the visualization of exogenous iron) Histochemistry. 1980;66:239–244. doi: 10.1007/BF00495737. [DOI] [PubMed] [Google Scholar]
- 17.Petiet A, Hedlund L, Johnson GA. Staining methods for magnetic resonance microscopy of the rat fetus. J. Magn. Reson. Imaging. 2007;25:1192–1198. doi: 10.1002/jmri.20932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Faure A, Verret L, Bozon B, El Tannir El Tayara N, Ly M, Kober F, Dhenain M, Rampon C, Delatour B. Impaired neurogenesis, neuronal loss, and brain functional deficits in the APPxPS1-Ki mouse model of Alzheimer's disease. Neurobiol. Aging. 2010 doi: 10.1016/j.neurobiolaging.2009.03.009. in press. [DOI] [PubMed] [Google Scholar]
- 19.Le Cudennec C, Faure A, Ly M, Delatour B. One-year longitudinal evaluation of sensorimotor functions in APP751SL transgenic mice. Genes Brain Behav. 2008;7(Suppl 1):83–91. doi: 10.1111/j.1601-183X.2007.00374.x. [DOI] [PubMed] [Google Scholar]
- 20.Delesse MA. Procédé mécanique pour déterminer la composition des roches [Mechanical process to evaluate the coomposition of rocks] Comptes rendus de l'Académie des Sciences (Paris) 1847;25:544–545. [Google Scholar]
- 21.Masuda T, Kasai T, Satodate R. Quantitative measurement of hemosiderin deposition in tissue sections of the liver by image analysis. Ana.l Quant. Cytol. Histol. 1993;15:379–382. [PubMed] [Google Scholar]
- 22.Turlin B, Loreal O, Moirand R, Brissot P, Deugnier Y, Ramee MP. Détection histochimique du fer hépatique [Histochemical detection of hepatic iron. A comparative study of four stains] Ann. Pathol. 1992;12:371–373. [PubMed] [Google Scholar]
- 23.Bizzi A, Brooks RA, Brunetti A, Hill JM, Alger JR, Miletich RS, Francavilla TL, Di Chiro G. Role of iron and ferritin in MR imaging of the brain: a study in primates at different field strengths. Radiology. 1990;177:59–65. doi: 10.1148/radiology.177.1.2399339. [DOI] [PubMed] [Google Scholar]
- 24.Hill JM, Switzer RC. The regional distribution and cellular localisation of iron in the rat brain. Neuroscience. 1984;11:595–603. doi: 10.1016/0306-4522(84)90046-0. [DOI] [PubMed] [Google Scholar]