Abstract
The stomach, a hallmark of gnathostome evolution, represents a unique anatomical innovation characterized by the presence of acid- and pepsin-secreting glands. However, the occurrence of these glands in gnathostome species is not universal; in the nineteenth century the French zoologist Cuvier first noted that some teleosts lacked a stomach. Strikingly, Holocephali (chimaeras), dipnoids (lungfish) and monotremes (egg-laying mammals) also lack acid secretion and a gastric cellular phenotype. Here, we test the hypothesis that loss of the gastric phenotype is correlated with the loss of key gastric genes. We investigated species from all the main gnathostome lineages and show the specific contribution of gene loss to the widespread distribution of the agastric condition. We establish that the stomach loss correlates with the persistent and complete absence of the gastric function gene kit—H+/K+-ATPase (Atp4A and Atp4B) and pepsinogens (Pga, Pgc, Cym)—in the analysed species. We also find that in gastric species the pepsinogen gene complement varies significantly (e.g. two to four in teleosts and tens in some mammals) with multiple events of pseudogenization identified in various lineages. We propose that relaxation of purifying selection in pepsinogen genes and possibly proton pump genes in response to dietary changes led to the numerous independent events of stomach loss in gnathostome history. Significantly, the absence of the gastric genes predicts that reinvention of the stomach in agastric lineages would be highly improbable, in line with Dollo's principle.
Keywords: stomach, gene loss, proton pump, pepsinogen, gene duplication, gnathostomes
1. Introduction
Gene duplication is a powerful event underscoring evolutionary change [1]. In effect, genome duplications in vertebrate history (2R and 3R) have been linked with the evolution of novel morphological and physiological traits [2–5]. In parallel, the impact of gene loss on phenotypic variation and diversity has been less explored, though gene extinction events are frequent in evolution and have phenotypic impacts [6–9].
The stomach, a specialized segment of the gut, is characterized by the occurrence of acid (HCl) and pepsin-producing gastric glands [10]. It represents a notable case of morphological variability, as first noted by Aristotle (345 BC) while commenting in his Historia Animalium about ‘the diversity of shapes’ [11]. Gastric glands first appeared approximately 450 Myr ago and represent a key functional innovation found exclusively in jawed vertebrates (figure 1) [10]. Invertebrate chordates such as amphioxus and ascidians lack a stomach, as do the jawless lampreys and hagfishes. Evolutionarily, the acquisition of an acidic luminal environment extended the spectrum of dietary protein sources. At low pH, long proteins are denaturated, facilitating the action of endopeptidases, which also evolved to operate optimally at these low pH levels [10,12]. The improvement by low pH of phosphate [13] and calcium [14,15] uptake, as well as its effect as a barrier against pathogen entry to the intestine, were also probably beneficial [10].
Figure 1.
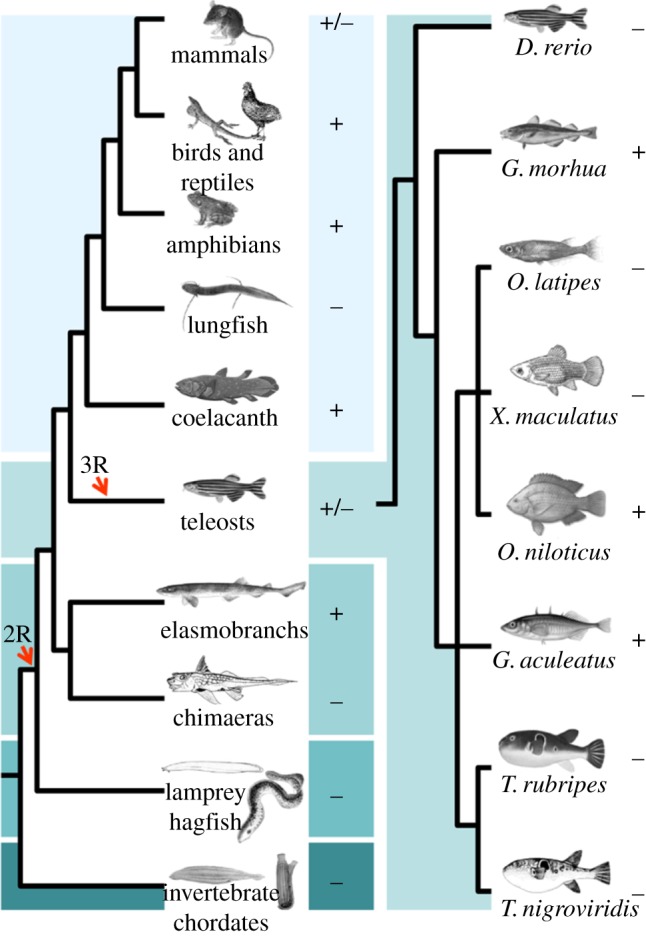
Phylogenetic relationships of extant chordate lineages. Plus and minus signs represent the presence or the absence of gastric glands, respectively. Timings of whole genome duplications (2R and 3R) are indicated. (Online version in colour.)
From the eighteenth to the twentieth century, a string of findings revealed the significance of gastric acid secretion in digestion, as well as the identification of its functional components, the pepsin and the gastric proton pump [16]. The gastric proton pump is an H+/K+-ATPase belonging to the P type ATPase IIc subfamily that is capable of pumping H+ against a 160 mM gradient [17]. It is a heterodimer composed of unrelated subunits: the HKα (alpha subunit), encoded by Atp4A, and the HKβ (beta subunit), encoded by Atp4B. Atp4A has been isolated and characterized in various vertebrate classes, including the Atlantic stingray, a cartilaginous fish [18,19]. The gastric Atp4B is unique among the P-type IIc family of ATPase beta subunit genes because it is the sole partner for Atp4A in vivo [17,20]. Fundamental to gastric function are the pepsinogens, such as Pepsinogen A (Pga), Pepsinogen B (Pgb), Pepsinogen F (Pgf), Progastricsin (Pgc) and Prochymosin (Cym) [12]. These are precursors of the pepsin enzymes, which are activated at low pH and digest proteins into smaller peptides, making them available for further digestion and absorption [12]. The diversity of pepsinogen gene families has been suggested to underscore differences in substrate specificity [12,21].
Surprisingly, acid–pepsinogen-producing glands are absent in several gnathostome lineages. The French zoologist Georges Cuvier (1805) [22], on the basis of gross morphology, first observed that some teleost groups (e.g. cyprinidae and labridae) lack a stomach. The number of observations of the agastric phenotype in teleost fishes has expanded greatly over the past 200 years. A conservative estimate indicates the occurrence of 15 independent loss events covering seven families and 20–27% of species [23]. Significantly, the lack of gastric acid secretion is not limited to the teleost lineage; Holocephali, dipnoids and monotremes also exhibit an agastric gut (figure 1) [24]. The molecular foundations for the occurrence of distinct stomach phenotypes in gnathostome lineages are still unclear. The lack of gastric glands in platypus coincides with the loss of the gene repertoire involved in gastric digestion [25]. Here, we test the hypothesis that absence of the gastric phenotype is correlated with absence of key gastric genes. We investigate whether the extensive distribution of the agastric phenotype in vertebrate history is paralleled by differences in gene complement, namely the pepsinogens and the Atp4A/Atp4B genes. Taking advantage of full genome sequences from all major vertebrate classes, we show that the lack of a stomach phenotype (acid secretion and pepsin activity) correlates with the targeted deletion or inactivation of the gene repertoire involved in the gastric function, in all of the examined species.
2. Material and methods
(a). Database searches and synteny analysis
The full coding sequences of the human proteins for ATP4A, ATP4B and pepsinogen genes were used as query for TBLASTN searches of the Ensembl (release 69) and GenBank databases. The genomes of 14 vertebrate species were investigated: Homo sapiens (human, gastric, GRCh37), Mus musculus (mouse, gastric, GRCm38), Monodelphis domestica (opossum, gastric, BROADO5), Gallus gallus (chicken, gastric, WASHUC2), Anolis carolinensis (anolis, gastric, AnoCar2.0), Xenopus tropicalis (western clawed frog, gastric, JGI_4.2), Gasterosteus aculeatus (stickleback, gastric, BROADS1), Oryzias latipes (medaka, agastric, MEDAKA1), Takifugu rubripes (pufferfish, agastric, FUGU4), Tetraodon nigroviridis (green-spotted pufferfish, agastric, TETRAODON8), Gadus morhua (cod, gastric, gadMor1), Oreochromis niloticus (Nile tilapia, gastric, Orenil1.0), Xiphophorus maculatus (platyfish, agastric, Xipmac v. 4.4.2) and Danio rerio (zebrafish, agastric, Zv. 9). The genomic location of each gene of interest was identified along with the two closest flanking genes. The orthology of each gene was verified through phylogenetics (not shown). The Callorhinchus milii genome was searched with TBLASTN at http://esharkgenome.imcb.a-star.edu.sg/Blast/.
(b). Alignment and phylogenetics
The retrieved protein sequences were aligned using MAFFT with the L-INS-i method [26]. The GenBank accession numbers or Ensembl codes for each sequence are indicated in electronic supplementary material S1. Gaps were removed from each alignment. The final alignment datasets were as follows: 21 alpha subunit sequences (768 amino acids length), 47 beta subunit sequences (207 amino acids length) and 42 aspartic protease sequences (283 amino acids length). To determine the best model of amino acid substitution ProtTest2 was run [27]. The selected models were LG + I + G + F (alpha subunit), LG + I + G (beta subunit) and WAG + I + G + F (aspartic proteases). The maximum-likelihood trees were reconstructed using PhyML online [28], and confidence in each node was assessed by 1000 bootstrap replicates of the data (presented as percentage of trees). The same aspartic protease amino acid alignment was used to reconstruct a neighbour-joining (NJ) tree [29] using standard settings with ClustalX v. 2.0.11 [30]. Trees were visualized with TreeFig. The Atp4A and Atp4B trees were rooted using Ciona intestinalis as an outgroup. The pepsinogen tree was rooted with CTSE-like sequences.
(c). Isolation of chicken Atp4A and catshark Atp4B
Chicken (G. gallus) specimens were obtained commercially in Portugal and killed by cervical dislocation prior to tissue sampling. Catsharks (Scyliorhinus canicula) were obtained locally and anaesthetized with tricaine methylsufonate (1 : 5000 MS-222 Aquapharm UK) pH 7.5 and killed by cervical transaction. Animals were treated in accordance with the Portuguese Animal Welfare Law (Decreto-Lei no.197/96) and animal protocols approved by CIIMAR/UP and DGV (Ministry of Agriculture). RNA was isolated from gastric samples with Illustra RNAspin mini kits with on column DNase I treatment (GE Healthcare, UK). Concentration and purity of RNA were assessed and 1 µg of total RNA converted into first strand cDNA with oligo-dT primer and SuperScript III Reverse transcriptase (Life Technologies, USA). The polymerase chain reaction (PCR) and rapid amplification of cDNA ends (RACE) methods (SMARTer RACE, Clontech, USA) were used to isolate complete ORF of Atp4A and Atp4B in chicken and catshark. The Phusion Flash hot start high fidelity polymerase mix was used for all PCR protocols with the manufacture recommended conditions (Life Technologies; reaction details and primer pairs in electronic supplementary material S1).
(d). Isolation and sequencing of the Atp4B locus in Callorhinchus milii
Our search of the 1.4× assembly C. milii genome identified partial sequences of Atp4B locus flanking genes Tmco3 (AAVX01006853.1) and Grk1 (AAVX01074983.1). PCR primers designed for these gene fragments were used to screen pooled DNA of a C. milii BAC library (IMCB_Eshark BAC library; B. Venkatesh 2013, unpublished data) comprising 92 160 clones in a three-step PCR screening. Two positive BAC clones were confirmed by PCR amplification of the gene fragment and sequencing the PCR amplicons. The intergenic space between Tmco3 and Grk1 was sequenced through a combination of BAC primer walk sequencing and BLAST of the 1.4× genome assembly of C. milii. Degenerate primers to the Atp4B second exon were also used to PCR from elephant shark genomic DNA (Atp4BFexon2 5′ GGATCTCCCTGTACTACGTGgcnttytaygt 3′ and Atp4BRexon2 5′ CGGTCCTGGTAGTCGggngyrtangg 3′).
3. Results
(a). Agastric species have lost Atp4A and Atp4B
We investigated the genome sequences of 14 species covering a wide range of evolutionary lineages, where the presence or the lack of a stomach phenotype is well established (figure 1). The analysed species included mammals, reptiles, birds, amphibians and teleosts. We further confirmed the presence/absence of gastric glands through histological analysis in some fish species (gastric: catshark S. canicula, stickleback G. aculeatus, armoured catfish Hypostomus plecostomus, mudskipper Periophthalmus barbarus; agastric: green-spotted pufferfish T. nigroviridis, zebrafish D. rerio, lungfish Protopterus annectens; electronic supplementary material S2).
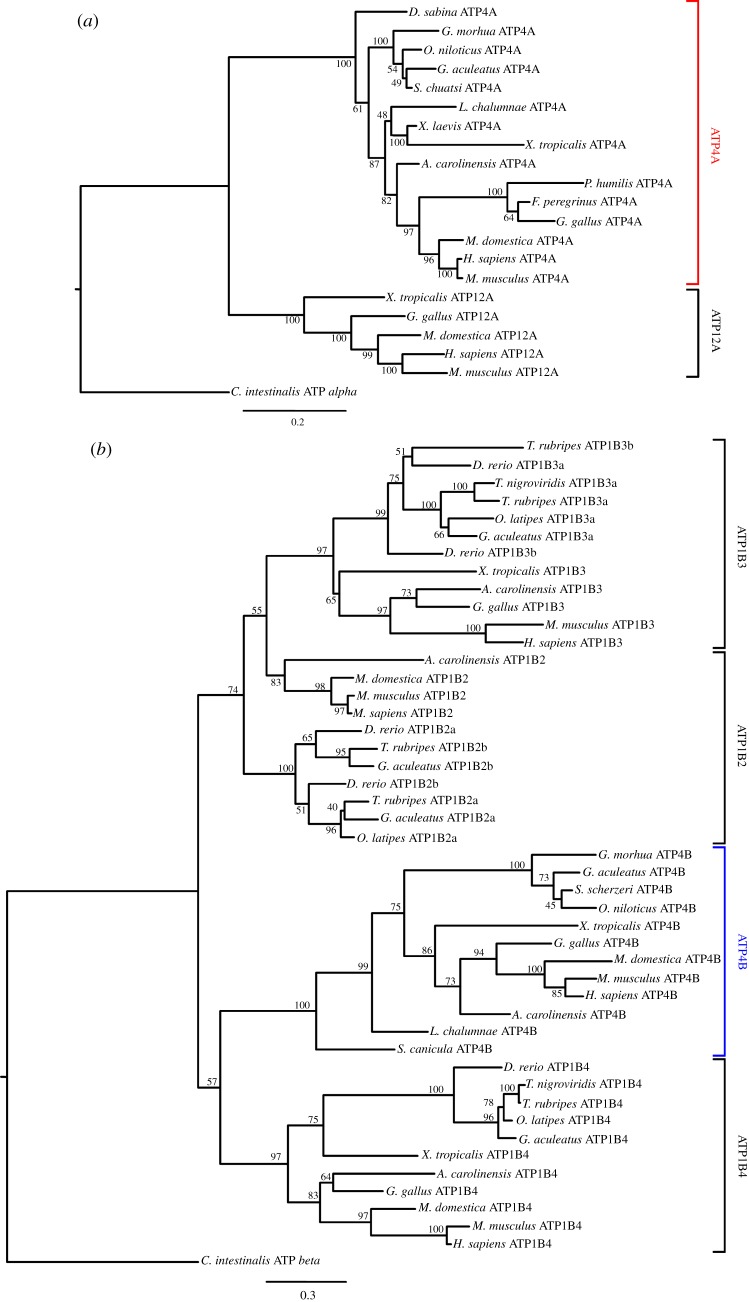
We started by establishing the phylogenetic distribution of Atp4A and Atp4B genes. Sequences with similarity to both gene families were found in anolis, western clawed frog and most mammals, with the exception of platypus, as was previously reported [25] (not shown). No Atp4A sequence was found in the chicken genome (see below). In teleosts, Atp4A and Atp4B genes were uniquely recovered in stickleback, cod and tilapia. Phylogenetic analyses clearly show that these sequences are orthologues of Atp4A and Atp4B (figure 2a,b). To investigate whether incomplete genome coverage or true gene loss was the cause behind the apparent absence of Atp4A and Atp4B, we next examined their respective genomic loci. The Atp4A locus displayed a strong degree of synteny in all the analysed tetrapod species (figure 3a). A different conserved gene set was found in the vicinity of the teleost Atp4A represented here by stickleback, cod and tilapia (figure 3a). In the agastric teleost species, even though a strongly conserved syntenic block was found, there was no evidence for an intervening Atp4A gene ORF. Surprisingly, we were unable to find in the current chicken genome assembly hits similar to Atp4A, or any of the flanking gene families (e.g. Tmem147). The finding was surprising since birds undoubtedly have HCl secretion [31]. To determine whether Atp4A was unsequenced in the chicken genome we undertook a PCR approach to isolate a complete Atp4A sequence. Our results indicate that the gene is present in the chicken genome (figure 2a). Independently, we also found Atp4A orthologue in two other birds: Falco peregrinus (peregrine falcon) and Pseudopodoces humilis (Tibetan ground-tit).
Figure 2.
Maximum-likelihood tree of (a) alpha and (b) beta H+/K+-ATPase subunit genes in vertebrate species with bootstrap values (percentage of 1000 replicates) shown at each node. Each phylogeny was rooted with a C. intestinalis sequence. (Online version in colour.)
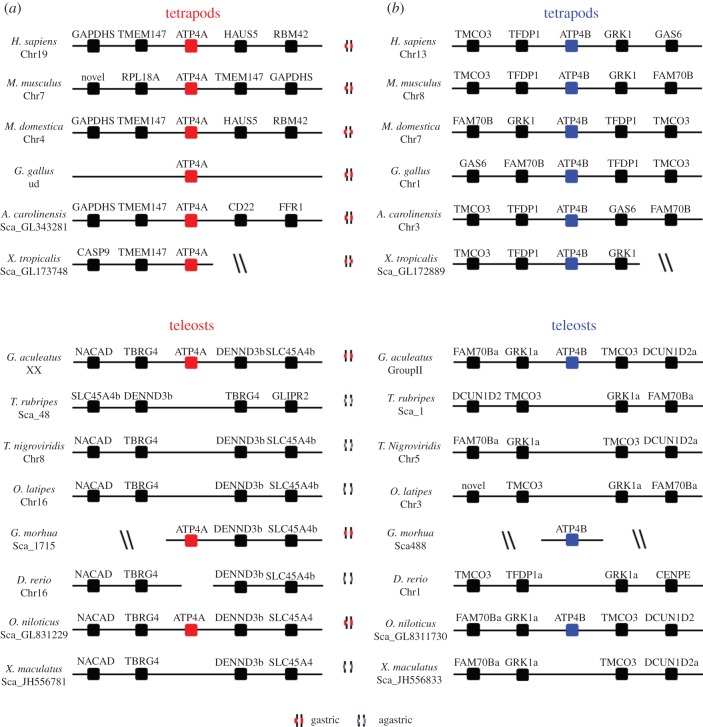
Figure 3.
Genomic loci of the (a) Atp4A and (b) Atp4B in vertebrate species. Chromosome or scaffold number is indicated. (Online version in colour.)
Regarding the Atp4B loci, a strongly conserved gene arrangement was found in both tetrapods and teleosts (figure 3b). Again, the agastric species have no intervening Atp4B ORF. Thus, we find an absolute correlation between the agastric phenotype and the absence of the two proton pump genes in their expected genomic loci. The preservation of the gene loci arrangements in combination with the phylogenetic analysis indicates that gene deletion has taken place in the genome of all four agastric teleosts, paralleling similar findings in the stomach-less mammal, the platypus [25].
(b). Duplication, pseudogenization and loss of pepsinogen genes in vertebrate species
Tetrapod aspartic proteases expressed almost uniquely in the stomach include Pga, Pgf, Pgc, Pgb and Cym [12,32–34]. In fishes, a distinct nomenclature is currently used and three pepsinogen gene families have been described: Pgc, Pga1 and Pga2 [35–38].
We began by determining the repertoire of Pga, Pgf and Cym in tetrapods. Pgc and Pgb were not examined here since we previously documented their history in tetrapods [38]. Both phylogenetic methods recovered similar tree topologies although with some differences (see electronic supplementary material S3). For example, Cym sequences form a monophyletic assembly in the NJ tree, while in the ML tree the mammalian and sauropsid sequences are separated. In general, higher bootstrap support was found in the NJ analysis, although the more basal nodes display lower values (see electronic supplementary material, S3). A single Cym gene was found in all of the examined tetrapod species, except in the western clawed frog and human. In the latter, a Cym pseudogene has been previously reported [39]. The number of Pga genes varies significantly between the studied species. Three clear paralogues are found in humans, although they represent an independent duplication. In chicken, we found two Pga-like genes, although the sequence of the second gene was very short and was not included in the phylogeny (not shown). Single-copy Pga orthologues were recovered in anolis and the western clawed frog. In contrast to previous suggestions [25], we found no indication for Pga or Pgf in the opossum. In the analysed species, we only identified Pgf in mammalian species. One sequence from the western clawed frog groups with mammalian Pgf, although with low statistical support to infer a proper orthology. This finding suggests that the Pga/Pgf duplication pre-dates tetrapod radiation.
We next inspected the gene loci of Pga, Pgf and Cym (figure 4). A strong conservation in gene arrangement was detected, with Pga and Pgf residing in the same locus. In the opossum, we were able to determine the existence of two pseudogenes with similarity to Pga/Pgf, which explains the conflicting results with Ordoñez et al. [25] (figure 4; electronic supplementary material S4a). The combination of phylogenetics and the analysis of the genomic loci supports the interpretation that Pgf was lost at least in humans and opossum, and probably in anolis and chicken.
Figure 4.
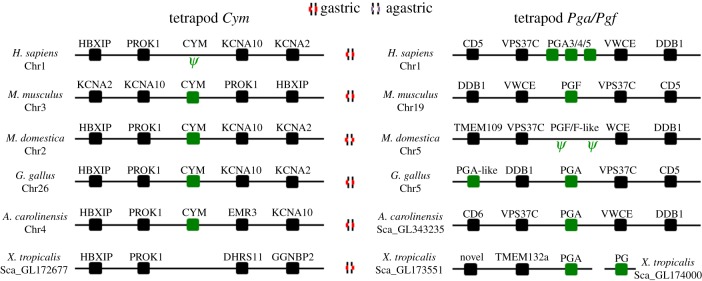
Genomic neighbourhood of the pepsinogen genes in tetrapod species. Chromosome or scaffold number is indicated. Ψ indicates a pseudogene. (Online version in colour.)
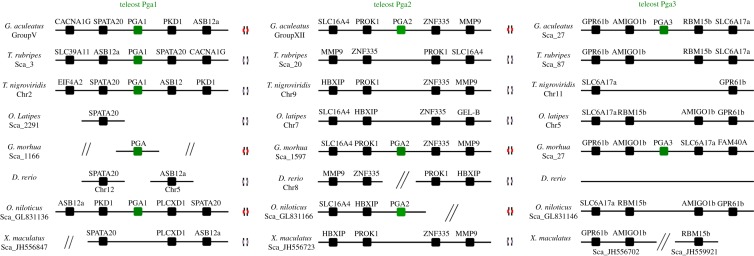
In the analysed teleost species, a substantial degree of variation was found with respect to the pepsinogen gene complement. The highest gene number was found in stickleback and cod, each with four pepsinogen genes. In tilapia, three pepsinogen-like sequences were recovered, while a single gene was found in pufferfishes (Takifugu and Tetraodon). In zebrafish, platyfish and medaka no relevant sequences with similarity to pepsinogen genes were discovered. The phylogenetic analysis provided clarification to the orthology of the retrieved sequences (see electronic supplementary material S3). Three of the sequences found in the stickleback and cod correspond to orthologues of Pgc, Pga1 and Pga2. The fourth sequence represents a novel Pga-like gene clade not previously reported, which we name Pga3. In tilapia, one sequence robustly groups with Pga2 clade, while the remaining groups with Pga1. The analysis of genomic loci of the various pepsinogen genes shows that these are mostly conserved between teleost species (figure 5). Surprisingly, even though teleost Pga genes form an independent clade, the gene families in the vicinity of Pga2 and Pga3 suggest that these are orthologues of tetrapod Cym and not Pga as previously reported [32]. Finally, the conservation of the loci in combination with the phylogenetic analysis provides support that extensive gene loss has taken place. With the exception of pufferfishes, no pepsinogen genes were found in the other agastric teleost lineages.
Figure 5.
Genomic neighbourhood of the pepsinogen genes in teleost species. Chromosome or scaffold is indicated. Gastric and agastric species are identified with appropriate symbols (figure 3). (Online version in colour.)
Previously, we also established that Pgc orthologues are absent in agastric teleosts such as medaka, zebrafish, Takifugu and Tetraodon, while present in the gastric stickleback [38]. Here, we extend the analysis to include two other teleost species: cod and tilapia (see electronic supplementary material S4b). We find that Pgc is present in cod but absent in tilapia. The lack of Pgc in tilapia is noticeable, as this species has a clear gastric phenotype [40], supported by our finding of Atp4A and Atp4B orthologues. Examination of the ‘Pgc’ locus in this species provides strong evidence for pseudogenization.
In summary, we find that the stomach phenotype (presence/absence of gastric glands) positively correlates with the complement of pepsinogen genes in the vertebrate species, although we also find gene retention in agastric species (e.g. Takifugu) and gene loss in gastric ones (e.g. tilapia).
(c). Atp4b is missing in the stomach-less chimaera Callorhinchus milii, but present in the gastric elasmobranch Scyliorhinus canicula
We next investigated the earliest branch of jawed vertebrates, the chondrichthyans (cartilaginous fishes), where gastric and agastric lineages have been documented (agastric Holocephali [41] and gastric Elasmobranchii [42]; figure 1). We started by searching the genome sequence of the Holocephali elephant shark C. milii [43]. No relevant hits were found for the Atp4A and Atp4B genes as well as pepsinogens (not shown). Nevertheless, the low coverage of the genome (1.4×) and the absence of synteny data impedes a firm conclusion. Thus, we decided to determine whether true gene loss justified our negative findings. We selected the Atp4B gene locus for analysis given the striking conservation of the flanking gene families among the analysed species. Typically, Grk1 and Tmco3 flank Atp4B in both teleosts and tetrapods (figure 3). We identified orthologues of these genes in the C. milii genome and isolated two BAC clones (25I21 and 132E7) that contained both genes. Sequencing and analysis of the intergenic sequence showed no evidence for the presence of the Atp4B gene between C. milii Tmco3 and Grk1 genes (see electronic supplementary material S5a). PCR with Atp4B degenerate primers using genomic C. milii DNA was also negative (data not shown). Additionally, to support our prediction of secondary loss in the Holocephali, we next attempted to isolate the Atp4B orthologue in the gastric S. canicula, a member of the sister group, the Elasmobranchii. Through a combination of PCR strategies, we isolated a gene sequence in S. canicula, which was confirmed to be an orthologue of Atp4B by phylogenetic analysis (figure 2b). Expression analysis shows that the isolated gene is expressed as predicted in the stomach, an indirect indication of its functionality (see electronic supplementary material S5b). Overall, the analysis of genes in fishes representing the two lineages of cartilaginous fishes, the Holocephali and Elasmobranchii, provided further support for the hypothesis that the absence of stomach function genes (e.g. Atp4B) correlates with the lack of gastric glands.
4. Discussion
Stomach acid-peptic digestion first evolved in gnathostome ancestry [10]. Nevertheless, the early findings of Cuvier [22] suggested that gastric glands are not a general anatomical trait among vertebrate species. In fact, the agastric phenotype is surprisingly common in gnathostomes, and gastric glands are absent from numerous teleost fish species, Chimaeriformes (extant Holocephali lineage), dipnoids and monotremes [23,25,41,44]. The distribution of both phenotypes and the phylogenetic relationships between extant vertebrate classes provides a clear indication that the loss of gastric glands represents a secondary event. Previously, it was shown that the agastric phenotype in the platypus correlates with the absence of the proton pump genes, Atp4A and Atp4B, and the pepsinogens [25]. Here, we investigated the repertoire of the gastric function genes in a diverse array of vertebrate species where gastric and agastric representatives are present. Using a combination of phylogenetics and comparative genomics, we show that the lack of stomach coincides with the recurrent targeted deletion of the proton pump genes (Atp4A and Atp4B) and the various pepsinogen gene families in all of the examined species. In the exceptional cases of the agastric Takifugu and platypus, pepsinogen genes have been retained only to serve non-digestive functions [25,35]. Although we have not investigated dipnoids, we expect that this lineage should also have undergone loss of gastric genes.
By providing a strong linkage between loss of the gastric phenotype and the loss of the functional genes for acid-peptic digestion, we give evidence for the irreversibility of stomach loss, consistent with Dollo's principle.1 Such gene loss precludes the reinvention of complex traits [45]. This contrasts with a number of other studies demonstrating the reinvention of complex traits (e.g. wings in stick insect, coiling in snails, limb development in reptiles [46]). However, in these cases, gene loss and recovery have not been demonstrated but rather the genes involved are retained for functioning at different development stages or in other processes. There is no evidence for any alternative acid-excreting mechanism being involved in gastric acidification, and the gastric proton pump is highly conserved [10]. A missing link, lacking the gastric phenotype but possessing the gastric acid-peptic genes, and thus potentially capable of re-inventing the stomach, has yet to be identified.
Why exactly did the stomach undergo so many secondary episodes of loss during vertebrate evolution is a priori difficult to establish, as the conditions experienced by the ancestor may no longer be present in the extant species of that lineage. Presumably, there is a high cost in maintaining a complex organ like the stomach; however, estimates of these costs from specific dynamic action measurements remain ambiguous [23,47–49]. Nevertheless, acid pumping by the H+/K+-ATPase, protection of the stomach lining from the acid and neutralization of the acid in the intestine will all contribute [23,47]. By contrast, the presence of gastric glands represents a selective advantage by extending and facilitating the digestion of dietary proteins through the action of pepsins. These endopeptidases are capable of hydrolysing peptide bonds at low pH and act at the first stage of protein digestion. They probably have had their evolution affected by the dietary peptide composition [12,32,50]. Moreover, relative rate ratio tests have identified that each round of pepsinogen gene duplication is characterized by adaptive evolution [32]. Positive selection has been documented also in four lysine residues found in the primate Pga specific gene lineage (designated Pga2), with anticipated modifications of the catalytic activity [50]. In effect, the diversity of pepsinogens is reflected in their hydrolytic specificities against different proteins and peptides [12,50–52]. For example, the hydrolytic activity of human Pgc towards haemoglobin is 100% and almost 0% for the porcine Pgb [12]. The Pgb from dog is also unable to hydrolyse haemoglobin, whereas it efficiently hydrolyses gelatin [53]. The cleavage pattern of the pepsins from the rice field eel against a set of proteins (e.g. cytochrome c) is similar for the three isomers (PG1, PG2 and PG3), but PG1 showed a higher cleavage efficiency [54]. Thus, protein dietary sources are prime selective drivers for functional pepsinogen diversification (positive selection). While new peptide components can lead to the retention and/or diversification of novel pepsinogen duplicates, the opposite is a possibility as well. As species colonize different environments, new peptide sources are integrated into the diet, while others disappear. Therefore, these alterations might render the activity of some pepsin enzymes ineffective or worthless, leading to the relaxation of negative selection. If so, mutational inactivation would accumulate in pepsinogen genes, leading to gene loss. We consistently found numerous cases of ongoing pseudogenization in gastric species (electronic supplementary material S6). For example, CYM is inactive in humans [39] as a possible consequence of the lower level of proteins in human milk [33], although it is suggested to be essential for neonatal milk digestion in mammals [55]. Pga and Pgf in the opossum, Pga and Pgb in the mouse, Pgc in tilapia, and a copy of Pgb in anolis are also undergoing processes of pseudogenization [38] (this study; electronic supplementary material S6). Given the complexity of dietary protein components no clear pattern emerges as to what exact component triggers gene loss (electronic supplementary material S6), and this is probably variable between species. Nevertheless, in mammals pepsin levels (and gene numbers) in the stomach have been suggested to be related to the type of diet, with higher levels in herbivores [50]. Taken together, the overall findings suggest that pepsinogen genes are under distinct selective pressures in response to variable protein dietary components. Under this scenario, when the full complement of pepsinogen genes is lost due to hydrolytic activities no longer being required, the need of an acidic gastric environment disappears, causing the loss of gastric function (figure 6). An alternative scenario involves the loss of acidification of the stomach lumen with the pseudogenization of the pump genes that could secondarily trigger the complete loss of pepsinogen genes since an acid environment is fundamental for activation and activity. Dietary and/or environmental factors may again have created the relaxation of purifying selective pressures or emergence of neutral selective pressures for loss of gastric acidification. Possible scenarios include a switch to a diet rich in calcareous organisms (high buffer capacity; e.g. coral feeding parrot fish Scaridae) or feeding on large amounts of barely digestible material (sediment and detritus, e.g. many Cyprinidae) that neutralize gastric acidification, rendering the pump genes superfluous [23,56].
Figure 6.
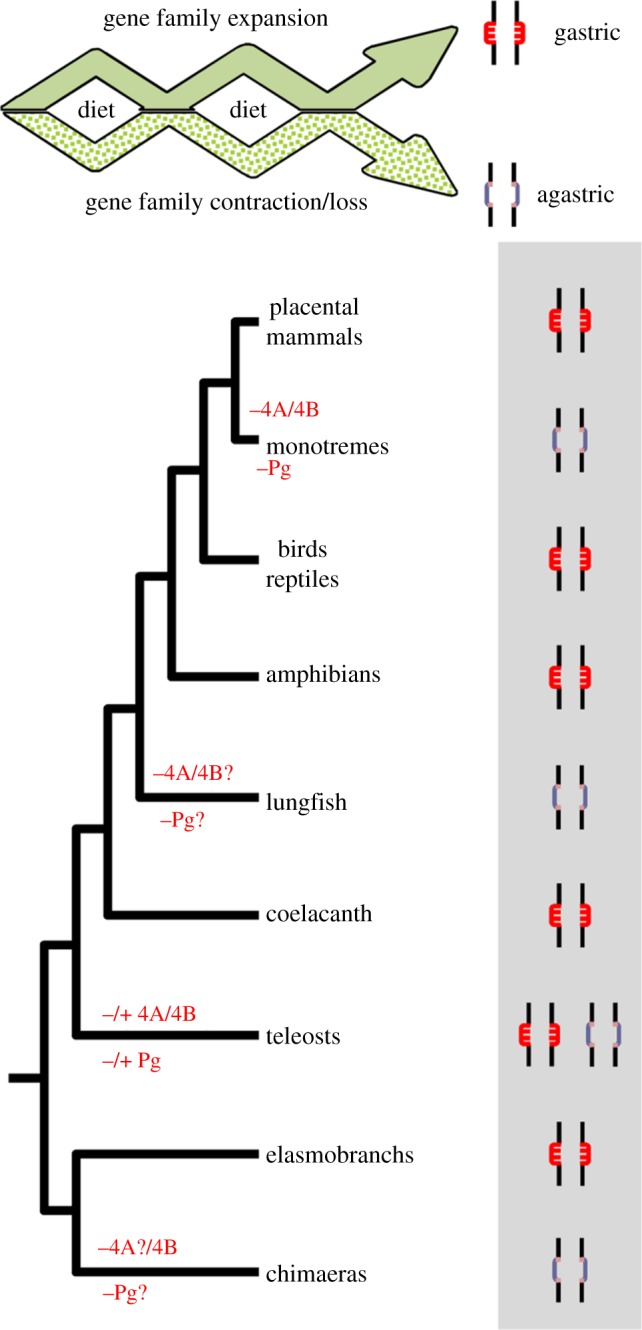
Evolutionary model of the stomach phenotype in gnathostomes. Plus and minus signs represent presence and absence of genes, respectively. Question mark indicates unknown. (Online version in colour.)
In conclusion, the findings reported clearly illustrate a remarkable case of the role of gene loss in phenotypic variability. We find that the simplification of the vertebrate gut, with the loss of gastric glands, has occurred multiple times in vertebrate evolution as a result of the loss of genes central to acid-peptic digestion.
Acknowledgements
We thank two anonymous reviewers for improving a previous version of the manuscript.
Endnote
The idea that complex characters that have been lost in evolution cannot be regained [45].
Funding statement
Work supported by Portuguese Foundation for Science and Technology (grant no. PTDC/MAR/98035/2008 and SFRH/BD/79821/2011). This research was partially supported by the European Regional Development Fund (ERDF) through the COMPETE—Operational Competitiveness Programme and national funds through FCT—Foundation for Science and Technology, under the project PEst-C/MAR/LA0015/2013. S.M. was supported by Région Bretagne (grant no. 079755 EVOVERT). Work in B.V.'s lab was supported by the Biomedical Research Council of A*STAR, Singapore.
References
- 1.Ohno S. 1970. Evolution by gene duplication. Berlin, Germany: Springer [Google Scholar]
- 2.Shimeld SM, Holland PW. 2000. Vertebrate innovations. Proc. Natl Acad. Sci. USA 97, 4449–4452 (doi:10.1073/pnas.97.9.4449) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Braasch I, Brunet F, Volff JN, Schartl M. 2009. Pigmentation pathway evolution after whole-genome duplication in fish. Genome Biol. Evol. 1, 479–493 (doi:10.1093/gbe/evp050) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Widmark J, Sundström G, Ocampo Daza D, Larhammar D. 2011. Differential evolution of voltage-gated sodium channels in tetrapods and teleost fishes. Mol. Biol. Evol. 28, 859–871 (doi:10.1093/molbev/msq257) [DOI] [PubMed] [Google Scholar]
- 5.Hoffmann FG, Opazo JC, Storz JF. 2012. Whole-genome duplications spurred the functional diversification of the globin gene superfamily in vertebrates. Mol. Biol. Evol. 29, 303–312 (doi:10.1093/molbev/msr207) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Opazo JC, Hoffmann FG, Storz JF. 2008. Differential loss of embryonic globin genes during the radiation of placental mammals. Proc. Natl Acad. Sci. USA 105, 12 950–12 955 (doi:10.1073/pnas.0804392105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuraku S, Kuratani S. 2011. Genome-wide detection of gene extinction in early mammalian evolution. Genome Biol. Evol. 3, 1449–1462 (doi:10.1093/gbe/evr120) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao H, Xu D, Zhang S, Zhang J. 2012. Genomic and genetic evidence for the loss of umami taste in bats. Genome Biol. Evol. 4, 73–79 (doi:10.1093/gbe/evr126) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hiller M, Schaar BT, Indjeian VB, Kingsley DM, Hagey LR, Bejerano G. 2012. A ‘forward genomics’ approach links genotype to phenotype using independent phenotypic losses among related species. Cell Rep. 2, 817–823 (doi:10.1016/j.celrep.2012.08.032) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koelz HR. 1992. Gastric acid in vertebrates. Scand. J. Gastroenterol. 27, 2–6 (doi:10.3109/00365529209095998) [DOI] [PubMed] [Google Scholar]
- 11.Wentworth-Thompson D. 1910. Historia animalium, p. 633. The works of Aristotle translated into English Oxford, UK: Clarendon Press [Google Scholar]
- 12.Kageyama T. 2002. Pepsinogens, progastricsins, and prochymosins: structure, function, evolution, and development. Cell Mol. Life Sci. 59, 288–306 (doi:10.1007/s00018-002-8423-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sugiura SH, Roy PK, Ferraris RP. 2006. Dietary acidification enhances phosphorus digestibility but decreases H+/K+-ATPase expression in rainbow trout. J. Exp. Biol. 209, 3719–3728 (doi:10.1242/jeb.02436) [DOI] [PubMed] [Google Scholar]
- 14.Pohl D, et al. 2008. Do we need gastric acid? Digestion 77, 184–197 (doi:10.1159/000142726) [DOI] [PubMed] [Google Scholar]
- 15.Bucking C, Wood CM. 2009. The effects of postprandial changes in pH along the gastrointestinal tract on the distribution of ions between solid and fluid phases of chyme in the rainbow trout. Aquacult. Nutr. 15, 282–296 (doi:10.1111/j.1365-2095.2008.00593.x) [Google Scholar]
- 16.Srodka A. 2003. The short history of gastroenterology. J. Physiol. Pharmacol. 54, 9–21 [PubMed] [Google Scholar]
- 17.Shin JM, Munson M, Vagin O, Sachs G. 2009. The gastric HK-ATPase: structure, function, and inhibition. Pflug Arch. Eur. J. Phys. 457, 609–622 (doi:10.1007/s00424-008-0495-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smolka AJ, Lacy ER, Luciano L, Reale E. 1994. Identification of gastric H,K-ATPase in an early vertebrate, the Atlantic stingray Dasyatis sabina. J. Histochem. Cytochem. 42, 1323–1332 (doi:10.1177/42.10.7930514) [DOI] [PubMed] [Google Scholar]
- 19.Choe KP, Verlander JW, Wingo CS, Evans DH. 2004. A putative H+-K+-ATPase in the Atlantic stingray, Dasyatis sabina: primary sequence and expression in gills. Am. J. Physiol. Reg1. 287, R981–R991 (doi:10.1152/ajpregu.00513.2003) [DOI] [PubMed] [Google Scholar]
- 20.Chow DC, Forte JG. 1995. Functional significance of the β-subunit for heterodimeric P-type ATPases. J. Exp. Biol. 198, 1–17 [DOI] [PubMed] [Google Scholar]
- 21.Narita Y, Oda S, Moriyama A, Kageyama T. 2002. Primary structure, unique enzymatic properties, and molecular evolution of pepsinogen B and pepsin B. Arch. Biochem. Biophys. 404, 177–185 (doi:10.1016/S0003-9861(02)00209-6) [DOI] [PubMed] [Google Scholar]
- 22.Cuvier G. 1805. Leçons d'anatomie comparée. Paris, France: Baudouin [Google Scholar]
- 23.Wilson JM, Castro LFC. 2010. In The multifunctional gut of fish, vol. 30 (eds Grosell M, Farrell AP, Brauner CJ.), pp. 1–55 London, UK: Academic Press [Google Scholar]
- 24.Stevens ED, Hume ID. 2004. Comparative physiology of the vertebrate digestive system, p. 400 Cambridge, UK: Cambridge University Press [Google Scholar]
- 25.Ordoñez GR, Hillier LW, Warren WC, Grützner F, López-Otín C, Puente XS. 2008. Loss of genes implicated in gastric function during platypus evolution. Genome Biol. 9, R81 (doi:10.1186/gb-2008-9-5-r81) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Katoh K, Kuma K, Toh H, Miyata T. 2005. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33, 511–518 (doi:10.1093/nar/gki198) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abascal F, Zardoya R, Posada D. 2005. ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21, 2104–2105 (doi:10.1093/bioinformatics/bti263) [DOI] [PubMed] [Google Scholar]
- 28.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321 (doi:10.1093/sysbio/syq010) [DOI] [PubMed] [Google Scholar]
- 29.Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425 [DOI] [PubMed] [Google Scholar]
- 30.Larkin MA, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 (doi:10.1093/bioinformatics/btm404) [DOI] [PubMed] [Google Scholar]
- 31.Koenig CS, Dabiké M, Bronfman M. 1987. Quantitative subcellular study of apical pole membranes from chicken oxyntic cells in resting and HCl secretory state . J. Cell Biol. 105, 2945–2958 (doi:10.1083/jcb.105.6.2945) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carginale V, Trinchella F, Capasso C, Scudiero R, Riggio M, Parisi E. 2004. Adaptive evolution and functional divergence of pepsin gene family. Gene 333, 81–90 (doi:10.1016/j.gene.2004.02.011) [DOI] [PubMed] [Google Scholar]
- 33.Kageyama T, Ichinose M, Tsukada-Kato S, Omata M, Narita Y, Moriyama A, Yonezawa S. 2000. Molecular cloning of neonate/infant-specific pepsinogens from rat stomach mucosa and their expressional change during development. Biochem. Biophys. Res. Commun. 267, 806–812 (doi:10.1006/bbrc.1999.2047) [DOI] [PubMed] [Google Scholar]
- 34.Szecsi PB. 1992. The aspartic proteases. Scand. J. Clin. Lab. Inv. Suppl. 210, 5–22 (doi:10.3109/00365519209104650) [PubMed] [Google Scholar]
- 35.Kurokawa T, Uji S, Suzuki T. 2005. Identification of pepsinogen gene in the genome of stomachless fish, Takifugu rubripes. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 140, 133–140 (doi:10.1016/j.cbpc.2004.09.029) [DOI] [PubMed] [Google Scholar]
- 36.Feng SZ, Li WS, Lin HR. 2008. Identification and expression characterization of pepsinogen A in orange-spotted grouper, Epinephelus coioides. J. Fish Biol. 73, 1960–1978 (doi:10.1111/j.1095-8649.2008.01999.x) [Google Scholar]
- 37.Tanji M, Yakabe E, Kubota K, Kageyama T, Ichinose M, Miki K, Ito H, Takahashi K. 2009. Structural and phylogenetic comparison of three pepsinogens from Pacific bluefin tuna: molecular evolution of fish pepsinogens. Comp. Biochem. Phys. B 152, 9–19 (doi:10.1016/j.cbpb.2008.08.006) [DOI] [PubMed] [Google Scholar]
- 38.Castro LFC, Lopes-Marques M, Gonçalves O, Wilson JM. 2012. The evolution of pepsinogen c genes in vertebrates: duplication, loss and functional diversification. PLoS ONE 7, e32852 (doi:10.1371/journal.pone.0032852) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ord T, Kolmer M, Villems R, Saarma M. 1990. Structure of the human genomic region homologous to the bovine prochymosin-encoding gene. Gene 91, 241–246 (doi:10.1016/0378-1119(90)90094-8) [DOI] [PubMed] [Google Scholar]
- 40.Osman AHK, Caceti T. 1991. Histology of the stomach of Tilapia nilotica (Linnaeus, 1758) from the River Nile. J. Fish Biol. 38, 211–223 (doi:10.1111/j.1095-8649.1991.tb03107.x) [Google Scholar]
- 41.Leydig F. 1851. Zur Anatomie und Histologie der Chimaera monstrosa. Arch. Anat. Physiol. 18, 241–271 [Google Scholar]
- 42.Sullivan MX. 1907. The physiology of the digestive tract of elasmobranchs. Bull. Bur Fish. 27, 1–27 [Google Scholar]
- 43.Venkatesh B, et al. 2007. Survey sequencing and comparative analysis of the elephant shark (Callorhinchus milii) genome. PLoS Biol. 5, e101 (doi:10.1371/journal.pbio.0050101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Purkerson ML, Jarvis JUM, Luse SA, Dempsey EW. 1975. Electron microscopy of the intestine of the African lungfish, Protopterus aethiopicus. Anat. Rec. 182, 71–90 (doi:10.1002/ar.1091820109) [DOI] [PubMed] [Google Scholar]
- 45.Marshall CR, Raff EC, Raff RA. 1994. Dollo's law and the death and resurrection of genes. Proc. Natl Acad. Sci. USA 91, 12 283–12 287 (doi:10.1073/pnas.91.25.12283) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Collin R, Miglietta MP. 2008. Reversing opinions on Dollo's Law. Trends Ecol. Evol. 23, 602–609 (doi:10.1016/j.tree.2008.06.013) [DOI] [PubMed] [Google Scholar]
- 47.Andrade DV, De Toledo LF, Abe AS, Wang T. 2004. Ventilatory compensation of the alkaline tide during digestion in the snake Boa constrictor. J. Exp. Biol. 207, 1379–1385 (doi:10.1242/jeb.00896) [DOI] [PubMed] [Google Scholar]
- 48.Secor SM. 2009. Specific dynamic action: a review of postprandial metabolic response. J. Comp. Physiol. B 179, 1–56 (doi:10.1007/s00360-008-0283-7) [DOI] [PubMed] [Google Scholar]
- 49.Yao X, Forte JG. 2003. Cell biology of acid secretion by the parietal cell. Annu. Rev. Physiol. 65, 103–131 (doi:10.1146/annurev.physiol.65.072302.114200) [DOI] [PubMed] [Google Scholar]
- 50.Narita Y, Oda S, Takenaka O, Kageyama T. 2010. Lineage-specific duplication and loss of pepsinogen genes in hominoid evolution. J. Mol. Evol. 70, 313–324 (doi:10.1007/s00239-010-9320-8) [DOI] [PubMed] [Google Scholar]
- 51.Kageyama H, Ueda H, Tezuka T, Ogasawara A, Narita Y, Kageyama T, Ichinose M. 2010. Differences in the P1’ substrate specificities of pepsin A and chymosin. J. Biochem. 147, 167–174 (doi:10.1093/jb/mvp158) [DOI] [PubMed] [Google Scholar]
- 52.Kageyama T. 2004. Role of S'1 loop residues in the substrate specificities of pepsin A and chymosin. Biochemistry 43, 15 122–15 130 (doi:10.1021/bi048440g) [DOI] [PubMed] [Google Scholar]
- 53.Kageyama T. 2006. Roles of Tyr13 and Phe219 in the unique substrate specificity of pepsin B. Biochemistry 45, 14 415–14 426 (doi:10.1021/bi061467a) [DOI] [PubMed] [Google Scholar]
- 54.Ahn J, Cao MJ, Yu YQ, Engen JR. 2013. Accessing the reproducibility and specificity of pepsin and other aspartic proteases. Biochim. Biophys. Acta 1834, 1222–1229 (doi:10.1016/j.bbapap.2012.10.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Foltmann B. 1992. Chymosin: a short review on foetal and neonatal gastric proteases. Scand. J. Clin. Lab. Invest. Suppl. 210, 65–79 (doi:10.3109/00365519209104656) [PubMed] [Google Scholar]
- 56.Kapoor BG, Smit H, Verighina IA. 1975. The alimentary canal and digestion in teleosts. Adv. Mar. Biol. 13, 109–239 (doi:10.1006/S0065-2881(08)60281-3) [Google Scholar]