Abstract
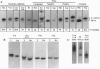
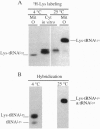
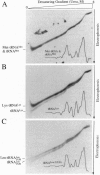
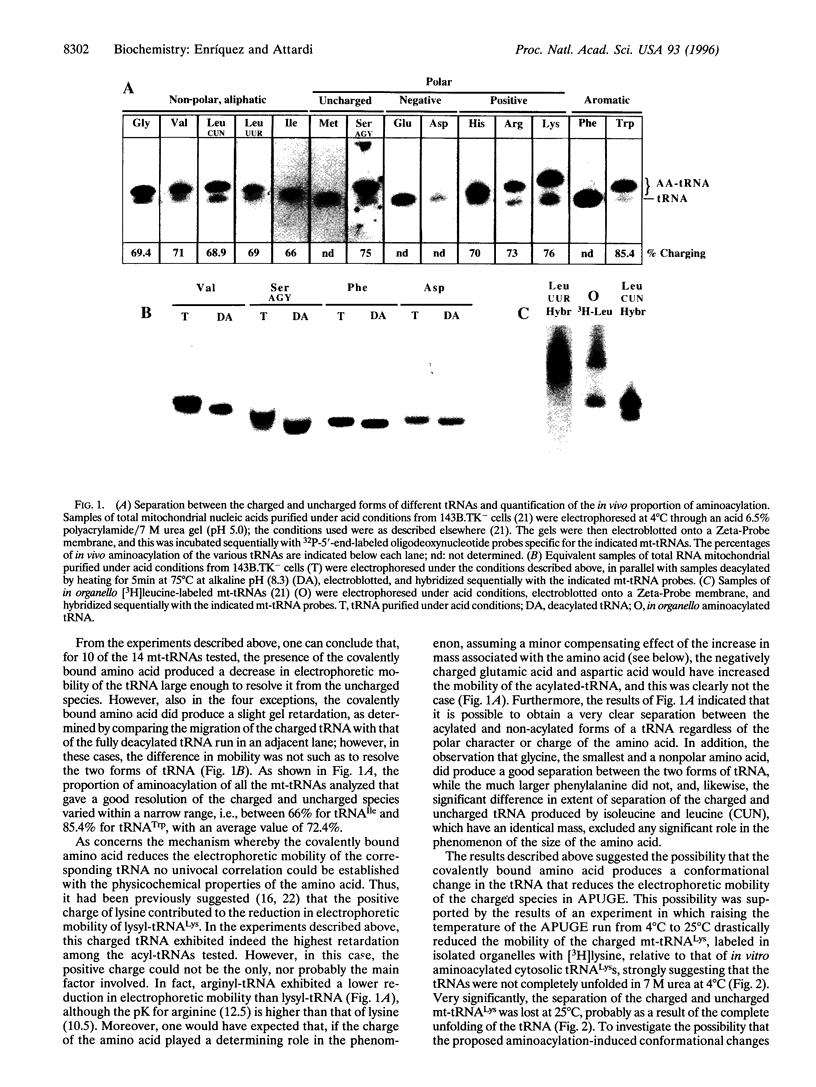
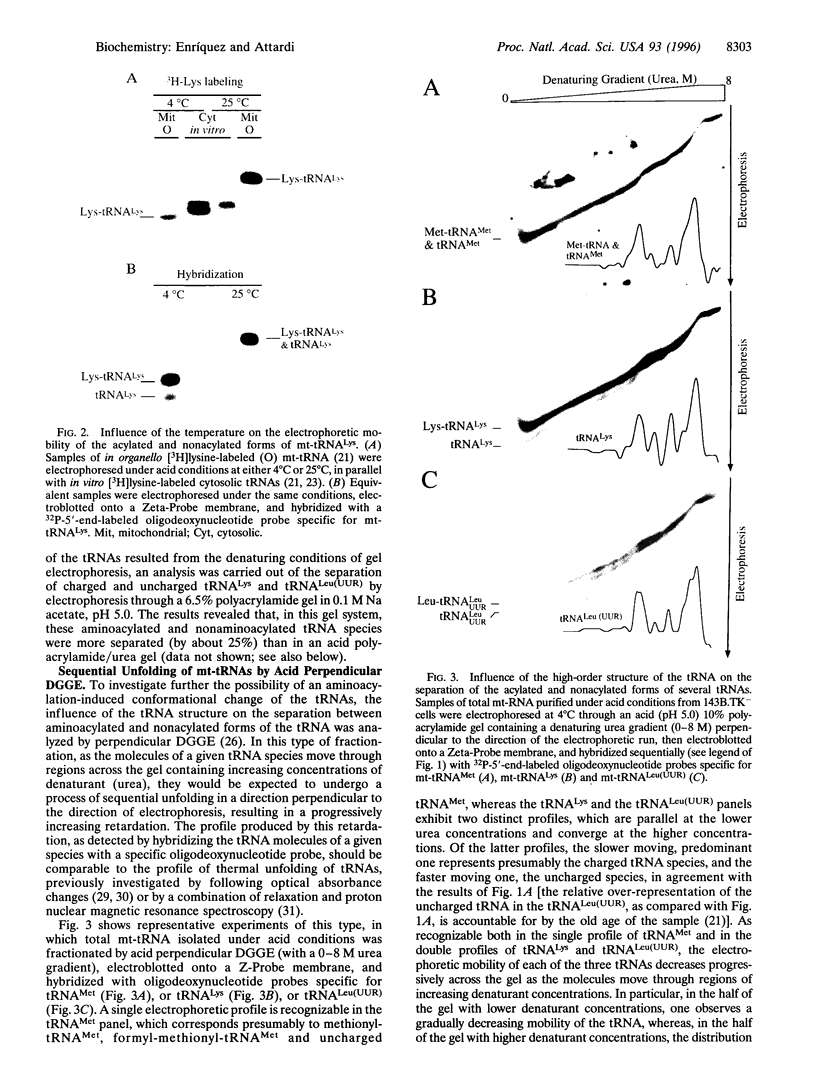
Analysis by acid polyacrylamide/urea gel electrophoresis of 14 individual mitochondrial tRNAs (mt-tRNAs) from human cells has revealed a variable decrease in mobility of the aminoacylated relative to the nonacylated form, with the degree of separation of the two forms not being correlated with the mass, polar character, or charge of the amino acid. Separation of the charged and uncharged species has been found to be independent of tRNA denaturation, being observed also in the absence of urea. In another approach, electrophoresis through a perpendicular denaturing gradient gel of several individual mt-tRNAs has shown a progressive unfolding of the tRNA with increasing denaturant concentration, which is consistent with an initial disruption of tertiary interactions, followed by the sequential melting of the four stems of the cloverleaf structure. A detailed analysis of the unfolding process of charged and uncharged tRNALys and tRNALeu(UUR) has revealed that the separation of the two forms of these tRNAs persisted throughout the almost entire range of denaturant concentrations used and was lost upon denaturation of the last helical domain(s), which most likely included the amino acid acceptor stem. These observations strongly suggest that the electrophoretic retardation of the charged species reflects an aminoacylation-induced conformational change of the 3'-end of these mt-tRNAs, with possible significant implications in connection with the known role of the acceptor end in tRNA interactions with the ribosomal peptidyl transferase center and the elongation factor Tu.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abrams E. S., Stanton V. P., Jr Use of denaturing gradient gel electrophoresis to study conformational transitions in nucleic acids. Methods Enzymol. 1992;212:71–104. doi: 10.1016/0076-6879(92)12006-c. [DOI] [PubMed] [Google Scholar]
- Adler A. J., Fasman G. D. Circular dichroism of valine and formylmethionine transfer RNA from Escherichia coli: effect of aminoacylation. Biochim Biophys Acta. 1970 Mar 19;204(1):183–190. doi: 10.1016/0005-2787(70)90501-0. [DOI] [PubMed] [Google Scholar]
- Amaldi F., Attardi G. Partial sequence analysis of ribosomal RNA from HeLa cells. I. Oligonucleotide pattern of 28 s and 18 s RNA after pancreatic ribonuclease digestion. J Mol Biol. 1968 May 14;33(3):737–755. doi: 10.1016/0022-2836(68)90317-3. [DOI] [PubMed] [Google Scholar]
- Anderson S., Bankier A. T., Barrell B. G., de Bruijn M. H., Coulson A. R., Drouin J., Eperon I. C., Nierlich D. P., Roe B. A., Sanger F. Sequence and organization of the human mitochondrial genome. Nature. 1981 Apr 9;290(5806):457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- Antosiewicz J., Porschke D. Effect of aminoacylation on tRNA conformation. Eur Biophys J. 1989;17(4):233–235. doi: 10.1007/BF00284730. [DOI] [PubMed] [Google Scholar]
- Chatterjee S. K., Kaji H. Conformational changes of transfer RNA on aminoacylations. Biochim Biophys Acta. 1970 Nov 12;224(1):88–98. doi: 10.1016/0005-2787(70)90623-4. [DOI] [PubMed] [Google Scholar]
- Cole P. E., Yang S. K., Crothers D. M. Conformational changes of transfer ribonucleic acid. Equilibrium phase diagrams. Biochemistry. 1972 Nov 7;11(23):4358–4368. doi: 10.1021/bi00773a024. [DOI] [PubMed] [Google Scholar]
- Crothers D. M., Cole P. E., Hilbers C. W., Shulman R. G. The molecular mechanism of thermal unfolding of Escherichia coli formylmethionine transfer RNA. J Mol Biol. 1974 Jul 25;87(1):63–88. doi: 10.1016/0022-2836(74)90560-9. [DOI] [PubMed] [Google Scholar]
- Delarue M., Moras D. The aminoacyl-tRNA synthetase family: modules at work. Bioessays. 1993 Oct;15(10):675–687. doi: 10.1002/bies.950151007. [DOI] [PubMed] [Google Scholar]
- Englander J. J., Kallenbach N. R., Englander S. W. Hydrogen exchange study of some polynucleotides and transfer RNA. J Mol Biol. 1972 Jan 14;63(1):153–169. doi: 10.1016/0022-2836(72)90527-x. [DOI] [PubMed] [Google Scholar]
- Enriquez J. A., Chomyn A., Attardi G. MtDNA mutation in MERRF syndrome causes defective aminoacylation of tRNA(Lys) and premature translation termination. Nat Genet. 1995 May;10(1):47–55. doi: 10.1038/ng0595-47. [DOI] [PubMed] [Google Scholar]
- Enriquez J. A., López-Pérez M. J., Montoya J. Saturation of the processing of newly synthesized rRNA in isolated brain mitochondria. FEBS Lett. 1991 Mar 11;280(1):32–36. doi: 10.1016/0014-5793(91)80197-b. [DOI] [PubMed] [Google Scholar]
- Enríquez J. A., Attardi G. Analysis of aminoacylation of human mitochondrial tRNAs. Methods Enzymol. 1996;264:183–196. doi: 10.1016/s0076-6879(96)64019-1. [DOI] [PubMed] [Google Scholar]
- Enríquez J. A., Ramos J., Pérez-Martos A., López-Pérez M. J., Montoya J. Highly efficient DNA synthesis in isolated mitochondria from rat liver. Nucleic Acids Res. 1994 May 25;22(10):1861–1865. doi: 10.1093/nar/22.10.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Förster C., Limmer S., Ribeiro S., Hilgenfeld R., Sprinzl M. Ternary complex between elongation factor Tu.GTP and Phe-tRNA(Phe). Biochimie. 1993;75(12):1159–1166. doi: 10.1016/0300-9084(93)90015-k. [DOI] [PubMed] [Google Scholar]
- Gantt R. R., Englander S. W., Simpson M. V. Hydrogen-exchange measurements on Escherichia coli transfer ribonucleic acid before, after, and during its aminoacylation. Biochemistry. 1969 Feb;8(2):475–482. doi: 10.1021/bi00830a003. [DOI] [PubMed] [Google Scholar]
- Goldman E., Jakubowski H. Uncharged tRNA, protein synthesis, and the bacterial stringent response. Mol Microbiol. 1990 Dec;4(12):2035–2040. doi: 10.1111/j.1365-2958.1990.tb00563.x. [DOI] [PubMed] [Google Scholar]
- Hashizume H., Imahori K. Circular dichroism and conformation of natural and synthetic polynucleotides. J Biochem. 1967 Jun;61(6):738–749. doi: 10.1093/oxfordjournals.jbchem.a128608. [DOI] [PubMed] [Google Scholar]
- Ho Y. S., Kan Y. W. In vivo aminoacylation of human and Xenopus suppressor tRNAs constructed by site-specific mutagenesis. Proc Natl Acad Sci U S A. 1987 Apr;84(8):2185–2188. doi: 10.1073/pnas.84.8.2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. H., Suddath F. L., Quigley G. J., McPherson A., Sussman J. L., Wang A. H., Seeman N. C., Rich A. Three-dimensional tertiary structure of yeast phenylalanine transfer RNA. Science. 1974 Aug 2;185(4149):435–440. doi: 10.1126/science.185.4149.435. [DOI] [PubMed] [Google Scholar]
- King M. P., Attardi G. Human cells lacking mtDNA: repopulation with exogenous mitochondria by complementation. Science. 1989 Oct 27;246(4929):500–503. doi: 10.1126/science.2814477. [DOI] [PubMed] [Google Scholar]
- King M. P., Attardi G. Post-transcriptional regulation of the steady-state levels of mitochondrial tRNAs in HeLa cells. J Biol Chem. 1993 May 15;268(14):10228–10237. [PubMed] [Google Scholar]
- Lee C. P., Dyson M. R., Mandal N., Varshney U., Bahramian B., RajBhandary U. L. Striking effects of coupling mutations in the acceptor stem on recognition of tRNAs by Escherichia coli Met-tRNA synthetase and Met-tRNA transformylase. Proc Natl Acad Sci U S A. 1992 Oct 1;89(19):9262–9266. doi: 10.1073/pnas.89.19.9262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. P., Mandal N., Dyson M. R., RajBhandary U. L. The discriminator base influences tRNA structure at the end of the acceptor stem and possibly its interaction with proteins. Proc Natl Acad Sci U S A. 1993 Aug 1;90(15):7149–7152. doi: 10.1073/pnas.90.15.7149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Kumar N. V., Varshney U., RajBhandary U. L. Important role of the amino acid attached to tRNA in formylation and in initiation of protein synthesis in Escherichia coli. J Biol Chem. 1996 Jan 12;271(2):1022–1028. doi: 10.1074/jbc.271.2.1022. [DOI] [PubMed] [Google Scholar]
- Lynch D. C., Attardi G. Amino acid specificity of the transfer RNA species coded for by HeLa cell mitochondrial DNA. J Mol Biol. 1976 Mar 25;102(1):125–141. doi: 10.1016/0022-2836(76)90077-2. [DOI] [PubMed] [Google Scholar]
- Nissen P., Kjeldgaard M., Thirup S., Polekhina G., Reshetnikova L., Clark B. F., Nyborg J. Crystal structure of the ternary complex of Phe-tRNAPhe, EF-Tu, and a GTP analog. Science. 1995 Dec 1;270(5241):1464–1472. doi: 10.1126/science.270.5241.1464. [DOI] [PubMed] [Google Scholar]
- Potts R. O., Ford N. C., Jr, Fournier M. J. Changes in the solution structure of yeast phenylalanine transfer ribonucleic acid associated with aminoacylation and magnesium binding. Biochemistry. 1981 Mar 17;20(6):1653–1659. doi: 10.1021/bi00509a038. [DOI] [PubMed] [Google Scholar]
- Puglisi E. V., Puglisi J. D., Williamson J. R., RajBhandary U. L. NMR analysis of tRNA acceptor stem microhelices: discriminator base change affects tRNA conformation at the 3' end. Proc Natl Acad Sci U S A. 1994 Nov 22;91(24):11467–11471. doi: 10.1073/pnas.91.24.11467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riesner D., Maass G., Thiebe R., Philippsen P., Zachau H. G. The conformational transitions in yeast tRNAPhe as studied with tRNAPhe fragments. Eur J Biochem. 1973 Jul 2;36(1):76–88. doi: 10.1111/j.1432-1033.1973.tb02887.x. [DOI] [PubMed] [Google Scholar]
- Robertus J. D., Ladner J. E., Finch J. T., Rhodes D., Brown R. S., Clark B. F., Klug A. Structure of yeast phenylalanine tRNA at 3 A resolution. Nature. 1974 Aug 16;250(467):546–551. doi: 10.1038/250546a0. [DOI] [PubMed] [Google Scholar]
- Rojiani M. V., Jakubowski H., Goldman E. Relationship between protein synthesis and concentrations of charged and uncharged tRNATrp in Escherichia coli. Proc Natl Acad Sci U S A. 1990 Feb;87(4):1511–1515. doi: 10.1073/pnas.87.4.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rould M. A., Perona J. J., Söll D., Steitz T. A. Structure of E. coli glutaminyl-tRNA synthetase complexed with tRNA(Gln) and ATP at 2.8 A resolution. Science. 1989 Dec 1;246(4934):1135–1142. doi: 10.1126/science.2479982. [DOI] [PubMed] [Google Scholar]
- Ruff M., Krishnaswamy S., Boeglin M., Poterszman A., Mitschler A., Podjarny A., Rees B., Thierry J. C., Moras D. Class II aminoacyl transfer RNA synthetases: crystal structure of yeast aspartyl-tRNA synthetase complexed with tRNA(Asp). Science. 1991 Jun 21;252(5013):1682–1689. doi: 10.1126/science.2047877. [DOI] [PubMed] [Google Scholar]
- Samaha R. R., Green R., Noller H. F. A base pair between tRNA and 23S rRNA in the peptidyl transferase centre of the ribosome. Nature. 1995 Sep 28;377(6547):309–314. doi: 10.1038/377309a0. [DOI] [PubMed] [Google Scholar]
- Varshney U., Lee C. P., RajBhandary U. L. Direct analysis of aminoacylation levels of tRNAs in vivo. Application to studying recognition of Escherichia coli initiator tRNA mutants by glutaminyl-tRNA synthetase. J Biol Chem. 1991 Dec 25;266(36):24712–24718. [PubMed] [Google Scholar]
- Varshney U., Lee C. P., RajBhandary U. L. From elongator tRNA to initiator tRNA. Proc Natl Acad Sci U S A. 1993 Mar 15;90(6):2305–2309. doi: 10.1073/pnas.90.6.2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickstrom E. Circular dichroism during deacylation of methionyl-tRNA met -f and formylmethionyl-tRNA met -f from E. coli. Biochem Biophys Res Commun. 1971 Jun 4;43(5):976–983. doi: 10.1016/0006-291x(71)90558-4. [DOI] [PubMed] [Google Scholar]
- Woo N. H., Roe B. A., Rich A. Three-dimensional structure of Escherichia coli initiator tRNAfMet. Nature. 1980 Jul 24;286(5771):346–351. doi: 10.1038/286346a0. [DOI] [PubMed] [Google Scholar]