Abstract
Motivation: With high-throughput DNA sequencing costs dropping <$1000 for human genomes, data storage, retrieval and analysis are the major bottlenecks in biological studies. To address the large-data challenges, we advocate a clean separation between the evidence collection and the inference in variant calling. We define and implement a Genome Query Language (GQL) that allows for the rapid collection of evidence needed for calling variants.
Results: We provide a number of cases to showcase the use of GQL for complex evidence collection, such as the evidence for large structural variations. Specifically, typical GQL queries can be written in 5–10 lines of high-level code and search large datasets (100 GB) in minutes. We also demonstrate its complementarity with other variant calling tools. Popular variant calling tools can achieve one order of magnitude speed-up by using GQL to retrieve evidence. Finally, we show how GQL can be used to query and compare multiple datasets. By separating the evidence and inference for variant calling, it frees all variant detection tools from the data intensive evidence collection and focuses on statistical inference.
Availability: GQL can be downloaded from http://cseweb.ucsd.edu/~ckozanit/gql.
Contact: ckozanit@ucsd.edu or vbafna@cs.ucsd.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
As sequencing costs drop, we envision a scenario where every individual is sequenced, perhaps multiple times in their lifetime. There is already a vast array of genomic information across various large-scale sequencing projects including the 1000 genome project (1000 Genomes Project Consortium et al., 2010) and the cancer genome atlas (TCGA) (Koboldt et al., 2012). In many of these projects, a re-sequencing strategy is applied in which whole genomes are sequenced redundantly with coverage between 4 and 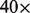 . The clone inserts (
. The clone inserts (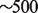 bp) and sequenced reads (
bp) and sequenced reads (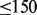 bp) are typically short and are not de novo assembled. Instead, they are mapped back to a standard reference to decipher the genomic variation in the individual relative to the reference. Even with advances in single-molecule sequencing and genomic assembly (Clarke et al., 2009), we are many years away from having finished and error-free assembled sequences from human donors. At least in the near to mid-term, we expect that the bulk of sequencing will follow the re-sequencing/mapping/variant calling approach (e.g. McKenna et al., 2010).
bp) are typically short and are not de novo assembled. Instead, they are mapped back to a standard reference to decipher the genomic variation in the individual relative to the reference. Even with advances in single-molecule sequencing and genomic assembly (Clarke et al., 2009), we are many years away from having finished and error-free assembled sequences from human donors. At least in the near to mid-term, we expect that the bulk of sequencing will follow the re-sequencing/mapping/variant calling approach (e.g. McKenna et al., 2010).
Mapped reads (represented by BAM files) from a single individual sequenced with 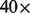 coverage are relatively inexpensive to generate, but they are storage intensive (
coverage are relatively inexpensive to generate, but they are storage intensive (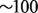 GB). As sequencing becomes more accessible, and larger numbers of individuals are sequenced, the amount of information will increase rapidly; this will pose a serious challenge to available community resources. Although raw archiving of large datasets is possible (Wheeler et al., 2008), the analysis of this huge amount of data remains a challenge. To facilitate access, some of the large datasets have been moved to commercially available cloud platforms. For example, the 1000 genome data are available on Amazon’s EC2 (1000genomescloud, 2012). The genomes on Amazon can be analyzed remotely using appropriate software frameworks like Galaxy [that allow for the pipelining/integration of multiple analysis tools (Goecks et al., 2010)], as well as tools like Genome Analysis Toolkit (GATK) (McKenna et al., 2010) and samtools (Li et al., 2009). The promise of this approach is that much of the analysis can be done remotely, without the need for extensive infrastructure on the user’s part.
GB). As sequencing becomes more accessible, and larger numbers of individuals are sequenced, the amount of information will increase rapidly; this will pose a serious challenge to available community resources. Although raw archiving of large datasets is possible (Wheeler et al., 2008), the analysis of this huge amount of data remains a challenge. To facilitate access, some of the large datasets have been moved to commercially available cloud platforms. For example, the 1000 genome data are available on Amazon’s EC2 (1000genomescloud, 2012). The genomes on Amazon can be analyzed remotely using appropriate software frameworks like Galaxy [that allow for the pipelining/integration of multiple analysis tools (Goecks et al., 2010)], as well as tools like Genome Analysis Toolkit (GATK) (McKenna et al., 2010) and samtools (Li et al., 2009). The promise of this approach is that much of the analysis can be done remotely, without the need for extensive infrastructure on the user’s part.
Even with these developments, a significant challenge remains. Each individual genome is unique, and the inference of variation, relative to a standard reference remains challenging. In addition to small indels and substitutions (the so-called single-nucleotide variations or SNVs), an individual might have large structural changes, including, but not limited to, insertions, deletions, inversions (Sharp et al., 2006), translocations of large segments (102–106 bp in size) (Giglio et al., 2001), incorporation of novel viral and microbial elements and recombination-mediated rearrangements (Perry et al., 2006). Further, many of these rearrangements may overlap leading to more complex structural variations. The detection of these variations remains challenging even for the simplest SNVs, and there is little consensus on the best practices for the discovery of more complex rearrangements. For large, remotely located datasets, it is often difficult to create a fully customized analysis. It is often desirable to download the evidence (reads) required for the detection of variation to a local machine, and experiment with a collection of analysis tools for the actual inference. In that case, we are back again to the problem of building a large local infrastructure, including clusters and large disks, at each analysis site in addition to the resources in the cloud.
As an example, we consider the use of paired-end sequencing and mapping (PEM) for identifying structural variation. In PEM, fixed length inserts are selected for sequencing at both ends, and the sequenced sub-reads are mapped to a reference genome. Without variation, the distance and orientation of the mapped reads match the a priori expectation. However, if a region is deleted in the donor relative to the reference, ends of the insert spanning the deleted region will map much further apart than expected. Similarly, the expected orientation of the read alignments for Illumina sequencing is (+,−). A (+,+) orientation is suggestive of an inversion event.
Using PEM evidence, different callers still use different inference mechanisms. GASV (Sindi et al., 2009) arranges overlapping discordantly mapping pair-end reads on the Cartesian plane and draws the grid of possible break point locations under the assumption that the discordancy is a result of a single SV. Breakdancer (Chen et al., 2009) finds all areas that contain at least two discordant pair-end reads, and it uses a Poisson model to evaluate the probability that those areas contain a SV as a function of the total number of discordant reads of each of those areas. VariationHunter (Hormozdiari et al., 2009) reports that regions of SV are the ones that minimize the total number of clusters that the pair ends can form. Given the complexity of the data, and the different inference methodologies, all of these methods have significant type 1 (false-positive), and type 2 (false-negative) errors. Further, as the authors of VariationHunter (Hormozdiari et al., 2009) point out, there are a number of confounding factors for discovery. For example, repetitive regions, sequencing errors, could all lead to incorrect mappings. At the same time, incorrect calls cannot be easily detected because tools need to be modified to re-examine the source data. In addition, the run time of the entire pipeline of those tools is not negligible given that they have to parse the raw data.
A starting point of our work is the observation all tools follow a two-step procedure, implicitly or explicitly, for discovery of variation. The first step—the evidence-step—involves the processing of raw data to fetch (say) the discordant pair-end reads; the second step—the inference-step—involves statistical inference on the evidence to make a variant call. Moreover, the evidence gathering step is similar and is typically the data-intensive part of the procedure.
For example, in SNV discovery, the evidence step is the alignment (‘pile-up’) of nucleotides to a specific location, whereas the inference step involves SNV estimation based on alignment quality and other factors. By contrast, for SVs such as large deletions, the evidence might be in the form of (a) length-discordant reads and (b) concordant reads mapping to a region; the inference might involve an analysis of the clustering of the length-discordant reads, and looking for copy-number decline and loss of heterozygosity in concordant reads.
In this article, we propose a Genome Query Language (GQL) that allows for the efficient querying of genomic fragment data to uncover evidence for variation in the sampled genomes. Note that our tool does not replace other variant calling tools, but it is complementary to existing efforts. It focuses on the collection of evidence that all inference tools can use to make custom inference. First, by providing a simple interface to extract the required evidence from the raw data stored in the cloud, GQL can free callers from the need to handle large data efficiently. Second, we show how existing tools can be sped up and simplified using GQL, with larger speed-ups possible through a cloud based parallel GQL implementation. Third, analysts can examine and visualize the evidence for each variant, independent of the tool used to identify the variant.
Software layers and interfaces for genomics: We also place GQL in the context of other genomics software. It is helpful to think of a layered, hourglass, model for genomic processing. At the bottom is the wide, instrument layer (customized for each instrument) for calling reads. This is followed by mapping/compression layers (the ‘narrow waist’ of the hourglass), and subsequently, multiple application layers. Some of these layers have been standardized. Many instruments now produce sequence data as ‘fastq’ format, which in turn is mapped against a reference genome using alignment tools, such as BWA (Li and Durbin, 2010) and MAQ (Li et al., 2008); further, aligned reads are often represented in the compressed, BAM format (Li et al., 2009) that also allows random access. Recently, more compressed alignment formats have come into vogue including SlimGene (Kozanitis et al., 2011) CRAM (Hsi-Yang Fritz et al., 2011) and others (Asnani et al., 2012; Cox et al., 2012; Popitsch and von Haeseler, 2013; Wan et al., 2012; Yanovsky, 2011) as well as compression tools for unmapped reads (Jones et al., 2012). At the highest level, standards such as VCF (VCF Tools, 2011) describe variants (the output of the inference stage of Fig. 1b).
Fig. 1.
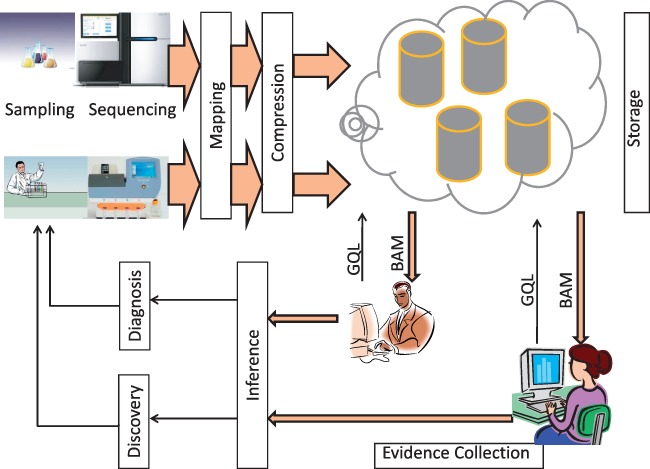
Abstraction and layering for genomics. The bottom (physical) layer is the instrumentation software for parsing raw data into sequences. Mapping against a known reference is the first level of abstraction of the data. Compression is used to reduce the storage requirements. Detection of variation involves an evidence layer to collect relevant reads, and an inference layer for statistical analysis. The inference results in variant calls (typically as VCF files) that can be used by other applications to make genetic discoveries
In this context, we propose additional layering. Specifically, we advocate the splitting of the processing below the application layer to support a query into an evidence layer(deterministic, large data movement, standardized) and an inference layer (probabilistic, comparatively smaller data movement, little agreement on techniques).
For evidence gathering, the closest tools are samtools (Li et al., 2009), BAMtools Barnett et al. (2011), BEDtools (Dale et al., 2011; Quinlan and Hall, 2010), BioHDF (Mason et al., 2010) and GATK (McKenna et al., 2010). Samtools consists of a toolkit and an API for handling mapped reads; together, they comprise the first attempt to hide the implications of raw data handling by treating datasets uniformly regardless of the instrument source. Samtools also provide quick random access to large files and provide a clean API to programmatically handle alignments. The tool combines index sorted BAM files with a lightweight and extremely efficient binning that clusters reads that map in neighboring locations. Thus, samtools can quickly return a set of reads that overlap with a particular location or create a pileup (i.e. all bases seen in reads that map to any reference locus). BAMtools is a C++ API built to support queries in a JSON format. BEDtools, closely aligned with samtools, allows interval-related queries through a clean unix and a python interface. Although powerful, these tools still require programmer-level expertise to open binary files, assign buffers, read alignments and manipulate various fields.
The GATK (McKenna et al., 2010) is built on top of samtools and reduces the programming complexity of data collection. GATK’s API provides two main iterator categories to an application programmer. The first iterator traverses individual reads; the second iterator walks through all genome loci, either individually or in adjacent groups. The toolkit, which is written based on the Map Reduce framework and thus easily parallelizable, is an excellent resource for developers of applications that need to determine local realignment, quality score re-calibration and genotyping (DePristo et al., 2011). The support of many of these tools for looking at paired-ends, and consequently for structural variation, is limited, depending (in GATK’s case) on the existence of the optional fields RNEXT and PNEXT of the SAM/BAM alignments (gatk-pairend, 2012).
The single biggest difference between our proposed tool, GQL and others is that GQL has a (SQL-like) declarative syntax in its own language, as opposed to a procedural syntax, designed to help programmers rather than the end user. Declarative languages, such as GQL and SQL, not only raise the level of abstraction of data access but also allow automatic data optimization without programmer intervention. By asking users to specify what data they want as opposed to how they want to retrieve it, we will show that GQL can facilitate automatic optimizations, such as the use of indices and caching; these seem harder to support in other tools without explicit programmer directives. Further, it is feasible to compile GQL queries to a distributed, cloud based, back-end.
Finally, GQL queries allow genomes to be browsed for variations of interest, allowing an interactive exploration of the data as we show in our results. Although the UCSC browser also allows genome browsing, it does only by position or string, which we refer to as syntactic genome browsing. By contrast, GQL allows browsing for all regions containing reads that satisfy a specified property (e.g. discrepant reads) and view the results on the UCSC browser. We refer to this as semantic genome browsing and give many examples in Section 2. Our performance results indicate that such browsing can be done interactively in minutes using a single cheap CPU.
1.1 An overview of GQL: language features and implementation
We chose to use an SQL-like syntax to express GQL because SQL is an accepted and popular standard for querying databases. Although our syntax is SQL-like, we need some special operators and relations for genomic queries that do not appear to fit well with existing off-the-shelf Relational Database Management Systems (RDBMS). Thus, we developed our own compiler to process GQL queries and translate them into C++ API calls (Section 4). Our compiler also allowed us the freedom to heavily optimize our GQL implementation (by several orders of magnitude), which would be harder to do with existing RDBMS.
We conceptualize the genomic fragment data as a database with some key relations. The first is a relation called READS that describes the collection of mapped fragment reads and all their properties but does not contain the actual bases or quality scores. For example, for paired-end sequencing, each entry in the READS table will contain information about a read, its mapping location and the location of its paired-end. The reads table is constructed by pre-processing a BAM file through a set of scripts that accompanies the source code that split, index and move the contents of the file to the appropriate directory that GQL can access; it contains fields such as the mapping location, the strand, the read length, the location of the pair-ends. In addition, GQL accepts a freely formatted Text table that can be any table that a user can define. Text table can, for example, be used to describe gene annotations.
GQL also accepts interval tables, which have three special fields (chromosome, and begin and end location within the chromosome) demarcating the interval. The user has the option of creating an interval table from any table by marking two of the attributes as begin and end; the chromosome field is updated automatically during the iteration through all chromosomes. The most interesting aspects of GQL semantics lie in allowing interval manipulation. In programming languages terminology, intervals are first-class entities in GQL. The Supplementary Information summarizes all GQL tables and the respective attributes of the language.
Language constructs. All GQL statements (like SQL) have the form select
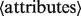 from
from
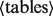 where
where
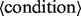 .
.
The from statement specifies the input tables to the statement.
The select statement corresponds to the projection operator in the relational calculus (Codd, 1970). It returns a subset of the attributes of the input table.
The where statement selects the subset of records of the input tables that satisfy the filter expression that follows the where statement.
The using intervals() expression optionally follows a table specifier in the from statement. It produces an interval for each entry of the corresponding table according to the specified expression. If the input table is of type READS the user has the ability to add the keyword both_mates as a third argument to the expression specified by using intervals to denote that a pair end is treated as a single interval. This expression does not return any table and can only be used with the create_intervals or mapjoin operations.
The create_intervals function constructs a table of intervals from the input table. When the function is called, the table in the from statement is followed by the statement using intervals(a,b) so that the function knows which fields to use as intervals.
The mapjoin statement takes two tables as input and concatenates any two entries of these tables whose corresponding intervals intersect. The user specifies intervals with the expression using intervals next to each input table.
The merge_intervals(interval_count op const) statement is a function whose input table needs to be of type Intervals. It creates an output table of intervals from the intervals in the input table that overlap with at least or at most the number of intervals specified inside the parenthesis. This statement uses op to specify at least or at most.
The implementation of GQL consists of a front-end that parses user input and a back-end that implements the remaining functionality of the system. The front-end is implemented using the flex (Flex, 1990) and Bison (Bison, 1988) tools and is based on the GQL grammar (Supplementary Information). It performs syntactic and semantic analysis and converts the GQL statements into a series of back-end procedure calls with the proper arguments. It also converts any user expressions, such as the ones found in the where and using intervals statements into customizable C++ files. These are compiled and run as executables on the back-end. The back-end implements the basic table types and all remaining GQL functionality (see Section 4 for details).
2 RESULTS AND DISCUSSION
We demonstrate the flexibility and efficiency of GQL by walking through some use cases that involve identifying donor regions with variations, and supplying the read evidence supporting the variation. We use the Yoruban trio (both parents and a child) from the Hapmap project (1000 Genomes Project Consortium et al., 2010) that was sequenced as part of the 1000 Genome Project. The genomes are labeled NA18507, NA18508 and NA18506 for the father, mother and the child, respectively. Each genome was sequenced to ∼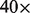 coverage (
coverage (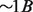 mapped reads) using
mapped reads) using 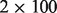 bp paired-end reads from 300 bp inserts. We use the large deletion detection problem for a case study. The corresponding input BAM file sizes are all in the range 73–100 GB.
bp paired-end reads from 300 bp inserts. We use the large deletion detection problem for a case study. The corresponding input BAM file sizes are all in the range 73–100 GB.
Large deletions on NA18506. Paired-end mapping provides an important source of evidence for identifying large structural variations (Bashir et al., 2007; Kidd et al., 2008). Length discordant clones (pairs of reads with uncharacteristically long alignment distance between the two mapped ends) are indicative of a deletion event. We start by using GQL to identify all genomic regions (in reference coordinates) that have an ‘abundance’ of length-discordant clones.
- Select inserts where both ends are mapped, and the insert size is at least 350 bp (i.e. the insert size exceeds the mean insert size by more than
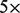 the standard deviation) and at most 1 Mb (Supplementary Information).
the standard deviation) and at most 1 Mb (Supplementary Information).
- Discordant = select * from READS
- where location ≥ 0
- and mate_loc ≥ 0
- and abs(mate_loc + length-location) > 350
- and abs(mate_loc + length-location) < 1 000 000)
- Create an interval table, with an interval for each discordant read (by specifying the begin and end coordinates). This changes the output data type from reads to intervals on the reference.
- Disc2Intrvl = select create_intervals() from Discordant
- using intervals(location, mate_loc, both_mates)
- We then merge overlapping intervals and identify maximal intervals that are overlapped by at least five clones. This set of intervals points to all regions of the reference with evidence of a deletion.
- Predicted_deletions = select merge_intervals (interval_ count > 4)
- from Disc2Intrvl
- To output the intervals, and the supporting discordant reads, we do a final MAPJOIN and print the result. This changes the output data type back to reads stored in a BAM file that can be used downstream by other software.
- out = select * from MAPJOIN Predicted_deletions, Discordant
- using intervals(location, mate_loc, both_mates)
- print out
Note that entire query is a few lines of GQL. Further, the query took 10 min to execute on the entire genome. All the results we report used a single i7 Intel CPU with 18 GB of random access memory and 2 TB of disk. Much of the execution time was spent on printing the large output (12 K regions, with 44 MB of supporting evidence).
These observations suggest that complex GQL queries could efficiently be performed in the cloud. Given that organizations can easily obtain 100 Mbps speeds today (Amazon’s Direct Connect even allows 1 Gbps speeds), the output of 44 MB can be retrieved interactively to a user desktop in <4 s. By contrast, downloading the original BAM files would take >2 h at 100 Mbps. Second, although we measured the query to take 10 min on a single processor, simple parallelism by chromosome (easily and cheaply feasible in the cloud) should provide a factor of 20 speed-up, leading to a query time of 30 s.
In addition, given the decreasing cost of random access memory, an implementation of GQL on a memory only database system, such as SAP HANA, can provide additional speed-up by eliminating disk accesses. Currently, the output products of each select statement are stored to disk and are loaded again by subsequent select statements. A memory only database will be able to remove this overhead. Further, writing output BAM files comprises a clear performance bottleneck caused by a large number of alternating read and write disk accesses, which can also be eliminated by a memory only database. Despite these advantages of memory only databases, cloud implementations are also useful because many current genomic archives are stored in the cloud, such as the 1000 genomes archive on EC2.
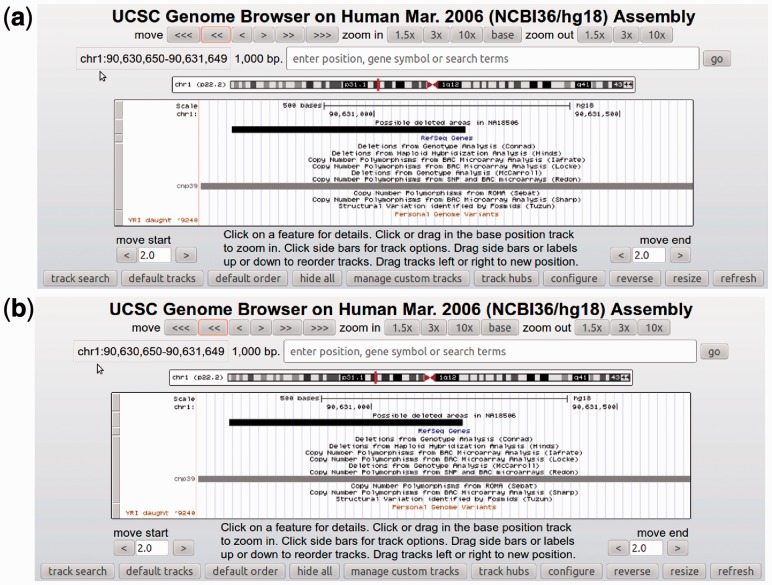
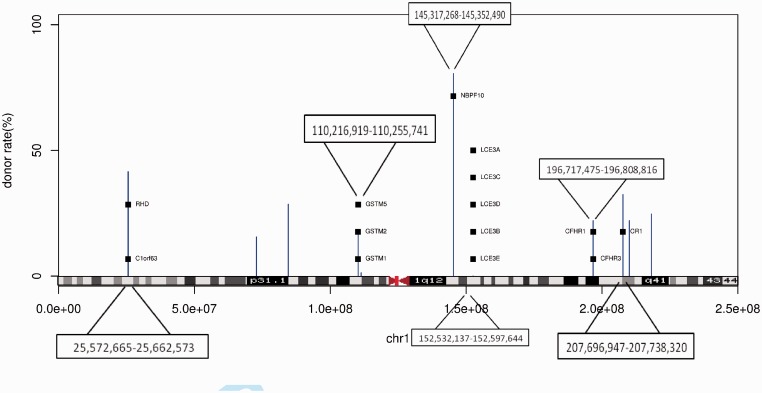
Further, we wrote a program to convert the intervals output by a GQL query to the BED format, which can then be uploaded to the UCSC genome browser for further investigation, including comparisons with other reference annotations. See Figure 2a and b for examples showing overlap between output regions, and known CNVs, and a previously reported Indel in NA18507 (Bentley et al., 2008), the father of NA18506.
Fig. 2.
UCSC browser snapshots from areas of NA18506 that are candidate deletions. (a) An area that overlaps with a known CNV site. (b) An area that overlaps with a known deletion of the father NA18507
Conrad et al. (2006) used array hybridization to identify deletions in a number of genomes, including NA18506. We wrote a GQL query to compare the two sets of predictions as follows: we created a table with all of Conrad’s predictions and performed a mapjoin with our predicted intervals. We then output the common regions that overlap and the corresponding discordant read evidence. This query ran in 7 min, and the total size of the produced BAM files was 164 KB. Our results overlap with 15 of the 23 findings of Conrad et al. (2006). To look at support for regions identified by Conrad et al. (2006), but not by us, we used mapjoin to identify all discordant and concordant reads that overlap with Conrad-only predictions using the following GQL query.
Discordant = select * from READS
where location ≥0 and mate_loc ≥ 0
and abs(mate_loc + length-location) > 350
and abs(mate_loc + length-location) < 1 000 000)
out = select * from mapjoin conr_only_intrvls, Discordant
using intervals(location, mate_loc, both_mates)
Interestingly, none of the eight Conrad-only predictions were supported by discordant-reads. Further, six of the eight Conrad-only regions had concordant read coverage exceeding 30; the other two had coverage exceeding 10, suggesting heterozygous deletion, at best. The GQL query to retrieve the entire evidence took 12 min, and a few lines of code.
To validate regions with discordant read evidence output by us, but not predicted by Conrad et al., we ran the same deletion-query in the parents of NA18506 to see whether they are reproduced in the parents. Three of the predicted deletions overlapped with discordant reads in both parents and nine in one parent. Only three of our predicted deletions do not appear in any of the parents. In each case, further statistical analysis can be used on the greatly reduced dataset to help validate the predicted deletions.
Inversions in the donor genome. To detect putative inversions, we locate regions that are covered by at least five different pairs of orientation discordant reads.
Discordant = select * from READS using intervals (location,mate_loc, both_mates)
where location ≥ 0 and mate_loc ≥ 0
and strand==mate_strand
and abs(mate_loc + length-location) > 270
and abs(mate_loc + length-location) < 1 000 000)
The query needs 8 min to run and identifies 366 regions of possible inversion events and returns 47 324 alignments, which are stored in BAM files of size 3 MB.
High CNV. The average coverage of reads in the dataset is 40. To identify high copy number regions (possible duplications), we locate regions with 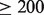 coverage.
coverage.
H1 = select create_intervals() from READS
where location ≥ 0
out = select merge_intervals(interval_coverage > 200) from H1
The query needs 30 min to run, and the evidence data of the output consist of 9.7 M reads stored in BAM files of size 664 MB. We contrast this with the original BAM file of size of 72 GB that a caller would have parsed had GQL not been used.
Reads mapping to specific intervals. Here, we output all reads that map to known genic regions. The query uses an external table that consists of all known genes based on the RefSeq database. The execution time is 288 min, limited by the huge output size (495 M reads).
mapped_reads = select * from reads
where location > 0
out = select *
from mapjoin Refseq_genes using intervals(txStart, txEnd),
mapped_reads using intervals (location, location + length, both_mates)
Efficacy of Mapjoin implementation. In most databases, Joins are typically the most time-expensive operation. Typical GQL queries use intermediate MapJoin operations extensively. We implement a special Lazy Join procedure to greatly improve the runtime, explained here with an example as ‘output all reads that overlap with genes whose transcriptional start is in a CpG island’. Tables describing CpG islands are available (e.g. Gardiner-Garden and Frommer, 1987) and can be downloaded from the UCSC genome browser. A non-expert user might write the following sub-optimal GQL code that applies the position restriction on the (large) output of the mapjoin between all reads and genes.
mapped_reads = select * from reads
where location > 0
reads_genes = select * from
mapjoin Refseq_genes using intervals(txStart, txEnd),
mapped_reads using intervals (location, location + length)
out = select * from mapjoin cpgIsland_hg18 using intervals(chromStart, chromEnd), reads_genes using intervals(location, location + length)
In the absence of lazy evaluation, the execution time of this snippet would be bounded by the extremely large execution time (288 min) of the data intensive query of the previous paragraph. Lazy evaluation, which allows us to join using virtual joins and bypasses intermediate data generation, provides the same result within 42 min for the entire genome.
Common deletions in the YRI population. Here, we extend queries from single donor to multiple donors. Kidd et al. (2008) validated deletions in eight geographically diverse individuals using a fosmid sub-cloning strategy, including NA18507. Of these deletions, the ones that overlap with genes suggest a phenotypically interesting deletion. Therefore, we ask how many of such Chr 1 deletions are prevalent in the entire HapMap YRI sub-population (77 individuals).
- Get intervals with at least four length-discordant reads.
- Disc = select * from Reads
- where 350 ≤ abs(location + mate_loc-length)
- and abs(location + mate_loc-length) ≤ 1 000 000
- Del = select merge_intervals(count > 4) from Disc
- MapJoin with validated intervals.
- Del_Overlapping = select * from MAPJOIN Del, Kidd_results
- using intervals (begin, end)
- Map Join with known genes.
- Gene_overlapping = select * from MAPJOIN Del, RefSeq_genes
- using intervals (txStart, txEnd)
The query takes 5 min to find the common regions of chr1 across the entire population and 30 min to print the accompanying reads that support the query. Figure 3 shows the rate according to which each validated deletion appears to other Yoruban individuals and the affected genes. Eight of the deletions are common in at least 30% of the individuals, two are common in at least 50% and one deletion is common in 80% of the YRI population. The information provides a quick first look at deletion polymorphisms in the Yoruban population. For example, 81% of the individuals have a deletion in 1q21.1 containing the Neuroblastoma Breakpoint gene family (NBPF), so called because of prevalent translocation event in neuroblastoma (Vandepoele et al., 2005). Also, 42% of the individuals have a deletion in 1p36.11, where the deletion removes a large part of the RHD gene, responsible for Rh group D antigen. Such deletions have previously been reported in other populations (Wagner and Flegel, 2000). We also find a common deletion (22%) involving complement factor H-related protein, which has been associated with age-related macular degeneration (AMD) (Sivakumaran et al., 2011). Other common deletions involving multiple genes are shown in Figure 3.
Fig. 3.
Frequencies of common deletions across the YRI population that match the results of Kidd et al. (2008) for the chromosome 1 of NA18507. For each genome, we find candidate deleted areas and we apply a mapjoin operation with the former deletions. The figure shows how many times each deletion of Kidd et al. (2008) overlaps with some deletion of other genomes, and it also shows the genes that are affected by said deletions
Integration with Variant Callers. In this experiment, we demonstrate the speed-up that the output of GQL can potentially provide to existing SV detection tools. Here, we use Breakdancer (Chen et al., 2009), which runs in two steps. The first, quick, step collects statistical information from the input BAM file, including read-coverage and distribution of insert sizes. The second (so-called Breakdancer_max) step involves the major processing of the input.
A normal run of Breakdancer_max with input being the NA18506 chr1 alignments (a BAM file of size 6 GB) takes 15 min and produces a collection of SVs, including deletion, inversion, insertion and intra-chromosomal translocation events.
We used GQL to filter the original BAM file to retain only reads (76 MB) that are needed by Breakdancer (Chen et al., 2009). Next, we ran Breakdancer_max again using the filtered input. This time the tool needed only 2:30 min, 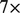 improvement in speed that can be attributed to the reduced data in the input file.
improvement in speed that can be attributed to the reduced data in the input file.
Note that the results are not identical because of the stochastic nature of the caller, but overlap strongly. We call an identified variant from the initial experiment consistent with the second run if it overlaps by at least 50% of its length with the latter. With this definition, we found that 947 of 948 of the deletions, all 204 intra-chromosomal translocations, 337 of 340 inversions and 397 of 461 of insertions are consistent.
These results were based on our estimation of the evidence used by Breakdancer found by reading the article. Even more accurate results could be obtained if the writers of the caller wrote the GQL query to gather the evidence they need. Although this is only one experiment, it supports the vision that writers of callers can concentrate on the hard problems of inference and leave the management and filtering of large genomic datasets to GQL.
3 CONCLUSIONS
The central message of this article is that a declarative query language such as GQL can allow interactive browsing of genome data that is hard to do with existing tools. We agree that in terms of functionality, especially with respect to interval calculus, there exist other tools with similar functionality, such as samtools, bedtools and others. However, the choice of a declarative syntax allows for richer syntax, including multiple join operations and operations on population data. Moreover, it separates the implementation from the query and allows for optimizations in the implementation that are transparent to the naïve user.
The results suggest that a cloud implementation of GQL can be efficient and fast. In particular, for most selective queries, the resulting output is small (MB) and can be retrieved in a few seconds across the network. Further, the query times we report are in the order of minutes using a cheap single processor for genome-wide scans. Simple map-reduce style parallel implementation should reduce this to seconds. However, one of the optimization relates to separating files by chromosomes, which effectively disallow querying for discordant paired-ends that map to different chromosomes. These queries will be added in a future iteration.
We note that we had to implement at least five non-trivial optimizations to reduce query processing times by at least three orders of magnitude. These include the use of a materialized view of the metadata inherent in reads, lazy joins, precompiled parsing of expressions, stack-based interval merging and interval trees. Although interval trees are commonly used in genomic processing, the other optimizations may be novel in a genomic context. These low-level optimizations will be described elsewhere.
Finally, GQL is compatible with existing genomic software. Existing callers can be re-factored to retrieve evidence from cloud repositories using GQL, thereby relegating large data handling to GQL with consequent speed-ups as we demonstrated for Breakdancer. GQL is also compatible with SNP calling because GQL produces smaller BAM files that can be input to SNP callers. We have chosen to focus on the use of GQL for structural variation analysis because SNP calling is well studied in the literature, and there are a number of tools already to provide the evidence needed for SNP calling. Further, the results of GQL queries can be viewed using the UCSC browser. In principle, we can also support ‘semantic browsing’ by properties of genomic regions in addition to browsing by position (syntactic browsing).
4 MATERIALS AND METHODS
The GQL pipeline starts with the parsing of a user’s GQL code (Supplementary Information) by the front-end. We developed a syntax checker and a parser to interpret GQL queries. We used the open source tool flex (Flex, 1990) to identify the keywords of GQL (Supplementary Information). Syntax checking code was created using the open source tool Bison (Bison, 1988) and checks the current syntax of GQL described by a context free grammar.
Next, we perform a semantic analysis of the code to understand the basic primitives. Appropriate keywords (Select, From and so forth) are recognized, checked in a specific order and used to make calls to back-end routines. The front-end also passes the algebraic part of the user statements (such as expressions) to the back-end by creating customized C++ files. This ‘precompiled parsing’ speeds up the back-end almost 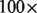 , which would otherwise have to interpret each expression when applied to each read in the input BAM file. The end of processing leads to a custom C++ queries that is automatically compiled and used to run GQL queries.
, which would otherwise have to interpret each expression when applied to each read in the input BAM file. The end of processing leads to a custom C++ queries that is automatically compiled and used to run GQL queries.
READS table. The Reads table is the abstraction by which GQL provides to a user with access to BAM files and its implementation considers that these files are so large that they do not necessarily fit into main memory. Without care, the disk traffic can slow down query processing that involves genome-wide scans. GQL chooses to speed-up most common structural variation queries by extracting as metadata a subset of fields (namely, the read length, a pointer to the pair-end and the mapping location and strand) from each read which is small enough to fit into main memory at least for a per-chromosome execution. Thus, a query that only uses a combination of the metadata fields does not have to access the raw BAM file at all.
This extraction needs to occur only once per genome during pre-processing, and it is highly efficient given that a BAM file follows our recommended formatting. We require that the input BAM files are sorted according to their alignment location, and all alignment locations are chromosome isolated: in other words, for every genome there should be a single file containing reads for each chromosome. Under these assumptions, a dataset of ∼90 M reads of size 6.5 GB from NA18507 that map to chr1 takes ∼6 min to extract the metadata.
Text tables. This type of table includes all tab-separated text files that a user uploads. As no assumptions can be made about the nature or the size of the contents of those tables, the main functions of the tables are simple. The evaluation of an expression on an entry of a text table fetches the appropriate fields and converts them from ASCII strings to the proper type according to the specification that the user supplies to the compiler. The selection on a file of a text table prints to the output the entries for which the evaluation of the provided boolean expression is True.
Interval Tables: creation and projection (merging). Recall the query for CNVs:
H1 = select create_intervals(…) from READS where location ≥ 0
out = select merge_intervals(interval_coverage > 200) from H1
We allow the user to create interval tables from any table, simply by identifying specific fields as begin and end. The function iterates over the entries of interest of the source table. It evaluates the user-provided interval expressions for begin and end on each entry. We discard entries of intervals whose end field is no less than begin.
The ‘merge-interval’ command operates by virtually projecting all intervals (using an efficient data-structure) to a reference and maintaining a count-vector.
Computing mapjoin. The mapjoin operation takes two interval tables as input, and outputs pairs of records where the intervals intersect. We allow for the joins to be applied to multiple tables, including user-defined ones. This significantly increases functionality, but it requires the use of ‘lazy-joins’ and interval trees for efficient implementation.
In evaluating a select operation on a mapjoin table, we simply evaluate the provided boolean expression on all tuples of the table and outputs those tuples that satisfy the expression.
Supplementary Material
ACKNOWLEDGEMENT
The authors thank Alin Deutsch, Lucila Ohno-Machado, Nitin Udpa, Anand Patel and Sangwoo Kim for useful discussions and support. GQL can be downloaded from http://cseweb.ucsd.edu/∼ckozanit/gql.
Funding: NIH (NIH 5R01-HG004962) sponsored iDASH project (Grant U54 HL108460) and a CSRO scholarship (to C.K.).
Conflict of Interest: none declared.
REFERENCES
- 1000 Genomes Project Consortium, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. [Google Scholar]
- 1000genomescloud. Using 1000 genomes data in the amazon web service cloud. 2012. http://www.1000genomes.org/using-1000-genomes-data-amazon-web-service-cloud (4 June 2013, date last accessed)
- Asnani H, et al. Lossy compression of quality values via rate distortion theory. ArXiv e-prints. 2012 [Google Scholar]
- Barnett DW, et al. BamTools: a C++ API and toolkit for analyzing and managing BAM files. Bioinformatics. 2011;27:1691–1692. doi: 10.1093/bioinformatics/btr174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashir A, et al. Optimization of primer design for the detection of variable genomic lesions in cancer. Bioinformatics. 2007;23:2807–2815. doi: 10.1093/bioinformatics/btm390. [DOI] [PubMed] [Google Scholar]
- Bentley DR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bison. Bison - GNU parser generator. 1988. http://www.gnu.org/software/bison/ (4 June 2013, date last accessed)
- Chen K, et al. BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nat. Methods. 2009;6:677–681. doi: 10.1038/nmeth.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke J, et al. Continuous base identification for single-molecule nanopore DNA sequencing. Nat. Nanotechnol. 2009;4:265–270. doi: 10.1038/nnano.2009.12. [DOI] [PubMed] [Google Scholar]
- Codd EF. A relational model of data for large shared data banks. Commun. ACM. 1970;13:377–387. [PubMed] [Google Scholar]
- Conrad D, et al. A high-resolution survey of deletion polymorphism in the human genome. Nat. Genet. 2006;38:75–81. doi: 10.1038/ng1697. [DOI] [PubMed] [Google Scholar]
- Cox AJ, et al. Large-scale compression of genomic sequence databases with the Burrows-Wheeler transform. Bioinformatics. 2012;28:1415–1419. doi: 10.1093/bioinformatics/bts173. [DOI] [PubMed] [Google Scholar]
- Dale RK, et al. Pybedtools: a flexible Python library for manipulating genomic datasets and annotations. Bioinformatics. 2011;27:3423–3424. doi: 10.1093/bioinformatics/btr539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flex. The Fast Lexical Analyzer. 1990. http://flex.sourceforge.net (4 June 2013, date last accessed)
- Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J. Mol. Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- gatk-pairend. Where does gatk get the mate pair info from bam files? 2012. http://gatkforums.broadinstitute.org/discussion/1529/where-does-gatk-get-the-mate-pair-info-from-bam-file (4 June 2013, date last accessed)
- Giglio S, et al. Olfactory receptor-gene clusters, genomic-inversion polymorphisms, and common chromosome rearrangements. Am. J. Hum. Genet. 2001;68:874–883. doi: 10.1086/319506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goecks J, et al. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010;11:R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hormozdiari F, et al. Combinatorial algorithms for structural variation detection in high-throughput sequenced genomes. Genome Res. 2009;19:1270–1278. doi: 10.1101/gr.088633.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsi-Yang Fritz M, et al. Efficient storage of high throughput DNA sequencing data using reference-based compression. Genome Res. 2011;21:734–740. doi: 10.1101/gr.114819.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DC, et al. Compression of next-generation sequencing reads aided by highly efficient de novo assembly. Nucleic Acids Res. 2012;40:e171. doi: 10.1093/nar/gks754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd JM, et al. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008;453:56–64. doi: 10.1038/nature06862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koboldt DC, et al. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozanitis C, et al. Compressing genomic sequence fragments using SlimGene. J. Comput. Biol. 2011;18:401–413. doi: 10.1089/cmb.2010.0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate long-read alignment with burrows-wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, et al. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason CE, et al. Standardizing the next generation of bioinformatics software development with BioHDF (HDF5) Adv. Exp. Med. Biol. 2010;680:693–700. doi: 10.1007/978-1-4419-5913-3_77. [DOI] [PubMed] [Google Scholar]
- McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry GH, et al. Hotspots for copy number variation in chimpanzees and humans. Proc. Natl Acad. Sci. USA. 2006;103:8006–8011. doi: 10.1073/pnas.0602318103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popitsch N, von Haeseler A. NGC: lossless and lossy compression of aligned high-throughput sequencing data. Nucleic Acids Res. 2013;41:e27. doi: 10.1093/nar/gks939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp A, et al. Structural variation of the human genome. Annu. Rev. Genomics Hum. Genet. 2006;7:407–442. doi: 10.1146/annurev.genom.7.080505.115618. [DOI] [PubMed] [Google Scholar]
- Sindi S, et al. A geometric approach for classification and comparison of structural variants. Bioinformatics. 2009;25:i222–i230. doi: 10.1093/bioinformatics/btp208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivakumaran TA, et al. A 32 kb critical region excluding Y402H in CFH mediates risk for age-related macular degeneration. PLoS One. 2011;6:e25598. doi: 10.1371/journal.pone.0025598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandepoele K, et al. A novel gene family NBPF: intricate structure generated by gene duplications during primate evolution. Mol. Biol. Evol. 2005;22:2265–2274. doi: 10.1093/molbev/msi222. [DOI] [PubMed] [Google Scholar]
- VCF Tools. Variant call format. 2011. http://vcftools.sourceforge.net/specs.html (4 June 2013, date last accessed)
- Wagner FF, Flegel WA. RHD gene deletion occurred in the Rhesus box. Blood. 2000;95:3662–3668. [PubMed] [Google Scholar]
- Wan R, et al. Transformations for the compression of FASTQ quality scores of next-generation sequencing data. Bioinformatics. 2012;28:628–635. doi: 10.1093/bioinformatics/btr689. [DOI] [PubMed] [Google Scholar]
- Wheeler DL, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2008;36:13–21. doi: 10.1093/nar/gkm1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanovsky V. ReCoil - an algorithm for compression of extremely large datasets of DNA data. Algorithms Mol. Biol. 2011;6:23. doi: 10.1186/1748-7188-6-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.