Abstract
Background
Glutamate decarboxylase 1 (GAD1), a rate-limiting enzyme in the production of γ-aminobutyric acid (GABA), is found in the GABAergic neurons of the central nervous system. Little is known about the relevance of GAD1 to oral squamous cell carcinoma (OSCC). We investigated the expression status of GAD1 and its functional mechanisms in OSCCs.
Methods
We evaluated GAD1 mRNA and protein expressions in OSCC-derived cells using real-time quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR) and immunoblotting analyses. To assess the critical functions of GAD1, i.e., cellular proliferation, invasiveness, and migration, OSCC-derived cells were treated with the shRNA and specific GAD1 inhibitor, 3-mercaptopropionic acid (3-MPA). GAD1 expression in 80 patients with primary OSCCs was analyzed and compared to the clinicopathological behaviors of OSCC.
Results
qRT-PCR and immunoblotting analyses detected frequent up-regulation of GAD1 in OSCC-derived cells compared to human normal oral keratinocytes. Suppression of nuclear localization of β-catenin and MMP7 secretion was observed in GAD1 knockdown and 3-MPA-treated cells. We also found low cellular invasiveness and migratory abilities in GAD1 knockdown and 3-MPA-treated cells. In the clinical samples, GAD1 expression in the primary OSCCs was significantly (P < 0.05) higher than in normal counterparts and was correlated significantly (P < 0.05) with regional lymph node metastasis.
Conclusions
Our data showed that up-regulation of GAD1 was a characteristic event in OSCCs and that GAD1 was correlated with cellular invasiveness and migration by regulating β-catenin translocation and MMP7 activation. GAD1 might play an important role in controlling tumoral invasiveness and metastasis in oral cancer.
Keywords: Glutamate acid decarboxylase 1, β-catenin, Matrix metalloproteinase-7, 3-mercaptopropionic acid, Metastasis, Oral squamous cell carcinoma
Background
Glutamate decarboxylase 1 (GAD1) catalyzes production of γ-aminobutyric acid (GABA) from L-glutamic acid, the principal inhibitory neurotransmitter in the brain [1,2]. GAD1 is associated with development of insulin-dependent diabetes mellitus [3] and many cases of the Stiff-Person syndrome [4]. The murine model of cleft palate also lacks GAD1 expression [5].
GAD1 is related closely to β-catenin expression by microarray analysis in ovarian endometrioid adenocarcinoma and Wilms’ tumor [6,7], whereas the functional interaction between GAD1 and β-catenin has not been demonstrated clearly. β-catenin is an essential component of both intercellular junctions and the canonical Wnt signaling pathway and connects the adherens junction complex with the actin cytoskeleton that binds directly to the intracellular domain of E-cadherin [8]. Disruption of β-catenin plays critical roles in the regulation of cellular invasiveness, proliferation, and migration [9-11].
The Wnt/β-catenin pathway is activated when the Wnt ligand binds to cell-surface receptors, the Frizzled family receptor, and the LRP 5/6 co-receptor. Activation of this pathway leads to inhibition of a complex comprising adenomatous polyposis coli, axis inhibition protein, and glycogen synthase kinase 3β. This complex has functions is involved in phosphorylation and degradation of β-catenin by the ubiquitin/proteosome system. After activation of the Wnt/β-catenin pathway, β-catenin translocates to the nucleus for binding to T-cell factor/lymphoid enhancer factor (TCF/LEF) and activates transcription of Wnt-targeting genes [12].
MMP7 is a Wnt-targeting gene that has been detected in several cancers, such as prostate, colon, stomach, lung, and breast [13-17] and degrades components of the extracellular matrix (ECM), including collagens (I, III, IV, and V), fibronectin, vitronectin, laminin, and elastin [18]. In the oral region, Chuang et al. demonstrated that MMP7 is closely related to invasion in OSCCs of buccal mucosa [19]. Therefore, MMP7 contributes significantly to the cellular invasiveness and metastasis of tumors.
The current study found that GAD1 is overexpressed frequently in OSCC-derived cell lines, and that GAD1 knockdown affects cellular invasiveness and migration. Based on this evidence, we proposed that GAD1 might be a therapeutic target to prevent metastasis in OSCCs.
Methods
Ethics statement
The Ethics Committee of the Graduate School of Medicine, Chiba University (approval number, 236) approved the study protocol, which was performed according to the tenets of the Declaration of Helsinki. All patients provided written informed consent.
OSCC-derived cell lines and tissue samples
RIKEN BRC (Ibaraki, Japan) provided the Sa3, HO-1-u-1, KOSC-2, Ca9-22, HO-1-N-1, HSC-2, and HSC-3 cell lines through the National Bio-Resource Project of the MEXT, Tokyo, Japan. Short tandem repeat profiles confirmed the cellular identity. Primary cultured human normal oral keratinocytes (HNOKs) were used as normal controls [20,21]. All cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) (Sigma, St. Louis, MO) supplemented with 10% fetal bovine serum (FBS) (Sigma) and 50 units/ml of penicillin and streptomycin (Sigma). Primary OSCCs and patient-matched normal oral epithelial samples were obtained during surgical resections of the tumors with simultaneous neck dissection at Chiba University Hospital. The average age of the patients was 64.6 years (range, 27–90 years). The mean follow-up time for all of the patients was 68.5 months (range, 24–98 months). The resected tissues were fixed in 10% buffered formaldehyde solution for pathological diagnosis and immunohistochemistry (IHC). Histopathological diagnosis of each tissue was performed according to the tumor-node-metastases classification of the International Union against Cancer.
Preparation of cDNA
Total RNA was isolated using TRIzol Reagent (Invitrogen, Carlsbad, CA). cDNA was generated from 5 μg of total RNA using Ready-To-Go You-Prime First-Strand Beads (GE Healthcare, Buckinghamshire, UK) and oligo (dT) primer (Sigma Genosys, Ishikari, Japan).
mRNA expression analysis
Real-time quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR) was performed using a Light Cycler 480 apparatus (Roche Diagnostics GmbH, Mannheim, Germany) to evaluate the expression levels of GAD1 mRNA in the seven OSCC-derived cell lines (HSC-2, HSC-3, Sa3, HO-1-u-1, HO-1-N-1, KOSC-2, and Ca9-22) and HNOKs. Primers were designed using the Probe Finder qRT-PCR assay design software (available at http://www.universalprobelibrary.com). The sequences of the gene-specific primers and universal probes were as follows: GAD1 forward, 5′-CCA TGG TCG TAC CTG ACT CC-3′ and reverse, 5′-CCT GGA ACT GGC TGA ATA CC-3′ (probe #78); MMP7 forward, 5′-TCT CCT CCG AGA CCT GTC C-3′ and reverse, 5′-GCT GAC ATC ATG ATT GGC TTT-3′ (probe #72); and β-catenin forward, 5′-GCT TTC AGT TGA GCT GAC CA-3′ and reverse, 5′-CAA GTC CAA GAT CAG CAG TCT C-3′ (probe #21). The PCR reactions were carried out in a final volume of 20 μl of a reaction mixture comprised of 10 μl of Light Cycler 480 Probes Master (Roche), 0.2 μl of universal probe (Roche), and 4 μM of the primers. The reaction mixture was loaded onto the PCR plate and subjected to an initial denaturation at 95°C (10 min), followed by 45 rounds of amplification at 95°C (10 sec) for denaturation, 60°C (30 sec) for annealing, and 72°C (1 sec) for extension, followed by a cooling step at 50°C for 30 seconds. The transcript amounts for the GAD1 and other genes were estimated from the respective standard curves and normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) forward, 5′-AGCCACATCGCTCAGACAC-3′ and reverse, 5′-GCCCAATACGACCAAATCC-3′ (probe #60) transcript amounts determined in corresponding samples.
Protein extraction
The cells were washed twice with cold phosphate buffered saline (PBS) and centrifuged briefly. The cell pellets were incubated at 4°C for 30 min in a lysis buffer (7 M urea, 2 M thiourea, 4% w/v CHAPS, and 10 mM Tris pH 7.4) with a proteinase inhibitor cocktail (Roche) to extract whole cell lysates. The protein concentrations of whole cell lysates were measured using the Bradford reagent (Bio-Rad, Richmond, CA). Cytoplasmic and nuclear fractions from cultured cells were isolated using the NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo, Rockford, IL). The protein concentrations were measured using the BCA Protein Assay Kit (Thermo).
Immunoblotting
Protein extracts were electrophoresed on 4% to 12% Bis-Tris gel, transferred to nitrocellulose membranes (Invitrogen), and blocked for 1 hr at room temperature with Blocking One (Nacalai Tesque, Inc., Kyoto, Japan). The membranes were washed three times with 0.1% Tween-20 in Tris-buffered saline and incubated with antibody for GAD1 (Santa Cruz Biotechnology, Dallas, TX) and β-catenin (Novus Biologicals, Littleton, CO) overnight at 4°C and GAPDH (Thermo) for 1 hr at room temperature. The membranes were washed again and incubated with a anti-rabbit or anti-mouse IgG horseradish peroxidase conjugate (Promega, Madison, WI) as a secondary antibody for 1 hr at room temperature. Finally, the membranes were detected using SuperSignal West Pico Chemiluminescent Substrate (Thermo), and immunoblotting was visualized by exposing the membranes to ATTO Light-Capture II (ATTO, Tokyo, Japan). Signal intensities were quantitated using the CS Analyzer version 3.0 software (ATTO).
IHC
IHC of 4-μm sections of paraffin-embedded specimens was performed using mouse anti-GAD1 monoclonal antibody (Santa Cruz Biotechnology). Briefly, after deparaffinization and hydration, the endogenous peroxidase activity was quenched by a 30-min incubation in a mixture of 0.3% hydrogen peroxide solution in 100% methanol, after which the sections were blocked for 2 hr at room temperature with 1.5% blocking serum (Santa Cruz Biotechnology) in PBS before reaction overnight with anti-GAD1 antibody (1:100 dilution) at 4°C in a moist chamber. Upon incubation with the primary antibody, the specimens were washed three times in PBS and treated with Envision reagent (DAKO, Carpinteria, CA) followed by color development in 3,3′-diaminobenzidine tetrahydrochloride (DAKO). The slides then were lightly counterstained with hematoxylin, dehydrated with ethanol, cleaned with xylene, and mounted. To avoid non-specific binding, an immunizing peptide blocking experiment was performed. As a negative control, triplicate sections were immunostained without exposure to primary antibodies. To quantify the status of the GAD1 protein expression in those components, we used an IHC scoring system to quantitatively evaluate the IHC staining, described previously [22-24]. We counted 300 cells/one field of vision. The staining intensity (0, negative; 1, weak; 2, moderate; 3, intense) and the number of positive cells in the field of vision then were multiplied to calculate the IHC score using the formula: IHC score = 0 × (number of negatively stained cells in the field) + 1 × (number of weakly stained cells in the field) + 2 × (number of moderately stained cells in the field) + 3 × (the highest score for normal tissue). Cases with a GAD1 IHC score exceeding 103 (maximal score within +3 standard deviations of the mean of normal tissues) were defined as GAD1-positive. Two independent pathologists, both masked to the patients’ clinical status, made these judgments.
Stable transfection of GAD1 shRNA
A total of 2 × 105 OSCC-derived cells (HSC2 and HSC3) were seeded into each well of 6-well plates in DMEM F-12 HAM (Sigma) containing 10% FBS (Sigma) without antibiotics. GAD1 shRNA (shGAD1, Santa Cruz Biotechnology) and the control shRNA (mock, Santa Cruz Biotechnology) vectors were transfected into OSCC-derived cells with Lipofectamine LTX (Invitrogen) and Plus Reagents (Invitrogen). After transfection, the cells were isolated using a culture medium containing 2 μg/mL Puromycin (Invitrogen). After 3 to 4 weeks, resistant cell clones were picked and transferred to 6-well plates and expanded gradually to 10-cm dishes. At 90% confluence, qRT-PCR and immunoblotting were performed to assess the efficiency of GAD1 knockdown.
3-Mercaptopropionic acid (3-MPA) treatment
To study the effect of decreased GAD1 activity, we used 3-MPA, a strong competitive inhibitor, at the active GAD1 site [25]. Because several studies has reported that the Ki of 3-MPA ranges from 2.7 to 5.1 μM [26-28], we used 3-MPA (Sigma) at a concentration of 5 μM for functional analyses.
Cellular growth
To evaluate the effect of GAD1 knockdown on cellular proliferation, we analyzed cellular growth in shGAD1 and mock cells. These transfectants were seeded in 6-well plates at a density of 1 × 104 viable cells/well. The experiments were carried out for 168 hr, and the cells were counted every 24 hr. At the indicated time point, the cells were trypsinized and counted using a hemocytometer in triplicate samples. We also performed a cellular growth assay using 3-MPA-treated cells.
Invasiveness assay
We evaluated the effect of GAD1 knockdown on cellular invasiveness. A total of 2.5 × 105 cells were seeded on a polyethylene terephthalate membrane insert with a pore size of 3 μm in a transwell apparatus (Becton-Dickinson Labware, Franklin Lakes, NJ). In the lower chamber, 1 ml of DMEM with 10% FBS was added. After the cells were incubated for 48 hr at 37°C, the insert was washed with PBS, and the cells on the top surface of the insert were removed with a cotton swab. Cells adhering to the lower surface of the membrane were fixed with methanol and stained with crystal violet. The numbers of cells invading the pores in five random fields were counted using a light microscope at × 100 magnification. We also performed the invasiveness assay using the 3-MPA-treated cells.
Migratory assay
shGAD1 and mock cells were seeded in a 6-well plate until they reached full confluence in a monolayer. One wound was created in the middle of each well using a micropipette tip. The plate was incubated at 37°C at 5% CO2. The results were visualized by measuring the wound spaces. The mean value was calculated from data obtained from three separate chambers. We also performed a migratory assay using 3-MPA-treated cells.
Casein zymography
The cells were cultured in serum-free DMEM for 48 hr. The cell culture media were then concentrated using Centrifugal Filter Units (Merck Millipore, Billerica, MA). The concentrated proteins were loaded on precast 12% Novex zymogram blue casein gels (Invitrogen) to measure MMP-7 proteolytic activity. After electrophoresis, the gels were renatured in Novex Zymogram Renaturing Buffer (Invitrogen) for 30 min at room temperature and then incubated at 37°C in Novex Zymogram Developing Buffer (Invitrogen) to allow degradation of the substrate in the gel matrix. Enzymatic activity was visualized as a clear band against a blue background [29-31].
Statistical analysis
Statistical significance was determined using Fisher’s exact test or the Mann–Whitney U test. P < 0.05 was considered significant. The data are expressed as the mean ± standard error of the mean (SEM).
Results
Evaluation of GAD1 expression in OSCC-derived cell lines
We performed qRT-PCR and immunoblotting using OSCC-derived cell lines (Sa3, HO-1-u-1, KOSC-2, Ca9-22, HO-1-N-1, HSC-2, and HSC-3) and HNOKs (Figure 1a, b). GAD1 mRNA was significantly (P < 0.05) up-regulated in all OSCC-derived cell lines compared with the HNOKs. Figure 1b shows representative results of immunoblotting analysis of GAD1 (67 kDa). All OSCC-derived cell lines had a significant (P < 0.05) increase in GAD1 protein expression compared with the HNOKs. Expression analyses indicated that both transcription and translation products of this molecule were highly expressed in OSCC-derived cell lines.
Figure 1.
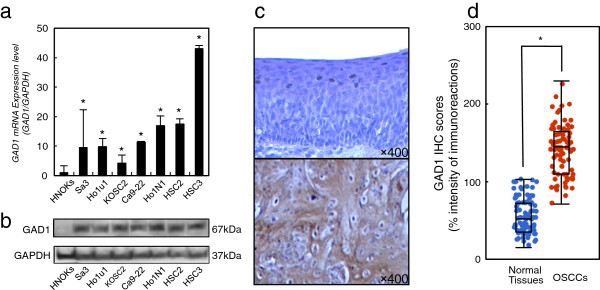
Evaluation of GAD1 expression in OSCC-derived cell lines. a Quantification of GAD1 mRNA levels in OSCC-derived cell lines by qRT-PCR analysis. All OSCC-derived cell lines have significant up-regulation of GAD1 mRNA compared with that in the HNOKs. Data are expressed as the mean ± SEM of values from three assays (*P < 0.05, Mann–Whitney U test). b Immunoblotting analysis of GAD1 protein in the OSCC-derived cell lines and HNOKs. GAD1 protein expressions are up-regulated in all OSCC-derived cell lines examined compared with that in the HNOKs. c Evaluation of GAD1 protein expression in primary OSCCs representative IHC results for GAD1 protein in normal tissue and primary OSCC. Original magnification, ×400. Scale bars, 10 μm. Strong GAD1 immunoreactivity is detected in primary OSCCs. Normal oral tissues show almost weak immunostaining. d The status of GAD1 protein expression in normal oral tissues and primary OSCCs (n = 80). The GAD1 IHC scores for normal oral tissues and OSCCs range from 15 to 103 (median, 52) and 71 to 230 (median, 145), respectively. GAD1 protein expression levels in OSCCs are significantly (*P < 0.001, Mann–Whitney U test) higher than in normal oral tissues.
Evaluation of GAD1 expression in primary OSCCs
We analyzed the GAD1 protein expression in primary OSCCs and paired normal oral tissues from 80 patients using the IHC scoring system. Figure 1c shows representative IHC results for GAD1 protein in normal oral tissues and primary OSCCs. Strong GAD1 immunoreactions were detected in the cytoplasm in the OSCCs. The GAD1 IHC scores for normal oral tissues and OSCCs ranged from 15 to 103 (median, 52) and 71 to 230 (median, 145), respectively. The GAD1 IHC score in primary OSCCs was significantly (P < 0.001) higher than in normal oral tissues (Figure 1d).
Establishment of GAD1 knockdown cells
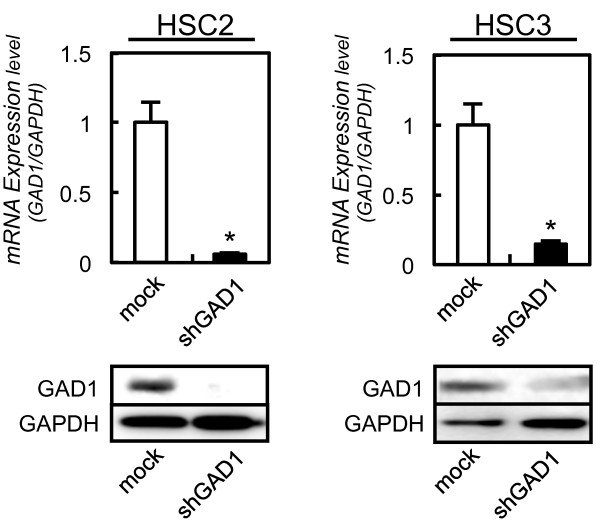
To assess the GAD1 functions in oral cancer, shRNA transfection was carried out in the OSCC-derived cells (HSC2 and HSC3). Expressions of GAD1 mRNA and protein in shGAD1 cells were significantly (P < 0.05) lower than in mock cells (Figure 2).
Figure 2.
Expression of GAD1 in GAD1 knockdown cells. qRT-PCR shows that GAD1 mRNA expression in the shGAD1 cells (HSC2 and HSC3-derived transfectants; 2 clones each) are significantly lower than that in the mock cells (*P < 0.05, Mann–Whitney U test). Immunoblotting analysis shows that the GAD1 protein levels in shGAD1 cells (HSC2 and HSC3-derivrd transfectants; 2 clones each) have decreased markedly compared with that in mock cells.
Functional analyses of GAD1 knockdown cells
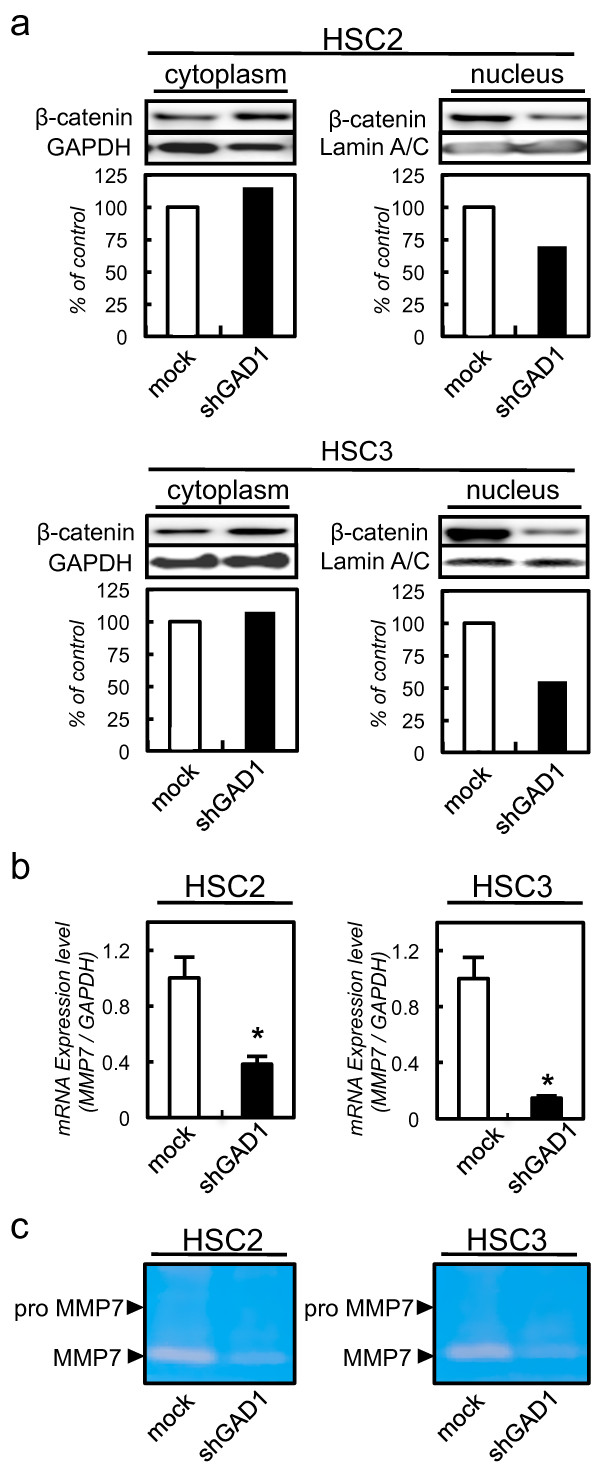
β-catenin, which is located along the cell membrane and cytoplasm in normal epithelial cells, is involved in cellular adhesion and migration [32]. In cancer epithelial cells, β-catenin is translocated into the nucleus, which activates oncogenes including MMP-7 [33]. To assess the translocation of β-catenin in shGAD1 cells, we performed immunoblotting analysis using shGAD1 and mock cells. The expression of β-catenin in the nucleus was suppressed in shGAD1 cells compared with mock cells. The expressions of β-catenin in the cytoplasm did not differ significantly between the shGAD1 and mock cells (Figure 3a). To evaluate the MMP7 mRNA expression, we also performed qRT-PCR using shGAD1 and mock cells. The expression of MMP7 mRNA decreased significantly in shGAD1 cells compared with mock cells (Figure 3b). Using casein zymography, we also detected secreted MMP7 in shGAD1 and mock cells. The MMP7 secretion was suppressed significantly (P < 0.05) in shGAD1 cells compared with mock cells (Figure 3c).
Figure 3.
GAD1 knockdown cells suppress translocation of β-catenin to the nucleus and MMP7 activation. a Immunoblotting analysis of β-catenin in the nucleus of shGAD1 cells. The expression of β-catenin in the nucleus of shGAD1 cells has decreased markedly compared with that in mock cells. β-catenin expression in the cytoplasm does not differ significantly between shGAD1 cells and mock cells. b Quantification of MMP7 mRNA levels in shGAD1 cells by qRT-PCR analysis. MMP7 mRNA is significantly down-regulated in shGAD1 cells compared with mock cells. c Casein zymography analysis of MMP7 activity in shGAD1. Cell culture media are collected and concentrated, MMP7 activity is analyzed by casein zymography. MMP7 secretion is decreased significantly in shGAD1 cells compared with mock cells.
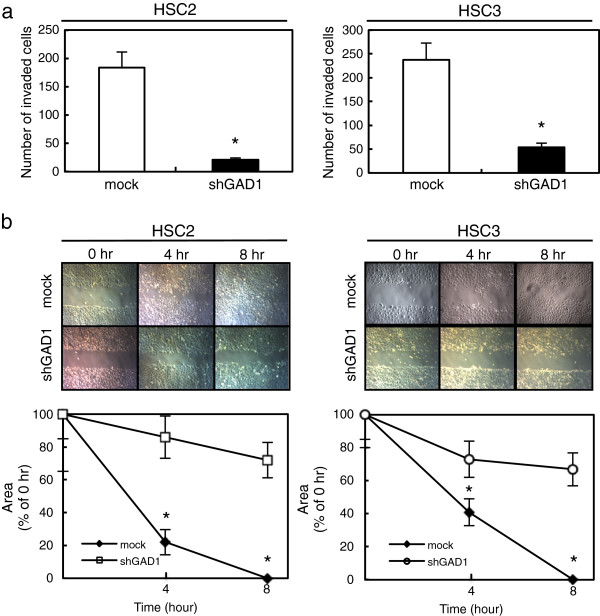
We also performed cellular proliferation, invasiveness, and migratory assays to evaluate the biologic effects of shGAD1 cells. A cellular proliferation assay showed similar growth curves for shGAD1 and mock cells, indicating that down-regulation of GAD1 did not affect cellular proliferation (data not shown). The invasiveness assay showed that the number of penetrating shGAD1 cells decreased compared with mock cells (Figure 4a). The migratory assay showed that the wounds in the shGAD1 cells closed later than in the mock cells when we visually monitored the area of uniform wounds in confluent cell cultures (Figure 4b).
Figure 4.
Functional analyses of GAD1 knockdown cells. a Invasiveness assay of the shGAD1 cells. After crystal violet staining, the numbers of cells invading the pores is counted (×100 magnification). The numbers penetrating the shGAD1 cells are significantly (*P < 0.05, Mann–Whitney U test) greater compared with mock cells. Scale bars, 100 μm. b Migratory assay of shGAD1 cells. The wound area has decreased significantly (*P < 0.05, Mann–Whitney U test) in the culture of mock cells after 8 hr, whereas there is still a gap in the shGAD1 cells. Original magnification, ×100. Scale bars, 100 μm.
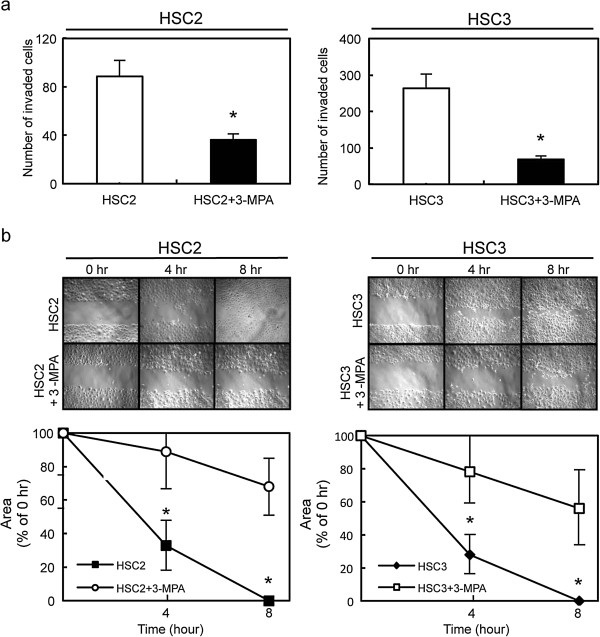
Functional analyses of 3-MPA-treated cells
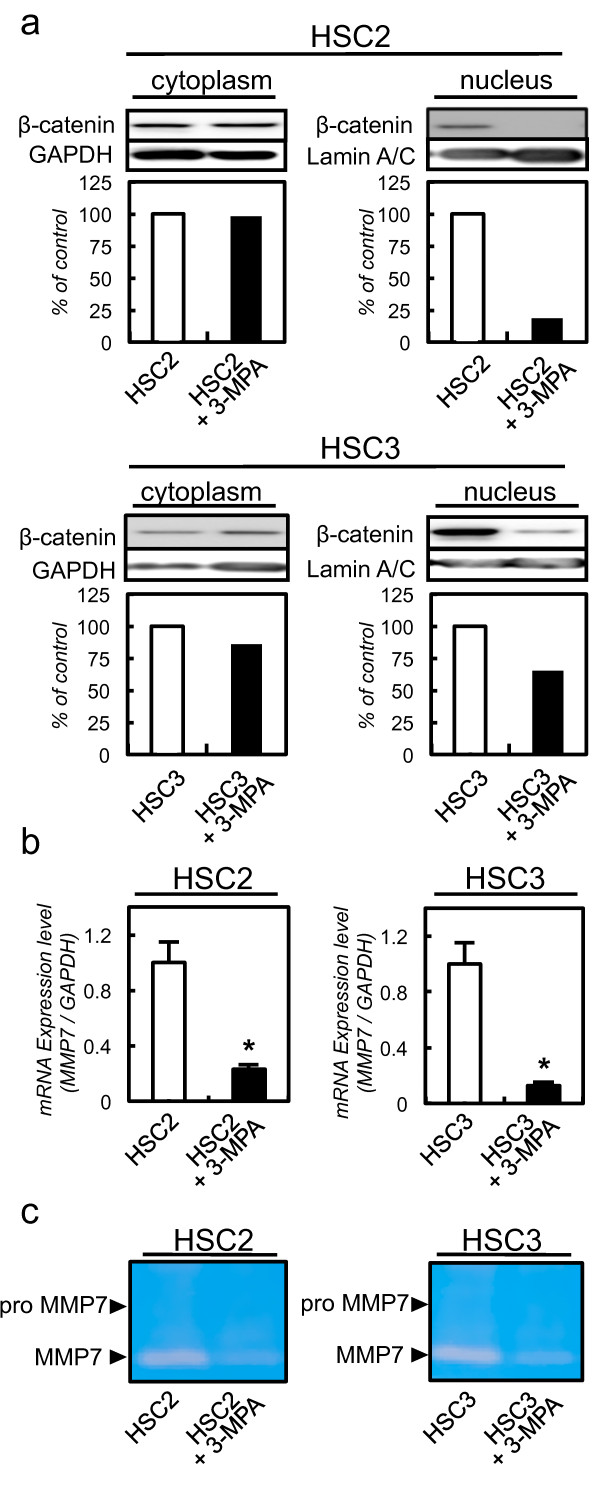
We also performed functional analysis using 3-MPA. To assess the translocation of β-catenin in 3-MPA-treated cells, we performed immunoblotting analysis using 3-MPA-treated and control cells. The expression of β-catenin in the nucleus was suppressed in 3-MPA-treated cells. The expression of β-catenin in the cytoplasm did not differ significantly between the 3-MPA-treated cells and control cells (Figure 5a). To evaluate the MMP7 mRNA expression, we also performed qRT-PCR using 3-MPA-treated and control cells. The MMP7 mRNA expression decreased significantly in the 3-MPA-treated cells compared with control cells (Figure 5b). We also detected MMP7 secreted by casein zymography in 3-MPA and control cells. The secretion of MMP7 was suppressed in 3-MPA-treated cells compared with control cells (Figure 5c).
Figure 5.
3-MPA-treated cells suppress translocation of β-catenin to the nucleus and MMP7 activation. a Immunoblotting analysis of β-catenin in the nuclei of 3-MPA-treated cells. β-catenin expression in the nucleus of 3-MPA-treated cells is decreased markedly compared with that in control cells. β-catenin expression in the cytoplasm does not differ significantly between 3-MPA-treated cells and control cells. b Quantification of MMP7 mRNA level in 3-MPA-treated cells by qRT-PCR analysis. MMP7 mRNA is significantly down-regulated in 3-MPA-treated cells compared with control cells. c Casein zymography analysis of MMP7 activity in 3-MPA-treated cells. Cell culture media are collected and concentrated, MMP7 activity is analyzed by casein zymography. MMP7 secretion is decreased significantly in 3-MPA-treated cells compared with control cells.
We performed cellular proliferation, invasiveness, and migratory assays to evaluate the biologic effects of 3-MPA-treated cells. The cellular proliferation assay showed similar growth curves for 3-MPA-treated and control cells, indicating that inhibition of GAD1 did not affect cellular proliferation (data not shown). The invasiveness assay showed that the number of penetrating 3-MPA-treated cells decreased compared with control cells (Figure 6a). The migratory assay showed that the wounds in the 3-MPA-treated cells closed later than in control cells (Figure 6b) when we visually monitored the area of uniform wounds in confluent cell cultures.
Figure 6.
Functional analysis of the 3-MPA-treated cells. a Invasiveness assay of the 3-MPA-treated cells. After crystal violet staining, the numbers of cells invading the pores are counted (×100 magnification). The numbers penetrating the 3-MPA-treated cells are significantly (*P < 0.05, Mann– Whitney U test) greater compared with control cells. Scale bars, 100 μm. b Migratory assay of 3-MPA-treated cells. The wound area is decreased significantly (*P < 0.05, Mann–Whitney U test) in the culture of control cells after 8 hr, whereas there is still a gap in the 3-MPA-treated cells. Original magnification, ×100. Scale bars, 100 μm.
Expression of GAD1 and clinicopathological variables of primary OSCCs
Table 1 shows the correlations between the clinicopathologic characteristics of patients with OSCC and the status of the GAD1 protein expression using the IHC scoring system. Among the clinical classifications, GAD1-positive OSCCs were significantly (P = 0.011) correlated with regional lymph node metastasis.
Table 1.
Correlation between GAD1 expression and clinical classification in OSCC
|
Clinical classification |
Total |
Result of immunostaining |
P
value |
||
|---|---|---|---|---|---|
| |
|
|
No. of patients (%) |
|
|
| GAD1 (high) | GAD1 (low) | ||||
| Age at surgery |
|
|
|
|
|
| |
< 60 |
17 |
15 (88%) |
2 (12%) |
0.527 |
| |
≧ 60 |
63 |
52 (83%) |
11 (17%) |
|
| Gender |
|
|
|
|
|
| |
Male |
34 |
29 (85%) |
5 (15%) |
0.401 |
| |
Female |
46 |
38 (83%) |
8 (17%) |
|
| T-primary tumor size |
|
|
|
|
|
| |
T1 |
8 |
6 (75%) |
2 (25%) |
0.263 |
| |
T2 |
48 |
38 (79%) |
10 (21%) |
|
| |
T3 |
14 |
13 (93%) |
1 (7%) |
|
| |
T4 |
10 |
10 (100%) |
0 (0%) |
|
| N-regional lymph node |
|
|
|
|
|
| |
N (negative) |
48 |
36 (75%) |
12 (25%) |
0.011* |
| |
N (positive) |
32 |
31 (97%) |
1 (3%) |
|
| Stage |
|
|
|
|
|
| |
I |
8 |
6 (75%) |
2 (25%) |
0.075 |
| |
II |
40 |
30 (75%) |
10 (25%) |
|
| |
III |
20 |
19 (95%) |
1 (5%) |
|
| |
IV |
12 |
12 (100%) |
0 (0%) |
|
| Histopathlogical type |
|
|
|
|
|
| |
Well |
54 |
46 (85%) |
8 (15%) |
0.441 |
| |
Moderately |
16 |
14 (88%) |
2 (12%) |
|
| |
Poorly |
10 |
7 (70%) |
3 (30%) |
|
| Tumor site |
|
|
|
|
|
| |
Gingiva |
20 |
18 (90%) |
2 (10%) |
0.243 |
| |
Tongue |
52 |
44 (85%) |
8 (15%) |
|
| |
Buccal mucosa |
4 |
2 (50%) |
2 (50%) |
|
| Oral floor | 4 | 3 (75%) | 1 (25%) | ||
GAD1(high), up-regulated GAD1; GAD1 (low), down-regulated GAD1; *P < 0.05.
Discussion
GAD1 was overexpressed in OSCC-derived cell lines and new functions of GAD1 were related closely to cellular invasiveness and migration in oral cancer. GAD1 knockdown and 3-MPA-treated cells had suppressed β-catenin levels in the nucleus and secretion of MMP7. Surprisingly, GAD1-positive OSCCs were significantly (P < 0.05) associated with regional lymph node metastasis (Table 1).
GAD isoforms, GAD1 and GAD2, are derived from a common ancestral gene [34]. GAD2 is localized to the nerve terminal and is reversibly bound to the membrane of synaptic vesicles, which has been linked with lower birth weights and additional risk for metabolic diseases [35], whereas GAD1 is a cytosolic enzyme distributed throughout the organs and central nervous system [36]. The enzymatic functions of GAD1 and GAD2 are almost similar; however, their functions remain unclear in cancer tissues [37]. Since our previous microarray data showed that GAD1 is up-regulated significantly in OSCCs [38], we focused on GAD1 in the current study.
β-catenin plays crucial and diverse roles in cadherin-mediated cell-cell adhesion, Wnt signal transduction, gene activation, and tumoral formation [39-41]. Although the interaction mechanism between GAD1 and β-catenin has not yet been reported, the current data suggested that GAD1 expression controls β-catenin localization. β-catenin in nuclei binds to the TCF/LEF in several types of cancers for transcriptional activation of downstream genes, such as MMP7, cyclinD1, and c-myc[42-46], which play important roles in carcinogenesis and metastasis.
We then investigated MMP7 secretion, a downstream candidate of GAD1/β-catenin interaction, because MMP7 often is overexpressed in human cancer tissues and associated with cancer cell invasiveness by proteolytic cleavage of the ECM substrates and degradation of basement membrane proteins [47-50]. Interestingly, we found that GAD1 knockdown and 3-MPA-treated cells inhibited MMP7 secretion by decreasing nuclear translocation of β-catenin. We speculated that the GAD1/β-catenin/MMP7 interaction affects cancer cell behaviors, such as cellular invasiveness and migration. In addition to the in vitro data that down-regulation of GAD1 led to low cellular invasiveness and migratory abilities, patients with GAD1-negative OSCC had a low risk of regional lymph node metastasis. Consistent with our hypothesis, the GAD1/β-catenin/MMP7 interaction is correlated closely with metastasis both in vitro and in vivo.
Conclusion
Our results showed that oral cancer carcinogenesis overexpression of GAD1 occurs frequently and that it might be closely associated with invasion and metastasis of OSCC by β-catenin translocation and MMP7 activation, while further studies are needed to research the GAD1/β-catenin/MMP7 interaction, the current data indicated that GAD1 is likely a molecular marker for early detection of lymph node metastasis and an efficacious treatment target for preventing cancer metastasis in OSCCs.
Abbreviations
GAD1: Glutamate acid decarboxylase 1); GABA: Gamma-aminobutyric acid; MMP7: Matrix metalloproteinase-7; 3-MPA: 3-mercaptopropionic acid.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
Conceived and designed the experiments: RK, AK, HT, UK. Performed the experiments: RK, AK, UK. Analyzed the data: RK, AK, UK. Contributed reagents/materials/analysis tools: RK, AK, TK, CF, YK, MH, YE-S, KO, MS, UK, HT. Wrote the paper: RK, AK, UK, HT. All authors read and approved the final manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Contributor Information
Ryota Kimura, Email: rkimura0131@yahoo.co.jp.
Atsushi Kasamatsu, Email: kasamatsua@faculty.chiba-u.jp.
Tomoyoshi Koyama, Email: koyamatomoyoshi@mail.goo.ne.jp.
Chonji Fukumoto, Email: chonji.lee@gmail.com.
Yukinao Kouzu, Email: kouzuy@faculty.chiba-u.jp.
Morihiro Higo, Email: higom@chiba-u.jp.
Yosuke Endo-Sakamoto, Email: end@faculty.chiba-u.jp.
Katsunori Ogawara, Email: katsunori.ogawara@faculty.chiba-u.jp.
Masashi Shiiba, Email: m.shiiba@faculty.chiba-u.jp.
Hideki Tanzawa, Email: tanzawap@faculty.chiba-u.jp.
Katsuhiro Uzawa, Email: uzawak@faculty.chiba-u.jp.
Acknowledgement
We thank Lynda C. Charters for editing this manuscript.
References
- Erlander MG, Tobin AJ. The structural and functional heterogeneity of glutamic acid decarboxylase: a review. Neurochem Res. 1991;16:215–226. doi: 10.1007/BF00966084. [DOI] [PubMed] [Google Scholar]
- Young SZ, Bordey A. GABA’s control of stem and cancer cell proliferation in adult neural and peripheral niches. Physiology (Bethesda) 2009;24:171–185. doi: 10.1152/physiol.00002.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambrand T, Pociot F, Rønningen KS, Rønningen K, Nerup J, Michelsen BK. Genetic markers for glutamic acid decarboxylase do not predict insulin-dependent diabetes mellitus in pairs of affected siblings. The Danish Study Group of Diabetes in Childhood. Hum Genet. 1997;99:177–185. doi: 10.1007/s004390050334. [DOI] [PubMed] [Google Scholar]
- Hijazi J, Bedat-Millet AL, Hannequin D. Stiff-person syndrome and other neurological disorders associated with anti-GAD antibodies. Rev Med Interne. 2010;31:23–28. doi: 10.1016/j.revmed.2009.01.012. [DOI] [PubMed] [Google Scholar]
- Condie BG, Bain G, Gottlieb DI, Capecchi MR. Cleft palate in mice with a targeted mutation in the gamma-aminobutyric acid-producing enzyme glutamic acid decarboxylase 67. Proc Natl Acad Sci U S A. 1997;94:11451–11455. doi: 10.1073/pnas.94.21.11451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CM, Kim CE, Margolin AA, Guo M, Zhu J, Mason JM, Hensle TW, Murty VV, Grundy PE, Fearon ER. et al. CTNNB1 mutations and overexpression of Wnt/beta-catenin target genes in WT1-mutant Wilms’ tumors. Am J Pathol. 2004;165:1943–1953. doi: 10.1016/S0002-9440(10)63246-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz DR, Wu R, Kardia SL, Levin AM, Huang CC, Shedden KA, Kuick R, Misek DE, Hanash SM, Taylor JM. et al. Novel candidate targets of beta-catenin/T-cell factor signaling identified by gene expression profiling of ovarian endometrioid adenocarcinomas. Cancer Res. 2003;63:2913–2922. [PubMed] [Google Scholar]
- Orsulic S, Huber O, Aberle H, Arnold S, Kemler R. E-cadherin binding prevents beta-catenin nuclear localization and beta-catenin/LEF-1-mediated transactivation. J Cell Sci. 1999;112(Pt 8):1237–1245. doi: 10.1242/jcs.112.8.1237. [DOI] [PubMed] [Google Scholar]
- Huang J, Xiao D, Li G, Ma J, Chen P, Yuan W, Hou F, Ge J, Zhong M, Tang Y. EphA2 promotes epithelial-mesenchymal transition through the Wnt/β-catenin pathway in gastric cancer cells. Oncogene. 2013;10:1–11. doi: 10.1038/onc.2013.238. [DOI] [PubMed] [Google Scholar]
- Howard S, Deroo T, Fujita Y, Itasaki N. A positive role of cadherin in Wnt/β-catenin signalling during epithelial-mesenchymal transition. PLoS One. 2011;6:e23899. doi: 10.1371/journal.pone.0023899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li VS, Ng SS, Boersema PJ, Low TY, Karthaus WR, Gerlach JP, Mohammed S, Heck AJ, Maurice MM, Mahmoudi T. et al. Wnt signaling through inhibition of β-catenin degradation in an intact Axin1 complex. Cell. 2012;149:1245–1256. doi: 10.1016/j.cell.2012.05.002. [DOI] [PubMed] [Google Scholar]
- Valenta T, Hausmann G, Basler K. The many faces and functions of β-catenin. EMBO J. 2013;31:2714–2736. doi: 10.1038/emboj.2012.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ougolkov AV, Yamashita K, Mai M, Minamoto T. Oncogenic beta-catenin and MMP-7 (matrilysin) cosegregate in late-stage clinical colon cancer. Gastroenterology. 2002;122:60–71. doi: 10.1053/gast.2002.30306. [DOI] [PubMed] [Google Scholar]
- Pajouh MS, Nagle RB, Breathnach R, Finch JS, Brawer MK, Bowden GT. Expression of metalloproteinase genes in human prostate cancer. J Cancer Res Clin Oncol. 1991;117:144–150. doi: 10.1007/BF01613138. [DOI] [PubMed] [Google Scholar]
- Yoshimoto M, Itoh F, Yamamoto H, Hinoda Y, Imai K, Yachi A. Expression of MMP-7 (PUMP-1) mRNA in human colorectal cancers. Int J Cancer. 1993;54:614–618. doi: 10.1002/ijc.2910540415. [DOI] [PubMed] [Google Scholar]
- Adachi Y, Itoh F, Yamamoto H, Matsuno K, Arimura Y, Kusano M, Endoh T, Hinoda Y, Oohara M, Hosokawa M. et al. Matrix metalloproteinase matrilysin (MMP-7) participates in the progression of human gastric and esophageal cancers. Int J Oncol. 1998;13:1031–1035. doi: 10.3892/ijo.13.5.1031. [DOI] [PubMed] [Google Scholar]
- Heppner KJ, Matrisian LM, Jensen RA, Rodgers WH. Expression of most matrix metalloproteinase family members in breast cancer represents a tumor-induced host response. Am J Pathol. 1996;149:273–282. [PMC free article] [PubMed] [Google Scholar]
- Samukange V, Yasukawa K, Inouye K. Interaction of 8-anilinonaphthalene 1-sulphonate (ANS) and human matrix metalloproteinase 7 (MMP-7) as examined by MMP-7 activity and ANS fluorescence. J Biochem. 2012;151:533–540. doi: 10.1093/jb/mvs025. [DOI] [PubMed] [Google Scholar]
- Chuang HC, Su CY, Huang HY, Huang CC, Chien CY, Du YY, Chuang JH. Active matrix metalloproteinase-7 is associated with invasion in buccal squamous cell carcinoma. Mod Pathol. 2008;21:1444–1450. doi: 10.1038/modpathol.2008.99. [DOI] [PubMed] [Google Scholar]
- Endo Y, Uzawa K, Mochida Y, Shiiba M, Bukawa H, Yokoe H, Tanzawa H. Sarcoendoplasmic reticulum Ca(2+) ATPase type 2 downregulated in human oral squamous cell carcinoma. Int J Cancer. 2004;110:225–231. doi: 10.1002/ijc.20118. [DOI] [PubMed] [Google Scholar]
- Kasamatsu A, Uzawa K, Nakashima D, Koike H, Shiiba M, Bukawa H, Yokoe H, Tanzawa H. Galectin-9 as a regulator of cellular adhesion in human oral squamous cell carcinoma cell lines. Int J Mol Med. 2005;16:269–273. [PubMed] [Google Scholar]
- Iyoda M, Kasamatsu A, Ishigami T, Nakashima D, Endo-Sakamoto Y, Ogawara K, Shiiba M, Tanzawa H, Uzawa K. Epithelial cell transforming sequence 2 in human oral cancer. PLoS One. 2010;5:e14082. doi: 10.1371/journal.pone.0014082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzu Y, Uzawa K, Koike H, Saito K, Nakashima D, Higo M, Endo Y, Kasamatsu A, Shiiba M, Bukawa H. et al. Overexpression of stathmin in oral squamous-cell carcinoma: correlation with tumour progression and poor prognosis. Br J Cancer. 2006;94:717–723. doi: 10.1038/sj.bjc.6602991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogoshi K, Kasamatsu A, Iyoda M, Sakuma K, Yamatoji M, Sakamoto Y, Ogawara K, Shiiba M, Tanzawa H, Uzawa K. Dickkopf-1 in human oral cancer. Int J Oncol. 2011;39:329–336. doi: 10.3892/ijo.2011.1046. [DOI] [PubMed] [Google Scholar]
- Crick EW, Osorio I, Bhavaraju NC, Linz TH, Lunte CE. An investigation into the pharmacokinetics of 3-mercaptopropionic acid and development of a steady-state chemical seizure model using in vivo microdialysis and electrophysiological monitoring. Epilepsy Res. 2007;74:116–125. doi: 10.1016/j.eplepsyres.2007.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netopilová M, Drsata J, Kubová H, Mares P. Differences between immature and adult rats in brain glutamate decarboxylase inhibition by 3-mercaptopropionic acid. Epilepsy Res. 1995;20:179–184. doi: 10.1016/0920-1211(94)00068-8. [DOI] [PubMed] [Google Scholar]
- Tunnicliff G. Action of inhibitors on brain glutamate decarboxylase. Int J Biochem. 1990;22:1235–1241. doi: 10.1016/0020-711X(90)90304-L. [DOI] [PubMed] [Google Scholar]
- Erdö SL. Postmortem increase of GABA levels in peripheral rat tissues: prevention by 3-mercapto-propionic acid. J Neural Transm. 1984;60:303–314. doi: 10.1007/BF01249102. [DOI] [PubMed] [Google Scholar]
- Yamamoto A, Kasamatsu A, Ishige S, Koike K, Saito K, Kouzu Y, Koike H, Sakamoto Y, Ogawara K, Shiiba M. et al. Exocyst complex component Sec8: a presumed component in the progression of human oral squamous-cell carcinoma by secretion of matrix metalloproteinases. J Cancer Res Clin Oncol. 2013;139:533–542. doi: 10.1007/s00432-012-1356-2. [DOI] [PubMed] [Google Scholar]
- Ho BY, Wu YM, Chang KJ, Pan TM. Dimerumic acid inhibits SW620 cell invasion by attenuating H2O2-mediated MMP-7 expression via JNK/C-Jun and ERK/C-Fos activation in an AP-1-dependent manner. Int J Biol Sci. 2011;7:869–880. doi: 10.7150/ijbs.7.869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snoek-van Beurden PA, Von den Hoff JW. Zymographic techniques for the analysis of matrix metalloproteinases and their inhibitors. Biotechniques. 2005;38:73–83. doi: 10.2144/05381RV01. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Park SY, Park OJ, Kim YM. Curcumin suppresses migration and proliferation of Hep3B hepatocarcinoma cells through inhibition of the Wnt signaling pathway. Mol Med Rep. 2013;8:282–286. doi: 10.3892/mmr.2013.1497. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Shimura T, Yajima T, Kubo N, Araki K. Transient gene silencing of galectin-3 suppresses pancreatic cancer cell migration and invasion through degradation of β-catenin. Int J Cancer. 2011;129:2775–2786. doi: 10.1002/ijc.25946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zik M, Arazi T, Snedden WA, Fromm H. Two isoforms of glutamate decarboxylase in Arabidopsis are regulated by calcium/calmodulin and differ in organ distribution. Plant Mol Biol. 1998;37:967–975. doi: 10.1023/A:1006047623263. [DOI] [PubMed] [Google Scholar]
- Meyre D, Boutin P, Tounian A, Deweirder M, Aout M, Jouret B, Heude B, Weill J, Tauber M, Tounian P. et al. Is glutamate decarboxylase 2 (GAD2) a genetic link between low birth weight and subsequent development of obesity in children? J Clin Endocrinol Metab. 2005;90:2384–2390. doi: 10.1210/jc.2004-1468. [DOI] [PubMed] [Google Scholar]
- Maddox DM, Condie BG. Dynamic expression of a glutamate decarboxylase gene in multiple non-neural tissues during mouse development. BMC Dev Biol. 2001;1:1. doi: 10.1186/1471-213X-1-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maemura K, Shiraishi N, Sakagami K, Kawakami K, Inoue T, Murano M, Watanabe M, Otsuki Y. Proliferative effects of gamma-aminobutyric acid on the gastric cancer cell line are associated with extracellular signal-regulated kinase 1/2 activation. J Gastroenterol Hepatol. 2009;24:688–696. doi: 10.1111/j.1440-1746.2008.05687.x. [DOI] [PubMed] [Google Scholar]
- Yamano Y, Uzawa K, Shinozuka K, Fushimi K, Ishigami T, Nomura H, Ogawara K, Shiiba M, Yokoe H, Tanzawa H. Hyaluronan-mediated motility: a target in oral squamous cell carcinoma. Int J Oncol. 2008;32:1001–1009. [PubMed] [Google Scholar]
- Kruck S, Eyrich C, Scharpf M, Sievert KD, Fend F, Stenzl A, Bedke J. Impact of an altered Wnt1/β-catenin expression on clinicopathology and prognosis in clear cell renal cell carcinoma. Int J Mol Sci. 2013;14:10944–10957. doi: 10.3390/ijms140610944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada G, Ueo H, Matsumura T, Uchi R, Ishibashi M, Mima K, Kurashige J, Takahashi Y, Akiyoshi S, Sudo T. et al. CHD8 is an independent prognostic indicator that regulates Wnt/β-catenin signaling and the cell cycle in gastric cancer. Oncol Rep. 2013;30:1137–1142. doi: 10.3892/or.2013.2597. [DOI] [PubMed] [Google Scholar]
- Abd El-Rehim D, Ali MM. Aberrant expression of β-catenin in invasive ductal breast carcinomas. J Egypt Natl Canc Inst. 2009;21:185–195. [PubMed] [Google Scholar]
- Usongo M, Li X, Farookhi R. Activation of the canonical WNT signaling pathway promotes ovarian surface epithelial proliferation without inducing β-catenin/Tcf-mediated reporter expression. Dev Dyn. 2013;242:291–300. doi: 10.1002/dvdy.23919. [DOI] [PubMed] [Google Scholar]
- Chen HJ, Hsu LS, Shia YT, Lin MW, Lin CM. The β-catenin/TCF complex as a novel target of resveratrol in the Wnt/β-catenin signaling pathway. Biochem Pharmacol. 2012;84:1143–1153. doi: 10.1016/j.bcp.2012.08.011. [DOI] [PubMed] [Google Scholar]
- Wang K, Li N, Yeung CH, Li JY, Wang HY, Cooper TG. Oncogenic Wnt/β-catenin signalling pathways in the cancer-resistant epididymis have implications for cancer research. Mol Hum Reprod. 2013;19:57–71. doi: 10.1093/molehr/gas051. [DOI] [PubMed] [Google Scholar]
- Lustig B, Jerchow B, Sachs M, Weiler S, Pietsch T, Karsten U, van de Wetering M, Clevers H, Schlag PM, Birchmeier W. et al. Negative feedback loop of Wnt signaling through upregulation of conductin/axin2 in colorectal and liver tumors. Mol Cell Biol. 2002;22:1184–1193. doi: 10.1128/MCB.22.4.1184-1193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima T, Akaike M, Yoshihara K, Shiozawa M, Yamamoto N, Sato T, Yamada R, Fujii S, Rino Y, Kunisaki C. et al. Clinicopathological significance of the gene expression of matrix metalloproteinase-7, insulin-like growth factor-1, insulin-like growth factor-2 and insulin-like growth factor-1 receptor in patients with colorectal cancer: insulin-like growth factor-1 receptor gene expression is a useful predictor of liver metastasis from colorectal cancer. Oncol Rep. 2008;20:359–364. [PubMed] [Google Scholar]
- Yamamoto H, Adachi Y, Itoh F, Iku S, Matsuno K, Kusano M, Arimura Y, Endo T, Hinoda Y, Hosokawa M. et al. Association of matrilysin expression with recurrence and poor prognosis in human esophageal squamous cell carcinoma. Cancer Res. 1999;59:3313–3316. [PubMed] [Google Scholar]
- Mimori K, Yamashita K, Ohta M, Yoshinaga K, Ishikawa K, Ishii H, Utsunomiya T, Barnard GF, Inoue H, Mori M. Coexpression of matrix metalloproteinase-7 (MMP-7) and epidermal growth factor (EGF) receptor in colorectal cancer: an EGF receptor tyrosine kinase inhibitor is effective against MMP-7-expressing cancer cells. Clin Cancer Res. 2004;10:8243–8249. doi: 10.1158/1078-0432.CCR-04-0849. [DOI] [PubMed] [Google Scholar]
- Fanelli MF, Chinen LT, Begnami MD. The influence of transforming growth factor-α, cyclooxygenase-2, matrix metalloproteinase (MMP)-7, MMP-9 and CXCR4 proteins involved in epithelial-mesenchymal transition on overall survival of patients with gastric cancer. Histopathology. 2012;61:153–161. doi: 10.1111/j.1365-2559.2011.04139.x. [DOI] [PubMed] [Google Scholar]
- Barros SS, Henriques Á, Pereira KM, de Medeiros AM, Galvão HC, Freitas RA. Immunohistochemical expression of matrix metalloproteinases in squamous cell carcinoma of the tongue and lower lip. Arch Oral Biol. 2011;56:752–760. doi: 10.1016/j.archoralbio.2010.11.022. [DOI] [PubMed] [Google Scholar]