Abstract
Most eukaryotic messenger RNA precursors (pre-mRNAs) undergo extensive maturational processing, including 3’-end cleavage and polyadenylation1–8. Despite the characterization of a large number of proteins that are required for the cleavage reaction, the identity of the endoribonuclease is not known4,9,10. Recent analyses suggested that the 73 kD subunit of cleavage and polyadenylation specificity factor (CPSF-73) may be the endonuclease for this and related reactions10–15, although no direct data confirmed this. Here we report the crystal structures of human CPSF-73 at 2.1 Å resolution, complexed with zinc ions and a sulfate that may mimic the phosphate group of the substrate, and the related yeast protein CPSF-100 (Ydh1p) at 2.5 Å resolution. Both CPSF-73 and CPSF-100 contain two domains, a metallo-β-lactamase domain and a novel β-CASP domain. The active site of CPSF-73, with two zinc ions, is located at the interface of the two domains. Purified recombinant CPSF-73 possesses endoribonuclease activity, and mutations that disrupt zinc binding in the active site abolish this activity. Our studies provide the first direct experimental evidence that CPSF-73 is the pre-mRNA 3’-end processing endonuclease.
Keywords: polyadenylation, metallo-β-lactamase, pre-mRNA processing, Artemis, V(D)J recombination, double-strand break repair
CPSF-73 belongs to the metallo-β-lactamase superfamily of zinc-dependent hydrolases11,12. Canonical metallo-β-lactamases contain five signature sequence motifs—Asp (motif 1), His-X-His-X-Asp-His (motif 2), His (motif 3), Asp (motif 4) and His (motif 5), most of which are ligands to the two zinc ions in their active site. Sequence conservation between CPSF-73 and the canonical metallo-β-lactamases is limited to these signature motifs. While the first four motifs can be identified in the N-terminal segment of CPSF-73 (Supplemental Fig. 1a, Supplemental Table 1), the fifth motif was uncertain, with three candidates, A (Asp or Glu), B (His), and C (His) (Supplemental Fig. 1a), in the so-called β-CASP motif12. Motif B was proposed to be equivalent to motif 5 in the canonical metallo-β-lactamases. Another subunit of CPSF, CPSF-100, shares sequence conservation (Supplemental Fig. 1b) and a similar domain architecture (Supplemental Fig. 1a) with CPSF-73 but lacks the putative Zn2+ binding residues.
To understand the roles of CPSF-73 and CPSF-100 in pre-mRNA 3’-end processing, we determined the structures of human CPSF-73 (residues 1–460), and yeast CPSF-100 (residues 1–720) (the crystallographic data are summarized in Supplemental Table 2). The two structures obtained for CPSF-73 were crystallized in the absence or presence of 0.5 mM zinc (although both structures contained zinc atoms; see below). We discovered serendipitously that in situ proteolysis by a fungal protease is crucial for the crystallization of yeast CPSF-10016.
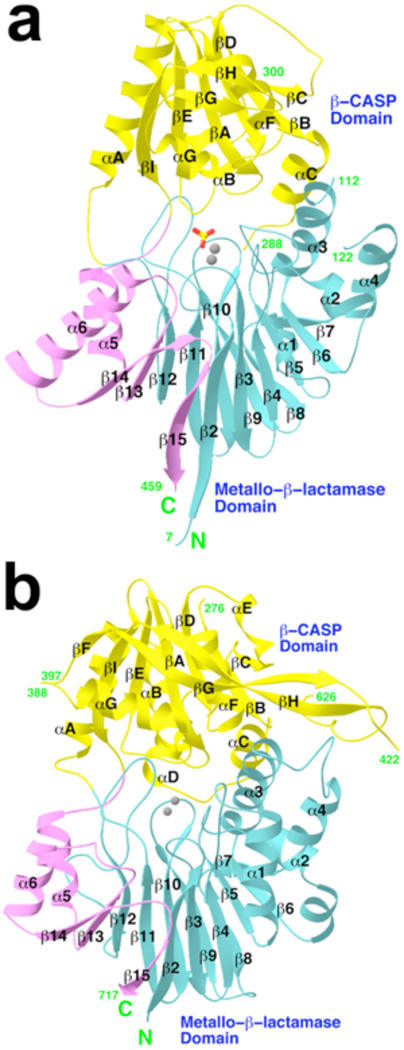
The structure of CPSF-73 can be divided into two domains (Fig. 1a). The N-terminal residues (amino acids 1–208) form a domain similar to the structure of canonical metallo-β-lactamases, with a four-layered αβ/βα architecture (Supplemental Fig. 2a). A close structural homolog of this domain is the L1 metallo-β-lactamase from S. maltophilia (Supplemental Fig. 2c)17, with a rms distance of 3.3 Å for 167 equivalent Cα atoms, calculated using the program Dali18. The sequence identity for these structurally equivalent residues is only 17%, however.
Figure 1. Structures of human CPSF-73 and yeast CPSF-100 (Ydh1p).
a, Schematic representation of the structure of human CPSF-73. The β-strands and α-helices are labeled, and the two zinc atoms in the active site are shown as gray spheres. The sulfate ion is shown as a stick model. b, Schematic representation of the structure of yeast CPSF-100. The zinc atoms in the CPSF-73 structure are shown for reference.
Despite the overall similarity to canonical metallo-β-lactamases, the structure of this domain of CPSF-73 contains several distinctive features. Most remarkably, residues 395–460, at the C-terminal end of the expression construct (Supplemental Fig. 1a), are also part of the metallo-β-lactamase domain (Fig. 1a). These residues add three strands (β13 through β15) to the central β-sandwich, and the last residue of the structure is located directly next to the first residue (Supplemental Fig. 2a). These additional β-strands in the metallo-β-lactamase fold are also observed in the structure of CPSF-100 (Fig. 1b, Supplemental Fig. 2b, see below) and RNase Z (Supplemental Fig. 2d)19,20, suggesting that they may be unique to RNA/DNA processing nucleases. This feature of the metallo-β-lactamase fold is crucial for the activity of these proteins, as the loop after strand β13 provides one of the ligands (motif C) to the zinc ions in the active site (Supplemental Fig. 2a, see below). The rms distance for 210 equivalent Cα atoms between CPSF-73 and RNase Z is 2.6 Å, but the sequence identity is only 18%. RNase Z is also distinct from CPSF-73 in that it lacks the second domain (Fig. 1a, Supplemental Fig. 2d).
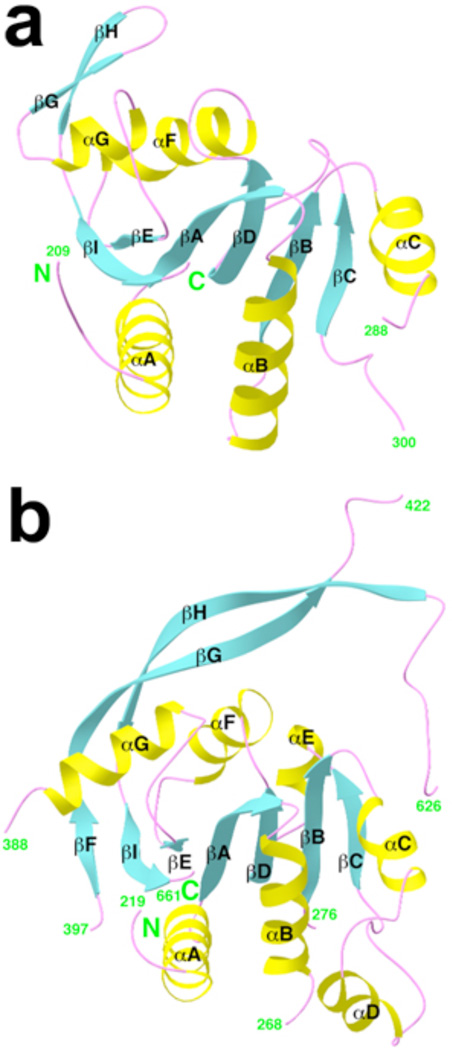
The second domain of CPSF-73 covers residues 209–394 (Supplemental Fig. 1a), and contains a central fully parallel six-stranded β-sheet that is surrounded by five α-helices on both faces (Fig. 2a). In addition, a small β-hairpin structure is on the surface of the domain. This second domain can be considered as a cassette inserted into the metallo-β-lactamase domain, between strand β12 and helix α5 (Fig. 1a). As it is related to the β-CASP motif identified earlier based on sequence analyses12, we have named it the β-CASP domain (Supplemental Fig. 1a).
Figure 2. The β-CASP domain of CPSF-73 and CPSF-100.
Schematic drawing of the β-CASP domain of human CPSF-73 (a) and yeast CPSF-100 (b).
The closest structural homolog of the β-CASP domain is the nucleotide binding fold (NBF, Supplemental Fig. 3), with a Z score of 6 from Dali18. However, the β-CASP domain lacks the Walker A motif for binding nucleotide phosphate groups (Supplemental Results and Supplemental Fig. 3)21. Therefore, the β-CASP domain appears to be a novel example of the NBF superfold but is unlikely to bind nucleotides. This domain is highly conserved among CPSF-73 proteins, and likely has important functions in regulating their activity (see below).
The overall structure of yeast CPSF-100 (Fig. 1b) is remarkably similar to that of human CPSF-73, despite the low degree of sequence conservation between them (Supplemental Fig. 1b). The rms distance for 235 equivalent Cα atoms in the metallo-β-lactamase domain (Supplemental Fig. 2b) of the two structures is 2.2 Å (with 21% sequence identity), and that for 154 equivalent Cα atoms in the β-CASP domain is 2.3 Å (with 17% identity).
There are, however, significant differences in the structures of CPSF-100 and CPSF-73 (Supplemental Fig. 4). Most importantly, motifs for zinc binding in the metallo-β-lactamase domain are missing in CPSF-100 (Supplemental Fig. 2b), and therefore this protein cannot bind zinc and is unlikely to possess nuclease activity. In addition, the β-CASP domain of CPSF-100 (Fig. 2b) is much larger than that of CPSF-73, but contains a highly flexible segment (Supplemental Results and Supplemental Fig. 5).
Clear electron density for two zinc ions (confirmed based on the anomalous scattering data) was observed in the CPSF-73 active site (Supplemental Fig. 6a), regardless of whether zinc was present in the crystallization solution. Even in the absence of added zinc, the zinc ions in the structure appear to have full occupancy as their temperature factor values are comparable to those of their protein ligands. This suggests that CPSF-73’s active site possesses extremely high affinity for zinc ions, making it impossible to remove the zinc from the protein, consistent with the known resistance of 3’ cleavage to relatively high concentrations of EDTA22. Two additional zinc ions were observed on the surface of CPSF-73 in the crystal grown in the presence of 0.5 mM zinc (Supplemental Results and Supplemental Fig. 7).
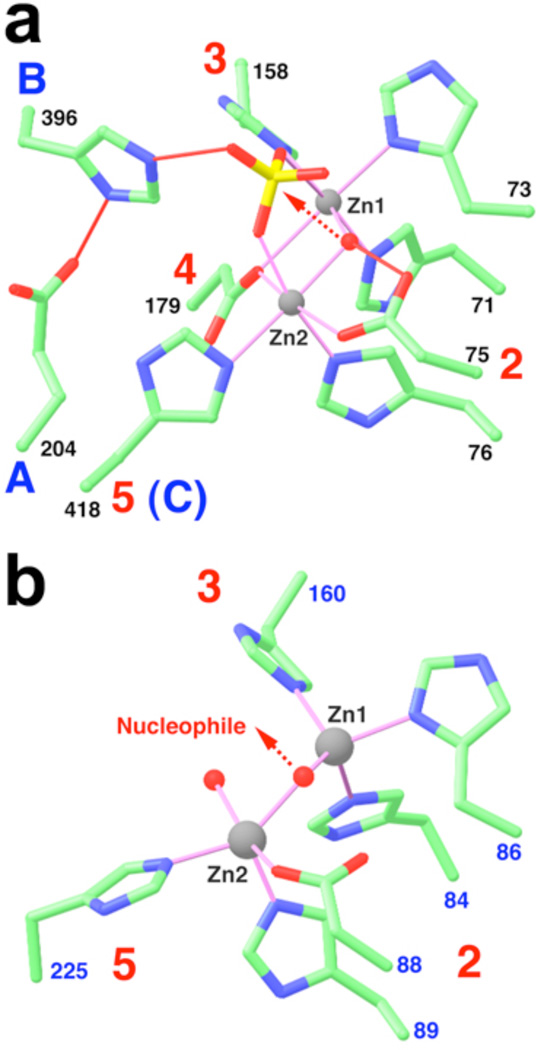
The two zinc atoms in the active site are each bound in an octahedral environment, with a hydroxide ion as one of the bridging ligands (Fig. 3a, Supplemental Fig. 6b). The other bridging ligand is the side chain carboxylate oxygen atom of Asp179 from motif 4 (Fig. 3a). His71, His73 (motif 2) and His158 (motif 3) provide three more ligands to the first zinc atom (Zn1), while Asp75, His76 (motif 2) and His418 (motif C, see below) are the ligands to Zn2 (Fig. 3a). Finally, the two zinc ions are coordinated by oxygen atoms from a sulfate ion (Fig. 3a), which was present at 300 mM concentration in the crystallization solution and has clear electron density (Supplemental Fig. 6a).
Figure 3. The active site of CPSF-73.
a, The zinc binding site for human CPSF-73. The motifs are labeled, and the bridging hydroxide ion is shown as a red sphere. Liganding interactions are indicated in thin magenta lines, and hydrogen-bonding interactions in thin red lines. The arrow indicates the nucleophilic attack from the hydroxide ion. b, The zinc binding site in L1 metallo-β-lactamase17.
The structure of the active site of CPSF-73 strongly implies that it possesses hydrolase/nuclease activity. In canonical metallo-β-lactamases, the hydroxide that bridges the two zinc ions is the nucleophile for the hydrolysis reaction, and the substrate is directly liganded to the zinc atoms23. In our structure, the hydroxide is placed directly below the sulfate group (Fig. 3a), which is likely a good mimic for the phosphate group of the pre-mRNA substrate at the cleavage site. Therefore, the hydroxide is located at the perfect position for an inline nucleophilic attack on the phosphate group to initiate the nuclease reaction (Fig. 3a).
Our analysis suggests that His396 (motif B, in the linker between the β-CASP domain and the metallo-β-lactamase domain, Supplemental Fig. 2a) is the general acid for catalysis. This residue is hydrogen-bonded to the side chain of Glu204 (motif A, last residue of strand β12 in the metallo-β-lactamase domain, Supplemental Fig. 2a) as well as an oxygen atom of the sulfate (Fig. 3a). This oxygen is located opposite from the direction of attack by the hydroxide (Fig. 3a), and therefore likely represents the leaving group in the hydrolysis reaction. After hydrolysis, the His396 side chain can function as the general acid to protonate the oxyanion in the reaction product. This pair of Glu-His residues is also observed in the structure of RNase Z, hydrogen-bonded to a phosphate group in the active site19. In yeast, mutation of either of these residues causes lethality13.
Although an earlier sequence analysis suggested that motif B was the best candidate for the zinc ligand in CPSF-7312, our structure shows that the ligand is the His residue from motif C. This residue is conserved among CPSF-73 proteins and its close homologs (RC-68 or Int11)24,25, but interestingly not in the DNA nuclease Artemis (Supplemental Fig. 1b)12,26. Therefore, the Zn2 atom may have a different coordination sphere in Artemis. The structure-based sequence alignment also indicates a change in the assignment of residues equivalent to motifs B and C in CPSF-100 (Supplemental Fig. 1b)12, although neither residue is His in CPSF-100.
The binding modes of the zinc ions in CPSF-73 (Fig. 3a) are similar to that in RNase Z but distinct from those in canonical metallo-β-lactamases (Fig. 3b, Supplemental Results). Another important difference between CPSF-73 and other metallo-β-lactamases is the presence of the β-CASP domain. While most metallo-β-lactamases (Supplemental Fig. 2c), and RNase Z (Supplemental Fig. 2d), have an open zinc binding site, the active site in CPSF-73 is located deep in the interface between the metallo-β-lactamase and the β-CASP domains (Fig. 1a). In fact, the β-CASP domain severely restricts access to the active site (Supplemental Fig. 8), and the scissile phosphate group probably cannot reach the zinc ions in the current structure. This suggests that a conformational change in the enzyme may be required for pre-mRNA binding (Supplemental Results).
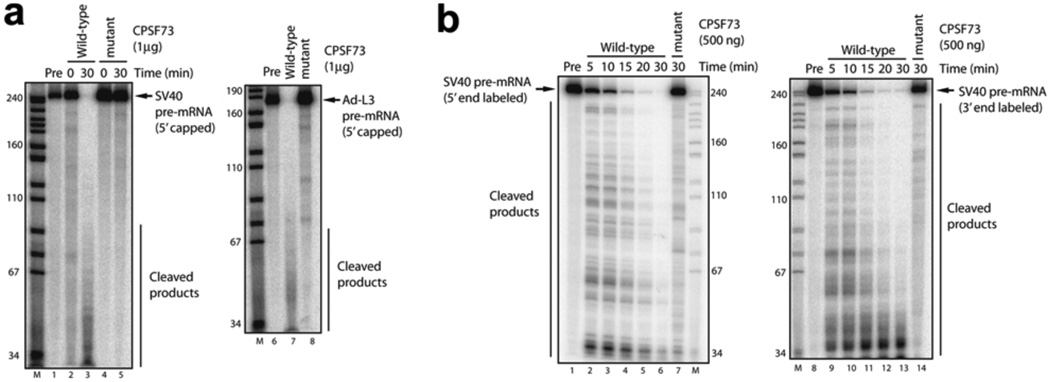
To demonstrate CPSF-73 endonuclease activity in vitro and to provide direct evidence for its role in 3’-end processing, we assessed the nuclease activity of the bacterially expressed CPSF-73, together with a derivative containing a double mutation (D75K/H76A) that destroys two of the zinc ligands in motif 2. We used a 5’ capped, uniformly labeled RNA containing the SV40 late polyadenylation site (SVL) as the substrate13,27. Using standard conditions for 3’ processing reactions, there was evidence for weak cleavage with the wild-type but not the mutant derivative (Supplemental Fig. 10a); a similarly prepared CPSF-100 derivative was also inactive (results not shown). This induced us to attempt to optimize conditions to obtain more efficient cleavage.
Among the possibilities tested to increase nuclease activity was preincubation with divalent cations. Our structural studies showed that there are binding sites for additional Zn2+ on the surface (Supplemental Fig. 7), and it was possible that (non-specific) binding of divalent cations to the protein surface could affect its activity. Preincubation with Zn2+ and all other divalent cations tested, except Ca2+, resulted in precipitation of the enzyme. Strikingly, after preincubation with Ca2+, the wild-type enzyme degraded the SVL substrate almost entirely to oligonucleotides, while the mutant was still inactive (Fig. 4a). A similar-sized RNA from adenovirus was also entirely degraded by wild-type but not mutant CPSF-73 (Fig. 4a). Preincubation with Ca2+, while required for enzyme activation (results not shown), was unnecessary for catalysis. It was diluted to 50 µM in the actual reactions and nuclease activity was not sensitive to the addition of 5 mM EGTA (results not shown). Furthermore, there is no evidence for a Ca2+ requirement in authentic 3’ processing, suggesting that its activating function is likely performed by other components of the polyadenylation machinery. At lower CPSF-73 enzyme levels, partially degraded, higher molecular weight intermediates were seen (Supplemental Fig. 10b). Significant cleavage required relatively high concentrations (10 ng/µl, or ~200 nM), which we attribute to only a small fraction of the enzyme being activated by the preincubation.
Figure 4. CPSF-73 possesses endoribonuclease activity.
a, Cleavage of 5’ capped SV40 late pre-mRNA (SVL) was performed with wild-type (lane 3) or mutant (lane 5) CPSF-73 at 30 °C for 30 min. 0 min incubations are shown in lanes 2 and 4. Cleavage of 5’ capped adenovirus L3 pre-mRNA (Ad-L3) is shown in lanes 7 and 8. Precursor RNAs (Pre) are shown in lanes 1 and 6. M, MspI-digested pBR322DNA markers. b, Time course analysis of RNA cleavage by CPSF-73. Cleavage of 5’ (lanes 2–6) and 3’ (lanes 9–13) end-labeled SVL pre-mRNAs was performed with wild-type CPSF-73 (500 ng); time points were 5, 10, 15, 20, 30 min. Mutant CPSF-73 (500 ng) did not exhibit ribonuclease activity (lanes 7 and 14).
While our data are consistent with purified CPSF-73 possessing non-specific endoribonuclease activity, they do not conclusively rule out that it may have exonuclease activity, as previously suggested14. Indeed, evidence has been presented that Artemis has exonuclease activity28. Although the presence of a 5’ cap on the RNAs analyzed above largely excludes 5’→3 exonuclease activity, we nonetheless compared the ability of CPSF-73 to degrade 5’ and 3’ end-labeled RNA (Fig. 4b). The two RNAs were degraded with comparable kinetics and generated similar patterns of intermediates, consistent only with endoribonuclease activity.
These results, together with previous data13, 14, establish CPSF-73 as a strong, sequence-independent endoribonuclease that functions with specificity in 3’-end processing of both polyadenylated and histone pre-mRNAs by interaction with other factors. In addition, our data strongly suggest that the close sequence homolog of CPSF-73, RC-68 or Int11, is the endoribonuclease in the 3’-end processing of small nuclear RNAs24.
Methods
Protein expression, purification and crystallization
Residues 1–460 of human CPSF-73 were sub-cloned into the pET28a vector (Novagen) and overexpressed in E. coli at 20 °C. The soluble protein was purified by nickel-agarose affinity chromatography and gel-filtration chromatography. Yeast CPSF-100 (YDH1p, residues 1–720) was expressed and purified by nickel-agarose affinity chromatography, anion exchange and gel-filtration chromatography.
Crystals of human CPSF-73 were obtained at room temperature by the sitting-drop vapor diffusion method. In an attempt to increase the occupancy of zinc ions, some crystals were grown from a reservoir solution that also contained 0.5 mM ZnCl2. Crystals of yeast CPSF-100 were obtained at 4 °C by the same protocol. The crystals were frozen in liquid propane for data collection at 100 K.
Data collection, structure determination and refinement
X-ray diffraction data were collected on an ADSC CCD at the X4A beamline of Brookhaven National Laboratory. The diffraction data were collected at the zinc absorption peak for crystals of CPSF-73, and at the gold absorption peak for a KAu(CN)2 derivative of CPSF-100. The structure of CPSF-73 was determined by the single-wavelength anomalous diffraction method29, using the anomalous signal of zinc. The structure of CPSF-100 was determined by the single-isomorphous replacement method, supplemented with anomalous diffraction. The crystallographic information are summarized in Supplemental Table 2. Figures 1–3 were produced with Ribbons30.
Pre-mRNA 3’-end cleavage assay
CPSF-73 was pre-incubated with 5 mM CaCl2 at 37 °C for 30 min. Cleavage assays were carried out in reaction mixture (10 µl) containing ~1 ng labeled RNA substrates, 10 mM HEPES (pH 7.9), 10% (v/v) glycerol, 50 mM KCl, 0.25 mM DTT, 0.25 mM PMSF, 0.125 mM EDTA, 50 µM CaCl2, RNase inhibitor (2 unit), 500 ng BSA and indicated amounts of recombinant CPSF-73. Cleaved RNAs were isolated and fractionated on 6% urea PAGE. The data were analyzed by Phospho-Imager.
Supplementary Material
Acknowledgements
We thank K. Ryan for discussions; B. Tweel for characterizing the fungus; R. Abramowitz, J. Schwanof and X. Yang for setting up the X4A beamline at the NSLS; J. Khan and Y. Shen for help with data collection at the synchrotron. This research is supported in part by grants from the National Institutes of Health.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
The atomic coordinates have been deposited at the Protein Data Bank (accession numbers 2I7T, 2I7V, and 2I7X).
The authors declare no competing financial interests.
References
- 1.Colgan DF, Manley JL. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 1997;11:2755–2766. doi: 10.1101/gad.11.21.2755. [DOI] [PubMed] [Google Scholar]
- 2.Wahle E, Ruegsegger U. 3'-end processing of pre-mRNA in eukaryotes. FEMS Microbiol. Rev. 1999;23:277–295. doi: 10.1111/j.1574-6976.1999.tb00400.x. [DOI] [PubMed] [Google Scholar]
- 3.Zhao J, Hyman L, Moore CL. Formation of mRNA 3' ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev. 1999;63:405–445. doi: 10.1128/mmbr.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Proudfoot NJ. New perspectives on connecting messenger RNA 3' end formation to transcription. Curr. Opin. Cell Biol. 2004;16:272–278. doi: 10.1016/j.ceb.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 5.Zorio DAR, Bentley D. The link between mRNA processing and transcription: communication works both ways. Exp. Cell Res. 2004;296:91–97. doi: 10.1016/j.yexcr.2004.03.019. [DOI] [PubMed] [Google Scholar]
- 6.Wilusz CJ, Wormington M, Peltz SW. The cap-to-tail guide to mRNA turnover. Nature Rev. Mol. Cell Biol. 2001;2:237–246. doi: 10.1038/35067025. [DOI] [PubMed] [Google Scholar]
- 7.Calvo O, Manley JL. Strange bedfellows: polyadenylation factors at the promoter. Genes Dev. 2003;17:1321–1327. doi: 10.1101/gad.1093603. [DOI] [PubMed] [Google Scholar]
- 8.Vinciguerra P, Stutz F. mRNA export: an assembly line from genes to nuclear pores. Curr. Opin. Cell Biol. 2004;16:285–292. doi: 10.1016/j.ceb.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 9.Shatkin AJ, Manley JL. The ends of the affair: capping and polyadenylation. Nature Struct. Biol. 2000;7:838–842. doi: 10.1038/79583. [DOI] [PubMed] [Google Scholar]
- 10.Wickens M, Gonzalez TN. Knives, accomplices, and RNA. Science. 2004;306:1299–1300. doi: 10.1126/science.1100137. [DOI] [PubMed] [Google Scholar]
- 11.Daiyasu H, Osaka K, Ishino Y, Toh H. Expansion of the zinc metallohydrolase family of the β-lactamase fold. FEBS Lett. 2001;503:1–6. doi: 10.1016/s0014-5793(01)02686-2. [DOI] [PubMed] [Google Scholar]
- 12.Callebaut I, Moshous D, Mornon J-P, de Villartay J-P. Metallo-β-lactamase fold within nucleic acids processing enzymes: the β-CASP family. Nucl. Acids Res. 2002;30:3592–3601. doi: 10.1093/nar/gkf470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ryan K, Calvo O, Manley JL. Evidence that polyadenylation factor CPSF-73 is the mRNA 3' processing endonuclease. RNA. 2004;10:565–573. doi: 10.1261/rna.5214404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dominski Z, Yang X-C, Marzluff WF. The polyadenylation factor CPSF-73 is involved in histone-pre-mRNA processing. Cell. 2005;123:37–48. doi: 10.1016/j.cell.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 15.Kolev NG, Steitz JA. Symplekin and multiple other polyadenylation factors participate in 3'-end maturation of histone mRNAs. Genes Dev. 2005;19:2583–2592. doi: 10.1101/gad.1371105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mandel CR, Gebauer D, Zhang H, Tong L. A serendipitous discovery that in situ proteolysis is required for the crystallization of yeast CPSF-100 (Ydh1p) Acta Cryst. 2006;F62:1041–1045. doi: 10.1107/S1744309106038152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ullah JH, et al. The crystal structure of the L1 metallo-β-lactamase from Stenotrophomonas maltophilia at 1.7Å resolution. J. Mol. Biol. 1998;284:125–136. doi: 10.1006/jmbi.1998.2148. [DOI] [PubMed] [Google Scholar]
- 18.Holm L, Sander C. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 19.de la Sierra-Gallay IL, Pellegrini O, Condon C. Structural basis for substrate binding, cleavage and allostery in the tRNA maturase RNase Z. Nature. 2005;433:657–661. doi: 10.1038/nature03284. [DOI] [PubMed] [Google Scholar]
- 20.Ishii R, et al. Crystal structure of the tRNA 3' processing endoribonuclease tRNase Z from Thermotoga maritima. J. Biol. Chem. 2005;280:14138–14144. doi: 10.1074/jbc.M500355200. [DOI] [PubMed] [Google Scholar]
- 21.Walker JE, Saraste M, Runswick MJ, Gay NJ. Distantly related sequences in the α- and β-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hirose Y, Manley JL. Creatine phosphate, not ATP, is required for 3' end cleavage of mammalian pre-mRNA in vitro. J. Biol. Chem. 1997;272:29636–29642. doi: 10.1074/jbc.272.47.29636. [DOI] [PubMed] [Google Scholar]
- 23.Garrity JD, Bennett B, Crowder MW. Direct evidence that the reaction intermediate of metallo-β-lactamase L1 is metal bound. Biochemistry. 2005;44:1078–1087. doi: 10.1021/bi048385b. [DOI] [PubMed] [Google Scholar]
- 24.Baillat D, et al. Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repeat of RNA polymerase II. Cell. 2005;123:265–276. doi: 10.1016/j.cell.2005.08.019. [DOI] [PubMed] [Google Scholar]
- 25.Dominski Z, Yang X-C, Purdy M, Wagner EJ, Marzluff WF. A CPSF-73 homologue is required for cell cycle progression but not cell growth and interacts with a protein having features of CPSF-100. Mol. Cell. Biol. 2005;25:1489–1500. doi: 10.1128/MCB.25.4.1489-1500.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moshous D, et al. Artemis, a novel DNA double-strand break repair/V(D)J recombination protein, is mutated in human severe combined immune deficiency. Cell. 2001;105:177–186. doi: 10.1016/s0092-8674(01)00309-9. [DOI] [PubMed] [Google Scholar]
- 27.Takagaki Y, Ryner LC, Manley JL. Separation and characterization of a poly(A) polymerase and a cleavage/specificity factor required for pre-mRNA polyadenylation. Cell. 1988;52:731–742. doi: 10.1016/0092-8674(88)90411-4. [DOI] [PubMed] [Google Scholar]
- 28.Ma Y, Pannicke U, Schwarz K, Lieber MR. Hairpin opening and overhand processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell. 2002;108:781–794. doi: 10.1016/s0092-8674(02)00671-2. [DOI] [PubMed] [Google Scholar]
- 29.Hendrickson WA. Determination of macromolecular structures from anomalous diffraction of synchrotron radiation. Science. 1991;254:51–58. doi: 10.1126/science.1925561. [DOI] [PubMed] [Google Scholar]
- 30.Carson M. Ribbon models of macromolecules. J. Mol. Graphics. 1987;5:103–106. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.