Abstract
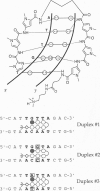
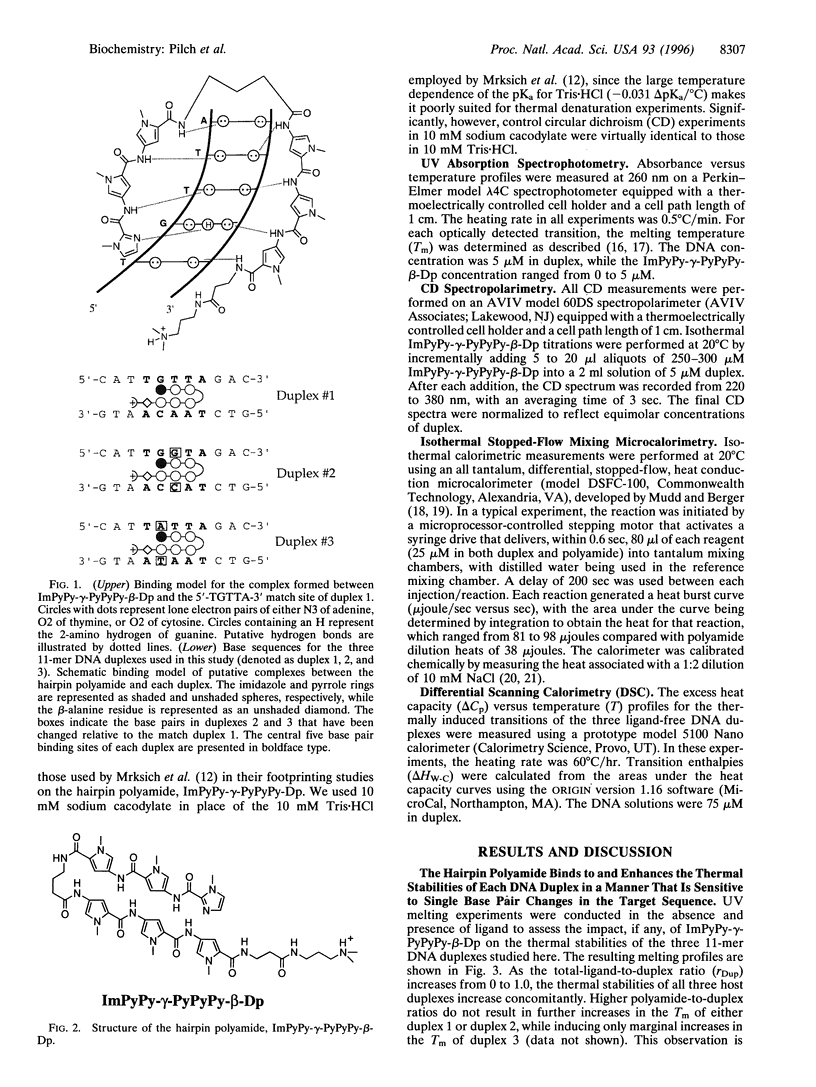
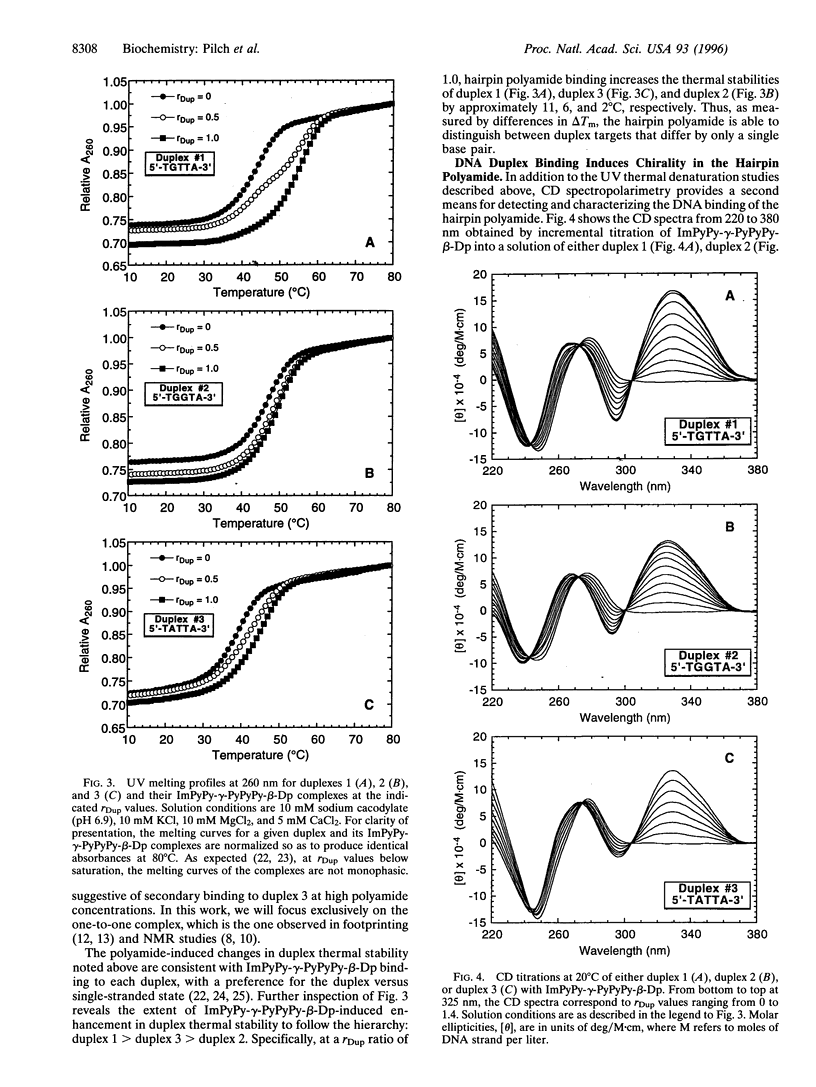
Hairpin polyamides are synthetic ligands for sequence-specific recognition in the minor groove of double-helical DNA. A thermodynamic characterization of the DNA-binding properties exhibited by a six-ring hairpin polyamide, ImPyPy-gamma-PyPyPy-beta-Dp (where Im = imidazole, Py = pyrrole, gamma = gamma-aminobutyric acid, beta = beta-alanine, and Dp = dimethylaminopropylamide), reveals an approximately 1-2 kcal/mol greater affinity for the designated match site, 5'-TGTTA-3', relative to the single base pair mismatch sites, 5'-TGGTA-3' and 5'-TATTA-3'. The enthalpy and entropy data at 20 degrees C reveal this sequence specificity to be entirely enthalpic in origin. Correlations between the thermodynamic driving forces underlying the sequence specificity exhibited by ImPyPy-gamma-PyPyPy-beta-Dp and the structural properties of the heterodimeric complex of PyPyPy and ImPyPy bound to the minor groove of DNA provide insight into the molecular forces that govern the affinity and specificity of pyrrole-imidazole polyamides.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Breslauer K. J. Extracting thermodynamic data from equilibrium melting curves for oligonucleotide order-disorder transitions. Methods Mol Biol. 1994;26:347–372. doi: 10.1007/978-1-59259-513-6_14. [DOI] [PubMed] [Google Scholar]
- Breslauer K. J., Freire E., Straume M. Calorimetry: a tool for DNA and ligand-DNA studies. Methods Enzymol. 1992;211:533–567. doi: 10.1016/0076-6879(92)11030-m. [DOI] [PubMed] [Google Scholar]
- Chalikian T. V., Plum G. E., Sarvazyan A. P., Breslauer K. J. Influence of drug binding on DNA hydration: acoustic and densimetric characterizations of netropsin binding to the poly(dAdT).poly(dAdT) and poly(dA).poly(dT) duplexes and the poly(dT).poly(dA).poly(dT) triplex at 25 degrees C. Biochemistry. 1994 Jul 26;33(29):8629–8640. doi: 10.1021/bi00195a003. [DOI] [PubMed] [Google Scholar]
- Chen X., Ramakrishnan B., Rao S. T., Sundaralingam M. Binding of two distamycin A molecules in the minor groove of an alternating B-DNA duplex. Nat Struct Biol. 1994 Mar;1(3):169–175. doi: 10.1038/nsb0394-169. [DOI] [PubMed] [Google Scholar]
- Chou W. Y., Marky L. A., Zaunczkowski D., Breslauer K. J. The thermodynamics of drug-DNA interactions: ethidium bromide and propidium iodide. J Biomol Struct Dyn. 1987 Oct;5(2):345–359. doi: 10.1080/07391102.1987.10506399. [DOI] [PubMed] [Google Scholar]
- Crothers D. M. Statistical thermodynamics of nucleic acid melting transitions with coupled binding equilibria. Biopolymers. 1971 Nov;10(11):2147–2160. doi: 10.1002/bip.360101110. [DOI] [PubMed] [Google Scholar]
- Drew H. R., Dickerson R. E. Structure of a B-DNA dodecamer. III. Geometry of hydration. J Mol Biol. 1981 Sep 25;151(3):535–556. doi: 10.1016/0022-2836(81)90009-7. [DOI] [PubMed] [Google Scholar]
- Geierstanger B. H., Jacobsen J. P., Mrksich M., Dervan P. B., Wemmer D. E. Structural and dynamic characterization of the heterodimeric and homodimeric complexes of distamycin and 1-methylimidazole-2-carboxamide-netropsin bound to the minor groove of DNA. Biochemistry. 1994 Mar 15;33(10):3055–3062. doi: 10.1021/bi00176a039. [DOI] [PubMed] [Google Scholar]
- Kopka M. L., Yoon C., Goodsell D., Pjura P., Dickerson R. E. The molecular origin of DNA-drug specificity in netropsin and distamycin. Proc Natl Acad Sci U S A. 1985 Mar;82(5):1376–1380. doi: 10.1073/pnas.82.5.1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marky L. A., Breslauer K. J. Calculating thermodynamic data for transitions of any molecularity from equilibrium melting curves. Biopolymers. 1987 Sep;26(9):1601–1620. doi: 10.1002/bip.360260911. [DOI] [PubMed] [Google Scholar]
- Marky L. A., Breslauer K. J. Origins of netropsin binding affinity and specificity: correlations of thermodynamic and structural data. Proc Natl Acad Sci U S A. 1987 Jul;84(13):4359–4363. doi: 10.1073/pnas.84.13.4359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhee J. D. Theoretical calculations of the helix-coil transition of DNA in the presence of large, cooperatively binding ligands. Biopolymers. 1976 Jul;15(7):1345–1375. doi: 10.1002/bip.1976.360150710. [DOI] [PubMed] [Google Scholar]
- Mrksich M., Wade W. S., Dwyer T. J., Geierstanger B. H., Wemmer D. E., Dervan P. B. Antiparallel side-by-side dimeric motif for sequence-specific recognition in the minor groove of DNA by the designed peptide 1-methylimidazole-2-carboxamide netropsin. Proc Natl Acad Sci U S A. 1992 Aug 15;89(16):7586–7590. doi: 10.1073/pnas.89.16.7586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudd C. P., Berger R. L. A computer-controlled all-tantalum stopped-flow microcalorimeter with microjoule resolution. J Biochem Biophys Methods. 1988 Nov;17(3):171–191. doi: 10.1016/0165-022x(88)90002-4. [DOI] [PubMed] [Google Scholar]
- Neidle S., Abraham Z. Structural and sequence-dependent aspects of drug intercalation into nucleic acids. CRC Crit Rev Biochem. 1984;17(1):73–121. doi: 10.3109/10409238409110270. [DOI] [PubMed] [Google Scholar]
- Pelton J. G., Wemmer D. E. Structural characterization of a 2:1 distamycin A.d(CGCAAATTGGC) complex by two-dimensional NMR. Proc Natl Acad Sci U S A. 1989 Aug;86(15):5723–5727. doi: 10.1073/pnas.86.15.5723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remeta D. P., Mudd C. P., Berger R. L., Breslauer K. J. Thermodynamic characterization of daunomycin-DNA interactions: microcalorimetric measurements of daunomycin-DNA binding enthalpies. Biochemistry. 1991 Oct 8;30(40):9799–9809. doi: 10.1021/bi00104a032. [DOI] [PubMed] [Google Scholar]
- Snyder J. G., Hartman N. G., D'Estantoit B. L., Kennard O., Remeta D. P., Breslauer K. J. Binding of actinomycin D to DNA: evidence for a nonclassical high-affinity binding mode that does not require GpC sites. Proc Natl Acad Sci U S A. 1989 Jun;86(11):3968–3972. doi: 10.1073/pnas.86.11.3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade W. S., Mrksich M., Dervan P. B. Binding affinities of synthetic peptides, pyridine-2-carboxamidonetropsin and 1-methylimidazole-2-carboxamidonetropsin, that form 2:1 complexes in the minor groove of double-helical DNA. Biochemistry. 1993 Oct 26;32(42):11385–11389. doi: 10.1021/bi00093a015. [DOI] [PubMed] [Google Scholar]