Abstract
To better understand the relationship between epitope variation and tumor escape from immune surveillance, SV40 T antigen transformed B6/K-0 cells were subjected to selection with individual CTL clones specific for the SV40 T antigen H-2Db-restricted epitopes I or V. CTL resistant populations were isolated from a majority of the selection cultures and substituted epitope sequences were identified within most of the resistant populations. T ag sequences deleted of all or portions of the selection-targeted epitope were identified, but in lower numbers compared to epitope sequences bearing single residue substitutions. Relatively few flanking residue substitutions were identified, and only in epitope I-targeted selections. The diversity (numbers and epitope residue locations) of substituted epitope residue positions varied between selections. These findings suggest that the scope of spontaneously occurring mutations that could allow for escape from individual CD8+ T cell clones is large.
Keywords: SV40 T ag, CTL clone, CTL Escape Variant, Epitope mutation, CTL Epitope, Cytotoxicity, CD8+ T Lymphocyte
Introduction
Multiple factors may influence the success of tumor control by cellular immunity (Dunn et al., 2002; Khong and Restifo, 2002). While general defects in the processing and presentation of MHC class I-restricted antigens may facilitate tumor progression, mutations that remove or alter a critical CD8+ T lymphocyte-restricted epitope may facilitate tumor escape under conditions where a relatively narrow, or oligoclonal cellular immune response focuses primarily on one immunodominant epitope within a single tumor antigen (Bai et al., 2003; Charini et al., 2001; Coulie et al., 2001; Dudley et al., 2002; Mortara et al., 1998; Pewe and Perlman, 1999; Turner and Carbone, 1998). Epitope loss may be achieved by deletion of sequences encoding the epitope (Akilesh et al., 2001; Dudley and Roopenian, 1996; Lill et al., 1992), or by amino acid substitutions that block processing of the intact determinant (Theobald et al., 1998; Yellen-Shaw et al., 1997). Single amino acid substitutions that alter primary or secondary MHC anchor residues, alter peptide conformation so as to prevent efficient MHC binding, or alter critical TCR contact residues within the determinant can prevent recognition (Bai et al., 2003; Lill et al., 1992; Lippolis et al., 1995; Mylin et al., 1995b). An alternative possibility is that production of an altered peptide ligand (APL) may functionally antagonize tumor-reactive cytotoxic T lymphocytes [CTL (Loftus et al., 1998)]. By providing a local shielding influence, such antagonist epitopes could interfere with recognition of wild-type peptide-MHC complexes on the same or neighboring cells. Examples of such escape mechanisms have been documented in viral systems (Borrow and Shaw, 1998; Klenerman, Wu, and Phillips, 2002; Mortara et al., 1998). Nonetheless, it remains unclear which mechanism(s) will favor escape from CTL surveillance for tumors where alteration of target epitopes within a chromosomally encoded oncoprotein is required.
To investigate mechanisms of tumor cell escape from oncoprotein-specific CTL, we have used a panel of Simian virus 40 large T antigen (SV40 T ag)-specific CTL clones (Campbell, Foley, and Tevethia, 1983; Mylin et al., 1995a; Tanaka et al., 1989; Tanaka et al., 1988) and SV40 T ag transformed B6/K-0 cells (Tanaka and Tevethia, 1988). The 708 residue SV40 T ag oncoprotein promotes cellular transformation in vitro and tumor progression in vivo by mechanisms which include dominant interference with cell cycle regulators (Fanning, 1992; Manfredi and Prives, 1994). The T ag induces a vigorous MHC class I-restricted response (Tevethia, Flyer, and Tjian, 1980). The SV40 T ag contains four CD8+ T lymphocyte epitopes, designated epitopes I, II/III, IV and V, which display varying immunological potencies in H-2b mice (Campbell, Foley, and Tevethia, 1983) (Deckhut, Lippolis, and Tevethia, 1992; Lippolis et al., 1995; Mylin et al., 1995a; Mylin et al., 1995b; Tanaka et al., 1989; Tanaka et al., 1988). Epitopes I, II/III and V are H-2Db-restricted, while epitope IV is presented by H-2Kb molecules. The hierarchy of CD8+ T cell responses directed at the individual epitopes has been defined by direct enumeration as IV>I>II/III>V (Mylin et al., 2000). Epitope V has been characterized as immunorecessive because epitope V-specific CD8+ T cells are undetectable following immunization with the wild type T ag (Fu et al., 1998; Mylin et al., 1995a; Mylin et al., 2000; Tanaka et al., 1989).
SV40 T ag-transformed B6/K-0 cells have been used previously to isolate CTL escape variants in vitro (Lill et al., 1992; Tanaka and Tevethia, 1988; Tanaka and Tevethia, 1990). B6/K-0 cells contain a single chromosomally integrated SV40 T ag gene and consistently express high levels of both cell surface H-2Db and H-2Kb MHC class I molecules (Tanaka and Tevethia, 1988). Previous in vitro selections of SV40 T ag transformed B6/K-0 cells with epitope I, II/III, IV, or V-specific CTL clones yielded CTL-resistant variant cell populations expressing T ags harboring substitution or deletion mutations affecting the respective, epitope-coding regions (Lill et al., 1992; Mylin et al., 1995b; Tanaka and Tevethia, 1988; Tanaka and Tevethia, 1990). Single residue substitutions, were identified in epitopes II/III, IV, or V (Lill et al., 1992; Mylin et al., 1995b). The results of analyses that utilized corresponding substituted synthetic peptides supported roles in TCR recognition or peptide-MHC binding for most of the affected residue positions (Lill et al., 1992; Mylin et al., 1995b). By contrast, escape variant T ags selected in the same study by the epitope I-specific CTL clone Y-1 contained only epitope I deletions (Lill et al., 1992). Because only a limited number of mutants were isolated and characterized in the initial study, it was important to conduct a more comprehensive analysis.
We report here the results of multiple, larger scale, in vitro CTL selections that individually targeted the H-2Db-restricted SV40 T ag epitopes I and V. While sharing limited amino acid similarity, epitopes I and V differ dramatically in immunogenicity and the half lives of cell surface MHC complexes bearing synthetic epitope I or V peptides differ dramatically (Fu et al., 1998; Mylin et al., 1995a; Mylin et al., 2000). Multiple CTL clones specific for each epitope have been established which express distinct T cell receptors (TCRs) (Mylin et al., 2000) and display distinct fine specificities (Deckhut et al., 1991; Lippolis et al., 1995; Tevethia et al., 1998) (Mylin and Tevethia, unpublished). By this approach, many additional naturally occurring substitutions and/or deletions were identified within (or surrounding) epitopes I and V. Our in vitro selection results imply that spontaneous mutations affecting target epitopes within non-essential regions of single copy tumor antigens may compromise immunotherapy strategies that target single epitopes with a limited number of TCR clonotypes.
Results
Analysis of epitope I escape variant sequences
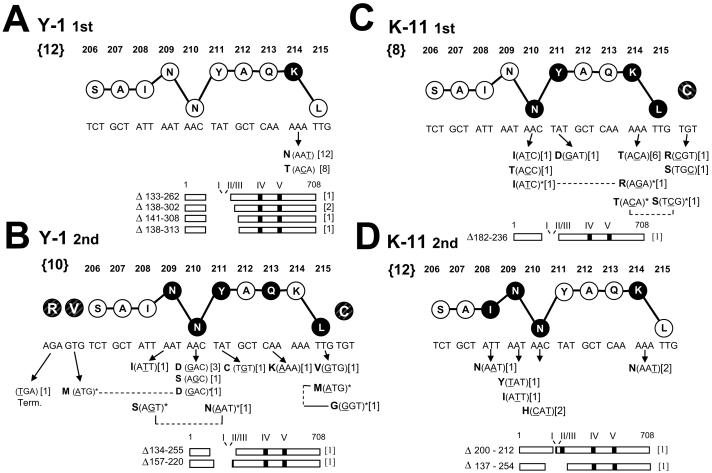
Selections for epitope I variants were conducted with the two epitope I specific CTL clones Y-1 and K-11 which express different TCRs that display distinct fine specificities [Table 1; (Tevethia et al., 1998)]. Two selections were performed using the CTL clone Y-1. Analysis of epitope I sequences amplified from the initial set of Y-1 resistant populations (41 - 52; Fig. 1) that were selected from B6/K-0 cells revealed that all harbored one or two nucleotide substitutions affecting codon 214 of epitope I (K214N or K214T; Fig. 2A). Residue K214 serves as a TCR contact residue for both CTL clones, and substitutions affecting residue 214 abrogate recognition by the CTL clones Y-1 or K-11 (Lippolis et al., 1995). Four different in-frame deletions also were identified in which a total of 130 (Δ133-262), 165 (Δ138-302), 168 (Δ141-308), or 176 (Δ138-313) codons had been lost, eliminating epitope I (Fig. 2A). These results demonstrate that the CTL clone Y-1 can select in vitro for epitope I variants bearing single amino acid substitutions as well as large deletions encompassing the epitope.
Table 1.
Epitope specificity, TCRβ usage and epitope substitutions that illustrate differences in fine specificity for SV40 T ag-specific CTL clones used in this study
| CTL clonea | Restriction | SV 40 T ag epitope specificity | TCRβb | Fine specificityc |
|---|---|---|---|---|
| Y-1 | H-2Db | I | 7 | S206T |
| A207G | ||||
| N209T | ||||
| K-11 | H-2Db | I | 10 | A212G/C |
| Q213H | ||||
| K-19 | H-2Db | II/III | 10 | NAd |
| Y-5 | H-2Db | V | 7 | |
| Q489A | ||||
| D495A | ||||
| H-1 | H-2Db | V | 9 |
References for the derivation and characterization of each of the CTL clones are provided in the text.
For additional information, see Mylin, et al. 2000.
Substituted epitope sequences which are differentially recognized by these pairs of CTL clones. For further information regarding the fine specificities of the epitope I-specific CTL clones Y-1 and K-11, see Lippolis, et al. 1995 and Tevethia, et al. 1998. The epitope V-specific CTL clones Y-5 and H-1 displayed differential recognition of alanine substituted synthetic peptides (Mylin and Tevehia, unpublished).
Not applicable.
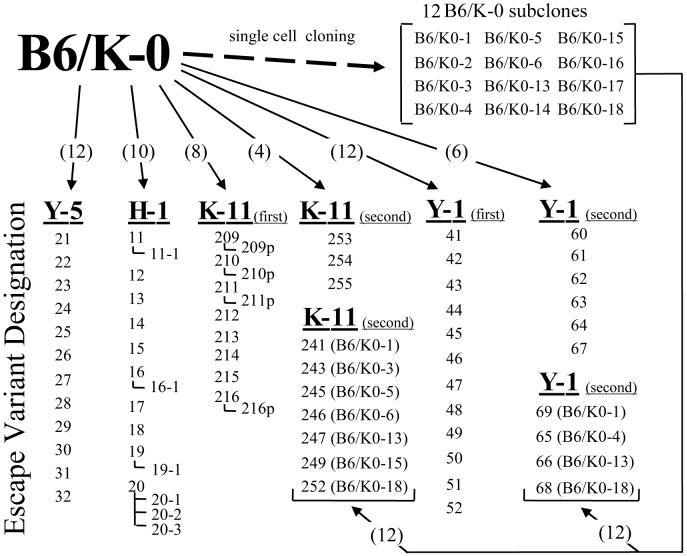
Fig.1.
Escape variant selections. Escape variant populations were selected from B6/K-0 cells or B6/K-0-derived subclones (arrows) by the SV40 T ag specific CTL clones H-1, Y-5, K-11 or Y-1. Two Y-1 selections and two K-11 selections were conducted (first, second). Numbers appearing in parentheses located on the arrows indicate, respectively, the total number of individual cultures used to begin each selection (multiple parallel cultures seeded with B6/K-0 cells, or cultures seeded with individual B6/K-0-derived subclone lines). The numbers or designations shown below a CTL clone represent designations given to the individual CTL resistant variant populations that emerged. Numbers set off by an angled line indicate a focus or island that was expanded from within a selection culture. The “p” designation for K-11 selected populations indicates that a secondary K-11 exposure (purge) was done for a portion of the resistant population following initial confluence of the primary selection culture. The identity of the B6/K-0 subclones used to seed selection cultures from which resistant populations emerged are indicated in parentheses beside the numeric designation for the corresponding resistant population.
Fig. 2.
Alterations affecting epitope I identified following selection with the CTL clones Y-1 (A, B) or K-11 (C, D). A, first Y-1 selection. B, second Y-1 selection. C, first K-11 selection. D, second K-11 selection. Nucleotide and amino acid sequences (single letter abbreviations) for an epitope I 10mer peptide are shown. The vertical location of each residue in the stick and ball representation implies function within the epitope peptide based on previous studies (Lippolis et al., 1995): residues implicated in peptide-MHC binding are lowered; residues involved in TCR recognition are raised. Darkened circles indicate residue positions at which substitutions were identified in this study; nucleotide alteration(s) and predicted amino acid substitutions are shown below the corresponding residue position (arrows). Deletions affecting epitopes are illustrated by diagrams of the 708 residue T ag, and the residues lost by each deletion are indicated. Numbers not within brackets indicate residue positions within the wild type T ag. Bold numbers within the “{}” brackets to the left of the wild type epitope sequences indicate the total number of selection cultures from which resistant populations arose. Numbers in brackets to the right of each substitution or deletion shown below the ball and stick epitope sequence indicate the number of selection cultures in which that substitution or deletion was detected. Positions of altered adjacent flanking residues (shaded and crosshatched, but not connected by lines to the epitope I residues 206-215) are included in (B) and (C). Residue positions altered in doubly mutated epitope sequences are connected by the dashed lines and indicated by asterisks. The epitope I mutations are also presented Figure 4 grouped by the variant populations in which they were identified.
A second selection was performed using the CTL clone Y-1 because only a limited number of different spontaneous substitutions were identified within epitope I in the first selection, and it was important to demonstrate that such epitope substitutions could arise independently. The second Y-1 selection was initiated with six cultures seeded with B6/K-0 cells and twelve cultures seeded with subclones derived from individual B6/K-0 cells. Substitutions isolated by selection from individual B6/K-0 subclones would represent independently occurring mutations. Y-1 resistant populations were obtained from each of the cultures seeded with B6/K-0 cells, and four of the twelve cultures seeded with the B6/K-0-derived subclones (Fig. 1). Interestingly, this second CTL clone Y-1 selection yielded a different set of variant mutations (compare Figs. 2B and 2A). Most notably, substitutions affecting TCR contact residue K214 were not detected in the second Y-1 selection (Fig. 2B). Instead, two substitutions, N201D and N210S affecting residue N210 which is absolutely required for efficient H-2Db binding by the epitope I peptide (Lippolis et al., 1995), were identified in three of six selection cultures seeded with B6/K-0 cells or in the resistant population selected from the B6/K-0-4 subclone. (See Fig. 1 for information about selection cultures in which variant populations arose, Fig. 2B for the locations of epitope I residues affected by those mutations, and Fig. 4B for epitope I sequences identified in each of the variant populations.) Two substitutions, L215V or L215M, altered the identity of the epitope I carboxyl terminal anchor residue position; such conservative substitutions would not be expected to abrogate MHC binding of the altered peptide (Lippolis et al., 1995). Substitutions were identified in three positions (209, 211 and 213) that have been previously implicated as TCR contact residues (Lippolis et al., 1995). Three doubly mutated epitope I-region sequences were also identified. Two of the double mutations included substitutions of the central anchor residue (N210D/V205M and N210S/Y211N; Fig. 2B) which by itself would be expected to abrogate Y-1 recognition (Lippolis et al., 1995). The third double mutation (L215M/C216G) included a conservative substitution (L215M) of the C terminal anchor residue combined with a substitution of residue 216 (C216G) which flanks the C terminus of epitope I (Fig. 2B). Two of the three double mutations included a substitution that affected an epitope I flanking residue (C216G or V205M; Fig. 2B); however, variant sequences containing single substitutions affecting only epitope I flanking residues were not identified in this Y-1 section. Two deletions that removed epitope I with a total of either 122 (Δ134-255) or 74 (Δ157-220) T ag residues were identified in the second Y-1 selection. These results reveal dramatic differences in the diversity and locations of substitution mutations, including TCR contact and MHC anchor residues, were identified in the two Y-1 selections.
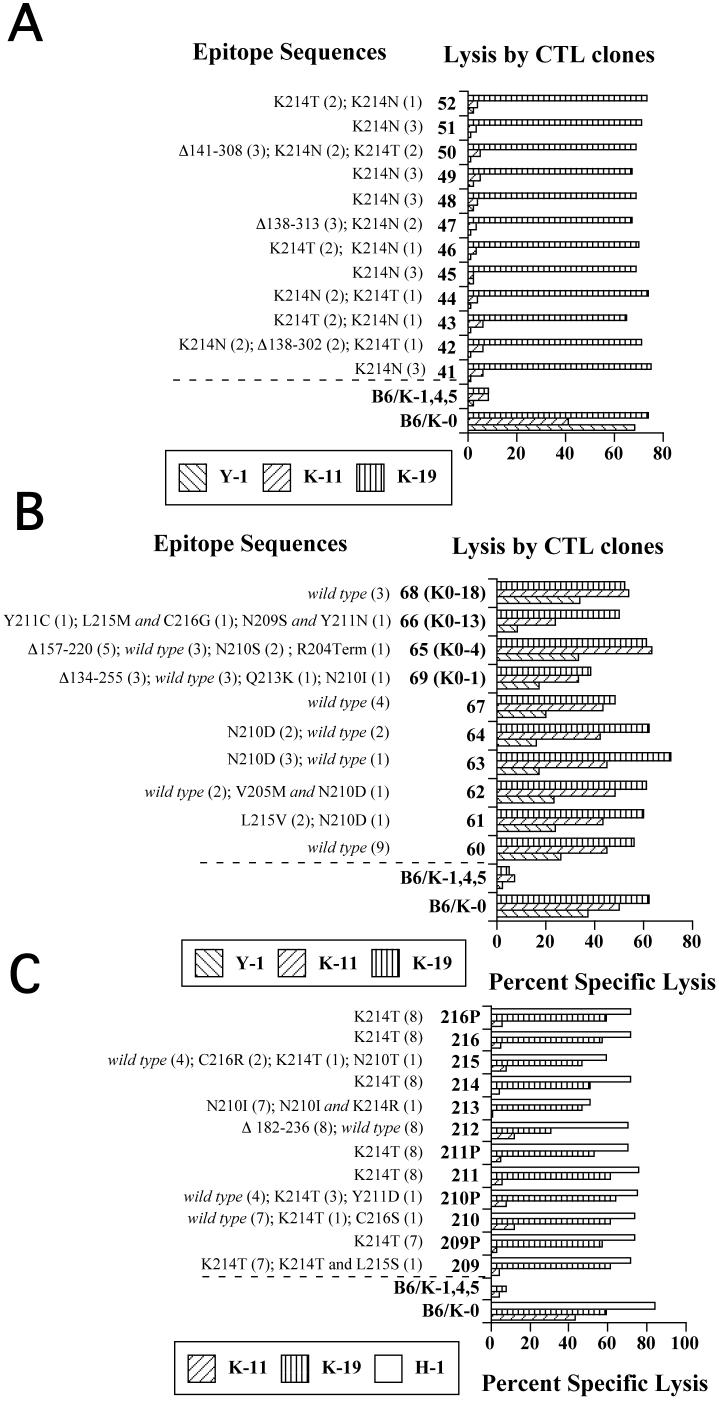
Fig. 4.
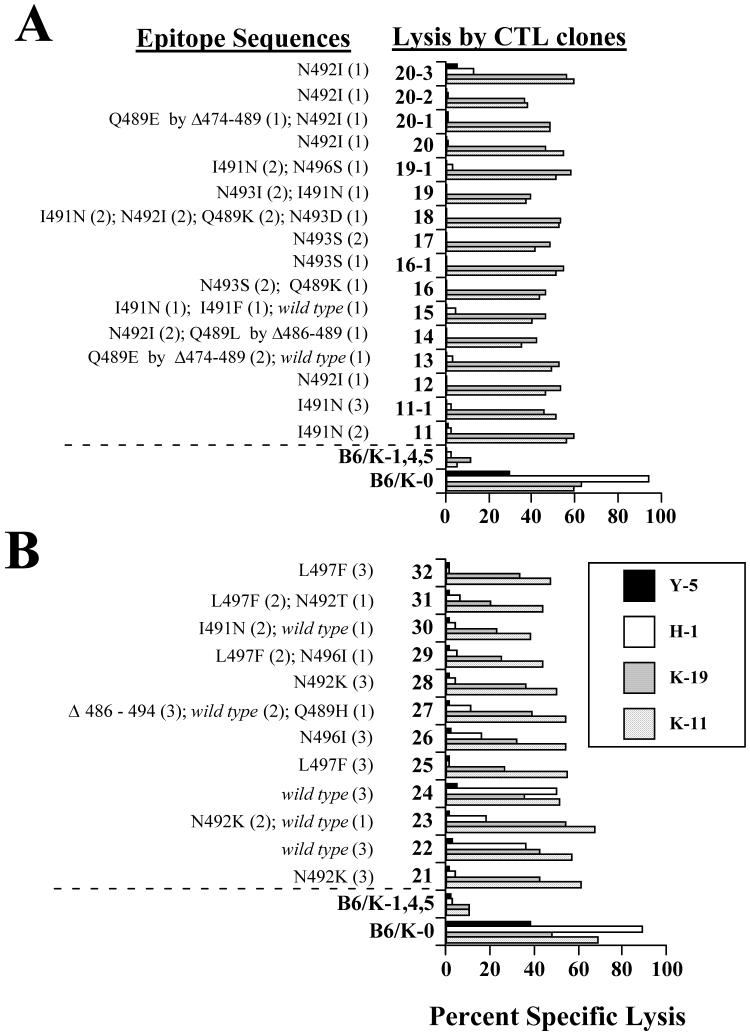
Epitope sequences and lysis of Y-1- or K-11-selected populations. Populations (A) obtained from the first Y-1 selection. B. Second Y-1 selection. C. First K-11 selection. Numbers (parentheses) and types of epitope sequences isolated from CTL-selected populations are shown. The CTL selected populations or control cell lines, B6/K-0 or B6/K-1,4,5, were combined with the indicated CTL clones (Y-1, K-11, or K-19 for A and B; K-11, K-19 or H-1 for C) in standard four hour cytotoxicity assays.
To investigate how TCR fine specificity may influence the diversity of escape variant mutations, additional selections of B6/K-0 cells and B6/K-0 subclones were conducted using a second epitope I-specific CTL clone, K-11 (Fig. 1; Table 1). K-11-resistant populations developed within most of the cultures seeded with B6/K-0 cells (209-216; 253-256) and approximately half of those seeded with individual B6/K-0 subclones (241 - 252). Mutations identified in the K-11 resistant populations included substitutions affecting epitope I positions 208, 209, 210, 211, 214 and 215 (Figs. 2C, 2D). A K214T substitution was detected in six of eight resistant populations in the first K-11 selection (Fig. 2C, Fig. 4C) while the K214N substitution, which was prevalent in the first Y-1 selection, was not detected in the first set of K-11-resistant populations (compare Figs. 2A and 2C). However, the K214N substitution was selected from B6/K-0 cells and the B6/K-0-3 subclone in the second K-11 selection (Fig. 2D). Large in-frame deletions that removed both epitopes I and II/III (Δ182-236, Δ137-254) were identified in two K-11-resistant populations (212, 254; Fig. 2C, 2D), while a third and smaller in-frame deletion was identified that inactivated epitope I but retained epitope II/III (Δ200-212; Fig. 2D). Epitope I sequences bearing two substitutions within the epitope sequence were identified in two K-11-resistant populations (209 and 213; Fig. 2C); both contained an alteration of the TCR contact residue position 214 in addition to a substitution affecting an anchor residue (K214T/L215S and N210I/K214R). Single substitutions affecting only the C terminal flanking residue (C216S, C216R) were identified in two K-11-resistant populations. Unlike the Y-1-selected flanking residue substitutions (see above; Fig. 2B), the position 216 substitutions selected by the CTL clone K-11 appeared as single mutations (Fig. 2C). Together, these results show that, under appropriate conditions, epitope I-specific CTL clones can select in vitro for epitope I variants containing single residue substitutions that occur within the epitope or flank the epitope in addition to variants that have been deleted of the epitope.
Analysis of epitope V escape variant sequences
SV40 T ag epitope V is immunorecessive (Mylin et al., 1995a; Mylin et al., 2000; Tanaka et al., 1989). Epitope V is presented on H-2Db MHC class I molecules on the surface of SV40 T ag transformed cells and cells infected by vaccinia virus recombinants encoding the full length T ag or epitope V minigenes (Fu et al., 1998). Nonetheless, epitope V remains nonimmunogenic within Tag derivatives that retain subsets of the immunodominant T ag epitopes (Fu et al., 1998; Mylin et al., 1995a; Mylin et al., 2000). Epitope V is also of interest due to its escape from negative selection in Tag transgenic mice where central tolerance leads to the deletion of CD8+ T cells specific to the dominant T ag epitopes due to thymic expression of Tag (Schell, 2004; Schell et al., 1999). Epitope V specific T cells can be activated in SV11 transgenic mice upon appropriate immunization (Ryan and Schell, 2006; Schell, 2004).
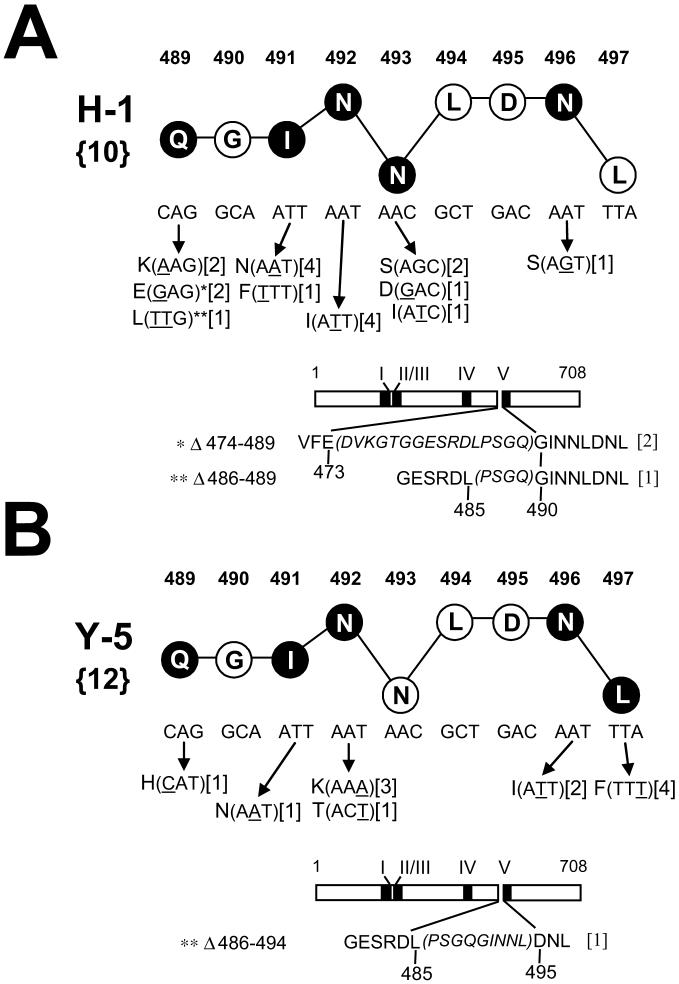
Variant selections were conducted with the two epitope V-specific CTL clones Y-5 and H-1 which express distinct TCRs [Table 1; (Mylin et al., 1995a; Mylin et al., 2000; Tanaka et al., 1989)]. All selection cultures initiated with the epitope V-specific CTL clones H-1 or Y-5 yielded resistant populations. Selection by the CTL clones Y-5 or H-1 revealed many variant sequences that as a group represented nucleotide substitutions or deletions affecting five of nine epitope V residues (Fig. 3A, B). Nucleotide substitutions affecting T ag amino acid residues 489, 491, 492, 496, and 497 were identified within variants selected by the CTL clone Y-5 (Fig. 3B), while substitutions affecting amino acid residues 489, 491, 492, 493, and 496 were identified within H-1 resistant populations (Fig. 3A). Therefore, each of the CTL clones Y-5 and H-1 selected for mutations affecting T ag residues 489, 491, 492, and 496. Even so, the nucleotide alterations and predicted amino acid substitutions affecting these four commonly substituted epitope V positions often differed between the populations selected by the two CTL clones (compare Fig. 3A, B). The diversity of variant epitope V sequences is further underscored by the fact that two or more different substitutions were selected at each of four epitope V residue positions by either CTL clone (codons 489, 491, 492, and 493; Fig. 3A, B). Although the majority of the predicted amino acid substitutions identified in the variant epitope V sequences affected non-MHC anchor residues, at least one anchor residue substitution was selected by Y-5 or H-1 (Fig. 3A, B). The CTL clone H-1 selected for substitutions affecting the central P5 anchor residue position (N493S, N493D, N493I) while the CTL clone Y-5 selected for one substitution affecting the CΩ anchor position (L497F). Therefore, an array of amino acid substitutions affecting epitope V were selected by two distinct SV40 T ag epitope V-specific CTL clones. Differences in TCR clonotype may have contributed to the selection of distinct sets of epitope variants by the CTL clones H-1 and Y-5.
Fig. 3.
Alterations affecting epitope V identified following selection with the CTL clones H-1 (A) or Y-5 (B). See legend of FIG. 2 for explanation of numbers and symbols. Deletions affecting epitope V are illustrated by diagrams of the 708 residue T ag; amino acid sequences retained in the deleted T ags are shown in plain text (below the T ag diagram) while T ag residues lost by the deletion are indicated in italic and within parentheses. (A) H-1-selected deletions Δ474-489 and Δ486-489, and the Q489E and Q489L substitutions, respectively, are highlighted by one or two asterisks because they represent the same altered epitope sequences. The epitope V mutations are also presented in Figure 5 grouped by the variant populations in which they were identified.
Short in-frame deletions affecting the epitope V region of T ag were selected by both Y-5 and H-1. These deletions removed one or more epitope V codons along with three or more amino terminal flanking codons (Fig. 3A, B). For two deletions selected by the CTL clone H-1, elimination of sequences 5’ to Gly 490 (the P2 residue) resulted in the replacement of the P1 Gln 489 codon of epitope V with codons specifying either a Glu or Leu residue (Fig. 3A). A small in frame deletion selected by the CTL clone Y-5 removed 6 of 9 epitope V codons and three amino terminal flanking residues (Fig. 3B). Variant sequences representing C-terminal deletions of epitope V were not detected. It is unclear if these deletions were not selected, or whether the annealing locations for the oligonucleotide primers used in this study to amplify the epitope V region of T ag, or reduced growth rate for cells expressing T ag genes bearing such deletions may have prevented detection of 3’ epitope V deletions (Kierstead and Tevethia, 1993). These results imply that a wealth of substitutions affecting TCR contact or residues within epitope V are readily selected by CTL clones in vitro.
Recognition of CTL-selected populations and altered epitope peptides
A large number of different epitope mutations were identified within the collection of variant populations selected in this study. It was important to determine the mechanism(s) by which the epitope alterations prevented destruction by the selecting CTL clones.
Variant cell populations arising from the first Y-1 selection (41 - 52), all of which contained either a substitution affecting residue 214 or were deleted of epitope I, were not lysed by either of the epitope I specific CTL clones (Y-1 and K-11). Each of the selected populations was lysed efficiently by the epitope II/III-specific CTL clone K-19 (Fig. 4A) suggesting that H-2Db-restricted processing and presentation remained intact in the escape variant populations and that epitope I-specific defects accounted for the resistance of these variant populations. By contrast, a majority of the populations derived by the second Y-1 selection (60 - 69) showed only marginally reduced levels of lysis by the CTL clone Y-1 (or near normal lysis by the CTL clone K-11) relative to that of unselected control B6/K-0 cells (Fig. 4B; K-19 reactivity towards the same Y-1 selected populations was comparable to that of the control B6/K-0 cells (Fig. 4B). The majority of substitutions identified in the same variant populations are known to affect residues that are critical for CTL recognition (Lippolis et al., 1995), but a significant number of unaltered (wild type) epitope I sequences were amplified from most of the same populations (Fig. 4B). The minor reduction in Y-1 (or K-11) lysis for these populations likely indicated that a significant number of cells that retained wild type epitope I sequences remained within the CTL selected populations.
K-11-resistant populations (209-216) were characterized for lysis by the CTL clones K-11, K-19 and H-1. The cytotoxicity assay results shown in Figure 4C revealed that these populations were no longer recognized by the CTL clone K-11. With the exception of population 212, loss of K-11 reactivity from the selected populations occurred with little apparent reduction in lysis by epitope II/III- or V-specific CTL clones. Specific reduction of K-11 lysis for the majority of the selected populations was consistent with the prevalence of substituted epitope I sequences within the same populations (Fig. 4C) although wild type epitope I sequences were detected in some populations.
Each of the epitope V-specific CTL clone H-1-selected populations was resistant to lysis by either of the CTL clones H-1 or Y-5 in standard cytotoxicity assays (Fig. 5A). The majority of CTL clone Y-5 selected populations showed resistance to lysis by Y-5 (Fig. 5B), while several of the Y-5-selected populations (e.g. 22 and 24) were recognized at varying efficiencies by the CTL clone H-1 (Fig. 5B). Lysis of the Y-5- or H-1-resistant populations by either epitope I or II/III-specific CTL clones was comparable to that of the wild type B6/K-0 cells (Fig. 5A, B). Consistent with the wealth of altered epitope V sequences amplified from variant populations, the cytotoxicity results suggest that alterations of epitope V were responsible for resistance of the H- 1- or Y-5-selected populations to lysis by the epitope V-specific CTL clones rather than reduced Tag expression or Tag-independent alteration(s) affecting MHC class I processing or presentation.
Fig. 5.
Epitope sequences and lysis of H-1- and Y-5-selected populations. A. Analyses for H-1-selected populations. B. Analyses for Y-5 -selected populations. Epitope sequences identified by sequencing recombinant clones representing each CTL-selected population are shown. The numbers in the parentheses indicate the number of each sequence type that was obtained for that population. Each of the CTL-selected populations or control cell lines (B6/K-0 or B6/K-1,4,5) were combined with the indicated CTL clones (K-11, K-19, H-1 or Y-5) in standard four hour cytotoxicity assays. Legend for A is as shown for B.
Analysis of substituted epitope I and V peptides
To determine the basis for loss of recognition for CTL escape variant epitope sequences identified in this study, substituted synthetic epitope peptides were used in standard cytotoxicity assays, and were evaluated for their ability to stabilize MHC class I molecules on the surface of TAP-defective RMA/s cells. Owing to the wealth of substitutions identified in this study, it was feasible to synthesize and study synthetic peptides corresponding to only a subset of the variant epitope sequences.
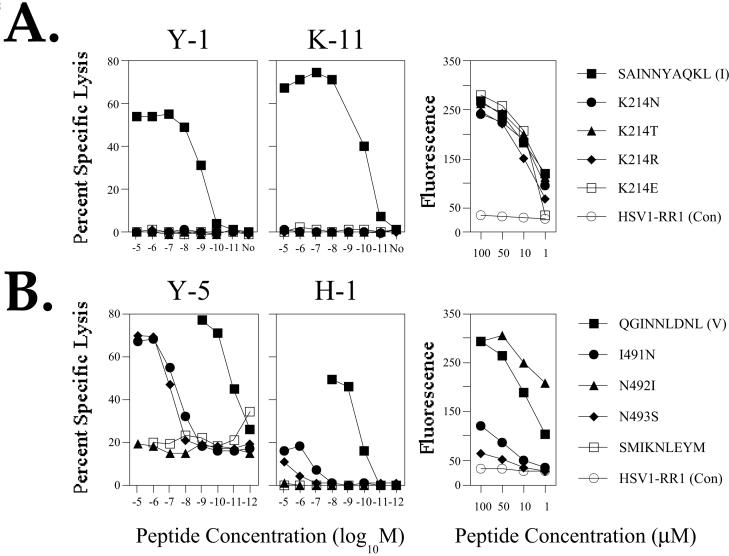
Due to the prevalence of K214 substitutions in CTL clone Y-1 and K-11 resistant populations, substituted synthetic epitope I peptide variants where asparagine (N), threonine (T), arginine (R), or glutamic acid (E) substitutions replaced Lysine (K) 214 were analyzed for recognition by the epitope I-specific CTL clones Y-1 and K-11. The results (Fig. 6A) indicated that each substitution at position 214 abolished recognition by the CTL clones Y-1 or K-11. Furthermore, recognition was not restored for either of the CTL clones when increased concentrations of the variant peptides were used. Analysis of H-2Db expression on the surface of MHC class I antigen presentation-deficient RMA/s cells incubated in the presence of synthetic epitope I peptides failed to reveal substantial differences in the ability of the residue 214-substituted epitope I peptide variants to stabilize H-2Db molecules relative to that of the wild type epitope I peptide (Fig. 6A). These results are consistent with those of previous studies in which position 214 substitutions have been studied (Deckhut and Tevethia, 1992; Lippolis et al., 1995). Therefore, the K214N and K214T epitope I substitutions selected in this study by the CTL clones Y-1 and K-11 alter a residue which appears to be critical for TCR engagement by both epitope I-specific CTL clones.
Fig. 6.
Target cell lysis by CTL clones and H-2Db stabilization for synthetic peptides corresponding to substituted epitope I or V sequences. RMA cells pulsed with substituted variant peptides at the concentrations indicated in the figure were combined with the CTL clones Y-1 or K-11 (A), or H-1 or Y-5 (B) in standard cytotoxicity assays (left two panels in A or B). No, no peptide added. The SMIKNLEYM peptide that efficiently binds H-2Db molecules, but is not recognized by T ag-specific CTL clones, was used as a control in assays utilizing the epitope V-specific CTL clones. RMA/s cells pulsed with varying concentrations of variant epitope I (A) or epitope V (B) peptides at the concentrations indicated in the figure were analyzed by flow cytometry following staining for cell surface H-2Db complexes using the conformation-sensitive monoclonal antibody 28-14-8. A peptide corresponding to an H-2Kb-restricted Herpes simplex virus epitope (HSVI-RR1) was used as a non-stabilizing control (Con).
Our analysis of the epitope V variants was confined to a set of peptides representing three of the more commonly detected substitutions (Fig. 3A, B). Synthetic epitope V peptide variants I491N, N492I, and N493S were analyzed in standard cytotoxicity assays using the CTL clones Y-5 and H-1 (Fig. 6B) and were compared to the unsubstituted epitope V peptide for their ability to stabilize H-2Db molecules on the surface of RMA/s cells (Fig. 6B). The results of the cytotoxicity assays revealed that the I491N (selected by H-1 or Y-5) and N493S (selected by H-1, but not by Y-5) substitutions dramatically reduced recognition by the CTL clone H-1 (Fig. 6B); the same substitutions reduced recognition by the CTL clone Y-5 as 1,000 fold higher concentrations of these peptides were required to trigger lysis by Y-5 that was comparable to lysis of targets pulsed with the unsubstituted epitope V peptide (Fig. 6B). Neither H-1 nor Y-5 lysed target cells pulsed with the N492I substituted variant (Fig. 6B; selected by H-1). Comparison of these peptides for H-2Db stabilization revealed that the N492I variant was more effective than the unsubstituted epitope V peptide (Fig. 6B). The I491N and N493S substituted epitope V peptide variants did not efficiently stabilize H-2Db complexes (Fig. 6B). These results suggest that the N492I substitution likely alters a TCR contact residue that is required for recognition by both of the CTL clones Y-5 and H-1, because the N492I-substituted peptide efficiently stabilized H-2Db molecules, but failed to trigger lysis by either CTL clone. By contrast, these results imply that the reduced lysis observed for the I491N- and N493S-substituted epitope V peptides likely results from reduced H-2Db binding and/or an altered conformation of the weakly bound peptide.
Discussion
The results of this study indicate that a large number of naturally occurring mutations (substitutions and deletions) affecting two distinct H-2Db-restricted SV40 Tag epitopes, I and V, can be selected from SV40 T ag transformed cells by CTL clones in vitro. While the number of different mutations identified and the fraction of epitope residue positions affected in the individual selections varied, the total number of mutations identified by the strategy used in this study was large and did include substitutions (in epitope V: I491F and N496I) and deletions (a variety that affected the epitope I region) that had been identified in our earlier variant selections (Lill et al., 1992; Tanaka and Tevethia, 1988; Tanaka and Tevethia, 1990). The presence of numerous mutant T ag genes within the B6/K-0 population was not unexpected as T ag immortalization does lead to increased genetic instability in rapidly dividing cells (Fanning, 1992; Hiscott, Murphy, and Defendi, 1980; Hiscott, Murphy, and Defendi, 1981; Manfredi and Prives, 1994; Wiesmuller, Cammenga, and Deppert, 1996) and the T ag can undergo mutation in many regions without the loss of immortalization function(Deckhut and Tevethia, 1992; Kierstead and Tevethia, 1993; Manfredi and Prives, 1994; Tevethia et al., 2001; Tevethia et al., 1998; Thompson et al., 1990). Further, cytotoxicity experiments employing H2b cells expressing constructed T ag mutants or the T ags of the papovaviruses JC or BK demonstrated that a wide variety of single residue substitutions can abrogate recognition of SV40 T ag epitopes I or V by CTL clones in vitro (Deckhut et al., 1991; Deckhut and Tevethia, 1992; Lippolis et al., 1995).
The strategy employed in this study was designed to primarily select for variants containing loss of function mutations affecting the epitope targeted by the selecting CTL clone. B6/K-0 cells contain a single integrated SV40 Tag gene (Tanaka and Tevethia, 1990). Therefore, escape due to mutations affecting the single copy Tag gene was expected to be favored relative to escape promoted by mutations affecting genes required for MHC class I-restricted epitope presentation. The requirement for T ag function to maintain immortalization of the B6/K-0 cells made it unlikely that rapidly growing variant cells would contain an inactivated T ag gene (complete antigen loss) as has been observed for other tumor associated antigens (Boon et al., 1994; Boon and van der Bruggen, 1996; Dudley and Roopenian, 1996). However, large portions of the T ag can be altered or lost without compromising immortalization function (Kierstead and Tevethia, 1993; Tevethia et al., 2001; Tevethia et al., 1998; Thompson et al., 1990). Specifically, T ag amino acids 127 - 250, the region containing epitope I (206-215), or amino acids which contain epitope V (489 - 497) can be deleted without compromising immortalization or tumorigenicity in immunocompromised mice. Because inactivation of one epitope within a single Tag gene could provide for escape from a given CTL clone, two classes of epitope variants could be expected: 1.) loss-of-recognition mutations which impaired epitope recognition due to epitope deletion, substitution of critical MHC or TCR contact residues, or alterations which severely reduced epitope processing; 2.) functional interference mutants where target cell recognition and lysis was compromised by substitutions which resulted in the production of TCR antagonist peptides. Based on the results of previous cytotoxicity studies involving T ag mutants and substituted T ag determinant peptides (Deckhut and Tevethia, 1992; Lill et al., 1992; Lippolis et al., 1995; Tevethia et al., 1998) and the analysis of a limited number of substituted peptides in this study, the majority of substitutions identified in the current study are likely to represent loss-of-recognition mutations.
The variety of variant epitope sequences selected by the SV40 T ag epitope I or V-specific CTL clones in this study agrees well with the results of previous studies in which altered T ag derivatives were analyzed for recognition by CTL clones. In the current study, single residue substituted epitope I variants were selected by both of the epitope I-specific CTL clones (Y-1 or K-11). Two Y-1-selections yielded substitutions affecting T ag epitope I residues 209, 210, 211, 213, 214, and 215, while the CTL clone K-11 selected for substitutions affecting residues 208, 209, 210, 211, 214 and 215 (Fig. 3). Based on previous studies, these residue positions have been assigned roles as either TCR contact or MHC anchor residues (Lippolis et al., 1995). Lippolis and coworkers (1995) have previously demonstrated that neither of the CTL clones Y-1 or K-11 lysed T ag cell lines engineered to express substitutions at epitope I positions 206, 207, 210, 211, or 214. Even so, not all substitutions at these positions are expected to abrogate recognition by epitope I-specific clones (Deckhut et al., 1991; Lippolis et al., 1995). Recognition of T ag transformed cell lines by Y-1 or K-11 can be differentially affected by substitution of residues 206, 207, 209, 212, or 213 [Table 1; (Lippolis et al., 1995; Tevethia et al., 1998)]. Therefore, with the exception of residue position 206, our larger scale selection strategy yielded substitutions in a majority of epitope I residue positions where substitutions had been previously shown to abrogate recognition of endogenously processed T ag epitopes by the CTL clones Y-1 or K-11.
Epitope V variant populations selected by either of the two CTL clones Y-5 or H-1 in the present study contained a wealth of substitutions that affected six different epitope V residue positions. The residues that occupy the altered positions appear to function as TCR contact, MHC anchor, or conformation maintaining spacer residues (see Fig. 2 legend) based on the analysis of substituted epitope V synthetic peptides [this study; (Lill et al., 1992); Mylin and Tevethia, data not shown)] and comparison with published H-2Db:peptide crystal structures (Young et al., 1994). Mutations previously selected by the CTL clone Y-5 [N496I in K-5; I491F in K-1,4,5; (Lill et al., 1992; Tanaka and Tevethia, 1990)] were also identified in this study. The Y-5 CTL clone did not re-select the I491F substitution in this study, but did select both I491N- and N496I-substitution variants. Therefore, the survival advantage provided by the I491F substitution is not unique to the TCR expressed by the CTL clone Y-5. Substitutions in a variety of epitope V positions therefore appear to promote survival during selection by epitope V-specific CTL clones in vitro.
The results of this study suggest that the role of flanking sequence variation in escape is minor, or is epitope dependent. Relatively few flanking sequence substitutions were identified, and only in the epitope I-targeted selections. Variant sequences bearing single substitutions affecting the epitope I carboxyl terminal flanking residue C216 (C216R, C216S) were identified only following selection with the CTL clone K-11. Two of the double mutations selected by the CTL clone Y-1 (second selection) included substitutions affecting flanking residues (C216G or V205M), however, individual flanking residue substitutions were not identified in either of the Y-1 selections. Due to the variety of substitutions that were identified in the epitope V-targeted selections, it was surprising that flanking sequence substitutions were not detected. Epitope context (flanking sequence variation) can, but does not universally appear to influence CTL clone recognition under the conditions of standard cytotoxicity assays (Bergmann et al., 1996; Brander et al., 1999; Del Val et al., 1991; Hahn et al., 1992; Lippolis et al., 1995; Mylin, 1999; Shastri, Serwold, and Gonzalez, 1995; Yellen-Shaw and Eisenlohr, 1997; Yellen-Shaw et al., 1997). Specifically, engineered substitutions affecting native SV40 T ag epitope I flanking residues did not appear to significantly impair recognition by the epitope I-specific CTL clones Y-1 or K-11 (Lippolis et al., 1995). However, flanking sequences did influence CTL recognition and immunogenicity for a single amino acid substituted epitope I variant (S206G) when it was located in the protein context normally occupied by epitope V (Mylin, 1999). Conditions used in this study to select CTL escape variants differ from traditional cytotoxicity assays in that lower effector to target ratios are used over days rather than hours. Minor differences in target cell recognition that are not apparent under the conditions of standard cytotoxicity assays may nonetheless provide a survival advantage under the in vitro selection conditions used in this study. Perhaps flanking sequence substitutions contribute to survival, but individually are not sufficient to provide for complete long-term resistance. It should be emphasized that single residue flanking substitutions were identified only as doubly mutated epitope sequences, or were found as single mutations within resistant populations in which additional substituted epitope sequences were identified.
The findings of this study in which CTL clones were used individually to select and enrich variant cells from monolayer cultures in vitro may be relevant to immune surveillance and the control of tumors in vivo. We note that CTL escape variants have been selected in vivo from transplanted tumors by others (Bai et al., 2003). In that study, substituted P1A epitope sequences were identified within CTL resistant tumor cells that grew out from tumors following transplantation into immunologically compromised mice and selection with adoptively transferred P1A-specific transgenic CD8+ T cells (Bai et al., 2003). Substituted P1A sequences expressed by the CTL resistant variants contained one of four single residue substitutions that affected residues located primarily in the carboxy terminal half of the epitope. The corresponding synthetic substituted peptides appeared to be compromised in TCR recognition and, in some cases, MHC association, but failed to demonstrate antagonist function in vitro (Bai et al., 2003). Escape variants were not obtained in that study when CTL populations representing a repertoire of naturally occurring P1A-specific clonotypes were used. It remains to be determined if the SV40 T ag escape variant substitutions identified in this study will promote survival under conditions of CTL selection in vivo.
Our results imply that the potential for tumor escape variation by cells transformed by a single copy of the SV40 T ag gene is high. Escape variants emerged following selections targeted at the stably-bound determinant, epitope I, or the more weakly-binding epitope, epitope V. Further studies will be required to determine whether biological properties of an epitope and/or the quality (avidity or aggressiveness) of tumor-specific CTL do influence the variety or frequency of escape variant mutations. Such considerations are relevant to immunological therapy of tumors in hosts where tumor-induced tolerance progressively eliminates or anergizes high avidity clonotypes reactive towards immunodominant determinants (Ryan and Schell, 2006; Schell, 2004; Schell, Knowles, and Tevethia, 2000; Schell et al., 1999; Schell and Tevethia, 2001; Staveley-O′Carroll et al., 2003). The results of this study imply that strategies that seek to recruit CD8+ T cells specific for weaker binding determinants, for which tumor-induced tolerance may be incomplete, may be frustrated by a high rate of escape variation in the targeted determinant. Alternately, it may be difficult to eliminate unmutated tumor cells when targeting weaker epitopes. Epitopes I and V within SV40 T ag will provide appropriate models to test such possibilities.
Materials and methods
Cell lines and viruses
The SV40-transformed cell lines B6/K-0 (Tanaka and Tevethia, 1988) and B6/WT-19 (Tevethia, 1984) were derived from the cells of C57BL/6 (H-2b) mice. The B6/K-1,4,5 cell line was derived by sequential rounds of selection of B6/K-0 cells with the SV40-T ag-specific CTL clones Y-1, Y-4, and Y-5 (Tanaka and Tevethia, 1988; Tanaka and Tevethia, 1990). For this study, twelve single-cell derived subclones were established from the B6/K-0 cell line (referred to as B6/K-0 population in the text) by limiting dilution plating. The B6/K-0 subclones were shown by flow cytometry to express levels of cell surface MHC class I molecules (H-2Db and H-2Kb) comparable to that expressed by the parental B6/K-0 line (not shown). The parental B6/K-0 line and the subclones were lysed with similar efficiencies by SV40 T ag specific CTL clones under the conditions of standard cytotoxicitsy assays (not shown). The RMA cell line and the antigen processing defective RMA/s derivative have been described (Karre et al., 1986; Ljunggren and Karre, 1985).
Growth and maintenance of CTL clones
The derivation and characterization of the SV40-T ag-specific H-2Db-restricted CTL clones Y-1, K-11, K-19, Y-5, and H-1 have been described (Campbell, Foley, and Tevethia, 1983; Mylin et al., 1995a; Mylin et al., 2000; Tanaka et al., 1989; Tanaka et al., 1988). Briefly, CTL clones were maintained in 12 well tissue culture plates (Costar 3513) and were subcultured twice weekly in RPMI 1640 medium (Gibco/Invitrogen) supplemented with either recombinant IL-2 (Amgen, Thousand Oaks, Calif.) or Rat T Stim Culture Supplement (Becton Dickson) and gamma-irradiated (10,000 rad) B6/WT-19 (Y-1, K-11, K-19) or B6/K-3,1,4 (Y-5, H-1) cells (Mylin et al., 1995a).
Selection and characterization of CTL clone-resistant variants
Multiple SV40 T ag transformed B6/K-0 kidney cell monolayers were individually subjected to selection by H-2Db-restricted, SV40 T antigen epitope V or I-specific CTL clones that express distinct T cell receptors (Mylin et al., 2000). Y-1 and K-11 selections were also performed using B6/K-0 cells in parallel with B6/K-0 subclones. The method used in this study for selection and molecular analysis of CTL clone-resistant cell populations from among B6/K-0 cells or B6/K-0 subclones was modified from that reported in a previous study (Tanaka and Tevethia, 1988; Tanaka and Tevethia, 1990). Selections were initiated using eight to twelve freshly confluent monolayers prepared in 75 cm2 flasks. Flasks were individually seeded with up to 107 B6/K-0 cells in Dulbeccos modified Eagle medium supplemented with 10% heat inactivated fetal bovine serum (HyClone, Logan, Utah), 100 U of penicillin per ml, 100 μg of streptomycin per ml, 2 mM L-glutamine, 100 μg of kanamycin per ml, 20 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid (HEPES), and 0.15% (wt/vol) sodium bicarbonate (DMEM10), and were incubated at 37 C with 5% CO2 in a humidified incubator from four hours to overnight. The plating medium was aspirated and replaced with 18 ml of fresh selection medium after which appropriate numbers of the respective CTL clone were added to each flask. Selections employing the CTL clones H-1, Y-5, K-19 and Y-1 were performed in RPMI 1640 medium (Gibco BRL, Gaithersburg, Md.) supplemented with 10% heat inactivated fetal bovine serum (HyClone), 2 mM L-glutamine, 5 × 10-5 M β-mercaptoethanol, 25 μg of pyruvic acid per ml, 100 U of penicillin per ml, 100 μg of streptomycin per ml, and 5 mM HEPES (CTL medium). K-11 selections were attempted using either CTL medium or DMEM10; no K-11-resistant variants were obtained following multiple attempts in which the RPMI-based CTL medium was used during the selection. CTL were harvested from culture wells using versene, pelleted, and resuspended in CTL medium to a concentration of 0.5 - 2 × 106 CTL, and volumes of CTL representing initial ratios of 0.1 to 0.3 effector CTL per B6/K-0 target cell were added to individual selection flasks. Following CTL addition, flasks were incubated at 37 C, 5% CO2 in a humidified incubator and agitated periodically for two to three days after which nonadherent cells and medium were aspirated from the flasks and replaced with 18 ml of DMEM10. Depending on the efficiency with which the target cell monolayer had been disrupted, individual flasks were incubated in medium alone overnight, or for several days (or weeks) before CTL addition was repeated. Because the CTL clone K-11 survived and proliferated in the selection cultures, it was necessary to routinely reduce the number of viable K-11 CTL in the selection cultures one or two days following addition of the CTL by replacing the selection culture medium with DMEM that contained 5% (v/v) FBS. CTL additions were routinely performed up to twice weekly over a period of three to seven weeks by which time a resistant monolayer (or large islands of resistant cells) had developed, or treatment on an individual selection culture was discontinued due to the absence of resistant fibroblast cells. Resistant cell monolayers were routinely harvested from the primary selection cultures by trypsinization, and were replated into fresh flasks for expansion prior to characterization and preservation. Following initial trypsinization and replating, most resistant populations were re-exposed to the selecting CTL clone to remove residual wild type B6/K-0 cells. CTL-selected populations were tested for uniform expression of the T ag by nuclear immunofluorescence using the monoclonal antibodies 419 and 901 (Harlow et al., 1981; Thompson et al., 1990) as described (Kierstead and Tevethia, 1993; Thompson et al., 1990) and were tested for recognition by T ag-specific CTL clones in standard cytotoxicity assays.
PCR amplification and DNA sequence analysis of variant epitopes
High molecular weight DNA was prepared from the CTL-selected populations or unselected B6/K-0 cells by phenol extraction and ethanol precipitation following overnight extraction of 1 - 3 × 106 trypsinized, PBS-washed cells in the presence of SDS and proteinase K (Schell et al., 1999). Aliquots representing less than 3% of the total nucleic acid yield were subjected to 30 cycles of PCR amplification using primers which flanked the region of SV40 T ag containing the epitope recognized by the selecting CTL clone using Perkin-Elmer models 9600 or 2400 Gene Amp or Techne Genius thermal cyclers and Taq DNA polymerase (Perkin Elmer or PGC Scientific). Multiple primer pairs were used for some amplifications because deletions encompassing the epitope and its flanking sequences were suspected, or preliminary PCR reactions performed with closely spaced primers produced no visible product. The primer pairs (sense and antisense) used to amplify sequences surrounding T ag epitopes I and II/III or epitope V and the sizes predicted for full sized amplification products were as follows: Epitopes I and/or II/III, STEV219 5′-TCCAACCTATGGAACTGATGAATG-3′ and STEV189 5′-TAGTTAATTGTAGGCTATCAACCCGC-3′ (820 bp), or JTEV27 5′-ATGGAAAATATTCTGTAACC-3′ and STEV02 5′-GCTCCTTTAACCCACCTGGC-3′ (238 bp); Epitope V, STEV335, 5′-GAATTATGTGGGGGGAAAGC-3′ and JTEV03, 5′-TCATGGTGACTATTCCAGGGGG-3′ (264 bp). Oligonucleotides used for PCR or DNA sequencing were synthesized by a MilliGen/Biosearch 7500 DNA synthesizer in the Macromolecular Core Facility of the Pennsylvania State University College of Medicine, and were routinely purified prior to use by NENsorb chromatography.
Products of the PCR reactions were visualized by ethidium bromide staining following electrophoresis on analytical 1 or 2% (w/v) agarose gels. The nucleotide sequences of PCR products resulting from a subset of epitope V selections were determined by direct sequencing following a second round of asymmetric amplification. The majority of individual variant sequences were identified by sequencing individual PCR products following their random isolation as recombinant subclones in the vectors pUC19 (Yanisch-Perron, Vieira, and Messing, 1985) or pGEM-T (Promega). PCR products of the predicted size (or shorter) were excised from preparative agarose gels, purified by the QIAEX II procedure (Qiagen), concentrated by ethanol precipitation, dissolved in a minimal volume (0.01 ml) of glass distilled water, and used for ligation. Candidate plasmids were screened by restriction digestion for the presence of appropriate sized inserts and were prepared for DNA sequence analysis by using the QIAwell8 (Qiagen) or High Pure Plasmid Isolation (Roche) procedures. DNA sequence analysis of plasmid DNAs was performed manually using 35S-labeled dATP and dideoxynucleotide triphosphate terminators (Sequenase, United States Biochemicals), or by an automated method utilizing fluorescent terminators that was performed by the Macromolecular Genetics Core Facility of the Penn State University School of Medicine. The primers JTEV27 or STEV335, respectively, were used to sequence recombinant plasmids bearing epitope I or epitope V region sequences. Sequencing reactions for deleted epitope regions were primed with either STEV219 or STEV189 (epitope I, II/II region) or universal primers complementary to flanking vector sequences.
MHC class I molecule stabilization assay
Synthetic peptides corresponding wild type or substituted SV40 T ag epitope sequences were compared for the ability to stabilize MHC class I molecules on the surface of MHC class I presentation-defective RMA/s cells by a flow cytometry assay as described previously (Fu et al., 1998; Lippolis et al., 1995; Mylin et al., 1995a) using the H-2Db-specific, conformation sensitive monoclonal antibody 28-14-8 (Potter et al., 1984). A peptide corresponding to the H-2Kb-restricted HSVI-RR1 epitope (Salvucci, Bonneau, and Tevethia, 1995) was used as a non-binding control.
Cytotoxicity assays
Conditions for the use of SV40 T ag-specific CTL clones in cytotoxicity assays have been described (Lippolis et al., 1995; Mylin et al., 1995a). CTL-selected fibroblast target populations were labeled with 51Cr, harvested with trypsin, washed, and combined with graded numbers of selecting or non-selecting CTL clone effectors. For most assays, fibroblast target cell populations were pre-treated with gamma interferon (Pharmacia) for two days to optimize MHC class I presentation (Lippolis et al., 1995; Mylin et al., 1995a). The conditions used to compare lysis of RMA cells incubated in the presence of varying concentrations of synthetic peptides have been described (Fu et al., 1998). The H-2Db-binding peptide, SMIKNLEYM (Gairin and Oldstone, 1992) that is not recognized by T ag-specific CTL (Mylin et al., 1995a) was used as a control. Synthetic peptides used in this study were synthesized at the Macromolecular Core Facility of The Pennsylvania State University College of Medicine by 9-fluorenylmethoxycarbonyl chemistry on an automated peptide synthesizer (9050 Milligen PepSynthesizer) or were obtained from Chiron (pin synthesis) and were routinely used without purification.
Acknowledgements
This study was supported by the Public Health Service Grant CA25000 from the National Cancer Institute and American Cancer Society Research Scholar Grant RSG04-059-01-LIB. Additional funding was provided through a grant to Messiah College (Project Inquiry) from The Whitaker Regional Foundation, and support from the Messiah College Department of Biological Sciences. We also acknowledge the contributions of Messiah College students Albert Siha, Sean Kelly, Jocelyn Megargell, Melissa Harris, Kevin Driver, Christian Boyer, Jeffery Siglin, Anuj Kalsy and Charles F. Rhoads III. We thank Dr. M. J. Tevethia for critical reading of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akilesh S, Dudley ME, Eden PA, Roopenian DC. Efficient chromosomal mapping of a methylcholanthrene-induced tumor antigen by CTL immunoselection. J. Immunol. 2001;167:5143–5149. doi: 10.4049/jimmunol.167.9.5143. [DOI] [PubMed] [Google Scholar]
- Bai XF, Liu J, Li O, Zheng P, Liu Y. Antigenic drift as a mechanism for tumor evasion of destruction by cytolytic T lymphocytes. J Clin Invest. 2003;111(10):1487–96. doi: 10.1172/JCI17656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann CC, Yao Q, Ho CK, Buckwold SL. Flanking residues alter antigenicity and immunogenicity of multi-unit CTL epitopes. J. Immunol. 1996;157:3242–3249. [PubMed] [Google Scholar]
- Boon T, Cerottini JC, Van den Eynde B, van der Bruggen P, Van Pel A. Tumor antigens recognized by T lymphocytes. Annu. Rev. Immunol. 1994;12:337. doi: 10.1146/annurev.iy.12.040194.002005. [DOI] [PubMed] [Google Scholar]
- Boon T, van der Bruggen P. Human tumor antigens recognized by T lymphocytes. J. Exp. Med. 1996;183:725–729. doi: 10.1084/jem.183.3.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrow P, Shaw GM. Cytotoxic T-lympocyte escape viral variants: how important are they in viral evasion of immune clearance in vivo? Immunol. Rev. 1998;164:37–51. doi: 10.1111/j.1600-065X.1998.tb01206.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brander C, Yang OO, Jones NG, Lee Y, Goulder P, Johnson RP, Trocha A, Colbert D, Hay C, Buchbinder S, Bergmann CC, Zweerink HJ, Wolinsky S, Blattner WA, Kalams SA, Walker BD. Efficient processing of the immunodominant HLA-A*0201-restricted human immunodeficiency virus type 1 cytotoxic T-lymphocyte epitope despite multiple variations in the epitope flanking sequences. J. Virol. 1999;73:10191–10198. doi: 10.1128/jvi.73.12.10191-10198.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell AE, Foley FL, Tevethia SS. Demonstration of multiple antigenic sites of the SV40 transplantation rejection antigen by using cytotoxic T lymphocyte clones. J. Immunol. 1983;130(1):490–492. [PubMed] [Google Scholar]
- Charini WA, Nuroda MJ, Schmitz JE, Beaudry KR, Lin W, Lifton MA, Krivulka GR, Necker A, Letvin NL. Clonally diverse CTL response to a dominant viral epitope recognizes potential epitope variants. J. Immunol. 2001;167:4996–5003. doi: 10.4049/jimmunol.167.9.4996. [DOI] [PubMed] [Google Scholar]
- Coulie PG, Karanikas V, Colau D, Lurquin C, Landry C, Marchand M, Dorval T, Brichard V, Boon T. A monoclonal cytolytic T-lymphocyte response observed in a melanoma patient vaccinated with a tumor-specific antigenic peptide encoded by gene MAGE-3. Proc Natl Acad Sci U S A. 2001;98(18):10290–5. doi: 10.1073/pnas.161260098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deckhut AM, Lippolis JD, Tevethia SS. Comparative analysis of core amino acid residues of H-2Db-restricted cytotoxic T-lymphocyte recognition epitopes in simian virus 40 T antigen. J. Virol. 1992;66(1):440–447. doi: 10.1128/jvi.66.1.440-447.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deckhut AM, Tevethia MJ, Haggerty S, Frisque RJ, Tevethia SS. Localization of common cytotoxic T lymphocyte recognition epitopes on Simian Papovavirus SV40 and human Papovavirus JC virus T antigens. Virol. 1991;183:122–132. doi: 10.1016/0042-6822(91)90125-u. [DOI] [PubMed] [Google Scholar]
- Deckhut AM, Tevethia SS. Effect of point mutations in the native Simian Virus 40 tumor antigen, and in synthetic peptides corresponding to the H-2Db-restricted epitopes, on antigen presentation and recognition by cytotoxic T lymphocyte clones. J. Immunol. 1992;148(10):3012–3020. [PubMed] [Google Scholar]
- Del Val M, Schlicht HJ, Ruppert T, Reddehase MJ, Kosinowski UH. Efficient processing of an antigenic sequence for presentation by MHC class I molecules depends on its neighboring residues in the protein. Cell. 1991;66:1145–1153. doi: 10.1016/0092-8674(91)90037-y. [DOI] [PubMed] [Google Scholar]
- Dudley ME, Roopenian DC. Loss of a unique tumor antigen by cytotoxic T lymphocyte immunoselection from a 3-methylcholanthrene-induced mouse sarcoma reveals secondary unique and shared antigens. J. Exp. Med. 1996;184:441–447. doi: 10.1084/jem.184.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley ME, Wunderlich JR, Robbins PF, Yang JC, Hwu P, Schwartzentruber DJ, Topalian SL, Sherry R, Restifo NP, Hubicki AM, Robinson MR, Raffeld M, Duray P, Seipp CA, Rogers-Freezer L, Morton KE, Mavroukakis SA, White DE, Rosenberg SA. Cancer regression and autoimmunity in patients after clonal repopulation with antitumor lymphocytes. Science. 2002;298(5594):850–4. doi: 10.1126/science.1076514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn GP, Bruce AT, Ikeda H, Old LJ, Schreiber RD. Cancer immunoediting: from immunosurveillance to tumor escape. Nature Immunol. 2002;3(11):991–998. doi: 10.1038/ni1102-991. [DOI] [PubMed] [Google Scholar]
- Fanning E. Simian virus 40 large T antigen; the puzzle, the pieces, and the emerging picture. J. Virol. 1992;66:1289–1293. doi: 10.1128/jvi.66.3.1289-1293.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu T-M, Mylin LM, Schell TD, Bacik I, Russ G, Yewdell JW, Bennink JR, Tevethia SS. An endoplasmic reticulum targeting signal sequence enhances the immunogenicity of an immunorecessive simian virus 40 T antigen cytotoxic T lymphocyte epitope. J. Virol. 1998;72:1469–1481. doi: 10.1128/jvi.72.2.1469-1481.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gairin JE, Oldstone MBA. Design of high-affinity major histocompatibility complex-specific antagonist peptides that inhibit cytotoxic T-lymphocyte activity: implications for control of viral disease. J. Virol. 1992;66:6755–6762. doi: 10.1128/jvi.66.11.6755-6762.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn YS, Hahn CS, Braciale VL, Braciale TJ, Rice CM. CD8+ T cell recognition of an endogenously processed epitope is regulated primarily by residues within the epitope. J. Exp. Med. 1992;176:1335–1341. doi: 10.1084/jem.176.5.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow E, Crawford LV, Pim DC, Williamson NM. Monoclonal antibodies specific for simian virus 40 tumor antigens. J. Virol. 1981;39:861–869. doi: 10.1128/jvi.39.3.861-869.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiscott J, Murphy D, Defendi V. Amplification and rearrangement of integrated SV40 DNA sequences accompany the selection of anchorage-independent transformed mouse cells. Cell. 1980;22(2 Pt 2):535–43. doi: 10.1016/0092-8674(80)90363-3. [DOI] [PubMed] [Google Scholar]
- Hiscott JB, Murphy D, Defendi V. Instability of integrated viral DNA in mouse cells transformed by simian virus 40. Proc Natl Acad Sci U S A. 1981;78(3):1736–40. doi: 10.1073/pnas.78.3.1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karre K, Ljunggren HG, Piontek G, Kiessling Selective rejection of H-2-deficient lymphoma variants suggests alternative immune defense strategy. Nature (London) 1986;319:675–678. doi: 10.1038/319675a0. [DOI] [PubMed] [Google Scholar]
- Khong HT, Restifo NP. Natural selection of tumor variants in the generation of “tumor escape” phenotypes. Nature Immunol. 2002;3(11):999–1005. doi: 10.1038/ni1102-999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kierstead TD, Tevethia MJ. Association of p53 binding and immortalization of primary C57BL/6 mouse embryo fibroblasts by using Simian Virus 40 T-antigen mutants bearing internal overlapping deletion mutations. J. Virol. 1993;67(4):1817–1829. doi: 10.1128/jvi.67.4.1817-1829.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klenerman P, Wu Y, Phillips R. HIV: current opinion in escapology. Curr. Opin. Microbiol. 2002;5:408. doi: 10.1016/s1369-5274(02)00339-9. [DOI] [PubMed] [Google Scholar]
- Lill NL, Tevethia MJ, Hendrickson WG, Tevethia SS. Cytotoxic T lymphocytes (CTL) against a transforming gene product select for transformed cells with point mutations within sequences encoding CTL recognition epitopes. J. Exp. Med. 1992;176:449–457. doi: 10.1084/jem.176.2.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippolis JD, Mylin LM, Simmons DT, Tevethia SS. Functional analysis of amino acid residues encompassing and surrounding two neighboring H-2Db-restricted cytotoxic T-lymphocyte epitopes in simian virus 40 tumor antigen. J. Virol. 1995;69(5):3134–3146. doi: 10.1128/jvi.69.5.3134-3146.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljunggren HG, Karre K. Host resistance directed selectively against H-2 deficient lymphoma variants. Analysis of mechanism. J. Exp. Med. 1985;162:1745–1759. doi: 10.1084/jem.162.6.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loftus DJ, Squarcina P, Nielsen MB, Geisler C, Castelli C, Odum N, Appella E, Parmiani G, Rivoltini L. Peptides derived from self-proteins as partial agonists and antagonists of human CD8+ T-cell clones reactive to melanoma/melanocyte epitope MART1(27-35) Cancer Res. 1998;58:2433–2439. [PubMed] [Google Scholar]
- Manfredi JJ, Prives C. The transforming activity of simian virus 40 large tumor antigen. Biochem. Biophys. Acta. 1994;1198:65–83. doi: 10.1016/0304-419x(94)90006-x. [DOI] [PubMed] [Google Scholar]
- Mortara L, Letourneur F, Gras-Masse H, Venet A, Guillet JG, Bourgault-Villada I. Selection of virus variants and emergence of virus escape mutants after immunization with an epitope vaccine. J. Virol. 1998;72:1403–1410. doi: 10.1128/jvi.72.2.1403-1410.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mylin LM. Context-dependent immunogenicity of an S206G-substituted H-2Db-restricted Simian virus 40 large T antigen epitope I variant. J. Immunol. 1999;162:2171–2179. [PubMed] [Google Scholar]
- Mylin LM, Bonneau RH, Lippolis JD, Tevethia SS. Hierarchy among multiple H-2b-restricted cytotoxic T-lymphocyte epitopes within simian virus 40 T antigen. J. Virol. 1995;69(a)(11):6665–6677. doi: 10.1128/jvi.69.11.6665-6677.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mylin LM, Deckhut AM, Bonneau RH, Kierstead TD, Tevethia MJ, Simmons DT, Tevethia SS. Cytotoxic T lymphocyte escape variants, induced mutations, and synthetic peptides define a dominant H-2Kb-restricted determinant in simian virus 40 tumor antigen. Virology. 1995;208(b):159–172. doi: 10.1006/viro.1995.1139. [DOI] [PubMed] [Google Scholar]
- Mylin LM, Schell TD, Roberts D, Epler M, Boesteanu A, Collins EJ, Frelinger JA, Joyce S, Tevethia SS. Quantitation of CD8+ T-lymphocyte responses to multiple epitopes from Simian virus 40 (SV40) large T antigen in C57BL/6 mice immunized with SV40, SV40 T-antigen transformed cells, or vaccinia virus recombinants expressing full-length T antigen or epitope minigenes. J. Virol. 2000;74:6922–6934. doi: 10.1128/jvi.74.15.6922-6934.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pewe L, Perlman S. Immune response to the immunodominant epitope of mouse hepatitis virus is polyclonal, but functionally monospecific in C57Bl/6 mice. Virology. 1999;255:106–116. doi: 10.1006/viro.1998.9576. [DOI] [PubMed] [Google Scholar]
- Potter TA, Boyer C, Verhulst AM, Golstein P, Rajan TV. Expression of H-2Db on the cell surface in the absence of detectable b-2 microglobulin. J. Exp. Med. 1984;160:317–322. doi: 10.1084/jem.160.1.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan CM, Schell TD. Accumulation of CD8+ T cells in advanced-stage tumors and delay of disease progression following secondary immunization against an immunorecessive epitope. J. Immunol. 2006;177:255–267. doi: 10.4049/jimmunol.177.1.255. [DOI] [PubMed] [Google Scholar]
- Salvucci LA, Bonneau RH, Tevethia SS. Polymorphism within the herpes simplex virus type I (HSV-1) ribonucleotide reductase large subunit (ICP6) confers type specificity for recognition by HSV-1 specific cytotoxic T lymphocytes. J. Virol. 1995;69:1122–1131. doi: 10.1128/jvi.69.2.1122-1131.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schell TD. In vivo expansion of the residual tumor antigen-specific CD8+ T lymphocytes that survive negative selection in simain virus 40 T-antigen-transgenic mice. J. Virol. 2004;78:1751–1762. doi: 10.1128/JVI.78.4.1751-1762.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schell TD, Knowles BB, Tevethia SS. Sequential loss of cytotoxic T lymphocyte responses to Simian virus 40 large T antigen epitopes in T antigen transgenic mice developing osteosarcomas. Cancer Res. 2000;60:3002–3012. [PubMed] [Google Scholar]
- Schell TD, Mylin LM, Georgoff I, Teresky AK, Levine AJ, Tevethia SS. Cytotoxic T-lymphocyte epitope immunodominance in the control of choroid plexus tumors in simian virus 40 large T antigen transgenic mice. J. Virol. 1999;73:5981–5993. doi: 10.1128/jvi.73.7.5981-5993.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schell TD, Tevethia SS. Control of advanced choroid plexus tumors in SV40 T antigen transgenic mice following priming of donor CD8+ T lymphocytes by the endogenous tumor antigen. J. Immunol. 2001;167:6947–6956. doi: 10.4049/jimmunol.167.12.6947. [DOI] [PubMed] [Google Scholar]
- Shastri N, Serwold T, Gonzalez F. Presentation of endogenous peptide/MHC complexes is profoundly influenced by specific C-terminal flanking residues. J. Immunol. 1995;155:4339–4352. [PubMed] [Google Scholar]
- Staveley-O′Carroll K, Schell TD, Jimenez M, Mylin LM, Tevethia MJ, Schoenberger SP, Tevethia SS. In vivo ligation of CD40 enhances priming against the endogenous tumor antigen and promotes CD8+ T cell effector function in SV40 T antigen transgenic mice. J Immunol. 2003;171(2):697–707. doi: 10.4049/jimmunol.171.2.697. [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Anderson RW, Maloy WL, Tevethia SS. Localization of an immunorecessive epitope on SV40 T antigen by H-2Db-restricted cytotoxic T-lymphocyte clones and a synthetic peptide. Virology. 1989;171:205–213. doi: 10.1016/0042-6822(89)90527-8. [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Tevethia MJ, Kalderon D, Smith AE, Tevethia SS. Clustering of antigenic sites recognized by cytotoxic T lymphocyte clones in the amino terminal half of SV40 T antigen. Virology. 1988;162:427–436. doi: 10.1016/0042-6822(88)90483-7. [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Tevethia SS. In vitro selection of SV40 T antigen epitope loss variants by site-specific cytotoxic T lymphocyte clones. J. Immunol. 1988;140(12):4348–4354. [PubMed] [Google Scholar]
- Tanaka Y, Tevethia SS. Loss of immunorecessive cytotoxic T lymphocyte determinant V on SV40 T antigen following cocultivation with site-specific cytotoxic T lymphocyte clone Y-5. Intervirology. 1990;31:197–202. doi: 10.1159/000150154. [DOI] [PubMed] [Google Scholar]
- Tevethia MJ. Immortalization of primary mouse embryo fibroblasts with SV40 virions, viral DNA, and a subgenomic DNA fragment in a quantitative assay. Virol. 1984;137:414–421. doi: 10.1016/0042-6822(84)90234-4. [DOI] [PubMed] [Google Scholar]
- Tevethia SS, Beachy T, Schell T, Lippolis J, Newmaster R, Mylin L, Tevethia MJ. Role of CTL host responses and their implication for tumorigenicity testing and the use of tumour cells as vaccine substrate. Dev. Biol. Stand. 2001;106:109–121. [PubMed] [Google Scholar]
- Tevethia SS, Flyer DC, Tjian R. Biology of Simian Virus 40 (SV40) transplantation antigen (TrAg). VI. Mechanism of induction of SV40 transplantation immunity in mice by purified SV40 T antigen (D2 protein) Virology. 1980;107:13–23. doi: 10.1016/0042-6822(80)90268-8. [DOI] [PubMed] [Google Scholar]
- Tevethia SS, Mylin L, Newmaster R, Epler M, Lednicky JA, Butel JS, Tevethia MJ. Cytotoxic T lymphocyte recognition sequences as markers for distinguishing among tumour antigens encoded by SV40, BKV, and JCV. Dev. Biol. Stand. 1998;94:329–339. [PubMed] [Google Scholar]
- Theobald M, Ruppert T, Kuckelhorn U, Hernzndez J, Haussler A, Ferreira EA, Liewer U, Biggs J, Levine AJ, Huber C, Kozinowski UH, Kloetzel PM, Sherman LA. The sequence alteration associated with a mutational hotspot in p53 protects cells from lysis by cytotoxic T lymphocytes specific for a flanking peptide epitope. J. Exp. Med. 1998;188:1017–1028. doi: 10.1084/jem.188.6.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson DL, Kalderon D, Smith AE, Tevethia MJ. Dissociation of Rb-binding and anchorage-independent growth from immortalization and tumorigenicity using SV40 mutants producing N-terminally truncated large T antigens. Virol. 1990;178:15–34. doi: 10.1016/0042-6822(90)90375-2. [DOI] [PubMed] [Google Scholar]
- Turner SJ, Carbone FR. A dominant V beta bias in the CTL response after HSV-1 infection is determined by peptide residues predicted to also interact with the TCR beta-chain CDR3. Mol. Immunol. 1998;35:307–316. doi: 10.1016/s0161-5890(98)00051-0. [DOI] [PubMed] [Google Scholar]
- Wiesmuller L, Cammenga J, Deppert WW. In vivo assay of p53 function in homologous recombination between Simian virus 40 chromosomes. J. Virol. 1996;70:737–744. doi: 10.1128/jvi.70.2.737-744.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Yellen-Shaw AJ, Eisenlohr LC. Regulation of class I-restricted epitope processing by local or distal flanking residues. J. Immunol. 1997;158:1727–1733. [PubMed] [Google Scholar]
- Yellen-Shaw AJ, Wherry EJ, Dubois GC, Eisenlohr LC. Point mutation flanking a CTL epitope ablates in vitro and in vivo recognition of a full-length viral protein. J. Immunol. 1997;158:3227–3234. [PubMed] [Google Scholar]
- Young AC, Zhang W, Sacchettini JC, Nathenson SG. The three-dimensional structure of H-2Db at 2.4 A resolution: implications for antigen-determinant selection. Cell. 1994;76(1):39–50. doi: 10.1016/0092-8674(94)90171-6. [DOI] [PubMed] [Google Scholar]