Abstract
Background/Aim
Geographic gradients in breast cancer incidence and mortality suggest that vitamin D may reduce risk. The enzyme 25-hydroxyvitamin D 24-hydroxylase (CYP24A1), which degrades the active form of vitamin D, and the vitamin D receptor (VDR) are both found in breast tissue. We investigated six polymorphisms in CYP24A1 and two in the VDR gene in association with breast cancer risk.
Materials and Methods
We conducted a case--control study within the nationwide U.S. Radiologic Technologists cohort, including 845 controls and 484 incident breast cancer cases. Associations of polymorphic variants and ecologic and personal measures of sun exposure with breast cancer risk were assessed using unconditional logistic regression.
Results
Two polymorphisms in CYP24A1 were associated with increased breast cancer risk (rs34043203, Ptrend = 0.03; rs2762934, Ptrend = 0.005) and one with reduced breast cancer risk (rs1570669, Ptrend=0.048). Risk was inversely associated with minor alleles for the VDR Bsm1 polymorphism (rs1544410, Ptrend = 0.05) but not Fok1 (rs2228570). Sunlight measures were not associated with breast cancer risk, however significant interactions between time outdoors in the teen years and three unlinked genotypes were found for VDR (rs1544410, rs2228570) and CYP24A1 (rs1570669).
Conclusion
In this nation-wide breast cancer case--control study, we found the vitamin D pathway was involved in disease etiology and further suggest that reduced cancer risk in association with sunlight may depend on timing of exposure and genetic background. These findings merit further investigation.
Keywords: Vitamin D, sunlight, polymorphisms, breast cancer, gene, case—control
Observations of North-South gradients in breast cancer incidence and mortality have led investigators to hypothesize that vitamin D, which is synthesized in sunlight-exposed skin, may play a role in breast cancer prevention (1, 2). A large body of experimental evidence supports the hypotheses that vitamin D plays an important role as a regulator of cell proliferation and differentiation in the breast and that vitamin D insufficiency can contribute to the pathogenesis of breast cancer (3).
Geographic and behavioral factors play important roles in determining exposure to ambient ultraviolet (UV) radiation and can therefore influence the ability to synthesize the vitamin D precursor 7-dehydrocholesterol cutaneously (4, 5). Genetic factors may also play an important role in determining systemic and tissue levels of vitamin D and the health consequences of vitamin D insufficiency (6).
Circulating 25-hydroxyvitamin D reflects UV exposure and dietary vitamin D intake, and is the precursor to the active form of vitamin D: 1,25-hydroxyvitamin D. The effects of vitamin D are mediated through an interaction of 1,25-hydroxyvitamin D with the vitamin D receptor (VDR). The enzyme 25-hydroxyvitamin D 24-hydroxylase (CYP24A1) catalyzes an irreversible and rate-limiting step in the degradation of 1,25-hydroxyvitamin D. CYP24A1 and the VDR are found in normal breast tissue and in breast tumors (3). Because CYP24A1 is strongly up-regulated by VDR signaling, it provides a negative feedback mechanism to vitamin D and its expression and activity are important determinants of the availability of activated vitamin D in breast and other target tissues. CYP24A1 has been observed to be overexpressed in breast tumors, where its association with increased cancer risk is presumed to act through suppression of vitamin D signaling (7).
Single nucleotide polymorphisms (SNPs) in vitamin D-related genes could represent risk factors for breast cancer, and thus potentially identify susceptible subgroups which might benefit from elevated and/or sustained levels of vitamin D. While many studies have focused on candidate polymorphisms in VDR and risk of breast cancer (8, 9), variants in CYP24A1 have not been studied extensively with respect to breast cancer risk. In this case--control study, we investigated associations of breast cancer risk with six polymorphisms in CYP24A1 and two variants in VDR. We also investigated whether these genetic variants modify the associations of ecologic and individual measures of sunlight exposure in early and later life, with breast cancer risk.
Materials and Methods
Study population
In 1982, the U.S. National Cancer Institute, in collaboration with the University of Minnesota and the American Registry of Radiologic Technologists, initiated a study of cancer incidence and mortality among 146,022 (106,953 female) U.S. radiologic technologists (USRT) who had been certified for at least two years between 1926 and 1982. Surveys were mailed to all eligible cohort members at study baseline in 1984-89 and again in 1993-98; each questionnaire included queries about work history as a radiologic technologist, family history of cancer, reproductive history, height, weight, other cancer risk factors (such as alcohol and tobacco use), and queries regarding health outcomes, including breast cancer. Seventy-one percent (69,524 out of 98,233) and 74% (69,998 out of 94,508) of female technologists known to be alive at the time of the first and second survey responded to the questionnaires, respectively [for questionnaires, see http://radtechstudy.nci.nih.gov; for other study participation details, see (10)]. This study has been approved annually by the Human Subjects Review Boards of the National Cancer Institute and the University of Minnesota.
Case and control recruitment
Cases were recruited from the set of living female technologists reporting a primary breast cancer (ductal carcinoma in situ or invasive breast cancer) on either the first or second questionnaire; diagnoses were confirmed based on pathology or medical records. In December 1999 when biospecimen collection began, there were 1386 living prevalent breast cancer cases that had been diagnosed between 1955-1998. By the end of December 2003, 874 (63%) breast cancer cases had provided informed consent and a blood sample.
Female controls were selected from the set of USRT participants who responded to either the first and/or the second questionnaire and had not reported a breast cancer at the time of blood collection. Controls were frequency-matched to cases (ratio 1.5:1) by birth year in 5-year strata. Of 2,268 living controls identified, 1,094 (48%) provided informed consent and a blood sample, and completed a telephone interview. Details on participation, characteristics of responders and non-responders, and comparisons with decedents have been previously published (11). These comparisons did not reveal any meaningful differences between participants and non-participants.
Because variation in vitamin D pathway genes could affect both risk of breast cancer and survival after diagnosis, the present analysis was designed as an incident breast cancer case-control study. Therefore, we selected participants who responded to both the first and second surveys and reported no history of breast cancer at baseline. Of 1,094 enrolled controls, 845 met these criteria. Of 874 cases in the series, 484 met these criteria and also reported an incident, primary breast cancer diagnosis occurring in the intervening period between the first and second surveys.
Sample handling
After venipuncture, whole blood samples were shipped overnight with a temperature stabilizing pack to the processing laboratory in Frederick, MD, USA. Blood components were separated and DNA was extracted using Qiagen mini-kits (Qiagen Inc., Valencia, CA, USA). Samples were tracked by a unique ID code, and laboratory investigators were blinded to case--control status.
Selection of candidate SNPs and sample genotyping
The selected polymorphic variants are listed in Table I. We based our selection strategy on whether the variants had been studied in previous epidemiological studies and if they were considered potentially functional based on their locations in promoter regions or splice sites. We chose six variants in the CYP24A1 gene (rs2248137, rs2296237, rs2762934, rs1570669, rs1977297, rs34043203) and two variants in the VDR gene (Bsm1, which is rs1544410 and Fok1, which is rs2228570). Samples were genotyped using standard TaqMan or MGB Eclipse assays. Genotyping methods for specific SNPs can be found online (16). There were 115 quality control samples embedded randomly in the sample trays, composed of between nine and 14 replicate samples from the same 10 individuals. All laboratory personnel were blinded to the location of the replicates. Of the replicated samples for the eight assays, there were no discrepancies. For the various SNP assays, completion success ranged between 96.2% -99.1%; with an average of 98.0%.
Table I. Selected polymorphic variants in the vitamin D receptor (VDR) and 25-hydroxyvitamin D 24-hydroxylase (CYP24A1) genes.
| Gene/SNP | Description | Chromosome | Base position* | Minor allele frequency | Function |
|---|---|---|---|---|---|
| CYP24A1 | |||||
| rs2248137 | IVS1-105C>G | 20 | 52789743 | 0.37 | Non-coding |
| rs34043203 | IVS2-341G>A | 20 | 52788550 | 0.11 | Non-coding |
| rs2296237 | IVS8+204T>C | 20 | 52775292 | 0.49 | Non-coding |
| rs1977297 | IVS9+146G>A | 20 | 52774479 | 0.07 | Non-coding |
| rs1570669 | IVS9+198T>C | 20 | 52774427 | 0.23 | Non-coding |
| rs2762934 | Ex12+40T>C | 20 | 52771261 | 0.29 | 3′ untranslated region |
|
| |||||
| VDR | |||||
| rs1544410 (Bsm1) | IVS9+283C>T | 12 | 48239835 | 0.48 | Non-coding |
| rs2228570 (Fok1) | Ex2+4T>C | 12 | 48272895 | 0.17 | Missense |
Per genome build 37.2
Measures of sun exposure
Daily measures of sunlight were made at ground level at National Weather Service stations across the United States from 1977-80 using meters developed by Robertson and Berger (RB) which were calibrated to capture action spectra pertinent to erythemal exposures (wavelengths of 280-330 nm) (12); one RB unit corresponds to approximately 0.35 joules per square meter. This measure is a proxy for the spectra associated with vitamin D production (280-315 nm), although it may somewhat overestimate winter exposures (13). Average annual UVB radiation at Earth's surface was estimated for each state in the U.S. using a regression equation which used latitude, altitude, and cloud cover to explain up to 97% of variability in this data (14).
The state of residence at baseline was ascertained for each participant based upon the mailing address from the first survey; the state of birth was available from the American Registry of Radiologic Technologists records. Time spent outdoors in the summer during childhood (ages <13 years), and the teen years (13-19 years) were ascertained in a third survey, conducted 2001-2005, and so are available only for a subset of cases and controls in the genetic analyses presented here who went on to participate at this later time point [n=372 cases (77%) and n=691 controls (82%)]. Cut-off points for measures of average annual UVB exposure in RB units for each state were determined based upon quartiles of RB values, in controls, for the state of residence at the time of the first survey. Quartiles for hours spent outdoors during the summer in childhood and adolescence were assigned based upon their respective distributions in controls.
Data analysis
A chi-square test was used to test whether allelic distributions among controls departed from expectation based on Hardy-Weinberg equilibrium; no significant deviations were noted. Associations between SNPs and breast cancer risk were evaluated while adjusting for birth year (in 5-year groups) using unconditional logistic regression (SAS v. 9.1; SAS Institute, Cary, NC, USA).
For each SNP, the common allele among controls was considered the referent. Tests for trend were conducted assuming a log additive model for genotype. Our decision to include only incident cases of breast cancer resulted in different age distributions for cases and controls. We used two analytic strategies to account for this problem; firstly, all regression models adjusted for the matching factor (year of birth, in 5-year intervals); secondly, we compared these results with results obtained when a subset of controls was selected randomly from age strata to match the distribution of selected cases. Since risk associations were similar in magnitude and direction using both approaches, we present the results of analyses using the larger set of cases and controls, adjusted for age.
Unconditional logistic regression models were also used to assess trends across categories of sunlight exposure measures. Finally, in exploratory analyses, for each polymorphism we assessed whether the trend in disease risk across categories of sunlight differed significantly in women homogeneous for the common allele vs. those who had at least one rare allele by adding the measure of sunlight and an interaction term to the logistic models. Because participants with some of the sunlight measures represent a subset of all those included in the present report, we also assessed whether genotypes were associated with the probability of having participated in the third survey, at which time spent outdoors in childhood and teen years were queried; none of these associations were statistically significant.
We assessed the following breast cancer risk factors as potential confounders: age at menarche; age at menopause; number of full-term pregnancies and age at first birth, as reported on the baseline survey, and family history of breast cancer reported at the time of blood collection. None of these factors modified regression coefficients associated with the number of rare alleles by >10%, so we present models with adjustment for age only (15).
Haploview was used to estimate linkage disequilibrium (LD) for each pair of polymorphisms (16). A haplotype block involving 4 SNPs in the CYP24A1 gene was identified (Figure 1). Haplostats for R was used to estimate haplotype frequencies and assess associations between genotypes and breast cancer risk (17).
Figure 1.
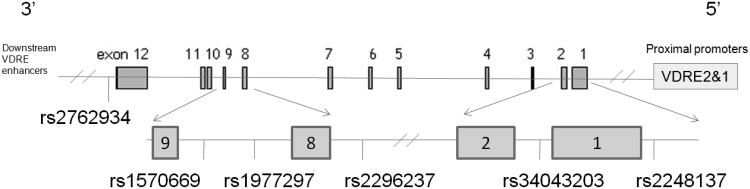
Relative positions of selected polymorphic variants in 25-hydroxyvitamin D 24-hydroxylase (CYP24A1).
The gene CYP24A1 is located at 20q13.
P-Values <0.05 were considered statistically significant given the relatively high priors and small number of variants tested. Tests for interactions were evaluated using cross-product terms, assumed departure from a multiplicative expectation, and due to their exploratory nature were assessed as significant for p-values <0.05. All statistical tests were two-sided.
Results
Of 494 incident breast cancer cases, 397 (80%) had invasive breast cancer, while the rest had in situ tumors. The median time from breast cancer diagnosis to sample collection was nine years. Baseline characteristics of study participants are presented in Table II. Because prevalent cases were excluded from these analyses, study controls, who had been matched to the entire case series on year of birth, were significantly older than remaining cases (P=0.002) and also had a higher body mass index (BMI) (P=0.002). Cases were more likely than controls to be nulliparous (23% vs. 18.5%, P=0.04) and to report a family history of breast cancer (20.5% vs. 16.3%, p=0.05). Compared to parous controls, parous cases reported significantly fewer live births (2.4 vs. 2.6, P =0.04) and later ages at first birth (25.6 years vs. 25.0 years, P =0.04).
Table II.
Baseline characteristics of breast cancer cases and controls from the U.S. Radiologic Technologists cohort.
| Controls (n=845) | Cases (n=494) | P-Value | |
|---|---|---|---|
|
| |||
| Mean (S.D.) | Mean (S.D.) | ||
| Age (years) | 45.2 (10.6) | 43.4 (10.0) | 0.002 |
| Body mass index (kg/m2) | 24.6 (4.3) | 23.9 (4.2) | 0.002 |
| Age at menarche (years) | 12.7 (1.7) | 12.5 (1.5) | 0.10 |
| Age at first birth (years)* | 25.0 (4.2) | 25.6 (4.1) | 0.04 |
| Number of live births* | 2.6 (1.2) | 2.4 (1.2) | 0.04 |
| Age at menopause (years)** | 43.1 (7.9) | 42.2 (8.2) | 0.25 |
| Years from diagnosis to specimen collection (years) | --- | 9 (3) | --- |
|
| |||
| % | % | ||
|
| |||
| Caucasian | 97.0 | 98.1 | 0.63 |
| Postmenopausal | 42.2 | 46.1 | 0.03 |
| Nulliparous | 18.5 | 23.2 | 0.04 |
| Positive family history of breast cancer | 16.3 | 20.5 | 0.05 |
| Ever smoker | 50.0 | 50.6 | 0.95 |
| Invasive breast cancer | --- | 80.4 | --- |
Among 371 parous cases and 687 parous controls.
Among 119 postmenopausal cases and 265 postmenopausal controls.
In Table III, we present associations of individual polymorphisms with breast cancer risk. Two polymorphisms in CYP24A1 showed statistically significant associations with breast cancer risk. Breast cancer risk increased significantly with the number of rare alleles of rs34043203 at CYP24A1 [per allele odds ratio (OR)=1.27, 95% confidence interval (CI)= 1.02-1.59, Ptrend=0.02]. A significant trend in risk was seen with increasing number of rare alleles at rs2762934 in the CYP24A1 gene (per allele OR=1.35, 95% CI=1.09-1.67, Ptrend=0.005). Results for two moderately correlated SNPs (D′=0.50) had marginal associations with risk; SNP rs1977297 of CYP24A1 showed an increasing trend with risk by number of rare alleles (per allele OR=1.25, 95% CI=0.97-1.64, Ptrend=0.08) while those for rs1570669 suggested an inverse association (per allele OR=0.84, 95% CI=0.71-1.00, Ptrend=0.048). Risk associations were similar in magnitude and direction when only invasive breast cancer cases were included (data not shown).
Table III. Risk of breast cancer* by 25-hydroxyvitamin D 24-hydroxylase (CYP24A1) and vitamin D receptor (VDR) genotype in U.S. Radiologic Technologists cohort.
| Va riants | Cases/Controls | Odds ratio | 95% CI | Ptrend** | |
|---|---|---|---|---|---|
| CYP24A1 | |||||
| rs2248137 | |||||
| GG | 174/325 | 1.00 | - | - | |
| CG | 224/374 | 1.12 | 0.87 | 1.44 | |
| CC | 77/144 | 1.00 | 0.71 | 1.40 | |
| CG+CC | 1.08 | 0.86 | 1.38 | ||
| Per allele | 1.02 | 0.87 | 1.20 | 0.79 | |
|
| |||||
| rs34043203 | |||||
| CC | 345/631 | 1.00 | - | - | |
| TC | 118/191 | 1.14 | 0.87 | 1.48 | |
| TT | 18/13 | 2.81 | 1.35 | 5.87 | |
| TC+TT | 1.25 | 0.97 | 1.61 | ||
| Per allele | 1.27 | 1.02 | 1.59 | 0.02 | |
|
| |||||
| rs2296237 | |||||
| CC | 296/508 | 1.00 | - | - | |
| TC | 155/290 | 0.92 | 0.72 | 1.18 | |
| TT | 23/43 | 0.93 | 0.55 | 1.59 | |
| TC+TT | 0.92 | 0.73 | 1.17 | ||
| Per allele | 0.94 | 0.78 | 1.14 | 0.54 | |
|
| |||||
| rs1977297 | |||||
| GG | 372/691 | 1.00 | - | - | |
| GA | 73/143 | 1.22 | 0.91 | 1.63 | |
| AA | 7/7 | 2.10 | 0.72 | 6.09 | |
| GA+AA | 1.27 | 0.96 | 1.69 | ||
| Per allele | 1.25 | 0.97 | 1.64 | 0.08 | |
|
| |||||
| rs1570669 | |||||
| TT | 209/327 | 1.00 | - | - | |
| TC | 214/399 | 0.83 | 0.65 | 1.05 | |
| CC | 53/114 | 0.73 | 0.50 | 1.05 | |
| TC+CC | 0.82 | 0.65 | 1.03 | ||
| Per allele | 0.84 | 0.71 | 1.00 | 0.048 | |
|
| |||||
| rs2762934 | |||||
| CC | 298/566 | 1.00 | - | - | |
| TC | 160/219 | 1.36 | 1.06 | 1.75 | |
| TT | 17/18 | 1.79 | 0.90 | 3.54 | |
| TC+TT | 1.39 | 1.09 | 1.78 | ||
| Per allele | 1.35 | 1.09 | 1.67 | 0.005 | |
|
| |||||
| VDR | |||||
| rs1544410 (Bsm1) | |||||
| GG | 185/290 | 1.00 | - | - | |
| GA | 214/404 | 0.81 | 0.63 | 1.04 | |
| AA | 67/144 | 0.69 | 0.49 | 0.97 | |
| GA+AA | 0.78 | 0.61 | 0.98 | ||
| Per allele | 0.82 | 0.70 | 0.97 | 0.02 | |
|
| |||||
| rs2228570 (Fok1) | |||||
| CC | 201/323 | 1.00 | - | - | |
| CT | 206/385 | 0.85 | 0.67 | 1.09 | |
| TT | 70/134 | 0.83 | 0.59 | 1.17 | |
| CT+TT | 0.85 | 0.67 | 1.07 | ||
| Per allele | 0.90 | 0.76 | 1.06 | 0.20 | |
Odds ratio (OR) were estimated using unconditional logistic regression with models adjusted for the frequency matching variable year of birth.
Ptrend reflects the significance level associated with the number of rare alleles (0, 1, or 2) in a log additive model.
SNP markers within CYP24A1 studied here were in moderate LD (D′ between each pair of variants range from 0.01-0.50) and results of haplotype analysis yielded results consistent with, and weaker than, associations observed for individual polymorphisms (data not shown).
We observed an inverse trend in breast cancer risk associated with number of rare alleles for the Bsm1 variant (per allele OR=0.82, 95% CI=0.70-0.97, Ptrend=0.02) in VDR. No significant association with risk was observed for the Fok1-related polymorphism.
We next considered associations of sunlight measures with risk of breast cancer. Breast cancer cases and controls showed no statistically significant differences with respect to geographic region of residence at study baseline (Table IV). No statistically significant trend in risk was noted across categories of average annual ambient sunlight for the state of residence at baseline, however, a marginally significant trend (P =0.07) was observed across categories of ambient sunlight for the state of birth. There were no statistically significant trends across categories of time spent outdoors in the summer during childhood, or during the teen years.
Table IV.
Residential and personal measures of sun exposure in breast cancer cases and controls from the U.S. Radiologic Technologists cohort.
| Controls n (%) | Cases n (%) | Estimates of association | ||
|---|---|---|---|---|
| Odds ratio (95% confidence interval) | Ptrend | |||
| Geographic region at study baseline (%) | 0.90 | |||
| Northwestern U.S. (including Alaska) | 179 (21.2) | 105 (21.7) | 1.00 (ref) | |
| Northeastern U.S. | 289 (34.2) | 160 (33.1) | 0.89 (0.63-1.25) | |
| Southwestern U.S. (including Hawaii) | 200 (23.7) | 108 (22.3) | 0.84 (0.58-1.22) | |
| Southeastern U.S. | 177 (21.0) | 111 (22.9) | 1.04 (0.71 -1.51) | |
|
| ||||
| Annual ambient sunlight in state of residence at study baseline in Robertson-Berger units (RB, joules/m2) | 0.46 | |||
| ≤ 109 | 203 (25.0) | 122 (26.1) | 1.00 (ref) | |
| 110 – 118 | 212 (26.1) | 121 (25.9) | 1.00 (0.70-1.43) | |
| 118 – 146 | 195 (24.0) | 121 (25.9) | 1.04 (0.73-1.49) | |
| >147 | 202 (24.9) | 103 (22.1) | 0.85 (0.58-1.22) | |
|
| ||||
| Annual ambient sunlight in state of residence at birth (RB, joules/m2) | 0.07 | |||
| ≤ 109 | 242 (29.8) | 167 (35.5) | 1.00 (ref) | |
| 110 – 118 | 267 (32.8) | 140 (29.8) | 0.84 (0.61 - 1.15) | |
| 118 – 146 | 150 (18.5) | 94 (20.0) | 0.85 (0.59-1.22) | |
| >147 | 154 (18.9) | 69 (14.7) | 0.69 (0.47-1.01) | |
|
| ||||
| Weekly hours spent outdoors during summer in childhood (age <13 years) | 0.74 | |||
| ≤ 12 | 166 (25.0) | 98 (26.9) | 1.00 (ref) | |
| 12-24.5 | 252 (38.0) | 144 (39.6) | 0.86 (0.60 - 1.24) | |
| 24.5-34.5 | 85 (12.8) | 34 (9.3) | 0.60 (0.35-1.02) | |
| >34.5 | 161 (24.3) | 88 (24.2) | 0.97 (0.65-1.45) | |
|
| ||||
| Weekly hours spent outdoors during summer in adolescence (age 13 to 19 years) | 0.60 | |||
| ≤ 10.5 | 209 (31.5) | 121 (33.5) | 1.00 (ref) | |
| 10.6-18.4 | 124 (18.7) | 59 (16.3) | 0.78 (0.53-1.14) | |
| 18.5-28.5 | 211 (31.8) | 124 (34.4) | 1.01 (0.73-1.38) | |
| ≥28.6 | 120 (18.1) | 57 (15.8) | 0.82 (0.56-1.21) | |
In exploratory analyses, we next assessed whether measured genotypes modified the associations between measures of sun exposure and breast cancer risk (Table V). Among women with rare alleles for the Fok1-related polymorphism in the VDR gene, risk declined with increasing time spent outdoors during summers in the teen years (Ptrend=0.05), while among those homozygous for the common allele, risk estimates did not trend towards reduced breast cancer risk (P for interaction=0.01). Associations between time spent outdoors during the summer and breast cancer risk were similarly modified by the Bsm1 variant of the VDR (P for interaction=0.02), and by a polymorphic variant in CYP24A1 (rs1570669, P for interaction=0.003).
Table V.
Modification of associations between weekly hours spent outdoors in summer during the teen years (13-19 years) and breast cancer risk by vitamin D-related genotypes in the U.S. Radiologic Technologists cohort.
| Homozygous for the common allele | One or more rare alleles | |||||||
|---|---|---|---|---|---|---|---|---|
| Time outdoors (hours per week) | Cases/Controls | Estimates of association | Cases/Controls | Estimates of association | P for interaction | |||
| Polymorphism | Odds ratio (95% confidence interval) | P trend | Odds Ratio (95% confidence interval) | Ptrend | ||||
| VDR Fok1 (rs2228570) | CC | CT+TT | 0.01 | |||||
| ≤10.5 | 45/85 | 1.00 (ref) | 0.09 | 74/123 | 1.00 (ref) | 0.05 | ||
| 10.6-18.4 | 19/55 | 0.62 (0.33-1.18) | 40/69 | 0.92 (0.57-1.51) | ||||
| 18.5-28.5 | 58/75 | 1.43 (0.87-2.36) | 63/135 | 0.79 (0.52-1.21) | ||||
| ≥28.6 | 29/41 | 1.36 (0.74 -2.47) | 28/79 | 0.60 (0.36 -1.01) | ||||
|
| ||||||||
| VDR Bsm1 (rs1544410) | GG | GA+AA | 0.02 | |||||
| ≤10.5 | 38/85 | 1.00 (ref) | 0.18 | 79/120 | 1.00 (ref) | 0.06 | ||
| 10.6-18.4 | 20/37 | 1.11 (0.56-2.17) | 38/86 | 0.61 (0.38-1.00) | ||||
| 18.5-28.5 | 54/60 | 1.95 (1.14-3.33) | 67/150 | 0.68 (0.45-1.03) | ||||
| ≥28.6 | 23/43 | 1.12 (0.59-2.13) | 30/76 | 0.64 (0.38-1.07) | ||||
|
| ||||||||
| CYP24A1 (rs1570669) | TT | TC+CC | 0.003 | |||||
| ≤10.5 | 49/93 | 1.00 (ref) | 0.08 | 72/115 | 1.00 (ref) | 0.02 | ||
| 10.6-18.4 | 22/48 | 0.79 (0.43-1.47) | 36/75 | 0.72 (0.44-1.19) | ||||
| 18.5-28.5 | 53/79 | 1.30 (0.79-2.13) | 68/130 | 0.83 (0.55-1.27) | ||||
| ≥28.6 | 32/39 | 1.59 (0.88-2.87) | 24/81 | 0.47 (0.28-0.83) | ||||
Discussion
In this case--control study, which drew participants from a national cohort, SNPs in genes in the vitamin D pathway were found to be associated with breast cancer risk. Of six polymorphisms in CYP24A1, two variants found in independent regions of the gene were significantly associated with breast cancer risk. In VDR, rare alleles for Bsm1 were significantly associated with breast cancer risk.
Germline polymorphisms in CYP24A1 have been the subject of only a few recent epidemiologic studies of cancer risk (18-21). The variants studied here are found in two regions of the gene (Figure 1): two near the proximal promoter region which includes two vitamin D response elements (VDRE) (22), and four near the 3′ end of the gene, where additional VDRE act as downstream enhancers of expression (23). While most of the selected SNPs are intronic; rs2762934 is found in the 3′ untranslated region of the CYP24A1 gene.
Three studies have investigated associations of CYP24A1 polymorphisms with risk of breast cancer (19-21). In a nested case--control study of postmenopausal breast cancer, which included measures of only one polymorphism in CYP24A1, rs2296241, a well-conserved variant in the fourth exon was not associated with breast cancer risk; this SNP was not among those selected for the present analysis. Recently, Anderson et al. conducted a large breast cancer case-control study in Ontario, Canada; they assessed four SNPs in CYP24A1 and found no statistically significant associations with risk among studied variants (21); of these polymorphisms none are examined or in linkage disequilibrium with the variants studied here.
In a population-based breast cancer case--control study in Shanghai, Dorjgochoo et al. measured 59 SNPs in CYP24A1 in addition to 500 variants in other genes in the vitamin D pathway (20). They found three markers in CYP24A1 with nominal statistical significance (P<0.05) but these did not survive correction for multiple comparisons, as was the case for all other SNPs under study, suggesting that genetic variation in the vitamin D pathway does not play an important role in breast cancer etiology in this population. Variants studied by Dorjgochoo et al. do not coincide with those in the present study however the SNP rs6097809, found to be modestly associated with breast cancer risk with a per allele OR of 0.90, (95% CI=0.83-0.98, Ptrend=0.02) in their study, is in LD with the variant rs1570669 (r2=0.25), for which we found effects of similar direction and magnitude (OR=0.84 per rare allele, 95% CI=0.71-1.00, Ptrend=0.05).
Wang et al. conducted a genome wide association study to find SNPs associated with vitamin D insufficiency among men and women of European descent (6). They found that SNP rs6013897, in the intergenic region downstream of CYP24A1, achieved genome-wide statistical significance as a predictor of vitamin D insufficiency in the pooled sample; in HapMap data this SNP is in weak LD with two SNPs we studied here (rs1570669, rs2762934, with r2=0.09 and 0.12, respectively), found to be associated with significantly reduced, and significantly increased breast cancer risk, respectively. However, it is not clear that these results are comparable, since vitamin D insufficiency for the analysis of Wang et al. was based on circulating 25-hydroxyvitamin D, while we have suggested that CYP24A1 polymorphisms might impact breast cancer risk through effects on the availability of its downstream metabolite, 1,25 hydroxyvitamin D. Genome-wide association studies of breast cancer have not identified loci in CYP24A1, although achieving genome-wide significance may exclude some true risk genes and it may be that additional studies conducted to evaluate interaction may be required to find these loci.
We recorded reduced risk of breast cancer in association with the number of rare alleles for rs1570669 in CYP24A1. This polymorphism is located in a haplotype block identified in our data that includes polymorphisms in the 8th, 9th and 12th introns. Previous research has shown differences among cancer cell lines in enzyme activities of induced and constitutive CYP24A1 resulting from differential splicing of transcripts between exons 9 and 11 (24). Investigators have speculated that germline polymorphisms in the ninth intron could influence splicing but this has not yet been shown (25).
Finally, in exploratory analyses, we assessed potential interactions between the genetic variants and a number of different measures of sunlight exposure, including average annual sunlight in the state of residence at study baseline and at birth, and retrospectively collected data on time spent outdoors during summers in childhood (<13 years) and during the teen years (13-19 years). Several interactions were observed, each indicating that among women carrying rare alleles of genotypes in the VDR and CYP24A1 genes, greater time spent outdoors during summers in the teen years was associated with reduced risk of breast cancer; in contrast, no inverse associations in association with time outdoors were observed in women homozygous for the same genotypes. While a large number of comparisons were made, the clustering of statistically significant interactions suggest that sun exposure and vitamin D may be most important for women who are genetically susceptible and at a time of life when breast tissue is more susceptible to insult.
Polymorphic variation in the CYP24A1 gene has not been extensively studied but may be important as a determinant of vitamin D availability in tissue. In the breast, available 1,25-hydroxyvitamin D is determined not only by the availability of substrate (25-hydroxyvitamin D), but also by the relative expression and activity of the hydroxylases (CYP27B1 and CYP24A1) which activate and degrade vitamin D. Paracrine mechanisms to regulate expression of these enzymes are complex, tissue-specific, and as yet, poorly understood (26). SNP variants in CYP24A1 could result in differences in gene expression, transcript stability, or enzyme activity, and so could have direct effects on tissue levels of 1,25-hydroxyvitamin D. Variation in the gene could also affect co-regulation and so modulate the ability to maintain homeostatic tissue levels of activated vitamin D when circulating 25-hydroxyvitamin D undergoes seasonal fluctuations (27).
A number of polymorphisms in the VDR have been studied previously in association with breast cancer risk, however results are not consistent. Epidemiological studies of Bsm1 and breast cancer risk have inconsistent findings and both a meta-analysis (9) and a pooled study of six prospective studies (8) found no statistically significant association for this polymorphism. The Bsm1–related variant has not been associated with VDR expression or function but is in LD with a known poly (A) variant in the 3′ untranslated region of the gene which is thought to modulate the stability of RNA transcripts. Limited data has suggested that differences across populations in patterns of LD near this variant may explain the heterogeneity seen in association studies using this genetic marker (28).
Variation in the VDR genotypes for Fok1 suggests that the rare allele results in compromised efficiency of signal transduction. Results of a recent pooled analysis of six prospective studies [8] suggest a modest increase in breast cancer risk associated with the rare allele for the Fok1-related polymorphism (combined OR for two rare alleles =1.16, 95% CI=1.04-1.28 and Ptrend=0.006), however, there was significant heterogeneity across studies (P=0.03). In our study, no significant main effect was seen for this polymorphism and in fact measures of association for this locus tended towards an inverse association.
While several previous studies have looked at interactions of dietary intake of vitamin D and its interactions with polymorphic variants in the VDR and CYP24A1, only one has looked at interactions of genotypes with exposure to sunlight, which has a greater impact on circulating vitamin D than diet. In a study by Anderson et al. (21), investigators report that they assessed interactions of variables related to sunlight exposure in adulthood, including geographic location, skin tone and sun-protective behaviors, and found no significant interactions with genetic polymorphisms in predicting breast cancer risk. Animal models of breast cancer and some epidemiological data suggest that prenatal, childhood, and adolescent periods are periods of greater susceptibility to carcinogenic insult since breast tissue remains undifferentiated until the first pregnancy (29). While exploratory, our findings of significant interactions with time spent outdoors in the teen years suggest that vitamin D exposure may be particularly important for later risk of breast cancer.
Our study has several unique features including the availability of genetic material and detailed information about reproductive, demographic and lifestyle factors derived from interviews of all subjects. Study participants came from states across the U.S. A limitation of the present study is retrospective identification of cases, which required survival from the time of breast cancer diagnosis to blood collection; however, analysis of allelic frequencies in cases with varying lengths of time from diagnosis to blood sampling did not suggest that any SNP was a correlate of survival (data not shown). Comparisons of demographic characteristics did not reveal any significant differences between participating cases and those who did not participate (11).
In summary, we have identified a number of variants in the gene that codes for the 1,25-dihydroxyvitamin D3 24-hydroxylase enzyme associated with breast cancer risk. These findings need to be confirmed in other studies. Future studies should include more genes in the vitamin D pathway and greater coverage of these genes, particularly the CYP24A1 gene, and measures of sunlight exposure in early life and adulthood to permit an examination of whether genetic variants interact with vitamin D levels to influence cancer risks.
Acknowledgments
We are grateful to the radiologic technologists who participated in the U. S. Radiologic Technologists Study; Jerry Reid of the American Registry of Radiologic Technologists for continued support of this study; Diane Kampa and Allison Iwan of the University of Minnesota for data collection and study coordination; Laura Bowen of Information Management Systems for biomedical computing statistical support. This research was supported by the Intramural Research Program of the Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health under Contract No. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
References
- 1.Boscoe FP, Schymura MJ. Solar ultraviolet-B exposure and cancer incidence and mortality in the United States, 1993-2002. BMC cancer. 2006;6:264. doi: 10.1186/1471-2407-6-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garland FC, Garland CF, Gorham ED, Young JF. Geographic variation in breast cancer mortality in the United States: a hypothesis involving exposure to solar radiation. Prev Med. 1990;19:614–622. doi: 10.1016/0091-7435(90)90058-r. [DOI] [PubMed] [Google Scholar]
- 3.Welsh J. Vitamin D and prevention of breast cancer. Acta Pharmacol Sin. 2007;28:1373–1382. doi: 10.1111/j.1745-7254.2007.00700.x. [DOI] [PubMed] [Google Scholar]
- 4.Webb AR, Kline L, Holick MF. Influence of season and latitude on the cutaneous synthesis of vitamin D3: exposure to winter sunlight in Boston and Edmonton will not promote vitamin D3 synthesis in human skin. J Clin Endocrinol Metab. 1988;67:373–378. doi: 10.1210/jcem-67-2-373. [DOI] [PubMed] [Google Scholar]
- 5.Kimlin MG. Geographic location and vitamin D synthesis. Mol Aspects Med. 2008;29:453–461. doi: 10.1016/j.mam.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 6.Wang TJ, Zhang F, Richards JB, Kestenbaum B, van Meurs JB, Berry D, Kiel DP, Streeten EA, Ohlsson C, Koller DL, Peltonen L, Cooper JD, O'Reilly PF, Houston DK, Glazer NL, Vandenput L, Peacock M, Shi J, Rivadeneira F, McCarthy MI, Anneli P, de Boer IH, Mangino M, Kato B, Smyth DJ, Booth SL, Jacques PF, Burke GL, Goodarzi M, Cheung CL, Wolf M, Rice K, Goltzman D, Hidiroglou N, Ladouceur M, Wareham NJ, Hocking LJ, Hart D, Arden NK, Cooper C, Malik S, Fraser WD, Hartikainen AL, Zhai G, Macdonald HM, Forouhi NG, Loos RJ, Reid DM, Hakim A, Dennison E, Liu Y, Power C, Stevens HE, Jaana L, Vasan RS, Soranzo N, Bojunga J, Psaty BM, Lorentzon M, Foroud T, Harris TB, Hofman A, Jansson JO, Cauley JA, Uitterlinden AG, Gibson Q, Jarvelin MR, Karasik D, Siscovick DS, Econs MJ, Kritchevsky SB, Florez JC, Todd JA, Dupuis J, Hypponen E, Spector TD. Common genetic determinants of vitamin D insufficiency: a genome-wide association study. Lancet. 2010;376:180–188. doi: 10.1016/S0140-6736(10)60588-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Albertson DG, Ylstra B, Segraves R, Collins C, Dairkee SH, Kowbel D, Kuo WL, Gray JW, Pinkel D. Quantitative mapping of amplicon structure by array CGH identifies CYP24 as a candidate oncogene. Nat Genetics. 2000;25:144–146. doi: 10.1038/75985. [DOI] [PubMed] [Google Scholar]
- 8.McKay JD, McCullough ML, Ziegler RG, Kraft P, Saltzman BS, Riboli E, Barricarte A, Berg CD, Bergland G, Bingham S, Brustad M, Bueno-de-Mesquita HB, Burdette L, Buring J, Calle EE, Chanock SJ, Clavel-Chapelon F, Cox DG, Dossus L, Feigelson HS, Haiman CA, Hankinson SE, Hoover RN, Hunter DJ, Husing A, Kaaks R, Kolonel LN, Le Marchand L, Linseisen J, McCarty CA, Overvad K, Panico S, Purdue MP, Stram DO, Stevens VL, Trichopoulos D, Willett WC, Yuenger J, Thun MJ. Vitamin D receptor polymorphisms and breast cancer risk: results from the National Cancer Institute Breast and Prostate Cancer Cohort Consortium. Cancer Epidemiol Biomarkers Prev. 2009;18:297–305. doi: 10.1158/1055-9965.EPI-08-0539. [DOI] [PubMed] [Google Scholar]
- 9.Tang C, Chen N, Wu M, Yuan H, Du Y. Fok1 polymorphism of vitamin D receptor gene contributes to breast cancer susceptibility: A meta-analysis. Breast Cancer Res Treat. 2009;117:391–399. doi: 10.1007/s10549-008-0262-4. [DOI] [PubMed] [Google Scholar]
- 10.Sigurdson AJ, Doody MM, Rao RS, Freedman DM, Alexander BH, Hauptmann M, Mohan AK, Yoshinaga S, Hill DA, Tarone R, Mabuchi K, Ron E, Linet MS. Cancer incidence in the US radiologic technologists health study, 1983-1998. Cancer. 2003;97:3080–3089. doi: 10.1002/cncr.11444. [DOI] [PubMed] [Google Scholar]
- 11.Bhatti P, Struewing JP, Alexander BH, Hauptmann M, Bowen L, Mateus-Pereira LH, Pineda MA, Simon SL, Weinstock RM, Rosenstein M, Stovall M, Preston DL, Linet MS, Doody MM, Sigurdson AJ. Polymorphisms in DNA repair genes, ionizing radiation exposure and risk of breast cancer in U.S. Radiologic technologists. Int J Cancer. 2008;122:177–182. doi: 10.1002/ijc.23066. [DOI] [PubMed] [Google Scholar]
- 12.Berger D, Robertson DF, Davies RE, Urbach F. Impacts of Climatic Change of the Biosphere. Springfield, VA: Department of Transportation, NTIS; 1975. Field measurements of biologically effective UV radiation. [Google Scholar]
- 13.Pennello G, Devesa S, Gail M. Association of surface ultraviolet B radiation levels with melanoma and nonmelanoma skin cancer in United States blacks. Cancer Epidemiol Biomarkers Prev. 2000;9:291–297. [PubMed] [Google Scholar]
- 14.Scotto J, Fears TR, Fraumeni JF., Jr . Solar radiation. In: Schottenfeld D, Fraumeni JF Jr, editors. Cancer Epidemiology and Prevention. New York: Oxford University Press; 1996. pp. 355–372. [Google Scholar]
- 15.Greenland S, Rothman KJ. Fundamentals of data analysis. In: Rothman KJ, Greenland S, Lash TL, editors. Modern Epidemiology. Philadelphia, PA: Lippincott Williams & Wilkins; 2008. [Google Scholar]
- 16.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 17.Sinnwell JP, Schaid D. Haplo Stats manual (version 1.5.0) Statistical Methods for Haplotypes When Linkage Phase is Ambiguous. 2011 URL http://mayoresearch.mayo.edu/mayo/research/schaid_lab/software.cfm.
- 18.Holick CN, Stanford JL, Kwon EM, Ostrander EA, Nejentsev S, Peters U. Comprehensive association analysis of the vitamin D pathway genes, VDR, CYP27B1, and CYP24A1, in prostate cancer. Cancer Epidemiol Biomarkers Prev. 2007;16:1990–1999. doi: 10.1158/1055-9965.EPI-07-0487. [DOI] [PubMed] [Google Scholar]
- 19.McCullough ML, Stevens VL, Diver WR, Feigelson HS, Rodriguez C, Bostick RM, Thun MJ, Calle EE. Vitamin D pathway gene polymorphisms, diet, and risk of postmenopausal breast cancer: a nested case-control study. Breast Cancer Res. 2007;9:R9. doi: 10.1186/bcr1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dorjgochoo T, Delahanty R, Lu W, Long J, Cai Q, Zheng Y, Gu K, Gao YT, Zheng W, Shu XO. Common genetic variants in the vitamin D pathway including genome-wide associated variants are not associated with breast cancer risk among Chinese women. Cancer Epidemiol Biomarkers Prev. 2011;20:2313–2316. doi: 10.1158/1055-9965.EPI-11-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anderson LN, Cotterchio M, Cole DE, Knight JA. Vitamin D-related genetic variants, interactions with vitamin D exposure, and breast cancer risk among Caucasian women in Ontario. Cancer Epidemiol Biomarkers Prev. 2011;20:1708–1717. doi: 10.1158/1055-9965.EPI-11-0300. [DOI] [PubMed] [Google Scholar]
- 22.Chen KS, DeLuca HF. Cloning of the human 1 alpha,25-dihydroxyvitamin D-3 24-hydroxylase gene promoter and identification of two vitamin D-responsive elements. Biochim Biophys Acta. 1995;1263:1–9. doi: 10.1016/0167-4781(95)00060-t. [DOI] [PubMed] [Google Scholar]
- 23.Meyer MB, Goetsch PD, Pike JW. A downstream intergenic cluster of regulatory enhancers contributes to the induction of CYP24A1 expression by 1alpha,25-dihydroxyvitamin D3. J Biol Chem. 2010;285:15599–15610. doi: 10.1074/jbc.M110.119958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muindi JR, Nganga A, Engler KL, Coignet LJ, Johnson CS, Trump DL. CYP24 splicing variants are associated with different patterns of constitutive and calcitriol-inducible CYP24 activity in human prostate cancer cell lines. J Steroid Biochem Mol Biol. 2007;103:334–337. doi: 10.1016/j.jsbmb.2006.12.060. [DOI] [PubMed] [Google Scholar]
- 25.Dong LM, Ulrich CM, Hsu L, Duggan DJ, Benitez DS, White E, Slattery ML, Farin FM, Makar KW, Carlson CS, Caan BJ, Potter JD, Peters U. Vitamin D related genes, CYP24A1 and CYP27B1, and colon cancer risk. Cancer Epidemiol Biomarkers Prev. 2009;18:2540–2548. doi: 10.1158/1055-9965.EPI-09-0228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Milani C, Welsh J, Katayama ML, Lyra EC, Maciel MS, Brentani MM, Folgueira MA. Human breast tumor slices: a model for identification of vitamin D regulated genes in the tumor microenvironment. J Steroid Biochem Mol Biol. 2010;121:151–155. doi: 10.1016/j.jsbmb.2010.03.051. [DOI] [PubMed] [Google Scholar]
- 27.Vieth R. How to optimize vitamin D supplementation to prevent cancer, based on cellular adaptation and hydroxylase enzymology. Anticancer Res. 2009;29:3675–3684. [PubMed] [Google Scholar]
- 28.Ingles SA, Haile RW, Henderson BE, Kolonel LN, Nakaichi G, Shi CY, Yu MC, Ross RK, Coetzee GA. Strength of linkage disequilibrium between two vitamin D receptor markers in five ethnic groups: implications for association studies. Cancer Epidemiol Biomarkers Prev. 1997;6:93–98. [PubMed] [Google Scholar]
- 29.Ruder EH, Dorgan JF, Kranz S, Kris-Etherton PM, Hartman TJ. Examining breast cancer growth and lifestyle risk factors: early life, childhood, and adolescence. Clin Breast Cancer. 2008;8:334–342. doi: 10.3816/CBC.2008.n.038. [DOI] [PMC free article] [PubMed] [Google Scholar]


