Abstract
N -acetylgalactosamine-4-sulfatase (Arylsulfatase B; ARSB) is the enzyme that removes sulfate groups from the N-acetylgalactosamine-4-sulfate residue at the non-reducing end of chondroitin-4-sulfate (C4S) and dermatan sulfate (DS). Previous studies demonstrated reduction in cell-bound high molecular weight kininogen in normal rat kidney (NRK) epithelial cells when chondroitin-4-sulfate content was reduced following overexpression of ARSB activity, and chondroitinase ABC produced similar decline in cell-bound kininogen. Reduction in the cell-bound kininogen was associated with increase in secreted bradykinin. In this report, we extend the in vitro findings to in vivo models, and present findings in Dahl salt-sensitive (SS) rats exposed to high (SSH) and low salt (SSL) diets. In the renal tissue of the SSH rats, ARSB activity was significantly less than in the SSL rats, and chondroitin-4-sulfate and total sulfated glycosaminoglycan content were significantly greater. Disaccharide analysis confirmed marked increase in C4S disaccharides in the renal tissue of the SSH rats. In contrast, unsulfated, hyaluronan-derived disaccharides were increased in the rats on the low salt diet. In the SSH rats, with lower ARSB activity and higher C4S levels, cell-bound, high-molecular weight kininogen was greater and urinary bradykinin was lower. ARSB activity in renal tissue and NRK cells declined when exogenous chloride concentration was increased in vitro. The impact of high chloride exposure in vivo on ARSB, chondroitin-4-sulfation, and C4S-kininogen binding provides a mechanism that links dietary salt intake with bradykinin secretion and may be a factor in blood pressure regulation.
Keywords: Bradykinin, Chondroitin, Disaccharide, Kininogen, Sulfatase, Sulfate
Introduction
In this report, we have considered how the interactions among the enzyme Arylsulfatase B (ARSB), chondroitin-4-sulfation, and high-molecular weight kininogen, might contribute to bradykinin activation in rat renal tubules. The enzyme ARSB (N-acetylgalactosamine-4-sulfatase) removes the 4-sulfate group from the non-reducing end of chondroitin-4-sulfate (C4S) and dermatan sulfate (DS) and is crucial for their degradation [1–3]. Previously, we reported how changes in expression of ARSB affected high molecular weight kininogen (HKK) and bradykinin (BK) in normal rat renal tubular epithelial (NRK) cells. Following ARSB overexpression, C4S and cell-bound HKK declined and BK secretion into the spent media increased [4]. These results supported the underlying rationale that changes in ARSB activity, which affect C4S degradation impact upon the production of BK from HKK by altering the binding of HKK with C4S. Other reports have also demonstrated interactions between kininogen and glycosaminoglycans (GAGs) [5, 6].
ARSB was previously regarded as only a lysosomal enzyme, deficiency of which causes Mucopolysaccharidosis VI (Maroteaux-Lamy Syndrome). However, recent studies have demonstrated the presence of ARSB activity in cell membranes of epithelial and endothelial cells, and decline in ARSB has been associated with cystic fibrosis, asthma, malignancy, and metastasis [7–11]. The extent of chondroitin-4-sulfation has been recognized as critically important in protection against attachment of malarial parasites to placenta and endothelium [12, 13]. Considerations about other functional consequences related to ARSB activity and chondroitin-4-sulfation were addressed with regard to secretion vs. cell sequestration of Interleukin-8 [14].
The current study was designed to determine if variation in dietary exposure to sodium chloride might impact upon the relationships among ARSB, C4S, HKK, and BK in vivo, since chloride has been reported to affect ARSB activity in vitro [15]. Dahl salt-sensitive (SS) rats were fed high salt (SSH) or low salt (SSL) diets, and the renal tissue ARSB, total sulfated GAGs, C4S, HKK, and C4S-, DS-, and hyaluronan-derived disaccharides and the urine BK and sulfate were compared between the high and low salt models. This work extends previous cell-based observations about the impact of ARSB activity and chondroitin-4-sulfation on cellular binding of high molecular weight kininogen and release of bradykinin, and demonstrates how physiological effects might arise in vivo due to changes in GAG sulfation.
Materials and methods
Dietary salt intake in rat models
Ten week-old Dahl salt-sensitive rats (Harlan Laboratories, Indianapolis, IN) were maintained on rat chow with either high salt (8 % NaCl) (SSH; n=5) or low salt (0.12 % NaCl) (SSL; n=5) content for 3 weeks, and kidneys were harvested and promptly frozen at the time of euthanasia. 24-hour urine samples for sulfate and creatinine were collected from other rats fed high (n=6) or low (n=6) salt diet for similar duration. All animals were treated in accordance with approved IACUC procedures for animal care at the University of Illinois at Chicago or the Jesse Brown VA Medical Center, Chicago, IL.
Normal rat kidney cell cultures
Normal rat kidney epithelial cells (NRK-E52; ATCC, Manassas, VA) were grown in DMEM with 10 % FBS, at 37 °C, 5 % CO2 and 95 % humidity, as previously described [4]. When cells reached 65–70 % confluence, silencing or over-expression of arylsulfatase B (ARSB) was performed (see below). Cells were harvested after 24 h by scraping and then frozen, pending further analysis. Also, spent media from the untreated control or treated cells were collected and frozen (−20 °C), pending further analysis. Some cell preparations were grown in phenol-red free media, in order to detect sulfate (see below).
Silencing and overexpression of ASB
Small interfering RNA (siRNA) to silence ARSB was obtained (Qiagen, Valencia, CA) and tested for specific effect on ARSB activity, as previously [14, 16]. The siRNA sequences for ARSB (NM_000046) silencing were: sense: 5 - GGGUAUGGUCUCUAGGCAtt - 3 anti-sense: 5 - UUGCCUAGAGACCAUACCCtt - 3.
ARSB and control plasmids in pCMV6-XL4 vector (Origene) were overexpressed in NRK cells by transient transfection using 2 μg of the plasmid and Lipofectamine™ 2000 (Invitrogen, CA) [4, 16] Controls included untransfected cells and cells transfected with vector only. Cells were incubated in humidified, 37 °C, 5 % CO2 environment and then harvested 24 h after initiation of silencing or overexpression.
N-acetylgalactosamine-4-sulfatase (arylsulfatase B; ARSB) activity assay
ARSB measurements were performed on whole kidney tissue extracts from the salt-sensitive rats exposed to either low or high salt diets, using the substrate 4-methylumbelliferyl sulfate, as previously [17]. Protein content of the homogenates was measured using BCA™ Protein Assay Kit (Pierce, Rockford, IL).
Measurement of sulfated glycosaminoglycans (sGAG) and chondroitin-4-sulfate
Total sulfated glycosaminoglycans (sGAG), including chondroitin 4-sulfate (C4S), chondroitin 6-sulfate, keratan sulfate, dermatan sulfate, heparan sulfate, and heparin, were measured in renal tissues by sulfated GAG assay (Blyscan™, Biocolor Ltd, Newtownabbey, Northern Ireland) [4, 16]. This assay uses the cationic dye 1,9-dimethylmethy-lene blue, which combines with the sulfate groups of the sulfated GAG to produce a dye-GAG complex. The dye measures the sulfated polysaccharide component of proteoglycans and protein-free sulfated GAG chains, but not degraded disaccharide fragments or hyaluronan. For this assay, the ratio of the GAG dye-binding capability to the C4S sulfation level is reported as 1.0/1.0 for C4S from bovine trachea. Absorbance maximum of 1,9-dimethylmethylene blue is at 656 nm. Concentration is expressed as μg/mg protein of cell lysate.
To measure chondroitin-4-sulfate, renal tissue lysates from frozen SSH and SSL were prepared using RIPA buffer (50 mmol/l Tris–HCl containing 0.15 mol/l NaCl, 1 % Non-idet P40, 0.5 % deoxycholic acid and 0.1 % SDS, pH7.4). C4S antibody (4D1; Santa Cruz Biotechnology, Santa Cruz, CA or Abnova, Walnut, California) was used to detect native C4S. The C4S (4D1) antibody (Abnova, Littleton, CO) detected native C4S, and the yield of C4S from immunoprecipitation, tested by recovery of pure C4S following immunoprecipitation with the C4S antibody (1 μg), was 93.3±2.7 % [16]. Cross-reactivity of the anti-C4S with CS-E or C6S was excluded by similar tests [16]. Immuno-precipitation with C4S antibody was performed as described previously [4, 16].
Assay of cellular high molecular weight kininogen bound to C4S
Methods to detect high molecular weight kininogen bound to C4S were reported previously and demonstrated the specificity of the kininogen binding to C4S by competition with exogenous C4, as well as treatments with chondroitinase ABC and keratanase [4]. Briefly, a 96-well ELISA plate was coated overnight at room temperature with C4S antibody at a concentration of 4 μg/ml, wells were washed three times with wash buffer (PBS with Tween-20 0.05 %), and then blocked for 1 h at room temperature with blocking buffer (1 % BSA in PBS). 100 μl of renal tissue lysates from high and low salt exposed animals were added to the coated wells, the plate was incubated for 2 h at room temperature, and then wells were washed three times. High molecular weight kininogen (HMWK) antibody (rabbit polyclonal IgG; H-70, SCBT), which recognizes the epitope from AA:261–330 was added (100 μl at a concentration of 1 μg/ml) and the plate was incubated for 2 h at room temperature. Then, goat anti-rabbit IgG-HRP (100 μl at a concentration of 1:1000) was added to each well, and the plate was then incubated for 1 h at room temperature. Color was developed by adding 100 μl of hydrogen peroxide/TMB substrate, the reaction was stopped by 2 N HCl, and the developed color was read at 450 nm in a microplate reader (BMG).
Chondroitin sulfate (CS)/dermatan sulfate (DS) and hyaluronan disaccharide analysis
Unsaturated disaccharide standards of CS/DS (ΔDi-0S: ΔUA-GalNAc, ΔDi-4S: ΔUA-GalNAc4S, ΔDi-6S: ΔUA-GalNAc6S, ΔDi-2S: ΔUA2S-GalNAc, ΔDi-diSB: ΔUA2S-GalNAc4S, ΔDi-diSD: ΔUA2S-GalNAc6S, ΔDi-diSE: ΔUA-GalNAc4S6S, ΔDi-triS: ΔUA2SGal-NAc4S6S) and of HA (ΔDi-UA-GlcNAc) were obtained from Seikagaku Corporation (Japan). All other chemicals were of reagent grade. The molecular weights (MW) of the CS disaccharides are: Di-0S (UA-GalNAc) 379; Di-4S(UA-GalNAc4S) 459; Di-6S(UAGalNAc6S) 459; Di-2S(UA2S-GalNAc) 459;Di-diSB(UA2S-GalNAc4S) 539; Di-diSD (UA2S-GalNAc6S) 539; Di-diSE(UA-GalNAc4S6S) 539; and Di-triS(UA2SGalNAc4S6S) 637 [18].
GAGs from the renal tissue samples from high and low salt exposed SS rats were isolated and purified as previously described [19, 20]. The individual samples were defatted and subjected to proteolysis at 55 °C with 10 % actinase E (20 mg/mL) for 20 h. After proteolysis, dry urea and dry CHAPS were added to each sample (2 wt.% in CHAPS and 8 M in urea), particulates were removed from the resulting solutions by passing each through a syringe filter containing a 0.22 μm membrane, and the GAGs were recovered as previously detailed [18, 21–23]. The GAGs were depolymerized by polysaccharide lyases and freeze-dried. Later, the GAG-derived disaccharides were added to 10 μl of a 0.1 M 2-aminoacridone (AMAC) solution in acetic acid/dimethylsulfoxide (3:17, v/v) as described [18]. The AMAC-tagged disaccharides were diluted using 50 % DMSW and subjected to LC-MS analyses as previously described [18, 21–23].
ARSB immunohistochemistry
Longitudinal frozen sections of the kidneys from rats fed low-salt and high-salt diets were obtained, and slides were fixed in 3.8 % formaldehyde for 20 min. Exogenous peroxidase was quenched with hydrogen peroxide buffer, and blocking was done with fetal bovine serum. Slides were then incubated with rabbit ARSB antibody (Open Biosystems, Huntsville, AL) at a dilution of 1:100 overnight at 4 °C and with secondary goat anti-rabbit HRP-conjugated antibody for 1 h at room temperature. Color was developed with 3,3′-Diaminobenzidine (DAB) for 10 min, and sections were counterstained with hematoxylin. Images were obtained using QCapture software (QImaging) at 10x magnification, and background color neutralization was done with GIMP Portable software (Portable Apps).
Effect of sodium chloride and sodium acetate on ARSB activity
ARSB determinations were performed on renal tissue lysates from the SSH and SSL rats and on NRK cells, using concentrations of NaCl and Na-acetate ranging from 1 to 100 mmol/l. ARSB was measured as previously described [17].
Rat urine collection and measurements of electrolytes, creatinine, and bradykinn
Urine samples were obtained by bladder puncture at the time of euthanasia or by 24-hour collection in rats housed in metabolic cages. Samples were promptly frozen, and at the time of assay, the samples were thawed and centrifuged at 2000×g for 10 min to remove cells and particulate matter. Electrolytes and creatinine were analyzed in a multichannel auto analyzer (Dimension® RxL Max® System, Siemens Corporation, Deerfield, IL), and measurements were expressed as mg/dL.
Bradykinin in the urine was determined by competitive EIA (BACHEM, Torrance, CA) in which color development is inversely proportional to the bradykinin content of the samples. 50 μl of samples or of standards, ranging from 10 pg to 10 ng, 25 μl of biotinylated bradykinin tracer, and 25 μl of rabbit polyclonal antisera to bradykinin were combined in wells of a 96-well plate that were coated with goat anti-rabbit IgG. Samples were incubated at room temperature for 2 h, washed three times, and streptavidin-horseradish peroxidase (HRP) conjugate (100 μl) was added to each well for 1 h. Color was developed by hydrogen peroxide/3,3′,5,5′-tetramethylbenzidine (TMB) substrate, and the reaction was stopped by 2 N HCl. Color intensity was detected at 450 nm in a plate reader (FLUOstar, BMG Labtech, Cary NC), and the concentration of bradykinin in the samples was extrapolated from the standard curve and expressed as pg per mg of creatinine from the 24-hour urine collection.
Measurement of sulfate in urine and spent media
Sulfate concentrations were measured by QuantiChrom™ Sulfate Assay (BioAssay Systems, Hayward, CA), which is linear in the detection range from 0.19 mg/dL to 19.2 mg/dL. Rat urine samples were diluted 1:20 or more, de-proteinized, and centrifuged at 14,000 RPM for 5 min. NRK cells were grown in phenol-red free media, and the spent media were de-proteinized. Samples were further processed as instructed by the manufacturer, and the OD was read at 540 nm, indicating the extent of formation of insoluble barium sulfate from the samples.
Statistics
Results are the mean ± standard deviation (S.D.) of at least three biological determinations with two technical replicates of each, except for the disaccharide analysis which was performed with two independent kidney tissue samples from high or low salt exposed animals. Statistical significance was determined by unpaired t-test, two-tailed, unless otherwise stated, and performed by one-way ANOVA followed by Tukey-Kramer post-test to correct for multiple comparisons using InStat3 software (GraphPad, San Diego, CA). A p-value of <0.05 is considered statistically significant. In the figures, three asterisks denote p≤0.001; two asterisks denote p≤0.01, and one asterisk indicates p≤0.05.
Results
Sulfatase activity in rat renal tissue following high or low chloride exposure
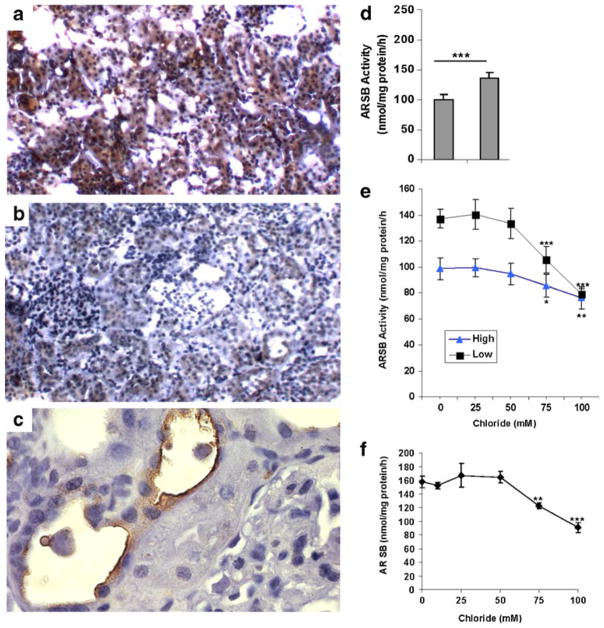
Differences in salt intake between the salt-sensitive rats fed high (SSH) or low (SSL) salt diet were confirmed by measurements of urinary sodium and chloride. Urinary sodium was 177±76 mEq/L in the SSH rats and 30±12 mEq/L in the SSL rats. Urinary chloride was 247±47 mEq/L in the SSH rats and 143±25 mEq/L in the SSL rats, consistent with the difference in diet. Immunohistochemistry demonstrated more intense and more extensive ARSB immunostaining in the SSL rat kidney (Fig. 1a) compared to the SSH kidney (Fig. 1b). ARSB immunostaining was evident at the apical cell surface in the renal tubular cells (Fig. 1c), as well as diffusely throughout the cytoplasm of the proximal and distal tubular epithelium. ARSB activity was significantly lower in the SSH rats than in the SSL rats, 98.3±8.6 nmol/mg protein/h vs. 137.1±7.4 nmol/mg protein/h (p<0.0001; unpaired t-test, two-tailed; n=10 for SSH and SSL) (Fig. 1d), consistent with the differences in immunostaining.
Fig. 1.
Reduced ARSB activity in vivo following higher salt exposure. a,b,c. Immunohistochemistry of representative renal tissue from SSL and SSH rats demonstrated increased intensity and extent of positive staining for ARSB in the epithelial cells of the proximal and distal tubules in the SSL renal tissue (a), compared to the SSH (b) renal tissue. Findings are consistent with reduced ARSB activity following exposure to high salt in the salt-sensitive rats (original magnification 10x). Prominent apical membrane staining of the tubular cells is present (c). (Original magnification 20x). d. ARSB activity was significantly lower in the SSH rats (n=5), than in the SSL rats (n=5) (p< 0.0001, unpaired t-test, two-tailed). e. With increasing concentrations of NaCl in the buffer, the ARSB activity in the rat renal tissue declined in the SSH rats (n=5) and the SSL rats (n=5), to nearly the same value. For concentrations of 75 mM and 100 mM, the declines were significantly different from the baseline for the SSH rats (p<0.05, p<0.01; unpaired t-test, two-tailed)) and for the SSL rats (p<0.001, p<0.001; unpaired t-test, two-tailed). In contrast to the profound inhibitory effect of chloride on ARSB activity, exposure to varying concentrations of Na-acetate (from 0 to 100 mmol/l) had no effect on the ARSB activity. f. Increase in exogenous NaCl exposure produced significant declines in the ARSB activity of the NRK cells, from 158±8.5 to 122.2± 4.3 nmol/mg protein/h (75 mM chloride) and to 79.2±4.6 nmol/mg protein/h (100 mM chloride) (p=0.003, and p=0.0005, unpaired t-test, two-tailed). [ARSB = arylsulfatase B; SSL = salt-sensitive on low salt diet; SSH = salt-sensitive on high salt diet; NRK = normal rat kidney]
The ARSB activity was determined using the exogenous substrate 4-methylumbelliferylsulfate (4-MUS). As the exogenous sodium chloride concentration increased from 0 to 100 mmol/l, the ARSB activity in the SSH and SSL renal tissue lysates declined, to 75.7±8.2 nmol/mg protein/h in the SSH rats and to 79.0±3.8 nmol/mg protein/h in the SSL rats (Fig. 1e). In contrast, increased concentration of sodium acetate from 1 to 100 mmol/l had no impact on the ARSB activity (data not shown).
In the NRK cells, increase in exogenous chloride produced 50 % decline in the ARSB activity (Fig. 1f). These in vitro findings are consistent with the inhibitory effect of chloride on ARSB activity observed in the renal tissue of the SSH rats, compared to the SSL rats.
Sulfated glycosaminoglycans (GAGs), chondroitin-4-sulfate (C4S), and sulfated disaccharides increased by high salt intake
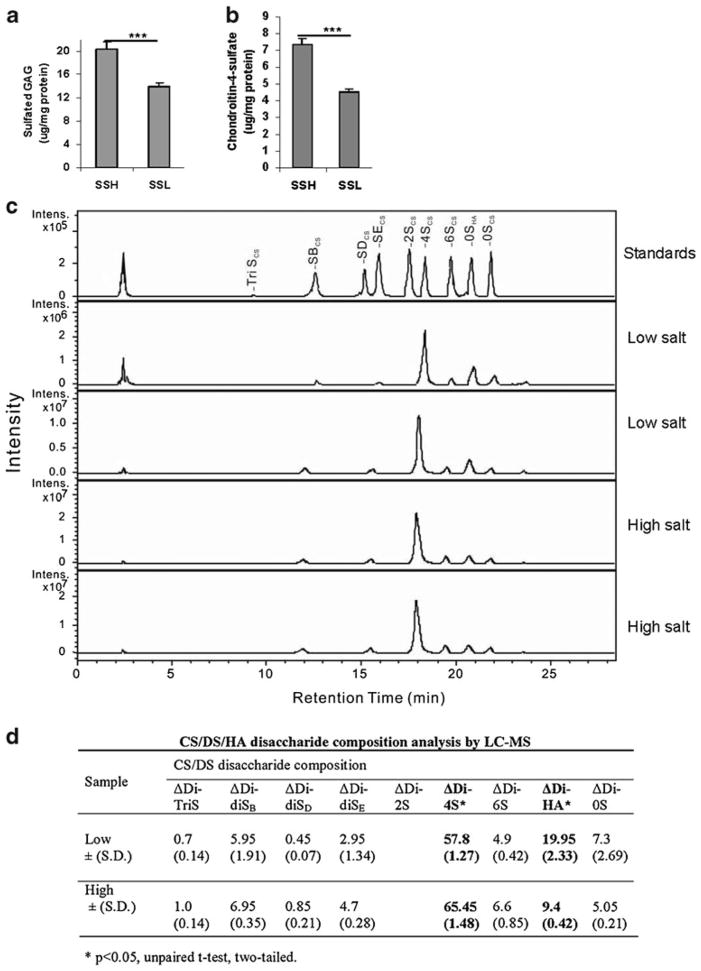
Consistent with the reduction in ARSB activity, the total cellular sulfated glycosaminoglycans (GAGs) (Fig. 2a) and the C4S (Fig. 2b) were significantly greater in the SSH rats than in the SSL rats. Total sGAG declined from 21.0± 1.3 μg/mg protein to 14.0±0.6 μg/mg protein, and C4S declined from 7.4±0.4 μg/mg protein to 4.5±0.2 μg/mg protein in SSH vs. SSL rats (p<0.0001, p<0.0001).
Fig. 2.
Increased sulfated GAGs, C4S, and sulfated disaccharides in renal tissue following higher salt exposure. a. sGAG in the rat kidney tissue following high salt was significantly greater in the SSH than in the SSL rats (p<0.0001, unpaired t-test, two-tailed; n=10 for SSH and for SSL). b. Similarly, the content of C4S in the rat kidney tissue was greater in the SSH rats with lower ARSB activity than in the SSL rat tissue (p< 0.0001; n=5 for SSH and n=5 for SSL). [ARSB = arylsulfatase B; GAGs = glycosaminoglycans; C4S = chondroitin-4-sulfate; SSH = salt-sensitive on high salt diet; SSL = salt-sensitive on low salt diet] c. 2-Aminoacridone (AMAC)-labeled chondroitin sulfate (CS)/dermatan sulfate (DS)/hyaluronan (HA) disaccharide analysis was performed by LC/MS. Chromatogram depicts the intensity of the disaccharides obtained following isolation, purification, and depolymerization of the glycosaminoglycans from the SSH and SSL renal tissue, and in comparison to standards. d. The disaccharide composition indicates marked differences between the average disaccharide content of the high and low salt renal tissue. The hyaluronan-derived disaccharide (ΔDi-HA) and the C4S-derived disaccharide (ΔDi-4S) are both present in high concentration and differ significantly between the high and low salt tissues (p=0.02 for ΔDi-HA and p=0.03 for ΔDi-4S, unpaired t-test, two-tailed). [CS = chondroitin sulfate; DS = dermatan sulfate; HA = hyaluronan; LC = liquid chromatography; MS = mass spectrometry; S = sulfate; UA = uronic acid; GalNac = N-acetylgalactosamine; GlcNAc = N-acetylglucos-amine; ΔDi-TriS = ΔUA2S-GlcNS6S; ΔDi-0S = ΔUA-GalNAc, ΔDi-4S = ΔUA-GalNAc4S, ΔDi-6S = ΔUA-GalNAc6S, ΔDi-UA2S = ΔUA2S-GalNAc, ΔDi-diSB = ΔUA2S-GalNAc4S;ΔDi-diSD =ΔUA2S-GalNAc6S; ΔDi-diSE = ΔUA-GalNAc4S6S; ΔDi = HA = ΔUA-GlcNAc; Intens = intensity]
Chromatogram depicts the peaks for the disaccharides obtained following digestion of the sulfated GAGs in the renal tissue from the SSH and SSL rats (Fig. 2c). Significant increase in the Δdi4S disaccharide (65.4±1.5 % vs. 57.8± 1.3 %; p=0.03, unpaired t-test, two-tailed) is present for the SSH renal tissue, consistent with an increase in C4S and reduced ARSB activity (Fig. 2d). In contrast, for the SSL renal tissue, significant increase in the ΔdiHA (hyaluronan) disaccharide is present (20.0±2.3 % vs. 9.4±0.4 %; p= 0.02), indicating an increased abundance of the un-sulfated GAGs. In addition, all of the sulfated disaccharide values are greater in the SSH renal tissue, and the 0S disaccharide is increased in the SSL renal tissue.
Cell-bound high molecular weight kininogen increased, and bradykinin and sulfate secretion reduced by high salt diet
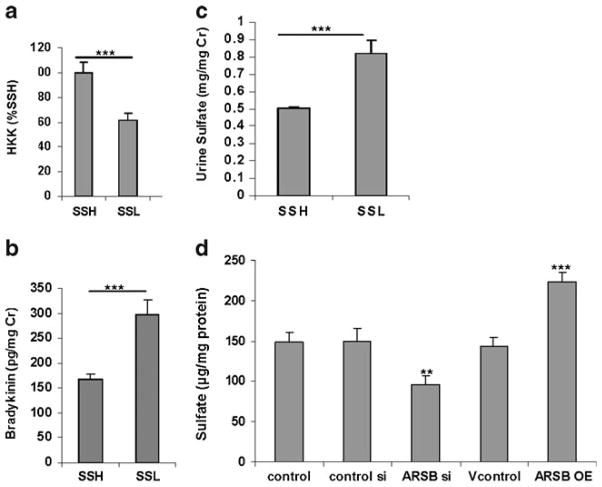
Previous work in NRK cells indicated that changes in ARSB activity and the associated changes in chondroitin-4-sulfation affected high molecular weight kininogen and bradykinin. Specifically, when ARSB was overexpressed, less high-molecular weight kininogen (HKK) was bound to C4S and more bradykinin was secreted [4]. To consider how the endogenous differences in ARSB activity following exposure to high or low salt affected kininogen and bradykinin in vivo, HKK in the rat renal tissue was determined by ELISA. In the SSL renal tissue with higher ARSB activity and lower C4S, the HKK was significantly less than in the SSH rat kidneys (p<0.0001) (Fig. 3a). This finding is consistent with the previous findings in the NRK cells.
Fig. 3.
Renal tissue kininogen increased, and urinary bradykinin and sulfate reduced following high salt. a. High-molecular weight kininogen (HKK) was measured by ELISA in the SSH and SSL renal tissue, and was significantly greater in the SSH tissue (p<0.0001; n=10 for SSH and SSL). b. Bradykinin was measured in the urine of the SSH and SSL rats on high and low salt diets, respectively. Bradykinin values were significantly less in the SSH group (p<0.0001; n=10 for SSH and SSL). c. Urinary sulfate was significantly less in the 24-hour urine of the SSH rats, compared to the SSL rats (p<0.0001; n=6 for SSH and n = 6 for SSL). d. Sulfate content in the spent media from the NRK cells declined following silencing of ARSB by siRNA, from a baseline value of 148±13 μg/mg protein to 96±11 μg/mg protein (p< 0.01, one-way ANOVA with Tukey-Kramer post-test) and increased following overexpression of ARSB to 223±12.5 μg/mg protein (p< 0.001, one-way ANOVA with Tukey-Kramer post-test). [HKK = high molecular weight kininogen; SSH = salt-sensitive on high salt; SSL = salt-sensitive on low salt diet; ARSB = arylsulfatase B; NRK = normal rat kidney]
To further address the impact of ARSB and chondroitin-4-sulfation on the HKK-bradykinin interaction, bradykinin was assayed in urine specimens from the SSH and SSL rats. Inverse to the HKK result, the urinary bradykinin was significantly lower in the SSH rats (p<0.0001) (Fig. 3b). These results indicate that in the salt sensitive rats, increased salt intake led to decline in urinary bradykinin which was associated with increased HKK binding to more highly sulfated chondroitin-4-sulfate.
Sulfate ion concentration was measured in urine samples from SSH and SSL rats (Fig. 3c). Sulfate was significantly higher in the SSL (p<0.0001) than in the SSH rats, consistent with higher ARSB activity. In addition to the sulfate determinations in the rat urine, measurements of sulfate in the spent media of normal rat kidney (NRK) cells following ARSB silencing and overexpression were performed (Fig. 3d). Sulfate declined by ~52 μg/mg cell protein when ARSB was silenced (p<0.01, one-way ANOVA with Tukey-Kramer post-test) and increased by ~72 μg/mg cell protein following ARSB overexpression (p<0.001, one-way ANOVA with Tukey-Kramer post-test).
Discussion
The current data support in vivo regulation by chloride of ARSB activity, with subsequent effects on chondroitin-4-sulfation, the interaction between HKK and C4S, and the release of BK from HKK. Lower ARSB activity in the SSH rats was associated with greater C4S, total sulfated glycosaminoglycans, C4S disaccharides, and cell-bound HKK, and with lower urinary BK and sulfate. These effects follow due to increased binding between kininogen and chondroitin-4-sulfate when chondroitin-4-sulfation is increased due to reduced ARSB activity. These relationships are presented schematically in Fig. 4. The depressed ARSB activity in the SSH rats is consistent with suppression of ARSB activity by chloride exposure in vivo. Chloride, as well as sulfate, sulfite, and phosphate, have been reported to reduce ARSB activity [7, 15, 24, 25], and increased renal tubular chloride following high salt intake may reduce ARSB activity in the SSH rat kidney. Interestingly, the rat Quantitative Trait Loci for high blood pressure and renal pathology suggest that the study findings of reduced activity of ARSB may be relevant to disease [26–30], since the ARSB gene on chromosome 2 is closely linked (~160,000 bases) to 5 genes for increased blood pressure.
Fig. 4.
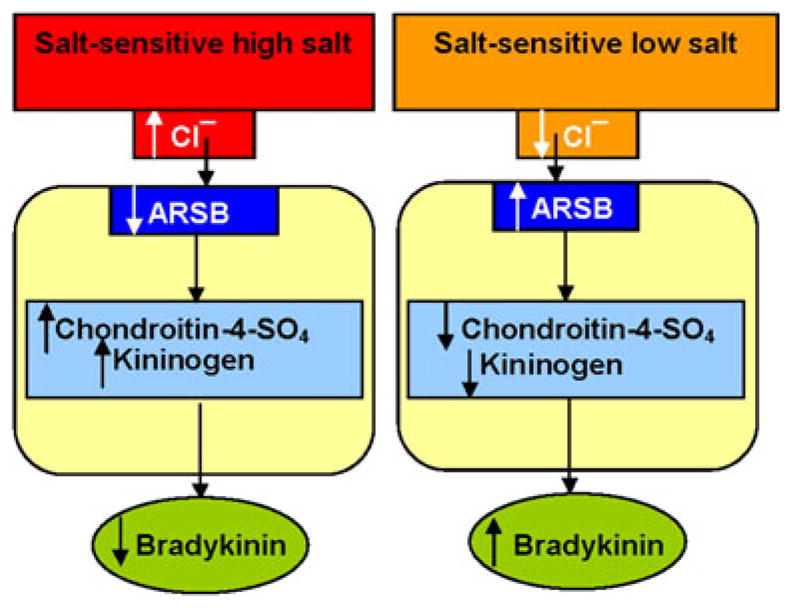
Schematic representation of the relationships between ARSB activity, chloride exposure, chondroitin-4-sulfation, kininogen, and bradykinin. The drawing depicts the impact of high vs. low salt exposure on the ARSB activity, chondroitin-4-sulfate content, cell-bound kininogen, and bradykinin in the rat kidney tissue. Higher chloride exposure leads to reduced ARSB activity, which leads to increase in chondroitin-4-sulfate (C4S) with increased binding of kininogen with C4S, producing a reduction in secreted bradykinin
Genetic differences that affect chloride channels or sensitivity may contribute to differences in ARSB activity in other rat models. Recently, effects on chloride homeostasis in the hypothalamus of spontaneously hypertensive rats due to upregulation of the Na(+)-K(+)-2CL(−) cotransporter-1 (NKCC1) and leading to increased sympathetic drive were identified [31]. Similarly, upregulation of the apical NKCC2 co-transporter was identified in the young Milan hypertensive strain (MHS) of salt-sensitive rats and was associated with increased salt reabsorption along the medullary thick ascending limb [32]. In the adult MHS model, luminal and basal transporters of sodium chloride were activated in the distal convoluted tubules, suggesting the potential effects of chloride reabsorption on ARSB activity beyond the proximal tubules. Disaccharide analysis has demonstrated a relative increase in the hyaluronan-derived disaccharides in the SSL rats, suggesting an increase in hyaluronan in the SSL rats. The implication of this finding is not clear at this time, although a hyaluronan binding serine protease has previously been reported to possess kallikrein-like activity and to release bradykinin from kininogen [33].
Future work will be needed to further elucidate the mechanisms responsible for some of the current findings. Additional studies to detect if there is variation in the responsiveness of the C4S-HKK-BK interactions with different levels of ARSB activity may help to clarify why the urine bradykinin varies between the SSH and SSL rats. The measurements of ARSB in this report were performed using an exogenous substrate (4-methylumbelliferyl sulfate) at pH 5.6; in vivo, it is possible that the activity of the ARSB may vary due to localized differences in pH. Cellular acid production is reported to be increased in SS rats [34], and this might increase the in vivo ARSB activity. However, the study measurements of C4S and disaccharides indicate significant differences in vivo, suggesting that any in vivo variation in pH is not so great as to equalize the ARSB activity.
The inhibition of ARSB activity in the NRK cells and in the renal tissue by exogenous chloride provides additional information about the relationship between ARSB activity and chloride. ARSB activity was reduced when cystic fibrosis transmembrane conductance regulator (CFTR) was defective in bronchial epithelial cells [17], and ARSB activity has been shown to be reduced in leukocytes of patients with cystic fibrosis [8, 9]. Mutations in the CFTR gene affect the regulation of chloride secretion, and non-functional CFTR is either unable to be translocated to its correct position on the cell membrane or, when localized at the cell membrane, unable to properly regulate the chloride channel activity [35, 36]. The mechanism by which ARSB activity is reduced when CFTR is non-functional is not yet established. It is pertinent to note that the formylglycine generating enzyme (FGE) and oxygen are required for ARSB activation [37, 38], and ARSB activity is reduced by hypoxia [39]. The FGE has a cleft occupied by oxygen which can also be occupied by chloride or other halides [40], suggesting a possible mechanism whereby increased chloride might inhibit ARSB activation. Increased attention to the function of ARSB and to its activation by the formylglycine generating enzyme and molecular oxygen may improve our fundamental understandings of the inter-relationship between chloride and ARSB activity, as well as of the interactions among chondroitin-4-sulfation, kininogen, and bradykinin.
Acknowledgments
The authors thank Robert Chanthimabha for his help with determinations of creatinine and electrolytes. Research was supported by VA Merit Awards to R.S. Danziger, M.D. and J.K. Tobacman, M.D. and NIDDK R21HL096031 to Dr. Danziger.
Contributor Information
Kumar Kotlo, University of Illinois at Chicago, Chicago, IL 60612, USA. Jesse Brown VA Medical Center, Chicago, IL 60612, USA.
Sumit Bhattacharyya, University of Illinois at Chicago, Chicago, IL 60612, USA. Jesse Brown VA Medical Center, Chicago, IL 60612, USA.
Bo Yang, Rensselaer Polytechnic University, Troy, NY, USA.
Leonid Feferman, University of Illinois at Chicago, Chicago, IL 60612, USA. Jesse Brown VA Medical Center, Chicago, IL 60612, USA.
Shah Tejaskumar, Rosalind Franklin University of Medicine and Science, North Chicago, IL 60064, USA.
Robert Linhardt, Rensselaer Polytechnic University, Troy, NY, USA.
Robert Danziger, University of Illinois at Chicago, Chicago, IL 60612, USA. Jesse Brown VA Medical Center, Chicago, IL 60612, USA.
Joanne K. Tobacman, Email: jkt@uic.edu, University of Illinois at Chicago, Chicago, IL 60612, USA. Jesse Brown VA Medical Center, Chicago, IL 60612, USA. Department of Medicine, University of Illinois at Chicago, 840 S. Wood St., CSN 440, M/C 718, Chicago, IL 60612, USA
References
- 1.De Sousa JF, Nader HB, Dietrich CP. Sequential degradation of chondroitin sulfate in mollusks. J Biol Chem. 1990;265:20150–20155. [PubMed] [Google Scholar]
- 2.Glaser JH, Conrad HE. Chondroitin SO4 catabolism in chick embryo chondrocytes. J Biol Chem. 1978;254:2316–2325. [PubMed] [Google Scholar]
- 3.Ingmar B, Wasteson B. Sequential degradation of a chondroitin sulfate trisaccharide by lysosomal enzymes from embryonic-chick epiphysial cartilage. Biochem J. 1979;179:7–13. doi: 10.1042/bj1790007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhattacharyya S, Kotlo K, Danziger RS, Tobacman JK. Arylsulfatase B regulates interaction of chondroitin-4-sulfate and kininogen in renal epithelial cells. Biochim Biophys Acta. 2010;1802(5):472–477. doi: 10.1016/j.bbadis.2010.01.014. [DOI] [PubMed] [Google Scholar]
- 5.Gozzo AJ, Nunes VA, Carmona AK, Nader HB, von Dietrich CP, Silveira VLF, Shimamoto K, Ura N, Sampaio MU, Sampaio CAM, Araujo MS. Glycosaminoglycans affect the action of human plasma kallikrein on kininogen hydrolysis and inflammation. Int Immunopharmacol. 2002;2:1861–1865. doi: 10.1016/s1567-5769(02)00145-5. [DOI] [PubMed] [Google Scholar]
- 6.Renné T, Schuh K, Müller-Esterl W. Local bradykinin formation is controlled by glycosaminoglycans. J Immunol. 2005;175:3377–3385. doi: 10.4049/jimmunol.175.5.3377. [DOI] [PubMed] [Google Scholar]
- 7.Bhattacharyya S, Tobacman JK. Steroid sulfatase, arylsulfatases A and B, galactose 6-sulfatase, and iduronate sulfatase in mammary cells and effects of sulfated and non-sulfated estrogens on sulfatase activity. J Steroid Biochem Mol Biol. 2007;103:20–34. doi: 10.1016/j.jsbmb.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 8.Ferrero GB, Pagliardini S, Veljkovic A, Porta F, Bena C, Tardivo I, Restagno G, Silengo MC, Bignamini D. In vivo specific reduction of arylsulfatase B enzymatic activity in children with Cystic Fibrosis. Mol Genet Metab. 2008;94:39. doi: 10.1016/j.ymgme.2007.10.010. [DOI] [PubMed] [Google Scholar]
- 9.Sharma G, Burke J, Bhattacharyya S, Sharma N, Katyal S, Park RL, Tobacman J. Reduced arylsulfatase B activity in leukocytes from cystic fibrosis patients. Pediatr Pulmonol. 2012 doi: 10.1002/ppul.22567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prabhu SV, Bhattacharyya S, Guzman-Hartman G, Macias V, Kajdacsy-Balla A, Tobacman JK. Extra-lysosomal localization of arylsulfatase B in human colonic epithelium. J Histochem Cytochem. 2011;59(3):328–335. doi: 10.1369/0022155410395511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bhattacharyya S, Tobacman JK. Arylsulfatase B regulates colonic epithelial cell migration by effects on MMP9 expression and RhoA activation. Clin Exp Metastasis. 2009;26(6):535–545. doi: 10.1007/s10585-009-9253-z. [DOI] [PubMed] [Google Scholar]
- 12.Achur RN, Valiyaveettil M, Gowda DC. The low sulfated chondroitin sulfate proteoglycans of human placenta have sulfate group-clustered domains that can efficiently bind Plasmodium falciparum-infected erythrocytes. J Biol Chem. 2003;278:11705–11713. doi: 10.1074/jbc.M211015200. [DOI] [PubMed] [Google Scholar]
- 13.Rogerson SJ, Brown G. Chondroitin sulfate A as an adherence receptor for Plasmodium falciparum-infected erythrocytes. Parasitol Today. 1997;13:70–75. doi: 10.1016/s0169-4758(96)10081-8. [DOI] [PubMed] [Google Scholar]
- 14.Bhattacharyya S, Solakyildirim K, Zhang Z, Linhardt RJ, Tobacman JK. Cell-bound IL-8 increases in bronchial epithelial cells following Arylsulfatase B silencing. Am J Respir Cell Mol Biol. 2010;42(1):51–61. doi: 10.1165/rcmb.2008-0482OC. [DOI] [PubMed] [Google Scholar]
- 15.Wòjczyk B. Lysosomal arylsulfatases A and B from horse blood leukocytes: purification and physico-chemical properties. Biol Cell. 1986;57:147–152. doi: 10.1111/j.1768-322x.1986.tb00471.x. [DOI] [PubMed] [Google Scholar]
- 16.Bhattacharyya S, Kotlo K, Shukla S, Danziger RS, Tobacman JK. Distinct effects of N-acetyl-galactosamine-4-sulfatase and galactose-6-sulfatase expression on chondroitin sulfates. J Biol Chem. 2008;283(15):9523–9530. doi: 10.1074/jbc.M707967200. [DOI] [PubMed] [Google Scholar]
- 17.Bhattacharyya S, Look D, Tobacman JK. Increased arylsulfatase B activity in cystic fibrosis cells following correction of CFTR. Clin Chim Acta. 2007;380(1–2):122–127. doi: 10.1016/j.cca.2007.01.021. [DOI] [PubMed] [Google Scholar]
- 18.Yang B, Chang Y, Weyers AM, Sterner E, Linhardt RJ. Disaccharide analysis of glycosaminoglycan mixtures by ultra-high-performance liquid chromatography-mass spectrometry. J Chromatog A. 2012;1225:91–98. doi: 10.1016/j.chroma.2011.12.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang B, Weyers A, Baik JY, Sterner E, Sharfstein S, Mousa SA, Zhang F, Dordick JS, Linhardt RJ. Ultra-performance ion-pairing liquid chromatography with on-line electrospray ion trap mass spectrometry for heparin disaccharide analysis. Anal Biochem. 2011;314:59–66. doi: 10.1016/j.ab.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang F, Sun P, Muñoz E, Chi L, Sakai S, Toida T, Zhang H, Mousa S, Linhardt RJ. Microscale isolation and analysis of heparin from plasma using an anion-exchange spin column. Anal Biochem. 2006;353:284–286. doi: 10.1016/j.ab.2006.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hernáiz MJ, Linhardt RJ. Degradation of chondroitin sulfate and dermatan sulfate with chondroitin lyases. Methods Mol Biol. 2001;171:363–371. doi: 10.1385/1-59259-209-0:363. [DOI] [PubMed] [Google Scholar]
- 22.Solakyildirim K, Zhang Z, Linhardt RJ. Ultraperformance liquid chromatography with electrospray ionization ion trap mass spectrometry for chondroitinal disaccharide analysis. Anal Biochem. 2010;397(1):24–28. doi: 10.1016/j.ab.2009.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang B, Bhattacharyya S, Linhardt R, Tobacman J. Exposure to common food additive carrageenan leads to reduced sulfatase activity and increase in sulfated glycosaminoglycans in human epithelial cells. Biochimie. 2012;94(6):1309–1316. doi: 10.1016/j.biochi.2012.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bhattacharyya S, Solakyildirim K, Zhang Z, Linhardt RJ, Tobacman JK. Chloroquine reduces arylsulphatase B activity and increases chondroitin-4-sulphate: implications for mechanisms of action and resistance. Malar J. 2009;8(1):303. doi: 10.1186/1475-2875-8-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rao GJ, Christe M. Inhibition of rabbit liver arylsulfatase B by phosphate esters. Biochim Biophys Acta. 1994;788:58–61. doi: 10.1016/0167-4838(84)90297-8. [DOI] [PubMed] [Google Scholar]
- 26.Bilusic M, Bataillard A, Tschannen MR, Gao L, Barreto NE, Vincent M, Wang T, Jacob HJ, Sassard J, Kwitek AE. Mapping the genetic determinants of hypertension, metabolic diseases, and related phenotypes in the Lyon hypertensive rat. Hypertension. 2004;44:695–701. doi: 10.1161/01.HYP.0000144542.57306.5e. [DOI] [PubMed] [Google Scholar]
- 27.Duong C, Charron S, Xiao C, Hamet P, Menard A, Roy J, Deng AY. Distinct quantitative trait loci for kidney, cardiac, and aortic mass dissociated from and associated with blood pressure in Dahl congenic rats. Mamm Genome. 2006;17(12):1147–1161. doi: 10.1007/s00335-006-0086-7. [DOI] [PubMed] [Google Scholar]
- 28.Garrett MR, Joe B, Dene H, Rapp JP. Identification of blood pressure quantitative trait loci that differentiate two hypertensive strains. J Hypertens. 2002;20(12):2399–2406. doi: 10.1097/00004872-200212000-00019. [DOI] [PubMed] [Google Scholar]
- 29.Garrett MR, Dene H, Walder R, Zhang QY, Cicila GT, Assadnia S, Deng AY, Rapp JP. Genome scan and congenic strains for blood pressure QTL using Dahl salt-sensitive rats. Genome Res. 1998;8(7):711–723. doi: 10.1101/gr.8.7.711. [DOI] [PubMed] [Google Scholar]
- 30.Schork NJ, Krieger JE, Trolliet MR, Franchini KG, Koike G, Krieger EM, Lander ES, Dzau VJ, Jacob HJ. A biometrical genome search in rats reveals the multigenic basis of blood pressure variation. Genome Res. 1995;5:164–172. doi: 10.1101/gr.5.2.164. [DOI] [PubMed] [Google Scholar]
- 31.Ye ZY, Li DP, Byun HS, Li L, Pan HL. NKCC1 upregulation disrupts chloride homeostasis in the hypothalamus and increases neuronal activity-sympathetic drive in hypertension. J Neurosci. 2012;32(25):8560–8568. doi: 10.1523/JNEUROSCI.1346-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trepiccione F, Zacchia M, Capasso G. The role of the kidney in salt-sensitive hypertension. Clin Exp Nephrol. 2012;16(1):68–72. doi: 10.1007/s10157-011-0489-y. [DOI] [PubMed] [Google Scholar]
- 33.Etscheid M, Beer N, Fink E, Seitz R, Johannes D. The hyaluronan-binding serine protease from human plasma cleaves HMW and LMW kininogen and releases bradykinin. Biol Chem. 2002;383(10):1633–1643. doi: 10.1515/BC.2002.184. [DOI] [PubMed] [Google Scholar]
- 34.Batlle D, Redon J, Gutterman C, LaPointe M, Saleh A, Sharma A, Rombola G, Ye M, Alsheikha W, Gomez L, Sobrero M. Acid–base status and intracellular pH regulation in lymphocytes from rats with genetic hypertension. J Am Soc Nephrol. 1994;5(5 Suppl 1):S12–S22. doi: 10.1681/ASN.V55s12. [DOI] [PubMed] [Google Scholar]
- 35.Cuthbert AW. New horizons in the treatment of cystic fibrosis: Br. J Pharmacol. 2011;163(1):173–183. doi: 10.1111/j.1476-5381.2010.01137.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sharma M, Benharounga M, Hu W, Lukacs GL. Conformation and temperature-sensitive stability defects of the ΔF508 cystic fibrosis transmembrane conductance regulator in post-endoplasmic reticulum compartments. J Biol Chem. 2001;276(12):8942–8950. doi: 10.1074/jbc.M009172200. [DOI] [PubMed] [Google Scholar]
- 37.Cosma MP, Pepe S, Annunziata I, Newbold RF, Grompe M, Parenti G, Ballabio A. The multiple sulfatase deficiency gene encodes an essential and limiting factor for the activity of sulfatases. Cell. 2003;113(4):445–456. doi: 10.1016/s0092-8674(03)00348-9. [DOI] [PubMed] [Google Scholar]
- 38.Roeser D, Preusser-Kunze A, Schmidt B, Gasow K, Wittmann JG, Dierks T, von Figura K, Rudolph MG. A general binding mechanism for all human sulfatases by the formylglycine-generating enzyme. Proc Natl Acad Sci U S A. 2006;103(1):81–86. doi: 10.1073/pnas.0507592102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bhattacharyya S, Tobacman JK. Hypoxia reduces arylsulfatase B activity and silencing arylsulfatase B replicates and mediates the effects of hypoxia. PLoS One. 2012;7(3):e33250. doi: 10.1371/journal.pone.0033250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roeser D, Schmidt B, Preusser-Kunze A, Rudolph MG. Probing the oxygen-binding site of the human formylglycine generating enzyme using halide ions. Acta Crystallog D Biol Crystallogr. 2007;63(Pt 5):621–627. doi: 10.1107/S0907444907009961. [DOI] [PubMed] [Google Scholar]