From yeast to humans, all spliceosomal snRNAs share structural similarities, but higher eukaryotes contain numerous post-transcriptional modifications with undefined roles. Using single-molecule fluorescence we show that several post-transcriptional modifications in human U2 snRNA modulate the dynamic equilibrium of a model U2–U6 complex by stabilizing the four-helix structure. The small magnitude of this effect, however, suggests a primary role in specific RNA–protein interaction mediation in vivo.
Keywords: splicing, human U2–U6 snRNAs, single-molecule FRET, structural dynamics, post-transcriptional modifications
Abstract
The spliceosome catalyzes precursor-mRNA splicing in all eukaryotes. It consists of over 100 proteins and five small nuclear RNAs (snRNAs), including U2 and U6 snRNAs, which are essential for catalysis. Human and yeast snRNAs share structural similarities despite the fact that human snRNAs contain numerous post-transcriptional modifications. Although functions for these modifications have been proposed, their exact roles are still not well understood. To help elucidate these roles in pre-mRNA splicing, we have used single-molecule fluorescence to characterize the effect of several post-transcriptional modifications in U2 snRNA on the conformation and dynamics of the U2–U6 complex in vitro. Consistent with yeast, the human U2–U6 complex reveals the presence of a magnesium-dependent dynamic equilibrium among three conformations. Interestingly, our data show that modifications in human U2 stem I modulate the dynamic equilibrium of the U2–U6 complex by stabilizing the four-helix structure. However, the small magnitude of this effect suggests that post-transcriptional modifications in human snRNAs may have a primary role in the mediation of specific RNA–protein interactions in vivo.
INTRODUCTION
Precursor mRNA (pre-mRNA) splicing is an essential process of eukaryotic gene expression, during which noncoding intron sequences are removed from primary transcripts to form mature mRNA. It consists of two transesterification steps that resemble analogous steps in group II intron splicing (Newman 1998; Sontheimer et al. 1999; Gordon et al. 2000). Splicing regulation is critical to maintain proper cellular function, and defects in splicing have been linked to neurodegenerative disorders and various cancers (Kalnina et al. 2005; Licatalosi and Darnell 2006; Pettigrew and Brown 2008).
Splicing is highly conserved in all eukaryotes (from yeast to humans) and is catalyzed by the spliceosome, a dynamic ribonucleoprotein (RNP) complex assembled from five small nuclear RNAs (U1, U2, U4, U5, and U6 snRNAs) and a large number of proteins (Stark and Luhrmann 2006; Ritchie et al. 2009; Wahl et al. 2009). In addition to catalyzing pre-mRNA splicing, the spliceosome plays an important central role in alternative splicing regulation (Saltzman et al. 2011). Following a highly orchestrated assembly, the activated spliceosome maintains only the U2, U5, and U6 snRNPs and releases U1 and U4 (Wahl et al. 2009). However, several lines of evidence have suggested that only U2 and U6 are critical for catalysis (Parker et al. 1987; Lesser and Guthrie 1993; O'Keefe et al. 1996; Segault et al. 1999; Valadkhan and Manley 2001, 2003). Furthermore, a recent crystal structure of a large Prp8 fragment (a U5 snRNP protein) further supports RNA-based catalysis (Galej et al. 2013). Although the U2–U6 complex is essential for pre-mRNA splicing, its structure and role in catalysis are still not well understood (Madhani and Guthrie 1992; Wolff and Bindereif 1993; Sun and Manley 1995; Sashital et al. 2004; Mefford and Staley 2009).
The U6 snRNA contains two highly conserved sequences that are involved in catalysis (Fig. 1): the 5′ splice site recognition sequence (ACAGAGA box) and the AGC triad (Lesser and Guthrie 1993; Hilliker and Staley 2004). In addition, it also contains the highly conserved U74 (analogous to U80 in yeast) in the U6 intramolecular stem–loop (U6 ISL), which is involved in metal ion binding and coordination for splicing (Huppler et al. 2002; Valadkhan et al. 2009). Based on in vivo genetic and in vitro NMR studies (Madhani and Guthrie 1992; Sun and Manley 1995; Sashital et al. 2004) two different secondary structures for the yeast U2–U6 (yU2–U6) snRNA complex have been proposed: a three- and a four-helix junction (Fig. 1A,B). The three-helix junction (Fig. 1B) comprises the so-called Helix Ib that forms between the highly conserved AGC triad in U6 and three residues in U2 snRNA (Madhani and Guthrie 1992; Mefford and Staley 2009). A recent NMR and SAXS study on the yU2–U6 complex has provided further evidence for the existence of a three-helix junction in solution (Burke et al. 2012). The resulting structural model positions the pre-mRNA recognition sites, the AGC triad, and U80 on the same face of the molecule.
FIGURE 1.
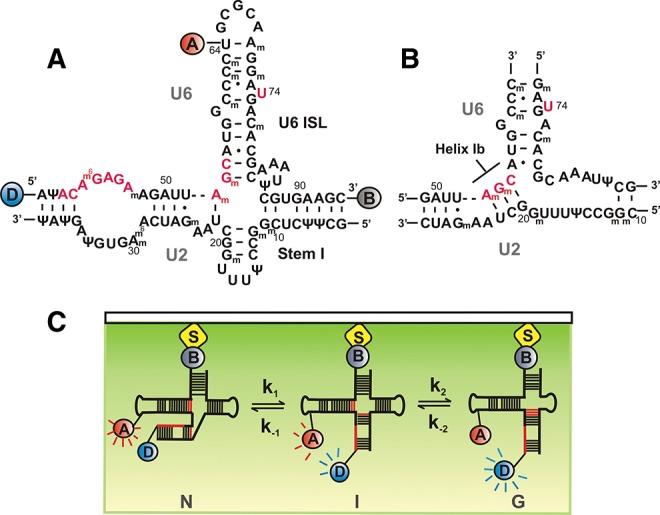
hU2–U6 snRNA complex and single-molecule experimental setup. (A) Proposed four-helix structure of the hU2–U6 complex with FRET donor-D (Cy3, blue), FRET acceptor-A (Cy5, red), and biotin-B (gray). Highly conserved regions of U6 snRNA (ACAGAGA, AGC triad, and U74) are shown in red. (B) Proposed three-helix structure of the hU2–U6 complex containing helix Ib. (C) Total internal reflection fluoresence (TIRF)-based single-molecule experimental setup. Flurophore-labeled RNA was surface immobilized via a biotin-streptavidin linkage and excited using a 532-nm laser beam. Fluoresence intensities of donor (blue) and acceptor (red) fluorophores are collected through the objective and detected using a CCD camera.
In the four-helix junction (Fig. 1A), the AGC triad forms three intramolecular base pairs within U6 snRNA that extend the U6 ISL (Sashital et al. 2004). Interestingly, compensatory mutational genetic studies have shown that Helix Ib residues from both U2 and U6 participate in intramolecular rather than intermolecular base-pairing in humans, suggesting that, in humans, the U2–U6 complex forms the four-helix junction (Wolff and Bindereif 1993; Sun and Manley 1995). Since then, several ensemble-averaged studies have provided evidence that support a hypothesis in which the U2–U6 complex can adopt multiple conformations corresponding to different activation states of the spliceosome (Rhode et al. 2006; Sashital et al. 2007). Recently, we have shown by single-molecule fluorescence that a protein-free U2–U6 snRNA complex from yeast can adopt at least three conformations, consistent with the previously proposed structures (Guo et al. 2009). Consistent with this notion, a recent NMR and enzymatic probing study has shown that a protein-free U2–U6 complex from humans can adopt primarily the four-helix junction structure but also the three-helix junction (Zhao et al. 2013).
Although the primary sequences of U2 and U6 are highly conserved from yeast to humans (Brow and Guthrie 1988), the human snRNAs contain numerous post-transcriptional modifications such as 2′-O-methyl groups and pseudouridines (Massenet et al. 1998; Yu et al. 1998; Sashital et al. 2007). The majority of these conserved modifications are distributed in functionally important regions of the snRNAs, where they can affect pre-mRNA splicing by regulating RNA–RNA and RNA–protein interactions (Newby et al. 2002b; Karijolich and Yu 2010). Out of the five snRNAs in humans, U2 is the most highly modified, containing 10 2′-O-methylated residues and 13 pseudouridines (Fig. 1A; Donmez et al. 2004). Recent NMR and UV melting studies have shown that although the human and yeast U2 stem I structures are nearly isosteric in vitro, post-transcriptional modifications in human U2 significantly increase its stability (Sashital et al. 2007). Since the formation of helix Ib requires the melting of base pairs in stem I, the increased stability of the modified U2 stem I may hinder formation of Helix Ib in human U2–U6 (hU2–U6).
Here, we have used single-molecule Förster resonance energy tranfer (smFRET) to characterize the structural dynamics of a protein-free U2–U6 from humans (Fig. 1A,C). smFRET experiments help dissect RNA folding pathways by revealing the presence of transient folding intermediates that can be hidden in ensemble-averaged experiments and by providing real-time dynamic information (Rueda and Walter 2005; Aleman et al. 2008; Karunatilaka and Rueda 2009; Zhao and Rueda 2009). Our data show that, similarly to the yeast complex, the hU2–U6 also adopts at least three conformations in dynamic equilibrium, and that Mg2+ ions modulate their relative stability. Furthermore, post-transcriptional modifications in the U2 stem I stabilize the four-helix structure. However, the relatively small energetic magnitude of this effect raises the interesting possibility that post-transcriptional modifications in U2 play a more important role in protein-specific recognition in vivo.
RESULTS
hU2–U6 snRNA adopts three dynamic conformations
To characterize the structural dynamics of the unmodified hU2–U6 complex, we have used a previously characterized labeling strategy (Fig. 1A,C; Guo et al. 2009). The U6 strand was labeled with a FRET pair (Cy3, Cy5) at the 5′ end and at uracil 64, respectively, and a 3′-biotin for surface immobilization. This labeling scheme enables monitoring of the dynamics of helix III relative to the U6 ISL, a motion that is thought to bring in close proximity the highly conserved residues involved in the first step of splicing (AGC triad, ACAGAGA loop, and U74) (Fig. 1A). Differing from our previous work, here we have used a single-stranded U6 snRNA, which simplifies the experiments and their interpretation.
We first monitored the folding dynamics of the surface-immobilized fluorophore-labeled construct in standard buffer (50 mM TRIS-HCl at pH 7.5, 100 mM NaCl, and 10 mM MgCl2) using smFRET (Fig. 2A). Similar to our previous work with yeast (Guo et al. 2009), the resulting time trajectories (Fig. 2A) clearly reveal the presence of three distinct FRET states (∼0.6, ∼0.4, and ∼0.2), corresponding to at least three different conformational states in dynamic equilibrium. A time-binned FRET histogram from 154 trajectories (Fig. 2B) confirms the presence of the three FRET states. Similar to the yeast construct, the histogram in 10 mM Mg2+ shows that the high FRET conformation is the least populated (13%), while the intermediate and low FRET conformations are approximately equally populated (43% and 44%, respectively).
FIGURE 2.
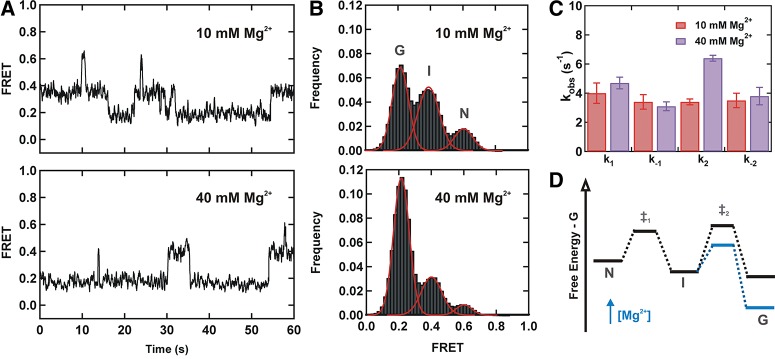
Magnesium stabilizes the low FRET conformation. (A) Typical FRET trajectories of the unmodified U2–U6 complex in 10 mM (top) and 40 mM Mg2+ (bottom). (B) FRET histograms of the unmodified U2–U6 complex obtained in 10 mM (top) and 40 mM Mg2+ (bottom). Both FRET trajectories and histograms exhibit three FRET states: low FRET (∼0.2, G), mid FRET (∼0.4, I), and high FRET (∼0.6, N). (C) Observed folding rate constants (k1, k−1, k2, and k−2) of the hU2–U6 complex without post-transcriptionally modified bases in 10 mM (red) and 40 mM Mg2+ (purple). (D) Effect of Mg2+ on the folding energy landscape of the hU2–U6 complex.
Based on the similarities between the human and yeast results (Guo et al. 2009), we propose a two-step folding pathway for the hU2-U6 snRNAs (Figs. 1C, 2D), where the high and low FRET conformations (N and G) may correspond to the four- and three-helix structures, respectively. As for the yeast construct, out of >350 observed trajectories, <2% exhibited direct transitions from high (G) to low (N) FRET states. Therefore, we assign the intermediate conformation (I) as an obligatory folding intermediate.
A dwell time analysis of the time trajectories yields the folding rate constants (k1, k−1, k2, and k−2) (Fig. 2C), which are in close agreement with those previously obtained with the yU2–U6 complex, except k2, which is ∼70-fold faster for the human construct. This difference may arise from the larger number of mismatched nucleotides in the human ISL, which may be less stable than the yeast ISL, and the presence of fewer base pairs in the human stem I. Overall, in the absence of post-transcriptional modifications, these data suggest that the hU2–U6 snRNA complex behaves similarly to the yeast complex, consistent with the idea that splicing is a highly conserved process amongst all eukaryotes (Will and Luhrmann 2011).
Characterization of the low FRET conformation
To support that the low FRET state corresponds to the three-helix structure with helix Ib formed, as observed for the yU2–U6 complex (Guo et al. 2009), we used a fourfold mutant that prevents helix Ib formation, while maintaining the junction structure of the four-helix structure (Fig. 3A). We flipped four bases in U2 stem I to prevent base-pairing with the catalytic AGC traid of U6. The resulting FRET trajectories and histogram (Fig. 3B,C) clearly show that the fourfold mutation significantly increases the mid FRET population while the low FRET population decreases, as expected. The FRET time trajectories confirm that excursions to the low FRET become less frequent and transient than for the wild-type complex (cf. Figs. 2A and 3B). These results are consistent with the formation of helix Ib in the low FRET conformation (Fig. 1B), similar to the yeast results. However, we cannot formally rule out alternative three-dimensional structures based on different coaxial and side-by-side stacking of helices in solution.
FIGURE 3.
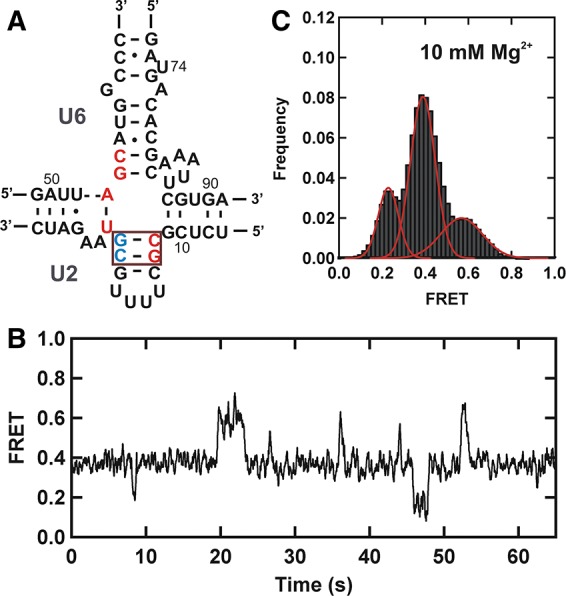
Low FRET state corresponds to the three-helix structure of the U2–U6 complex. (A) Schematic structure of the fourfold mutant hU2–U6 snRNA complex. Residues of U2 and U6 snRNAs participating in helix Ib formation are shown in red. Red box highlights flipped bases (5′ CG-red and 5′ CG-blue) in the U2 stem I. (B) Typical FRET trajectory of the fourfold mutant hU2–U6 snRNA complex. (C) FRET histogram showing the distribution of different FRET states of the mutant U2–U6 snRNA complex in 10 mM Mg2+.
Mg2+ ions modulate the U2–U6 conformational dynamics
Magnesium ions play an essential role during pre-mRNA splicing by facilitating RNA folding and catalysis (Yean et al. 2000; Butcher 2011). We have previously shown that Mg2+ ions stabilize the intermediate and low FRET conformation in the yU2–U6 construct. To test the role of Mg2+ ions on the unmodified hU2–U6 complex, we repeated the smFRET measurements in high [Mg2+] (40 mM). At this concentration the time trajectories (Fig. 2A) show that the molecules remain predominantly in the low FRET state and excursions to the intermediate and high FRET states are less frequent and more transient. A time-binned FRET histogram from 179 trajectories (Fig. 2B) confirms that, in high [Mg2+], the low FRET conformation becomes the most populated (69%), indicating that Mg2+ ions stabilize this conformation relative to the intermediate (26%) and the high (5%) FRET states.
A dwell time analysis of 99 and 108 trajectories at 10 and 40 mM, respectively, (Fig. 2C; Supplemental Fig. 1) shows that the rate constants k1, k−1, and k−2 do not change significantly (P>0.02) with increasing [Mg2+]. However, the rate constant k2 increases by approximately twofold when Mg2+ concentration increases from 10 mM to 40 mM (Fig. 2C). This result confirms that in the 10–40 mM range Mg2+ ions preferentially stabilize the low FRET conformation (Fig. 2D).
Taken together, these data indicate that in the absence of post-transcriptional modifications, a high concentration of divalent ions stabilizes the low FRET state, which we attribute to the three-helix conformation of the hU2–U6 complex similar to that of yeast snRNAs (Guo et al. 2009).
Post-transcriptional modifications modulate the U2–U6 conformational dynamics
Human U2–U6 snRNA complex contains 23 post-transcriptionally modified nucleotides in both strands (Fig. 1A), in contrast to the single modified nucleotide (pseudouridine at position 35 in U2) for the equivalent complex in yeast (Yu et al. 1998; Donmez et al. 2004; Sashital et al. 2007; Karijolich and Yu 2010). To understand the effect of these modifications on the structural dynamics of the hU2–U6 complex, we have focused on four modifications in U2 stem I (Fig. 4A) that have been shown to be important for splicing (Donmez et al. 2004; Karijolich and Yu 2010): 2′-O-methylguanosines (Gm) at positions 11, 12, 19, and a pseudouridine at position 15. Several studies have suggested that modifications in U2 snRNA are involved in spliceosomal assembly (Karijolich and Yu 2010). Specficially, 2′-O-methylations at positions 12 and 19 have been individually shown to be required for early spliceosomal E complex formation (Donmez et al. 2004).
FIGURE 4.
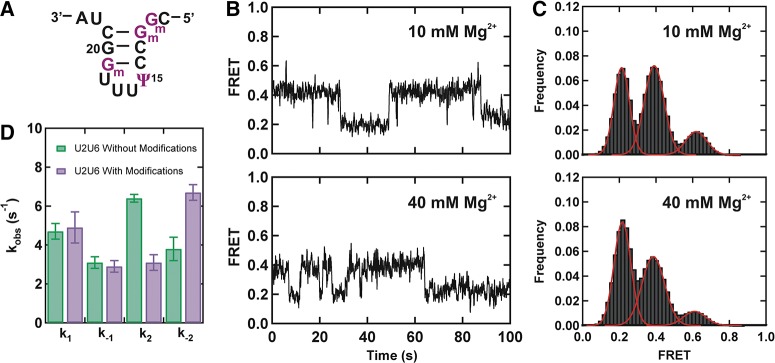
Modified bases affect structural dynamics of the hU2–U6 complex. (A) Schematic structure of the U2 stem I containing post-transcriptionally modified bases (purple). (B) FRET trajectories of the modified U2–U6 complex exhibiting three different FRET states in 10 and 40 mM Mg2+. (C) FRET histograms obtained from >100 trajectories in 10 and 40 mM Mg2+. (D) Observed folding rate constants (k1, k−1, k2, and k−2) of the hU2–U6 snRNA complex with (purple) and without (green) post-transcriptionally modified bases in 40 mM Mg2+.
The observed time-trajectories in 40 mM [Mg2+] with the fully modified U2 stem I reveal that the snRNA complex resides for less time in the low FRET state and more time in the intermediate FRET conformation (cf. Figs. 2A and 4B, bottom). A time-binned FRET histogram from 144 trajectories (Fig. 4C), confirms a significant population decrease in the low FRET state (from 69% to 47%) accompanied by a population increase in the intermediate FRET state (from 26% to 44%). Dwell time analysis of the time trajectories shows that while the presence of the modifications does not affect appreciably the rate constants k1 and k−1, they decrease k2 by approximately twofold and increase k−2 by approximately twofold (Fig. 4D). These effects in rate constants clearly explain the population decrease in the low FRET state and population increase in the intermediate FRET state. However, consistent with comparable rate constants of k1 and k−1, we did not observe a substantial change in the high FRET population with modifications (8% with modifications and 5% without modifications).
Collectively, these results indicate that the presence of post-transcriptional modifications destabilize the low FRET state that has been proposed as the three-helix conformation (helix Ib) and stabilize the intermediate conformation (stem I). Based on the FRET histograms, we estimate that these modifications stabilize the U2 stem I by ∼0.5 kcal/mol. These results are in qualitative agreement with previous UV melting studies that have shown a modification-mediated stabilization of the U2 stem I helix (Sashital et al. 2007). Lowering the [Mg2+] to 10 mM results in similar but less prominent effects (Fig. 4B,C).
To understand the effect of pseudouridine 15 and 11Gm on the structural dynamics of the U2–U6 complex, we performed single-molecule experiments with a U2–U6 snRNA construct containing only Gm at positions 12 and 19 (12Gm + 19Gm) (Fig. 5A; Supplemental Fig. 2). We used 40 mM Mg2+ because the stabilizing effect of the modifications is most pronounced at this concentration (Fig. 4D). A time-binned histogram from 102 FRET trajectories reveals almost identical distribution to the fully modified U2 construct (cf. Figs. 4C and 5A), indicating that the absence of pseudouridine 15 and 11Gm does not significantly affect the relative stability of the U2–U6 conformations. An estimate of the relative free-energy difference between states (Table 1, ΔΔG) confirms that most of the stabilization of the U2 stem I arises from 12Gm and 19Gm alone.
FIGURE 5.
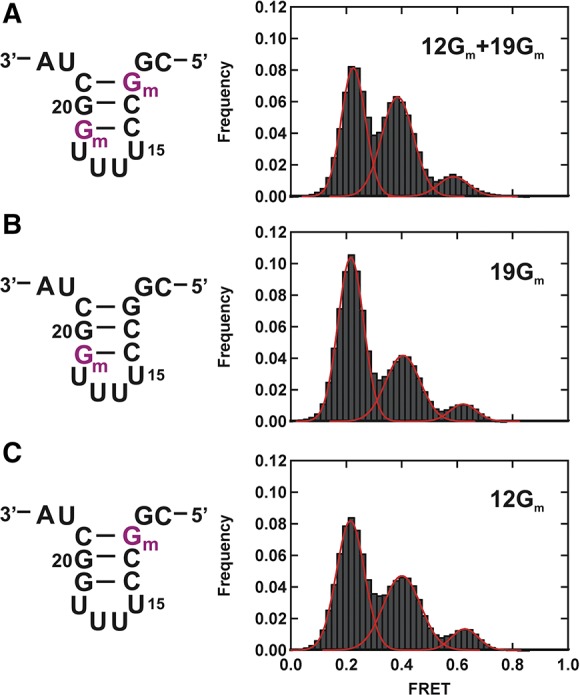
Charaterization of individual modifications. (A) Schematic structure of the U2 stem I containing 2′-O-methylguanosines (Gm) at positions 12 and 19 (left) and corresponding FRET histogram at 40 mM Mg2+ (right). (B) Schematic structure of the U2 stem I containing Gm at position 19 (left) and corresponding FRET histogram at 40 mM Mg2+ (right). (C) Schematic structure of the U2 stem I containing Gm at position 12 (left) and corresponding FRET histogram at 40 mM Mg2+ (right). All modified bases are shown in purple.
TABLE 1.
Post-transcriptional modifications destabilize the low FRET conformation (ψ = pseudouridine and Gm = 2′-O-methylguanosine)
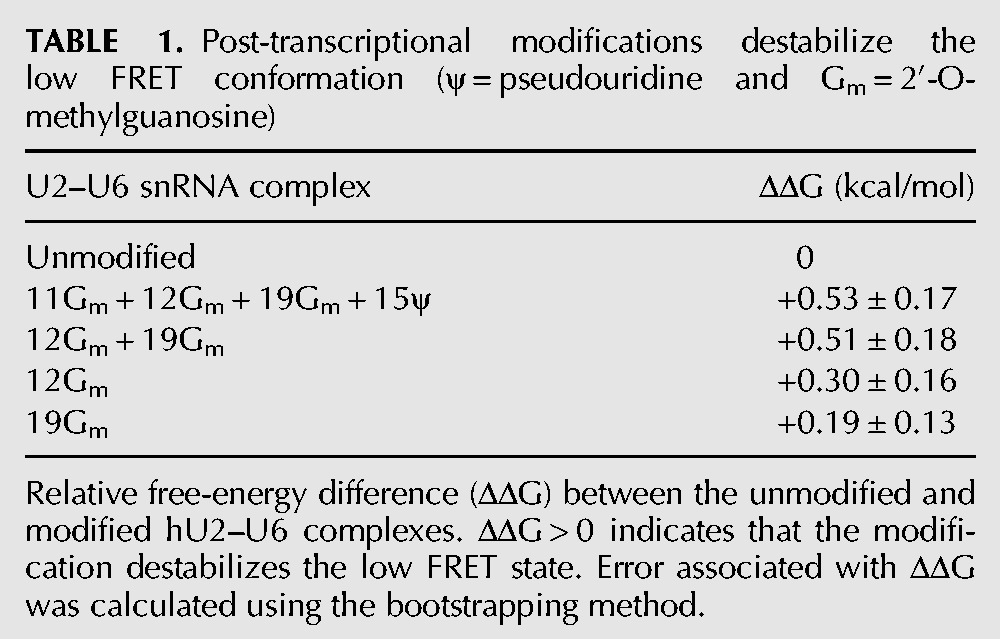
To assess the contribution from each of the remaining modifications (12Gm and 19Gm), we repeated the measurement with only one of them at a time (Fig. 5B,C). A time-binned histogram from 118 single-molecule trajectories of only 19Gm (Fig. 5B) reveals a population distribution that resembles the one obtained with the unmodified construct (cf. Figs. 2B and 5B), indicating that 19Gm plays a minor role in stabilizing the U2 stem I. An estimate of the relative free-energy difference between states (Table 1) shows that 0.19 of the 0.53 kcal/mol stabilization of the modified U2 stem I arise from 19Gm alone. In the presence of only 12Gm (Fig. 5C), a time-binned histogram from 151 single-molecule trajectories reveals a population distribution that is intermediate between the fully modified construct and the unmodified construct (cf. Figs 2B, 4C, and 5C), indicating that 12Gm plays a more important role in stabilizing the U2 stem I. An estimate of the relative free-energy difference between states (Table 1, ΔΔG) shows that 0.30 of the 0.53 kcal/mol stabilization of the U2 stem I arise from 12Gm alone. Together, these results show that the U2 stem I stabilization effect from each post-transcriptional modification is additive and not cooperative.
DISCUSSION
The spliceosomal RNA complex formed by U2 and U6 is one of the main components of the catalytic core of the eukaryotic spliceosome (Parker et al. 1987; Lesser and Guthrie 1993; O'Keefe et al. 1996; Segault et al. 1999; Valadkhan and Manley 2001, 2003). In the absence of proteins, the hU2–U6 snRNA complex can catalyze a splicing related reaction in vitro providing direct evidence for the catalytic ability of these spliceosomal snRNAs (Valadkhan and Manley 2001; Valadkhan et al. 2009). However, the specific conformation of the U2–U6 snRNA complex is not well understood. Here we have used single molecule fluorescence to characterize the structural dynamics of the hU2–U6 complex in real time.
Consistent with our previous studies on the yU2–U6 complex (Guo et al. 2009), the smFRET data shows that the hU2–U6 snRNA complex also adopts at least three distinct conformations in dynamic equilibrium. The relative stability of each conformation varies with the concentration of Mg2+ ions in solution, suggesting that the hU2–U6 is a Mg-dependent conformational switch. Based on sequence similarities between the human and the yeast complexes and the mutational data presented here, we have assigned the observed FRET states to the four-helix structure with an extended U6 ISL (FRET ∼0.6 and ∼0.4), as proposed by early NMR studies in yeast (Sashital et al. 2004), and more recently in the human U2–U6 complex (Zhao et al. 2013) and the three-helix structure with Helix 1B (FRET ∼0.2), as proposed by more recent NMR and SAXS studies in yeast (Burke et al. 2012).
Considering the positions of the FRET donor and acceptor fluorophores, we propose that in the high FRET state the highly conserved ACAGAGA sequence and the magnesium binding site U74 in U6 ISL are in close proximity. Therefore, the high FRET state resembles the four-helix structure with the extended U6 ISL containing the AGC triad. Furthermore, this structure supports the important properties of the active conformation exhibiting close association of the highly conserved regions that have been shown to participate in the splicing reaction.
In high magnesium concentrations, the U2–U6 complex prefers the low FRET state, which has two fluorophores far away from each other. As we previously observed in yeast, the hU2–U6 complex may also adopt the three-helix structure with helix Ib by rearranging the junction structure. The existence of the three-helix structure with helix Ib was further supported using a fourfold mutant containing flipped bases in the U2 stem I that prevent the formation of helix Ib. Although it can maintain the junction structure, the mutant U2–U6 construct greatly decreases the population of the low FRET state. Thus, smFRET data of the fourfold mutant provide evidence for the observed low FRET state as the previously proposed three-helix structure.
In addition to the conformations discussed above, smFRET data of U2–U6 snRNAs exhibited the existence of a previously unidentified folding intermediate with FRET ∼0.4. Since previous studies have shown that the stability of this conformation is independent of the mutations at U80 in yeast (comparable to U74 in humans) U6 snRNA (Guo et al. 2009), we propose this mid FRET state has the extended U6 ISL structure, but lacks the tertiary interactions between the ACAGAGA sequence and the U6 ISL. However, the role of this conformation in pre-mRNA splicing is not well understood in both humans and yeast. Although we have assigned the observed FRET states as the three-helix and the four-helix conformations, it is important to note that the hU2–U6 complex may fold into alternative three-dimensional structures based on different coaxial and side-by-side stacking of helices in solution.
Consistent with yeast smFRET data, the population of the low FRET state (FRET ∼0.2) increases with Mg2+ concentration, indicating a magnesium-mediated stabilization of the three-helix structure (Guo et al. 2009). As a result, under high Mg2+ concentrations, the hU2–U6 complex without modified bases showed a higher population of molecules in the three-helix structure than the four-helix structure. In contrast, the hU2–U6 snRNAs with post-transcriptionally modified bases in U2 stem I (11Gm, 12Gm, 19Gm, and pseudouridine at position 15) show a clear stabilization effect of the mid FRET state while lowering the population of the low FRET state corresponds to the three-helix structure. Interestingly, smFRET data obtained from the U2–U6 construct with 2′-O-methylguanosines at positions 12 and 19 (12Gm + 19Gm) exhibited folding dynamics similar to the fully modified construct. Therefore, these results suggest that 11Gm and pseudouridine at position 15 of U2 stem I do not exhibit an overall effect on the folding dynamics of the U2–U6 complex. This unique behavior can be due to either cancellation of opposite effects or due to lack of direct stabilization/destabilization effect from the two individual modifications. Furthermore, compared with the modification at position 19, the 2′-O-methylguanosines at position 12 showed a higher destabilization effect on the low FRET state of the hU2–U6 complex. This result is in agreement with previous measurements of Holliday Junction dynamics which show that the first base pair near the junction plays an important role in its stability and dynamics (McKinney et al. 2005).
Taken together, the smFRET data suggest that modified bases collectively affect the structural dynamics of the U2–U6 complex and may prevent the formation of the proposed three-helix structure consisting of helix Ib by stabilizing U2 stem I. 2′-O-methylations can alter the structural stabilization of U2 stem I by blocking sugar edge interactions and by reducing the reactivity of the RNA backbone (Auffinger and Westhof 1997; Helm 2006). In the case of pseudouridine, this may result from improved base stacking (Davis 1995) and from formation of a water-mediated hydrogen bond between the additional protonated imino group and adjacent phosphate residues, as reported in functionally important RNAs including tRNAs (Arnez and Steitz 1994; Newby et al. 2002a).
Although the modified bases destabilize the low FRET state, the effect of modifications on the stability of U2 stem I is less significant in the U2–U6 complex than in the U2 stem I alone, as shown in UV melting studies (Sashital et al. 2004). Therefore, we propose that post-transcriptionally modified residues may be involved in protein-dependent functions rather than direct stabilization of U2–U6 RNA structures in vivo. Hence, we suggest that modified bases in U2 snRNA may play a major role in protein recognition and interactions in cells. Thereby these modifications can directly participate in the spliceosome assembly process as suggested previously (Donmez et al. 2004). This hypothesis is further supported by the presence of a large number of spliceosomal proteins associating with human snRNAs, which are highly post-transcriptionally modified compared with the yeast system.
In summary, the hU2–U6 complex follows the minimal two-step folding pathway consisting of the previously proposed two structures and at least one obligatory intermediate conformation. Furthermore, RNA constructs without post-transcriptionally modified bases exhibited similar dynamics as yeast spliceosomal RNAs (Guo et al. 2009). Although modified bases can affect the structural dynamics of hU2–U6 snRNAs, the structural effects of the U2–U6 complex were less than that of U2 stem I alone, suggesting a protein-dependent role rather than direct stabilization of RNA structures in cells.
MATERIALS AND METHODS
RNA purification and labeling
5′-Cy3 labeled U6 RNA (5′-AUACAGAGAAGAUUAGCAUGGCCCCdTGCGCAAGGAUGACACGCAAAUUCGUGAAGC-3′ Biotin, where dT contains an internal, free primary amine for Cy5 labeling) and U2 RNA (5′-GCUUCUCGGCCUUUUGGCUAAGAUCAAGUGUAGUAU-3′) with or without modified bases (pseudouridine and 2′-O-methyl guanosines) were purchased from the Howard Hughes Medical Institution Biopolymer/Keck Foundation Biotechnology Resource Laboratory (Yale University). Synthetic RNAs were 2′-OH deprotected, labeled, and purified by denaturing 18% polyacrylamide gel electrophoresis and C8 reversed-phase HPLC as described (Steiner et al. 2008; Zhao and Rueda 2009). Internal dT of U6 was Cy5 labeled following the manufacturer's protocol and purified by reversed-phase HPLC.
Single-molecule FRET (smFRET)
Single-molecule FRET experiments were performed on a home-built, prism-based, total internal reflection microscope as described (Guo et al. 2009; Zhao and Rueda 2009). Briefly: Biotinylated and fluorophore-labeled U6 (1 µM) and U2 (10 µM) were heated (90°C for 45 sec) in standard buffer (50 mM TRIS-HCl at pH 7.5, 100 mM NaCl, and variable [Mg2+]) and annealed (15–20 min) at room temperature. The resulting U2–U6 RNA complex was diluted to ∼25 pM and surface immobilized onto a streptavidin-coated quartz slide via a biotin-streptavidin linkage. Unbound RNA was washed away with standard buffer. To reduce fluorophore photobleaching, an oxygen-scavenging system (OSS) consisting of 10% (w/v) glucose, 2% (v/v) 2-mercaptoethanol, 50 µg/mL glucose oxidase, and 10 µg/mL catalase was used in all experiments. All TIRF-based single-molecule experiments were performed using a 532-nm laser with 33-msec time resolution at room temperature.
Data analysis
FRET and dwell times histograms were built by time binning >100 dynamic single-molecule time trajectories. Kinetic rate constants were obtained by fitting the resulting dwell times histograms to exponential decays as described (Guo et al. 2009). Differences between free-energy changes associated with the formation of the low FRET state for unmodified (U) and modified (M) U2–U6 snRNA constructs were calculated as ΔΔG = ΔGM − ΔGU, where ΔG = −RT ln(K), K = A0.2/A0.4 and A represents the area of a given FRET state in a FRET histogram. Error associated with ΔΔG was analyzed using the bootstrap method as described (Efron and Gong 1983).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank N. Greenbaum and S. Butcher for critically reading the manuscript. The Rueda lab is funded by grants from the NIH (GM085116) and a CAREER award from the NSF (MCB-0747285).
Author contributions: K.S.K. and D.R. designed research; K.S.K. performed research and analyzed data; K.S.K. and D.R. wrote the paper.
REFERENCES
- Aleman EA, Lamichhane R, Rueda D 2008. Exploring RNA folding one molecule at a time. Curr Opin Chem Biol 12: 647–654 [DOI] [PubMed] [Google Scholar]
- Arnez JG, Steitz TA 1994. Crystal structure of unmodified tRNAGln complexed with glutaminyl-tRNA synthetase and ATP suggests a possible role for pseudo-uridines in stabilization of RNA structure. Biochemistry 33: 7560–7567 [DOI] [PubMed] [Google Scholar]
- Auffinger P, Westhof E 1997. Rules governing the orientation of the 2′-hydroxyl group in RNA. J Mol Biol 274: 54–63 [DOI] [PubMed] [Google Scholar]
- Brow DA, Guthrie C 1988. Spliceosomal RNA U6 is remarkably conserved from yeast to mammals. Nature 334: 213–218 [DOI] [PubMed] [Google Scholar]
- Burke JE, Sashital DG, Zuo X, Wang YX, Butcher SE 2012. Structure of the yeast U2/U6 snRNA complex. RNA 18: 673–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butcher SE 2011. The spliceosome and its metal ions. Met Ions Life Sci 9: 235–251 [DOI] [PubMed] [Google Scholar]
- Davis DR 1995. Stabilization of RNA stacking by pseudouridine. Nucleic Acids Res 23: 5020–5026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donmez G, Hartmuth K, Luhrmann R 2004. Modified nucleotides at the 5′ end of human U2 snRNA are required for spliceosomal E-complex formation. RNA 10: 1925–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efron B, Gong G 1983. A leisurely look at the bootstrap, the jackknife, and cross-validation. Am Stat 37: 36–48 [Google Scholar]
- Galej WP, Oubridge C, Newman AJ, Nagai K 2013. Crystal structure of Prp8 reveals active site cavity of the spliceosome. Nature 493: 638–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon PM, Sontheimer EJ, Piccirilli JA 2000. Metal ion catalysis during the exon-ligation step of nuclear pre-mRNA splicing: Extending the parallels between the spliceosome and group II introns. RNA 6: 199–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z, Karunatilaka KS, Rueda D 2009. Single-molecule analysis of protein-free U2–U6 snRNAs. Nat Struct Mol Biol 16: 1154–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helm M 2006. Post-transcriptional nucleotide modification and alternative folding of RNA. Nucleic Acids Res 34: 721–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilliker AK, Staley JP 2004. Multiple functions for the invariant AGC triad of U6 snRNA. RNA 10: 921–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppler A, Nikstad LJ, Allmann AM, Brow DA, Butcher SE 2002. Metal binding and base ionization in the U6 RNA intramolecular stem-loop structure. Nat Struct Biol 9: 431–435 [DOI] [PubMed] [Google Scholar]
- Kalnina Z, Zayakin P, Silina K, Line A 2005. Alterations of pre-mRNA splicing in cancer. Genes Chromosomes Cancer 42: 342–357 [DOI] [PubMed] [Google Scholar]
- Karijolich J, Yu YT 2010. Spliceosomal snRNA modifications and their function. RNA Biol 7: 192–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karunatilaka KS, Rueda D 2009. Single-molecule fluorescence studies of RNA: A decade's progress. Chem Phys Lett 476: 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesser CF, Guthrie C 1993. Mutations in U6 snRNA that alter splice site specificity: Implications for the active site. Science 262: 1982–1988 [DOI] [PubMed] [Google Scholar]
- Licatalosi DD, Darnell RB 2006. Splicing regulation in neurologic disease. Neuron 52: 93–101 [DOI] [PubMed] [Google Scholar]
- Madhani HD, Guthrie C 1992. A novel base-pairing interaction between U2 and U6 snRNAs suggests a mechanism for the catalytic activation of the spliceosome. Cell 71: 803–817 [DOI] [PubMed] [Google Scholar]
- Massenet S, Mougin A, Branlant C 1998. Posttranscriptional modifications in the U small nuclear RNAs. In The modification and editing of RNA. (ed. Grosjean H, Benne R), pp. 201–228 ASM Press, Washington, DC [Google Scholar]
- McKinney SA, Freeman AD, Lilley DM, Ha T 2005. Observing spontaneous branch migration of Holliday junctions one step at a time. Proc Natl Acad Sci 102: 5715–5720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mefford MA, Staley JP 2009. Evidence that U2/U6 helix I promotes both catalytic steps of pre-mRNA splicing and rearranges in between these steps. RNA 15: 1386–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newby MI, Greenbaum NL 2002a. Investigation of Overhauser effects between pseudouridine and water protons in RNA helices. Proc Natl Acad Sci 99: 12697–12702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newby MI, Greenbaum NL 2002b. Sculpting of the spliceosomal branch site recognition motif by a conserved pseudouridine. Nat Struct Biol 9: 958–965 [DOI] [PubMed] [Google Scholar]
- Newman A 1998. RNA splicing. Curr Biol 8: R903–R905 [DOI] [PubMed] [Google Scholar]
- O'Keefe RT, Norman C, Newman AJ 1996. The invariant U5 snRNA loop 1 sequence is dispensable for the first catalytic step of pre-mRNA splicing in yeast. Cell 86: 679–689 [DOI] [PubMed] [Google Scholar]
- Parker R, Siliciano PG, Guthrie C 1987. Recognition of the TACTAAC box during mRNA splicing in yeast involves base pairing to the U2-like snRNA. Cell 49: 229–239 [DOI] [PubMed] [Google Scholar]
- Pettigrew CA, Brown MA 2008. Pre-mRNA splicing aberrations and cancer. Front Biosci 13: 1090–1105 [DOI] [PubMed] [Google Scholar]
- Rhode BM, Hartmuth K, Westhof E, Luhrmann R 2006. Proximity of conserved U6 and U2 snRNA elements to the 5′ splice site region in activated spliceosomes. EMBO J 25: 2475–2486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie DB, Schellenberg MJ, MacMillan AM 2009. Spliceosome structure: Piece by piece. Biochim Biophys Acta 1789: 624–633 [DOI] [PubMed] [Google Scholar]
- Rueda D, Walter NG 2005. Single molecule fluorescence control for nanotechnology. J Nanosci Nanotechnol 5: 1990–2000 [DOI] [PubMed] [Google Scholar]
- Saltzman AL, Pan Q, Blencowe BJ 2011. Regulation of alternative splicing by the core spliceosomal machinery. Genes Dev 25: 373–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sashital DG, Cornilescu G, McManus CJ, Brow DA, Butcher SE 2004. U2–U6 RNA folding reveals a group II intron-like domain and a four-helix junction. Nat Struct Mol Biol 11: 1237–1242 [DOI] [PubMed] [Google Scholar]
- Sashital DG, Venditti V, Angers CG, Cornilescu G, Butcher SE 2007. Structure and thermodynamics of a conserved U2 snRNA domain from yeast and human. RNA 13: 328–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segault V, Will CL, Polycarpou-Schwarz M, Mattaj IW, Branlant C, Luhrmann R 1999. Conserved loop I of U5 small nuclear RNA is dispensable for both catalytic steps of pre-mRNA splicing in HeLa nuclear extracts. Mol Cell Biol 19: 2782–2790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sontheimer EJ, Gordon PM, Piccirilli JA 1999. Metal ion catalysis during group II intron self-splicing: Parallels with the spliceosome. Genes Dev 13: 1729–1741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark H, Luhrmann R 2006. Cryo-electron microscopy of spliceosomal components. Annu Rev Biophys Biomol Struct 35: 435–457 [DOI] [PubMed] [Google Scholar]
- Steiner M, Karunatilaka KS, Sigel RK, Rueda D 2008. Single-molecule studies of group II intron ribozymes. Proc Natl Acad Sci 105: 13853–13858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun JS, Manley JL 1995. A novel U2–U6 snRNA structure is necessary for mammalian mRNA splicing. Genes Dev 9: 843–854 [DOI] [PubMed] [Google Scholar]
- Valadkhan S, Manley JL 2001. Splicing-related catalysis by protein-free snRNAs. Nature 413: 701–707 [DOI] [PubMed] [Google Scholar]
- Valadkhan S, Manley JL 2003. Characterization of the catalytic activity of U2 and U6 snRNAs. RNA 9: 892–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valadkhan S, Mohammadi A, Jaladat Y, Geisler S 2009. Protein-free small nuclear RNAs catalyze a two-step splicing reaction. Proc Natl Acad Sci 106: 11901–11906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Luhrmann R 2009. The spliceosome: Design principles of a dynamic RNP machine. Cell 136: 701–718 [DOI] [PubMed] [Google Scholar]
- Will CL, Luhrmann R 2011. Spliceosome structure and function. Cold Spring Harb Perspect Biol 3: a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff T, Bindereif A 1993. Conformational changes of U6 RNA during the spliceosome cycle: An intramolecular helix is essential both for initiating the U4–U6 interaction and for the first step of slicing. Genes Dev 7: 1377–1389 [DOI] [PubMed] [Google Scholar]
- Yean SL, Wuenschell G, Termini J, Lin RJ 2000. Metal-ion coordination by U6 small nuclear RNA contributes to catalysis in the spliceosome. Nature 408: 881–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu YT, Shu MD, Steitz JA 1998. Modifications of U2 snRNA are required for snRNP assembly and pre-mRNA splicing. EMBO J 17: 5783–5795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R, Rueda D 2009. RNA folding dynamics by single-molecule fluorescence resonance energy transfer. Methods 49: 112–117 [DOI] [PubMed] [Google Scholar]
- Zhao C, Bachu R, Popovic M, Devany M, Brenowitz M, Schlatterer JC, Greenbaum NL 2013. Conformational heterogeneity of the protein-free human spliceosomal U2–U6 snRNA complex. RNA 19: 561–573 [DOI] [PMC free article] [PubMed] [Google Scholar]


