The btuB riboswitch from E. coli is known to alter its conformation upon interaction with its ligand, coenzyme B12; however, little is known about the ligand-free form of the btuB riboswitch. This work highlights the Mg2+-induced prefolding of the btuB aptamer and indicates the importance of the prefolded aptamer for recognition of coenzyme B12. The extrapolation of the data to the class of coenzyme B12 riboswitches suggests that the prefolding of the aptamer could be a common phenomenon for coenzyme B12 riboswitches to successfully interact with coenzyme B12.
Keywords: Mg2+, RNA, folding, riboswitch, coenzyme B12
Abstract
Mg2+ has been shown to modulate the function of riboswitches by facilitating the ligand-riboswitch interactions. The btuB riboswitch from Escherichia coli undergoes a conformational change upon binding to its ligand, coenzyme B12 (adenosyl-cobalamine, AdoCbl), and down-regulates the expression of the B12 transporter protein BtuB in order to control the cellular levels of AdoCbl. Here, we discuss the structural folding attained by the btuB riboswitch from E. coli in response to Mg2+ and how it affects the ligand binding competent conformation of the RNA. The btuB riboswitch notably adopts different conformational states depending upon the concentration of Mg2+. With the help of in-line probing, we show the existence of at least two specific conformations, one being achieved in the complete absence of Mg2+ (or low Mg2+ concentration) and the other appearing above ∼0.5 mM Mg2+. Distinct regions of the riboswitch exhibit different dissociation constants toward Mg2+, indicating a stepwise folding of the btuB RNA. Increasing the Mg2+ concentration drives the transition from one conformation toward the other. The conformational state existing above 0.5 mM Mg2+ defines the binding competent conformation of the btuB riboswitch which can productively interact with the ligand, coenzyme B12, and switch the RNA conformation. Moreover, raising the Mg2+ concentration enhances the ratio of switched RNA in the presence of AdoCbl. The lack of a AdoCbl-induced conformational switch experienced by the btuB riboswitch in the absence of Mg2+ indicates a crucial role played by Mg2+ for defining an active conformation of the riboswitch.
INTRODUCTION
Riboswitches are highly conserved RNA elements present in the noncoding regions of various mRNAs exerting their response on genetic regulation by binding small metabolites (Mironov et al. 2002; Nahvi et al. 2002; Winkler et al. 2002; Sudarsan et al. 2003a). Metabolite binding to the aptamer region of the riboswitch alters its conformation, accompanied by the structural changes in the downstream expression platform (Mandal and Breaker 2004). These conformational changes regulate the gene expression either at the transcriptional or translational level (Winkler and Breaker 2003; Soukup and Soukup 2004), due to intron splicing (Kubodera et al. 2003), activation of self cleavage (Winkler et al. 2004), or due to trans regulation (Loh et al. 2009; Bastet et al. 2011). The btuB riboswitch of Escherichia coli is placed in the 5′ UTR of the btuB gene encoding the cobalamine transporter BtuB, an outer membrane protein (Reynolds et al. 1980; Gudmundsdottir et al. 1988). At high cellular concentration, the ligand AdoCbl interacts with the btuB aptamer, inducing thereby a structural switch in the RNA (Nahvi et al. 2002). The coordinated conformational switch in the aptamer and the expression platform of the riboswitch can result in the attenuation of transcription or inhibition of translation of the btuB mRNA to maintain adequate levels of AdoCbl in the cytosol (Nou and Kadner 2000; Mandal and Breaker 2004). Apart from AdoCbl, this riboswitch also interacts with Vitamin B12 and other cobalamine derivatives, although with reduced affinity (Nahvi et al. 2002; Gallo et al. 2008).
To bind its ligand, a riboswitch needs to adopt its so-called binding competent conformation (Montange and Batey 2008; Haller et al. 2011b). In most cases, such a competent structure can be achieved by Mg2+- assisted folding of the riboswitch. Mg2+ favors the formation and stabilization of the tertiary structure of the polyanionic RNA by reducing the strong electrostatic repulsions experienced by the closely positioned phosphodiester groups (Pyle 2002; Draper 2004; Woodson 2005; Freisinger and Sigel 2007; Sigel and Pyle 2007; Chu et al. 2008; Sigel and Sigel 2013). Divalent metal ions like Mg2+ have been reported to be playing indispensable roles in the case of riboswitches in the formation of an active tertiary structure (Lemay et al. 2006; Noeske et al. 2007b; Serganov et al. 2009; Haller et al. 2011a; Jenkins et al. 2011; Ramesh et al. 2011). In some cases, Mg2+ also facilitates the interaction of the ligand with its riboswitch (Klein and Ferré-D'Amaré 2006; Serganov et al. 2006, 2009; Lipfert et al. 2010); e.g., the SAM I riboswitch (Heppell et al. 2011) requires both divalent metal ions and its ligand to induce a conformational alteration. In contrast, the binding of guanine to the xpt G riboswitch can take place at high monovalent ion concentration but in the absence of Mg2+, although 2 mM Mg2+ drives the complex formation to completion (Serganov et al. 2004).
Given the diverse activities attributed to Mg2+ for an efficient functioning of riboswitches, we focused our interest on the roles of Mg2+ in one of the most complex riboswitches, the btuB riboswitch from E. coli. Although the mechanism for AdoCbl binding to B12 riboswitches has been studied previously (Gallo et al. 2008, 2010; Johnson et al. 2012; Perdrizet et al. 2012; Peselis and Serganov 2012), the metal ion-induced folding and functioning of coenzyme B12 riboswitches remains unsolved.
Here, we describe the influence of Mg2+ on the preorganization of the binding competent structure for the btuB aptamer. At the same time, we deduce the riboswitch folding by establishing the affinity constants of different regions to Mg2+. Our results suggest that the formation of a binding competent conformation is crucial for the btuB aptamer to recognize AdoCbl. Furthermore, we have also defined the influence of Mg2+ in the AdoCbl-induced conformational switch of the aptamer. Our studies provide an insight into the Mg2+-dependent folding of the btuB riboswitch as well as indicate the relevance of Mg2+ in regulation of the conformational switch of the btuB RNA by AdoCbl.
RESULTS
Mg2+-dependent conformational states of the ligand-free btuB aptamer
To observe the dependence of the btuB riboswitch folding on Mg2+, we have used the well-established technique of in-line probing (Regulski and Breaker 2008; Choudhary et al. 2014). This technique has been used previously to distinguish the ligand-induced changes in various riboswitches (Nahvi et al. 2002; Winkler et al. 2002; Mandal et al. 2003; Sudarsan et al. 2003b). The in-line nucleophilic attack itself depends on the local geometry taken up by the involved nucleotides and thereby represents the overall RNA conformation (Soukup and Breaker 1999). Hence, to detect any metal ion-induced conformational alteration in the ligand-free RNA, we subjected the btuB aptamer to in-line probing by varying Mg2+ concentrations in the complete absence of AdoCbl.
As seen in Figure 1A, the btuB RNA is almost unstructured in water only. Addition of Tris-HCl (pH 8.3) that includes ∼25 mM K+ leads to first structuring, i.e., in the absence of further monovalent and divalent cations, some nucleotides show a marked protection from the cleavage (Fig. 1A, lanes 1, 2). The addition of 100 mM KCl induces distinct structural changes in some of the regions of the RNA as summarized in Figure 1B. With the addition of Mg2+ up to 0.1 mM, the cleavage pattern, and thus the structure, remain unchanged, indicating that the critical Mg2+ sites have KD values smaller than 100 μM. Generally, a rather unspecific and relatively high background cleavage at all positions can be observed at low Mg2+ concentration. This is true especially for the loop regions, indicating the absence of tertiary contacts.
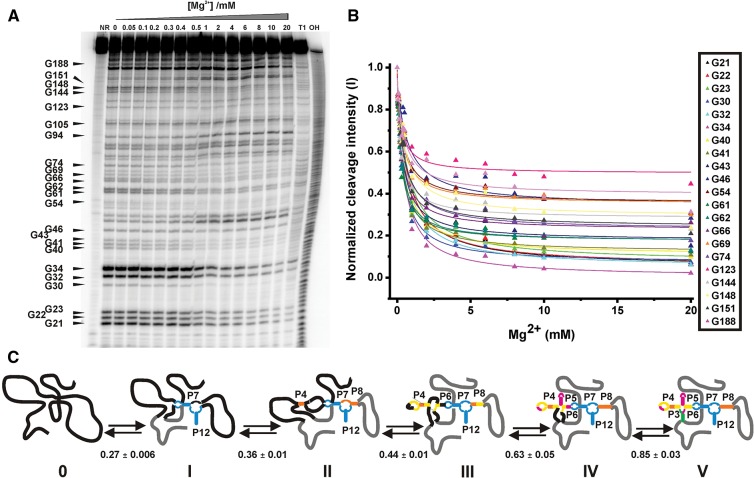
FIGURE 1.
Mg2+-induced conformational changes in the btuB aptamer. (A) In-line probing of the btuB aptamer in the absence of AdoCbl by varying the Mg2+ concentration from 0 to 20 mM in the absence and presence of 100 mM KCl. Lane 14 with 20 mM Mg2+ and 500 μM AdoCbl serves as a reference to map the AdoCbl interacting sites 1–8. Nucleotides 19–183 indicate the sites of the most visible changes at Mg2+ > 0.1 mM. (NR) btuB in plain water, (T1) RNase T1 ladder, (OH) alkaline hydrolysis ladder. Structural changes detected by in-line probing experiments in the absence of AdoCbl are mapped on the secondary structure of the btuB aptamer in the absence (B) and in the presence of 2 mM Mg2+ (C). Shown are the nucleotides undergoing strong cleavage (green), intermediate cleavage (orange), and complete protection (black). Nucleotides in gray could not be mapped from the gel. Black arrows in C indicate the sites modulated by AdoCbl.
Upon increase in Mg2+ concentration above 0.5 mM, the change in the geometry is noticeable at several nucleotides (Fig. 1C). In the region encompassing nucleotides 16–30, most of the residues exhibit a relative protection from the cleavage except for nucleotides U19, G23, and U26 (Fig. 1C). Further changes can be seen at loop L4 which, after addition of >0.5 mM Mg2+, displays less cleavage. Interestingly, some of the nucleotides in this loop like U31, A35, and A36 were found to be conserved in the consensus sequence of the coenzyme B12 aptamer (Nahvi et al. 2004), adding further to its importance in the function of the riboswitch. Moreover, the L4 interacts with an internal loop between P6 and P7 (T-loop-T-loop motif) in the crystal structure of TteAdoCbl riboswitch (Johnson et al. 2012). The btuB riboswitch probably undergoes a similar interaction, explaining the observed partial protection from the cleavage at L4.
At a Mg2+ concentration of 0.5 mM, nucleotides at the junction of P6/7, P7/8 (C52, G54,U58, A60), and in the region around stems P8–P11 (G106, A107, A112) show a decreasing susceptibility for cleavage as compared to the absence of Mg2+ (Fig. 1A). Another interesting change takes place at L11 that takes up more flexible conformation with increasing Mg2+ concentration. From the conserved B12 box situated between nucleotides 140–160, only G144 and G156 undergo a Mg2+-dependent structural transition toward a more rigid geometry.
Interesting to note are the changes seen at the nucleotides (sites 1–8) reported to be modulated upon interaction with AdoCbl (Nahvi et al. 2002; Gallo et al. 2008). As can be seen from Figure 1A, G23 (site 1) is less prone for in-line cleavage in the presence of up to 0.1 mM Mg2+, whereas at Mg2+ concentration >0.1 mM, along with monovalent cations, G23 takes up a dynamic geometry and undergoes a distinct cleavage. G23, therefore, attains two different geometries, one in the presence of 0–0.1 mM Mg2+ or at high Mg2+/AdoCbl and a second at 0.5–20 mM Mg2+. Site 2b (U77), site 3 (G87), site 4 (G106), and site 8 (U183) are relatively protected from cleavage at 0–0.1 mM Mg2+ as compared to 0.5–20 mM Mg2+ (Fig. 1A), whereas site 2 (U68), site 5 (G110), site 6 (U118), and site 7 (U167) do not change their geometry dramatically with increase in Mg2+ concentration. All of these sites (sites 1–8) switch their geometries in the presence of AdoCbl. The geometry displayed by sites 1–8 prior to ligand binding, therefore, could be important for the recognition by the ligand.
The structural changes observed between 0.1 and 0.5 mM Mg2+ persist when the Mg2+ concentration is raised to 20 mM. Although we could observe that the in-line nucleophilic cleavage is enhanced with Mg2+, the intensity of cleavage increases only at specific intra-nucleotide linkages keeping otherwise the pattern of cleavage and, thereby, the conformation of RNA the same from 0.5 to 20 mM Mg2+. It is important to note that the sites undergoing in-line cleavage may not correspond to specific Mg2+ binding sites of the RNA. Hence, the change in cleavage pattern of the btuB aptamer probably reflects the overall conformational change brought about by Mg2+. However, metal ion binding at certain sites of cleavage cannot be ruled out.
The btuB aptamer is structured in the absence of divalent cations
To investigate the extent to which Mg2+-introduced conformational alterations affect the stabilization of RNA, thermal denaturation studies of the btuB aptamer were carried out. One millimolar Mg2+ stabilizes the structure of the btuB RNA considerably as reflected by an increase of almost 10°C in the melting temperature of the RNA (Fig. 2A). Nevertheless, the transition seen in the absence of Mg2+ suggests that the btuB RNA adopts a specific secondary structure even in the absence of divalent cations. This is in agreement with our in-line probing data, as some of the nucleotides do not change their geometry regardless of Mg2+. CD spectra of the btuB aptamer (Fig. 2B) indicate that the RNA forms A-form helices already in the absence of Mg2+, as the addition of Mg2+ does not lead to any significant increase in the large positive ellipticity at 265 nm (Saenger 1984; Yamauchi et al. 2005), confirming the presence of secondary structural elements at high monovalent cation concentration.
FIGURE 2.
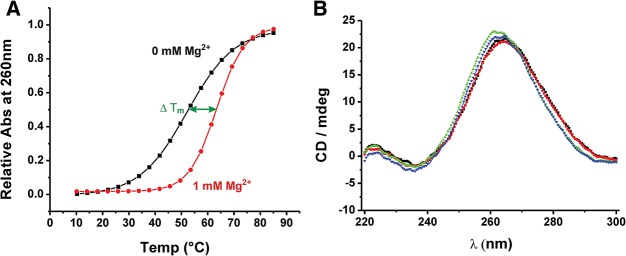
Structural changes in the btuB aptamer induced by K+ and Mg2+. (A) Mg2+-induced stabilization of the btuB riboswitch folded in 100 mM KCl as indicated by the increase of the melting temperature from 52.8 ± 0.13 (no Mg2+, black curve) to 63.3 ± 0.10 in the presence of 1 mM Mg2+ (red curve). (B) CD spectra of the btuB aptamer renatured in 100 mM KCl and 0 mM (black), 0.1 mM (red), 1.5 mM (green), or 20 mM (blue) MgCl2.
Mg2+ induces a hierarchical folding of the ligand-free btuB aptamer
As the in-line probing shows a structural transition in the btuB aptamer at Mg2+ concentrations between 0.1 and 0.5 mM, we determined the Mg2+ concentration needed for a folding transition of the btuB aptamer using RNase T1 (Rangan et al. 2003; Chauhan et al. 2005). This ribonuclease cleaves 3′ from unpaired guanine residues in RNA and hence, the folding of RNA by varying the Mg2+ concentrations should be reflected in the protection of specific guanine residues.
Most guanine residues were found to be sensitive to the increasing concentration of Mg2+ (Fig. 3A). Interestingly, the observed Mg2+ concentration required for structural transition of individual RNA domains varies with KD values ranging from 0.26 to 0.91 mM (Fig. 3B; Table 1). These distinct affinities exhibited by the different guanines suggest a stepwise folding of the aptamer into its complete secondary structure, as the regions having a lower dissociation constant fold before the other regions, as indicated in Figure 3C. Starting from a completely unfolded RNA, stems P7 and P12 undergo the first level of folding along with their associated loops. The second stage of folding is marked by the folding of stem P8 and the onset of folding for stem P4. In the next step, there is a complete folding of stem P4 along with its loop, stem P6, and probably some of the regions surrounding nucleotide G188. Stems P5 and P3 along with their associated regions undergo the fourth and fifth level of folding, respectively, due to their high KD for Mg2+.
FIGURE 3.
Mg2+-induced folding transition for the btuB aptamer. (A) Partial RNaseT1 digestion of the btuB aptamer subjected to renaturation in varying Mg2+ concentrations (0–20 mM). Arrows indicate the corresponding guanine residues. (NR) btuB in plain water, (T1) RNaseT1 ladder under denaturing conditions, (OH) alkaline hydrolysis ladder. (B) The relative intensity of cleavage as detected in the RNase T1 assay is plotted (▵) against the Mg2+ concentration, and the data points were fitted according to a 1:1 binding model (—) to obtain the respective dissociation constants KD (Sigel et al. 2000). The KD values for G94 and G105 are not considered due to the large error associated with the values. (C) The proposed folding pathway for the btuB aptamer as a function of the Mg2+ concentration indicated by the five stages of folding as I (blue), II (orange), III (yellow), IV (pink), and V (green). This pathway is based on categorizing the KD values of the individual guanine residues toward Mg2+ into the following five categories: 0.1–0.3 mM, 0.3–0.4 mM, 0.4–0.5 mM, 0.5–0.75 mM, 0.75–1 mM. The folding stages for the regions indicated in gray could not be predicted with certainty, e.g., for the 3′ tail, only the data for G188 could be fit (see also Table 1).
TABLE 1.
Dissociation constants KD of Mg2+ as determined by partial T1 digestion
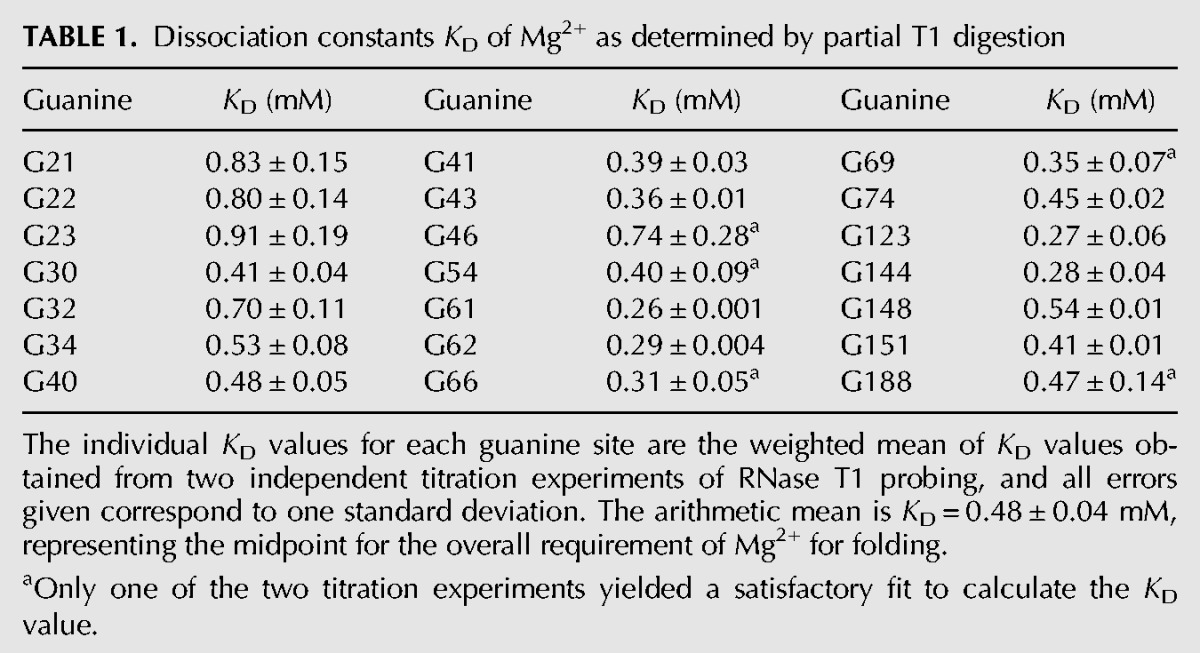
On average, 0.48 ± 0.04/ (∼0.5) mM Mg2+ is required for the guanine residues to undergo a transition in their geometries when the final RNA concentration is 1 μM. Considering the half-maximal Mg2+ concentration needed for a transition in folding (∼0.5 mM), it can be said that the btuB aptamer achieves its complete secondary structure at a Mg2+ concentration of ∼1.0 mM. The preorganization of the base-paired elements at the Mg2+ concentration of ∼1.0 mM might facilitate the rapid folding of the btuB RNA to its native form.
Mg2+ drives the switch in RNA conformation upon AdoCbl binding
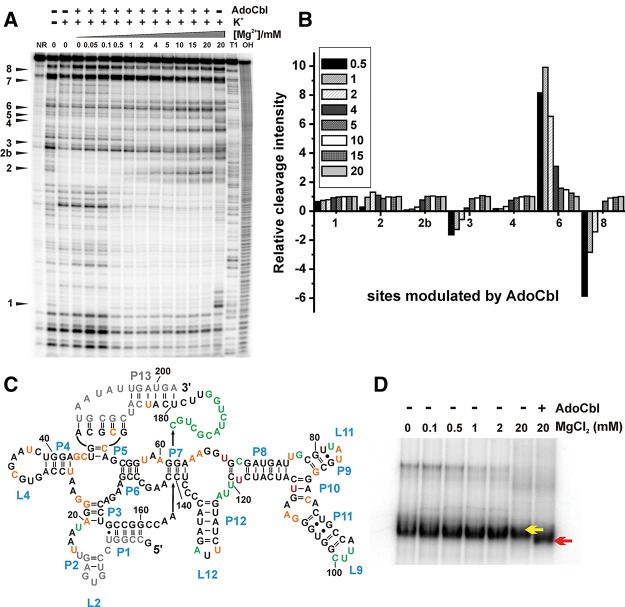
We defined the ligand-compatible structure of the btuB aptamer with the help of in-line probing done in the presence of AdoCbl by varying the Mg2+ concentrations (Fig. 4A). Up to concentrations of 0.1 mM Mg2+, the cleavage pattern in the presence of AdoCbl remains almost unchanged compared to the one in the absence of the ligand (Fig. 4A, lanes 3–6). Under these conditions, AdoCbl fails to switch the RNA, as evident from the absence of any intensity changes at the classical nine sites previously reported (Nahvi et al. 2002; Gallo et al. 2008). However, raising the Mg2+ concentration from 0.5 to 5 mM does lead to an enhanced conformational switch of the btuB aptamer in the presence of AdoCbl (Fig. 4A, lanes 7–14). The extent of switching for most of the sites modulated by AdoCbl at 5 mM Mg2+ appears similar to that at 20 mM Mg2+ (Fig. 4B), indicating that the proportion of the switched RNA is almost the same at these Mg2+ concentrations.
FIGURE 4.
Detection of the binding competent conformation of the btuB aptamer. (A) In-line probing of the btuB aptamer in the presence of 500 μM AdoCbl and 100 mM KCl by varying the Mg2+ concentration (0–20 mM). Sites 1–8 represent nucleotides modulated by AdoCbl. (NR) btuB in plain water, (T1) RNase T1 ladder, (OH) alkaline hydrolysis ladder. (B) Mg2+-dependent relative maximal changes at the sites (1–8) modulated by AdoCbl. The differences between the intensity changes in the presence and absence of AdoCbl at each Mg2+ concentration were normalized to the difference at 20 mM Mg2+. Data for site 5 and site 7 could not be evaluated. (C) Structural changes induced by AdoCbl along with Mg2+ (>0.1 mM). Superposition of structural changes detected by an in-line probing experiment in the presence of AdoCbl on the secondary structure of the btuB aptamer indicating nucleotides undergoing strong cleavage (green), intermediate cleavage (orange), or complete protection (black). Residues marked in red indicate the position of nucleotides undergoing enhanced cleavage by AdoCbl. Nucleotides in gray could not be mapped from the gel. (D) The native gel electrophoresis of the btuB riboswitch indicating the faster migration of the RNA species incubated with AdoCbl (100 μM) (red arrow) compared to the one in the absence of AdoCbl (yellow arrow).
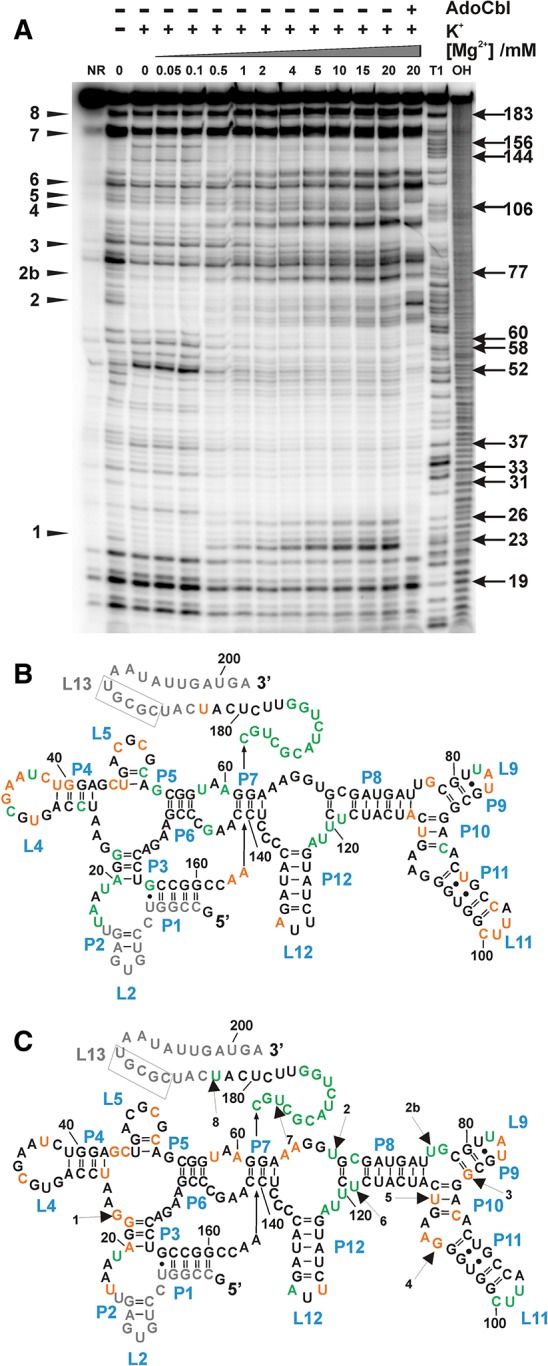
The cleavage pattern of the btuB aptamer with 100 mM KCl and in the absence of Mg2+ remains unaltered irrespective of the presence of AdoCbl, meaning that the secondary structure does not change much in the presence of AdoCbl (Fig. 4C). Both in the absence and presence of AdoCbl, we see the same influence of Mg2+ in folding the RNA only at a concentration higher than 0.1 mM. Above 0.5 mM Mg2+, the presence of AdoCbl affects the cleavage pattern at the AdoCbl modulated sites (sites 1–8) similar to an earlier observation with 20 mM Mg2+ (Nahvi et al. 2004; Gallo et al. 2008). On the contrary, binding of AdoCbl definitely makes the btuB aptamer more compact. This can be seen in native gel electrophoresis where the species of RNA in the presence of AdoCbl migrates faster compared to the one in the absence of the ligand (Fig. 4D; Supplemental Fig. S1): Also, native gel electrophoresis shows that both Mg2+ and AdoCbl add to the compaction of the btuB RNA and that the two ligands have a complementary effect as exhibited by compaction at lower Mg2+ concentrations in the presence of AdoCbl (Supplemental Fig. S1). Therefore, the formation of new tertiary interactions is likely to enable the RNA to adopt a more compact conformation in its switched form. Taking the recently published crystal structures of AdoCbl riboswitches into account (Johnson et al. 2012; Peselis and Serganov 2012), the ligand-induced compaction in the btuB aptamer can be a result of a kissing loop (KL) interaction between L5 and L13, the close packing of the coaxial stems P3-P6/P4-P5, and the specific tertiary interactions mediated by peripheral elements (P7–P12) with the ligand binding pocket. Indeed, the RNase T1 probing of the btuB aptamer in the presence of AdoCbl clearly indicates the new formation of the L5-L13 KL interaction as well as the possible interaction between J6/3 and the peripheral elements (Supplemental Fig. S2). The formation of the AdoCbl induced KL interaction between L5 and L13 is seen also in the 239-nt-long btuB riboswitch that includes the expression platform (Supplemental Fig. S3).
DISCUSSION
Pre-organization of the btuB aptamer with Mg2+
Mg2+: a driving factor for the binding competent conformation
The stabilization of the tertiary structure of a specific RNA by Mg2+ influences its activity as reported for many different RNAs, such as transfer RNA, catalytic ribozymes, or riboswitches (Lindahl et al. 1966; Cole et al. 1972; Pyle 1993; Cate et al. 1996; Baird et al. 2010). The btuB riboswitch is no exception and undergoes Mg2+-dependent folding. In the presence of monovalent cations, two conformations exist for the btuB aptamer with respect to its secondary structure (Supplemental Fig. S1)—one in the complete absence of Mg2+ or at Mg2+ concentrations of <0.5 mM and the second achieved at >0.5 mM Mg2+. The high monovalent cation concentration changes the geometry at some nucleotides. Interestingly, these nucleotides undergo further change in their geometries with Mg2+ concentrations above 0.5 mM. Such geometrical alterations brought about by Mg2+ have a great impact on the overall conformation of the RNA, as this conformation corresponds to the binding competent conformation of the btuB aptamer.
Folding of the btuB aptamer: a stepwise transition
It is possible that the Mg2+-induced structural transition of the btuB aptamer as seen in in-line probing represents an average conformation of the molecule on macroscopic scale. There could be transient folding intermediates which remain undetected by this method, taking into consideration the time frame of 40 h over which the cleavage reaction takes place. Based on the in-line probing cleavage pattern, we can observe only one transition in the conformational change of the btuB aptamer taking place at Mg2+ concentrations above 0.1 mM Mg2+. However, the existence of more than one folding intermediate cannot be excluded, as distinctive regions of the RNA exhibit different dissociation constants (KD) toward Mg2+ (Fig. 3). This allows the proposal of a folding pathway solely based on these KD values. Such a possible hierarchical folding of the btuB aptamer structural elements is, e.g., known for the aptamer of the pbuE adenine riboswitch (Greenleaf et al. 2008). In vivo, such a hierarchical folding model for the btuB riboswitch might be regulated by the transcriptional pausing of RNA polymerase as proposed recently (Perdrizet et al. 2012). The cotranscriptional folding model proposed for the btuB riboswitch suggests a coordinated folding of the aptamer and the expression platform driven by the RNA polymerase pause sites PA, PB, and PC (Supplemental Fig. S3A; Perdrizet et al. 2012; Souliere et al. 2013). The pausing by RNA polymerase ensures formation of the aptamer and the anti-aptamer regions depending on the presence and absence of AdoCbl (Perdrizet et al. 2012). The btuB riboswitch construct studied here allows L5-L13 KL formation and represents the folding pathway for the riboswitch with the RNA polymerase paused at site PB before transcription is continued to the expression platform. The observed AdoCbl induced L5-L13 KL interaction is also evident in a longer btuB riboswitch construct (239 nt) that resembles the riboswitch with the polymerase at pause site PC. These changes confirm the role of the regulatory L13-P13 in the presence of AdoCbl as reported previously (Johnson et al. 2012; Souliere et al. 2013).
Binding of AdoCbl to the btuB aptamer
No switching of the btuB riboswitch by AdoCbl in the absence of Mg2+
Our data provide strong evidence that the btuB riboswitch cannot be switched by its ligand in the absence of Mg2+. Based on our observations, we propose the following two possibilities regarding the recognition and switching of the btuB riboswitch by its ligand as shown in Figure 5.
A binding competent conformation of the btuB aptamer is needed for AdoCbl recognition, which can only be achieved at Mg2+ concentrations above ∼0.5 mM. Below 0.5 mM Mg2+, a binding-incompatible structure of the RNA is formed which fails to bind AdoCbl.
The aptamers of adenine binding (add A) and guanine binding (xpt G) riboswitches are known to bind their ligand independent of Mg2+ (Serganov et al. 2004). As in in-line probing experiments, AdoCbl binding is monitored by accompanied switching of the RNA, it is possible that AdoCbl binds to the btuB aptamer below 0.5 mM Mg2+ but is unable to switch its conformation. In this case, Mg2+ plays an important role in concert with the ligand to switch the structure of the riboswitch.
FIGURE 5.
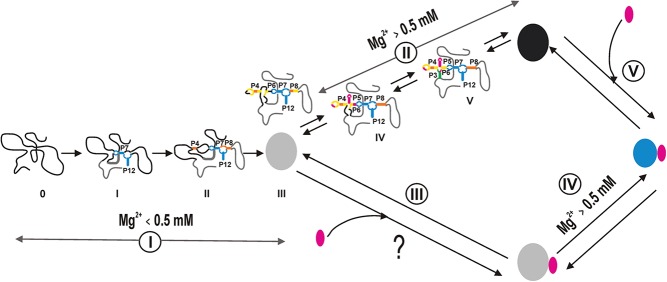
A model pathway for the folding and interaction between the btuB riboswitch and AdoCbl. The unfolded RNA begins to fold, leading to the formation of a binding-incompetent conformation (gray) at a Mg2+ concentration <0.5 mM (Pathway I). A binding-competent conformation of the RNA (black) is achieved at Mg2+ concentrations >0.5 mM (Pathway II). AdoCbl (pink) may or may not bind to the binding-incompetent conformation (gray) (Pathway III), but it does switch the RNA conformation at Mg2+ > 0.5 mM (Pathway IV). AdoCbl not only binds the binding-competent conformation but also switches its conformation, making it more compact (blue) (Pathway V).
If a specific RNA is preorganized, it can interact with its ligand in a classical induced fit manner, circumventing thereby the entropic cost to the free energy of binding (Noeske et al. 2007a). However, the different prefolded states observed for the btuB riboswitch suggest the possibility of a conformational ensemble of such prefolded states existing together with the binding-competent conformation. Therefore, the interaction between the btuB riboswitch and AdoCbl could also be explained by a synergism between the conformational selection model and the induced fit model (Boehr et al. 2009).
As the global architecture of the riboswitch achieved via RNA-RNA tertiary interactions is crucial for recognition of its ligand (Batey et al. 2004; Hampel and Tinsley 2006; Noeske et al. 2007b; Buck et al. 2010), we predict also for the btuB riboswitch that the Mg2+-induced folding transitions prepare the riboswitch for encountering its large and complex ligand. Although we do not know the exact nature of the tertiary interaction(s) accomplished by Mg2+ binding, the specific association of Mg2+ is crucial for an active btuB riboswitch. Moreover, the switched conformation (+ AdoCbl/Mg2+) of the btuB aptamer has a more compact conformation than the one in the absence of AdoCbl, indicating new or altered tertiary contacts induced by ligand binding rather than a drastic rearrangement of the secondary structural elements.
Has Mg2+ any role in the conformational switch of the btuB aptamer along with AdoCbl?
Our studies indicate a strong connection between the Mg2+ concentration and AdoCbl-induced conformational switching of the btuB RNA. The observed situation of Mg2+-aided switching of the RNA can be explained by two possibilities. As Mg2+ can assist the interaction of RNA with a ligand (Ferré-D'Amaré and Winkler 2011), it is probable that Mg2+ facilitates the binding of the complex AdoCbl to the btuB riboswitch. Increasing the Mg2+ concentration possibly can also increase the riboswitch's affinity for AdoCbl, a phenomenon observed earlier with TPP and guanine-binding riboswitches (Batey et al. 2004; Yamauchi et al. 2005). In both cases, the switching of the btuB riboswitch by AdoCbl could be a concerted effect induced by the ligand along with Mg2+. This concerted mechanism of switching by Mg2+ and ligand has been proposed also for the SAM I riboswitch (Heppell et al. 2011) and remains a topic for further investigation in the case of the btuB riboswitch. The mapping of Mg2+ binding sites from the in-line nucleophilic cleavage pattern can be indistinct, and therefore, detection of such metal ion binding sites in the btuB riboswitch will help in predicting their implication not only in folding of the riboswitch but also in switching the structure of the riboswitch upon ligand binding. However, there is a fair overlap between the nucleotides undergoing conformational changes in the presence of Mg2+ with those being part of the consensus sequence for AdoCbl riboswitches (Fig. 6; Supplemental Fig. S4). Consequently, the Mg2+-induced conformational changes in the btuB riboswitch core and peripheral regions containing conserved nucleotides are likely to occur also in other AdoCbl riboswitches.
FIGURE 6.
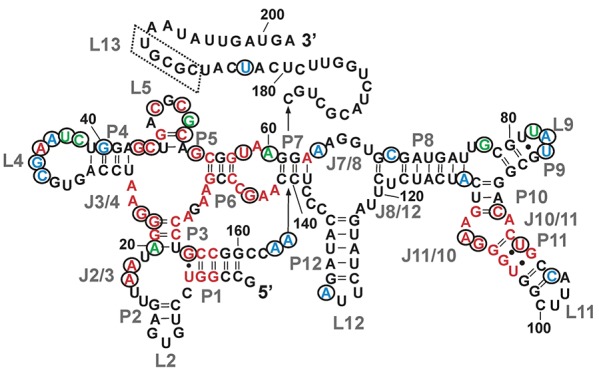
Mapping of Mg2+-induced changes on the AdoCbl aptamer consensus sequence depicted on the btuB sequence. The nucleotides undergoing Mg2+-induced conformational changes as observed in in-line probing experiments are indicated by black circles. The nucleotides in red are conserved in 90% of the representatives of AdoCbl aptamer sequences (Nahvi et al. 2004). The nucleotides in green represent the consensus sequence of the btuB aptamer and the AdoCbl riboswitches from the crystal structures (Johnson et al. 2012; Peselis and Serganov 2012). The nucleotides of the btuB aptamer shown in blue are common to one of the two sequences from the crystallized AdoCbl riboswitches (Johnson et al. 2012; Peselis and Serganov 2012).
Mg2+-dependent in-line probing and SHAPE on the btuB riboswitch
The recently published SHAPE (selective 2′-hydroxyl acylation analyzed by primer extension) study with NMIA (N-methylisatoic anhydride) as the chemical probing agent on the btuB riboswitch from E. coli does not indicate the distinct folding transitions with increasing Mg2+ concentration except at 15 mM Mg2+, where the KL interaction between L5 and L13 is predicted in the absence of ligand (Johnson et al. 2012). Our current data, in contrast, clearly indicate conformational changes in the RNA as a function of Mg2+ in the absence of AdoCbl. In the presence of ligand, however, the structural changes in the RNA are evident at a physiological Mg2+ concentration in NMIA as well as in in-line probing. The in-line probing and NMIA chemical probing vary mainly with respect to monovalent ions being 100 mM KCl or NaCl, respectively. In the absence of Mg2+, the two M+ ions might prefold the btuB aptamer differently, with varying requirements for further Mg2+ stabilization in the absence of AdoCbl. The presence of AdoCbl, however, might facilitate folding of the btuB aptamer toward a more stable conformation at a physiological Mg2+ concentration.
Altogether, we suggest that the complex btuB riboswitch from E. coli belongs to the group of riboswitches involving Mg2+ as a key player for riboswitch function. Therefore, the activity of the btuB riboswitch can be modulated by Mg2+ independently or along with its ligand, coenzyme B12.
MATERIALS AND METHODS
Materials
Nucleoside 5′-triphosphates (ATP, GTP, CTP) were purchased from GE Healthcare and Sigma-Aldrich (UTP). Coenzyme B12 (Sigma-Aldrich) was used without any further purification. Homemade T7 RNA polymerase was used for in vitro RNA transcription (Gallo et al. 2005). RNase T1 (Fermentas) 1000 units/μL was diluted to 0.05 units/μL in a buffer containing 50 mM Tris-HCl (pH 7.4) and 50% (v/v) glycerol. Denaturing polyacrylamide gels were prepared using Long Ranger gel solution (Lonza). Deionized water was further purified by Millipore-filtration and autoclaved before use. All the buffers, salt solutions, and gel solutions were filtered through 0.2-μm filters. All other chemicals were at least puriss p.a and were purchased from Sigma-Aldrich. Gels were analyzed by a Storm860 PhosphoImager and ImageQuant software (GE Healthcare).
Preparation of the RNA
The 202-nt-long btuB RNA was obtained by in vitro transcription from plasmid pPC1 by homemade T7 RNA polymerase (Gallo et al. 2005). The plasmid pPC1 is based on the plasmid pSG2 (Gallo 2008) that contains the natural 202-nt btuB aptamer sequence. In addition, pPC1 contains a newly introduced GGA sequence 5′ to the btuB aptamer sequence for efficient transcription (Gallo et al. 2005) and a GAGCUCG sequence at the 3′ end stemming from digestion with the restriction enzyme EcoR1 (Promega). These additional nucleotides have no effect on the aptamer's ability to switch (Supplemental Fig. S5). To obtain the 239-nt-long btuB riboswitch, the 202-nt-long btuB aptamer sequence was extended at its 5′ and 3′ ends using the forward primer (5′-GCC[AAGCTT]HindIII-[GCATGC]SphI-TAATACGACTCACTATAGGAGCCGGTCCTGTGAGTTAATAGG) and the reverse primer (5′-GCCCG[GAATTC]EcoRI-GTAAAGCATCCACAATAGAAGAAGGATGCC). The 5′ end of the construct includes the additional nucleotides GGA for better transcription yields (see above), whereas the 3′-terminal 37 nt correspond to the expression platform region. Both RNAs were purified by 10% denaturing PAGE after in vitro transciption, electroeluted, and concentrated using a Vivaspin concentrator (5000 MWCO) as described earlier (Gallo et al. 2005). The RNA was stored in water at −20°C.
In-line probing
In-line probing experiments were carried out using 32P-5′-labeled RNA at a final concentration of 7 nM. The RNA was incubated for 40 h at 25°C in 50 mM Tris-HCl (pH 8.3) and 100 mM KCl with varying concentrations of MgCl2. In the presence of AdoCbl, the incubation was done in the dark. The reaction was quenched by formamide loading buffer. An RNase T1 ladder and alkaline hydrolysis ladder were prepared as described (Regulski and Breaker 2008). The cleaved bands were separated by a 10% denaturing PAGE and were analyzed with ImageQuant.
UV melting experiments
First, 2.7 μM of RNA along with 100 mM KCl was heated to 90°C, followed by addition of the respective MgCl2 concentration. The sample was incubated at room temperature for 20 min and degassed prior to measurement. Then, the samples were transferred to a cuvette with a diameter of 1 mm and covered with a layer of 100 μL paraffin oil to prevent evaporation. The UV melting curves were recorded using a Cary 100 Bio UV-Visible spectrophotometer (Varian Inc.). The samples were subjected to thermal denaturation from 10°C–85°C at the rate of 0.5°/min. The data were fitted to the Boltzmann equation [y = A2 + (A1 − A2)/(1 + exp((x − x0)/dx))] to derive the melting temperature x0. y is the relative absorbance at 260 nm, x the temperature (°C), and A1 and A2 correspond to ymin and ymax, respectively. x0 is the point of inflection (i.e., Tm), and dx is a variable parameter.
Partial RNase T1 digestion
First, 6 nM of 32P-5′-labeled RNA and 1 μM of unlabeled RNA, along with 50 mM Tris-HCl (pH 7.5), were heated to 90°C for 1 min. The sample was then cooled at room temperature for 1 min, followed by the addition of 100 mM KCl and MgCl2 (0–20 mM). The sample was incubated at 37°C for 10 min, followed by addition of 1 μL of 0.05 units/μL of RNase T1 (Fermentas) and subsequent incubation of a further 10 min at 37°C. Ten microliters of formamide loading buffer (82% [v/v] Formamide, 0.16% [w/v] Xylene Cynol, 0.16% [w/v] Bromophenol blue, 10 mM EDTA [pH 8.0]) were added for quenching the reaction, and the samples were immediately loaded on 10% denaturing PAGE. The data were analyzed with ImageQuant.
Evaluation of data and calculation of KA
The relative cleavage intensity at the guanine residues is calculated according to Harris and Walter (2003). The normalized cleavage intensities were plotted against Mg2+ concentration, and the data points were fitted to a 1:1 binding model (Sigel et al. 2000) to derive the dissociation constant KD.
Native gel electrophoresis
First, 20 nM of 32P-5′-labeled RNA and 1 μM of unlabeled RNA, along with 50 mM Tris-HCl (pH 7.5) and 100 mM KCl, were denatured at 90°C for 1 min, followed by the addition of respective concentrations of MgCl2. The sample was then incubated at 37°C for 15 min. AdoCbl (100 μM) was then added to the sample, followed by subsequent incubation at 37°C for 30 min. Along with an equal volume of 60% glycerol, the samples were loaded on a 6% native gel and run in a buffer containing 3 mM MgOAc, 66 mM HEPES, and 34 mM Tris-HCl (pH 7.5) inside the refrigerator at 4°C.
CD spectroscopy
First, 0.2 μM RNA in 50 mM of Tris-HCl (pH 7.5) was denatured at 90°C for 1 min. The sample was then cooled to room temperature for 10 min, followed by the addition of 100 mM KCl and the respective amount of MgCl2 and incubated at 25°C for 30 min. The CD spectra were recorded using a Jasco J-810 spectropolarimeter at a 0.5-nm data pitch and a scanning speed of 100 nm/min. Three consecutive accumulations were collected for each sample.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank Sofia Gallo for careful reading and valuable comments on our manuscript as well as for providing the plasmid pSG2 for the cloning purpose. We also thank Fabio Steffen and Sofia Gallo for providing the native gel showing btuB folding. Financial support by the Swiss National Science Foundation (R.K.O.S.), the University of Zürich, the European Research Council (ERC starting grant to R.K.O.S.), as well as within the COST Action CM1105 is gratefully acknowledged.
REFERENCES
- Baird NJ, Kulshina N, Ferre-D'Amare AR 2010. Riboswitch function: Flipping the switch or tuning the dimmer? RNA Biol 7: 328–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastet L, Dube A, Masse E, Lafontaine DA 2011. New insights into riboswitch regulation mechanisms. Mol Microbiol 80: 1148–1154 [DOI] [PubMed] [Google Scholar]
- Batey RT, Gilbert SD, Montange RK 2004. Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine. Nature 432: 411–415 [DOI] [PubMed] [Google Scholar]
- Boehr DD, Nussinov R, Wright PE 2009. The role of dynamic conformational ensembles in biomolecular recognition. Nat Chem Biol 5: 789–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck J, Noeske J, Wöhnert J, Schwalbe H 2010. Dissecting the influence of Mg2+ on 3D architecture and ligand-binding of the guanine-sensing riboswitch aptamer domain. Nucleic Acids Res 38: 4143–4153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cate JH, Gooding AR, Podell E, Zhou K, Golden BL, Kundrot CE, Cech TR, Doudna JA 1996. Crystal structure of a group I ribozyme domain: Principles of RNA packing. Science 273: 1678–1685 [DOI] [PubMed] [Google Scholar]
- Chauhan S, Caliskan G, Briber RM, Perez-Salas U, Rangan P, Thirumalai D, Woodson SA 2005. RNA tertiary interactions mediate native collapse of a bacterial group I ribozyme. J Mol Biol 353: 1199–1209 [DOI] [PubMed] [Google Scholar]
- Choudhary PK, Gallo S, Sigel RKO 2014. Monitoring global structural changes and specific metal binding sites in RNA by in-line probing and Tb(III) cleavage. Methods Mol Biol 1086: 143–158 [DOI] [PubMed] [Google Scholar]
- Chu VB, Bai Y, Lipfert J, Herschlag D, Doniach S 2008. A repulsive field: Advances in the electrostatics of the ion atmosphere. Curr Opin Chem Biol 12: 619–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole PE, Yang SK, Crothers DM 1972. Conformational changes of transfer ribonucleic acid. Equilibrium phase diagrams. Biochemistry 11: 4358–4368 [DOI] [PubMed] [Google Scholar]
- Draper DE 2004. A guide to ions and RNA structure. RNA 10: 335–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferré-D'Amaré AR, Winkler WC 2011. The roles of metal ions in regulation by riboswitches. Met Ions Life Sci 9: 141–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freisinger E, Sigel RKO 2007. From nucleotides to ribozymes—a comparison of their metal ion-binding properties. Coord Chem Rev 251: 1834–1851 [Google Scholar]
- Gallo S 2008. “Investigations on the interaction between coenzyme B12 derivatives and the btuB riboswitch of E. coli.” PhD thesis, University of Zürich, Zürich [Google Scholar]
- Gallo S, Furler M, Sigel RKO 2005. In vitro transcription and purification of RNAs of different size. Chimia 59: 812–816 [Google Scholar]
- Gallo S, Oberhuber M, Sigel RK, Kräutler B 2008. The corrin moiety of coenzyme B12 is the determinant for switching the btuB riboswitch of E. coli. ChemBioChem 9: 1408–1414 [DOI] [PubMed] [Google Scholar]
- Gallo S, Mundwiler S, Alberto R, Sigel RKO 2010. The change of corrin-amides to carboxylates leads to altered structures of the B12-responding btuB riboswitch. Chem Commun 47: 403–405 [DOI] [PubMed] [Google Scholar]
- Greenleaf WJ, Frieda KL, Foster DA, Woodside MT, Block SM 2008. Direct observation of hierarchical folding in single riboswitch aptamers. Science 319: 630–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudmundsdottir A, Bradbeer C, Kadner RJ 1988. Altered binding and transport of vitamin B12 resulting from insertion mutations in the Escherichia coli btuB gene. J Biol Chem 263: 14224–14230 [PubMed] [Google Scholar]
- Haller A, Rieder U, Aigner M, Blanchard SC, Micura R 2011a. Conformational capture of the SAM-II riboswitch. Nat Chem Biol 7: 393–400 [DOI] [PubMed] [Google Scholar]
- Haller A, Soulière MF, Micura R 2011b. The dynamic nature of RNA as key to understanding riboswitch mechanisms. Acc Chem Res 44: 1339–1348 [DOI] [PubMed] [Google Scholar]
- Hampel KJ, Tinsley MM 2006. Evidence for preorganization of the glmS ribozyme ligand binding pocket. Biochemistry 45: 7861–7871 [DOI] [PubMed] [Google Scholar]
- Harris DA, Walter NG 2003. Probing RNA structure and metal-binding sites using terbium(III) footprinting. Curr Protoc Nucleic Acid Chem 10.1002/0471142700.nc0608s13 [DOI] [PubMed] [Google Scholar]
- Heppell B, Blouin S, Dussault AM, Mulhbacher J, Ennifar E, Penedo JC, Lafontaine DA 2011. Molecular insights into the ligand-controlled organization of the SAM-I riboswitch. Nat Chem Biol 7: 384–392 [DOI] [PubMed] [Google Scholar]
- Jenkins JL, Krucinska J, McCarty RM, Bandarian V, Wedekind JE 2011. Comparison of a preQ1 riboswitch aptamer in metabolite-bound and free states with implications for gene regulation. J Biol Chem 286: 24626–24637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JE Jr, Reyes FE, Polaski JT, Batey RT 2012. B12 cofactors directly stabilize an mRNA regulatory switch. Nature 492: 133–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein DJ, Ferré-D'Amaré AR 2006. Structural basis of glmS ribozyme activation by glucosamine-6-phosphate. Science 313: 1752–1756 [DOI] [PubMed] [Google Scholar]
- Kubodera T, Watanabe M, Yoshiuchi K, Yamashita N, Nishimura A, Nakai S, Gomi K, Hanamoto H 2003. Thiamine-regulated gene expression of Aspergillus oryzae thiA requires splicing of the intron containing a riboswitch-like domain in the 5′-UTR. FEBS Lett 555: 516–520 [DOI] [PubMed] [Google Scholar]
- Lemay JF, Penedo JC, Tremblay R, Lilley DM, Lafontaine DA 2006. Folding of the adenine riboswitch. Chem Biol 13: 857–868 [DOI] [PubMed] [Google Scholar]
- Lindahl T, Adams A, Fresco JR 1966. Renaturation of transfer ribonucleic acids through site binding of magnesium. Proc Natl Acad Sci 55: 941–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipfert J, Sim AY, Herschlag D, Doniach S 2010. Dissecting electrostatic screening, specific ion binding, and ligand binding in an energetic model for glycine riboswitch folding. RNA 16: 708–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh E, Dussurget O, Gripenland J, Vaitkevicius K, Tiensuu T, Mandin P, Repoila F, Buchrieser C, Cossart P, Johansson J 2009. A trans-acting riboswitch controls expression of the virulence regulator PrfA in Listeria monocytogenes. Cell 139: 770–779 [DOI] [PubMed] [Google Scholar]
- Mandal M, Breaker RR 2004. Gene regulation by riboswitches. Nat Rev Mol Cell Biol 5: 451–463 [DOI] [PubMed] [Google Scholar]
- Mandal M, Boese B, Barrick JE, Winkler WC, Breaker RR 2003. Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell 113: 577–586 [DOI] [PubMed] [Google Scholar]
- Mironov AS, Gusarov I, Rafikov R, Lopez LE, Shatalin K, Kreneva RA, Perumov DA, Nudler E 2002. Sensing small molecules by nascent RNA: A mechanism to control transcription in bacteria. Cell 111: 747–756 [DOI] [PubMed] [Google Scholar]
- Montange RK, Batey RT 2008. Riboswitches: Emerging themes in RNA structure and function. Annu Rev Biophys 37: 117–133 [DOI] [PubMed] [Google Scholar]
- Nahvi A, Sudarsan N, Ebert MS, Zou X, Brown KL, Breaker RR 2002. Genetic control by a metabolite binding mRNA. Chem Biol 9: 1043. [DOI] [PubMed] [Google Scholar]
- Nahvi A, Barrick JE, Breaker RR 2004. Coenzyme B12 riboswitches are widespread genetic control elements in prokaryotes. Nucleic Acids Res 32: 143–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noeske J, Buck J, Furtig B, Nasiri HR, Schwalbe H, Wöhnert J 2007a. Interplay of ‘induced fit’ and preorganization in the ligand induced folding of the aptamer domain of the guanine binding riboswitch. Nucleic Acids Res 35: 572–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noeske J, Schwalbe H, Wöhnert J 2007b. Metal-ion binding and metal-ion induced folding of the adenine-sensing riboswitch aptamer domain. Nucleic Acids Res 35: 5262–5273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nou X, Kadner RJ 2000. Adenosylcobalamin inhibits ribosome binding to btuB RNA. Proc Natl Acad Sci 97: 7190–7195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perdrizet GA 2nd, Artsimovitch I, Furman R, Sosnick TR, Pan T 2012. Transcriptional pausing coordinates folding of the aptamer domain and the expression platform of a riboswitch. Proc Natl Acad Sci 109: 3323–3328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peselis A, Serganov A 2012. Structural insights into ligand binding and gene expression control by an adenosylcobalamin riboswitch. Nat Struct Mol Biol 19: 1182–1184 [DOI] [PubMed] [Google Scholar]
- Pyle AM 1993. Ribozymes: A distinct class of metalloenzymes. Science 261: 709–714 [DOI] [PubMed] [Google Scholar]
- Pyle AM 2002. Metal ions in the structure and function of RNA. J Biol Inorg Chem 7: 679–690 [DOI] [PubMed] [Google Scholar]
- Ramesh A, Wakeman CA, Winkler WC 2011. Insights into metalloregulation by M-box riboswitch RNAs via structural analysis of manganese-bound complexes. J Mol Biol 407: 556–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangan P, Masquida B, Westhof E, Woodson SA 2003. Assembly of core helices and rapid tertiary folding of a small bacterial group I ribozyme. Proc Natl Acad Sci 100: 1574–1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regulski EE, Breaker RR 2008. In-line probing analysis of riboswitches. Methods Mol Biol 419: 53–67 [DOI] [PubMed] [Google Scholar]
- Reynolds PR, Mottur GP, Bradbeer C 1980. Transport of vitamin B12 in Escherichia coli. Some observations on the roles of the gene products of BtuC and TonB. J Biol Chem 255: 4313–4319 [PubMed] [Google Scholar]
- Saenger W 1984. Different hydration states associated with A-, B-, and C-DNA. In Principles of nucleic acid structure, pp. 370–372 Springer-Verlag, New York [Google Scholar]
- Serganov A, Yuan YR, Pikovskaya O, Polonskaia A, Malinina L, Phan AT, Höbartner C, Micura R, Breaker RR, Patel DJ 2004. Structural basis for discriminative regulation of gene expression by adenine- and guanine-sensing mRNAs. Chem Biol 11: 1729–1741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganov A, Polonskaia A, Phan AT, Breaker RR, Patel DJ 2006. Structural basis for gene regulation by a thiamine pyrophosphate-sensing riboswitch. Nature 441: 1167–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganov A, Huang L, Patel DJ 2009. Coenzyme recognition and gene regulation by a flavin mononucleotide riboswitch. Nature 458: 233–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigel RKO, Pyle AM 2007. Alternative roles for metal ions in enzyme catalysis and the implications for ribozyme chemistry. Chem Rev 107: 97–113 [DOI] [PubMed] [Google Scholar]
- Sigel H, Sigel RKO 2013. Metal ion interactions with nucleic acids and their constituents. In Comprehensive inorganic chemistry II (ed. Reedijk J, Poeppelmeier K), Vol. 3, pp. 623–661 Elsevier Ltd., Oxford, UK [Google Scholar]
- Sigel RKO, Freisinger E, Lippert B 2000. Effects of N7-methylation, N7-platination, and C8-hydroxylation of guanine on H-bond formation with cytosine: Platinum coordination strengthens the Watson-Crick pair. J Biol Inorg Chem 5: 287–299 [DOI] [PubMed] [Google Scholar]
- Soukup GA, Breaker RR 1999. Relationship between internucleotide linkage geometry and the stability of RNA. RNA 5: 1308–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soukup JK, Soukup GA 2004. Riboswitches exert genetic control through metabolite-induced conformational change. Curr Opin Struct Biol 14: 344–349 [DOI] [PubMed] [Google Scholar]
- Souliere MF, Haller A, Santner T, Micura R 2013. New insights into gene regulation—high-resolution structures of cobalamin riboswitches. Angew Chem Int Ed 52: 1874–1877 [DOI] [PubMed] [Google Scholar]
- Sudarsan N, Barrick JE, Breaker RR 2003a. Metabolite-binding RNA domains are present in the genes of eukaryotes. RNA 9: 644–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudarsan N, Wickiser JK, Nakamura S, Ebert MS, Breaker RR 2003b. An mRNA structure in bacteria that controls gene expression by binding lysine. Genes Dev 17: 2688–2697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler WC, Breaker RR 2003. Genetic control by metabolite-binding riboswitches. ChemBioChem 4: 1024–1032 [DOI] [PubMed] [Google Scholar]
- Winkler W, Nahvi A, Breaker RR 2002. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature 419: 952–956 [DOI] [PubMed] [Google Scholar]
- Winkler WC, Nahvi A, Roth A, Collins JA, Breaker RR 2004. Control of gene expression by a natural metabolite-responsive ribozyme. Nature 428: 281–286 [DOI] [PubMed] [Google Scholar]
- Woodson SA 2005. Metal ions and RNA folding: A highly charged topic with a dynamic future. Curr Opin Chem Biol 9: 104–109 [DOI] [PubMed] [Google Scholar]
- Yamauchi T, Miyoshi D, Kubodera T, Nishimura A, Nakai S, Sugimoto N 2005. Roles of Mg2+ in TPP-dependent riboswitch. FEBS Lett 579: 2583–2588 [DOI] [PubMed] [Google Scholar]