In this paper, the authors develop chemically synthesized pre-miRNAs (syn-pre-miRNAs) that retain the stem–loop structures for the simultaneous production of both guide (-5p) and passenger (-3p, or *) miRNA strands in transfected cells. The authors show that syn-pre-miRNAs fully recapitulate the properties of pre-miRNAs and that both 5p and 3p miRNAs can inhibit expression of sensors containing bona fide target sites.
Keywords: pre-miRNA, macrophages, TNF-α, miR-34a, miRNA biogenesis, 3p strand
Abstract
Functional microRNAs (miRNAs) are produced from both arms of their precursors (pre-miRNAs). Their abundances vary in context-dependent fashion spatiotemporarily and there is mounting evidence of regulatory interplay between them. Here, we introduce chemically synthesized pre-miRNAs (syn-pre-miRNAs) as a general class of accessible, easily transfectable mimics of pre-miRNAs. These are RNA hairpins, identical in sequence to natural pre-miRNAs. They differ from commercially available miRNA mimics through their complete hairpin structure, including any regulatory elements in their terminal-loop regions and their potential to introduce both strands into RISC. They are distinguished from transcribed pre-miRNAs by their terminal 5′ hydroxyl groups and their precisely defined terminal nucleotides. We demonstrate with several examples how they fully recapitulate the properties of pre-miRNAs, including their processing by Dicer into functionally active 5p; and 3p-derived mature miRNAs. We use syn-pre-miRNAs to show that miR-34a uses its 5p and 3p miRNAs in two pathways: apoptosis during TGF-β signaling, where SIRT1 and SP4 are suppressed by miR-34a-5p and miR-34a-3p, respectively; and the lipopolysaccharide (LPS)-activation of primary human monocyte-derived macrophages, where TNF (TNFα) is suppressed by miR-34a-5p indirectly and miR-34a-3p directly. Our results add to growing evidence that the use of both arms of a miRNA may be a widely used mechanism. We further suggest that syn-pre-miRNAs are ideal and affordable tools to investigate these mechanisms.
INTRODUCTION
MiRNAs are noncoding RNAs that regulate gene expression (Bartel 2009; Krol et al. 2010). Their biogenesis begins with transcription of a primary transcript (pri-miRNA). Processing of the pri-miRNA to an approximate 70-nucleotide (nt) pre-miRNA hairpin is performed by the microprocessor, a complex of the RNase III Drosha with the DiGeorge syndrome critical region gene 8 (DGCR8) (Denli et al. 2004; Gregory et al. 2004; Han et al. 2004). Pre-miRNAs are exported to the cytoplasm (Lund et al. 2004), where their terminal loops are excised by the RNase III Dicer as part of a complex with Argonaute proteins (Ago1–4), trans-activation RNA-binding protein (TRBP), and chaperone proteins (Hutvágner et al. 2001). This yields a stem composed of 5′ and 3′ strands representing 5p and 3p miRNAs, respectively. Human Dicer is a large protein composed of several domains, including a helicase/ATPase domain, a PAZ (Piwi, Argonaute, Zwille) domain, a dsRBD, and two RNase III domains (Zhang et al. 2004; Lau et al. 2012; Ma et al. 2012). The PAZ domain grips the termini of the pre-miRNA and positions it for double cleavage ∼22 nt into the stem, producing a double-stranded RNA (dsRNA) with 3′ overhangs (Macrae et al. 2006; Park et al. 2011). The RNase IIIA domain cleaves in the 3′ arm of the pre-miRNA, while the RNase IIIB domain cleaves in the 5′ arm (Gurtan et al. 2012). The overhang on the pre-miRNA plays an important role in cleavage, as blunt-ended stems show shifted cleavage patterns (Vermeulen et al. 2005). Dicer is assisted by TRBP, which affects its kinetics and can also facilitate production of miRNA isoforms, leading to changes in loading of the RNA-induced silencing complex (RISC) (Lee and Doudna 2012). The cleaved pre-miRNA forms part of a pre-miRISC in which one of the RNA strands (the guide) is subsequently maintained, while the other strand (the passenger or miRNA*) is discarded. Precisely how this occurs is not fully understood. Often the strand with the 5′ end engaged in the least stable base-pairing is selected for RISC incorporation: However, where the ends of the dsRNA exhibit similar stability, either miRNA can be taken into a RISC (Khvorova et al. 2003; Schwarz et al. 2003). Fully activated RISCs recognize sites in the 3′ UTR of mRNAs via the seed region at the 5′ end of the miRNA and induce mRNA degradation or translational inhibition (Mathonnet et al. 2007; Djuranovic et al. 2012).
Most of the early characterization of miRNAs focused on the guide strands (usually the 5p), because these were typically found to be more abundant than their miRNA* counterparts in humans (Hu et al. 2009). However, many miRNA* strands have conserved seed sequences and have been isolated from RISC complexes, therefore suggesting that they are functional (Okamura et al. 2008a; Yang et al. 2011). Indeed, the ratios of 5p to 3p strands from subsets of miRNAs vary in a cell-context and tissue-dependent fashion (Ro et al. 2007; Ruby et al. 2007; Azuma-Mukai et al. 2008; Okamura et al. 2008b; Chiang et al. 2010). In addition, there is increasing evidence of interplay between 5p and 3p strands from the same precursor. This is most prominent in arm-switching, where the dominant miRNA is switched from one arm of the precursor to the other (Griffiths-Jones et al. 2011), or where the abundance of both miRNAs from a human precursor is similar, in which case they are predicted to target overlapping sets of genes/functional classes (Marco et al. 2012).
Examples of miRNAs from which functional 5p and 3p miRNAs have been characterized include: miR-9 (Packer et al. 2008), miR-17 (Shan et al. 2009), miR-19 (Yang et al. 2011), miR-28 (Almeida et al. 2012), miR-30c (Byrd et al. 2012), miR-125a (Jiang et al. 2010), miR-142 (Wu et al. 2009), miR-155 (Zhou et al. 2010), miR-199 (Kim et al. 2008; Sakurai et al. 2011), miR-223 (Kuchenbauer et al. 2011), miR-342 (Montag et al. 2009), miR-2015 (Grimson et al. 2008), miR-18a (Tsang and Kwok 2009), and miR-582 (Uchino et al. 2013). In some cases the two strands of the miRNA may have opposing functions (miR-28 [Almeida et al. 2012], miR-125 [Jiang et al. 2010]). In others, for example, miR-199 (Sakurai et al. 2011), miR-155 (Zhou et al. 2010), and miR-582 (Uchino et al. 2013), the two arms were shown to function in a joint fashion, reinforcing a phenotype. It seems likely therefore that interplay between miRNAs from the same precursor is a reoccurring theme.
Deciphering the function of miRNAs has been aided by synthetic miRNA “mimics”—sometimes inappropriately named dsRNA reagents that are chemically modified in one strand to force only the desired 5p or 3p miRNA into RISC (Birmingham et al. 2007). However, with increasing evidence of the dual activity of 5p and 3p miRNAs, so the need for a convenient-to-use chemical mimic of the miRNA precursor (i.e., pre-miRNA mimic), which produces both 5p and 3p, has arisen.
Here we propose chemically synthesized pre-miRNAs (syn-pre-miRNAs) as a general class of pre-miRNA mimics. These are RNA hairpins with terminal hydroxyl groups, identical in sequence to naturally occurring pre-miRNAs. We demonstrate with examples how they fully recapitulate the properties of pre-miRNAs, including their processing by Dicer into simultaneously active 5p- and 3p-derived mature miRNAs. We use syn-pre-miRNAs to show that miR-34a uses both strands in two pathways: apoptosis during TGFβ signaling in HeLa cells, where SIRT1 and SP4 are suppressed by miR-34a-5p and miR-34a-3p, respectively; and the lipopolysaccharide (LPS) activation of primary human monocyte-derived macrophages, where TNF (TNFα) is suppressed by both miR-34a-5p and miR-34a-3p. Our results provide an example of a prominent miRNA using both strands to drive a pathway, and add to the growing evidence that the use of both arms of a miRNA may be a widely used mechanism. We further suggest that syn-pre-miRNAs are ideal tools to investigate such mechanisms, especially as they can be acquired from commercial vendors at similar cost to routine biological reagents.
RESULTS
TGF-β1 RNAi induces miR-34a-5p and miR-34a-3p
Recently we described how suppression of latent TGFB1 (TGF-β1) restores growth inhibitory TGF-β signaling through miRNAs in some cell types (Dogar et al. 2011). We screened a large number of miRNAs for their response to TGF-β1 RNAi. Whereas most miRNAs remained unchanged or were suppressed, miR-34a-5p was induced and caused apoptosis. Apoptosis was partly rescued by miR-34a-5p antimiR. Further examination of this mechanism showed that TGF-β1 RNAi in HeLa cells in fact caused more than fourfold increases in both miR-34a-5p and miR-34a-3p at the highest treatment concentration (Fig. 1A), suggesting that both arms of miR-34a were functional. Although the activity of miR-34a-5p is well documented (Chen and Hu 2012), the properties of miR-34a-3p are not well described. In one account miR-34c-3p was shown to induce caspase activity in cervical carcinoma cells, though no targets were identified (López and Alvarez-Salas 2011). In another, miR-34a-3p was found to inhibit XIAP, an inhibitor of apoptosis, in synovial fibroblasts (Niederer et al. 2011b). Evidence for an extensive activity of miR-34a-3p (as well as miR-34a-5p) was also seen in expression data from cells infected with a miR-34a expression construct (Yang et al. 2011).
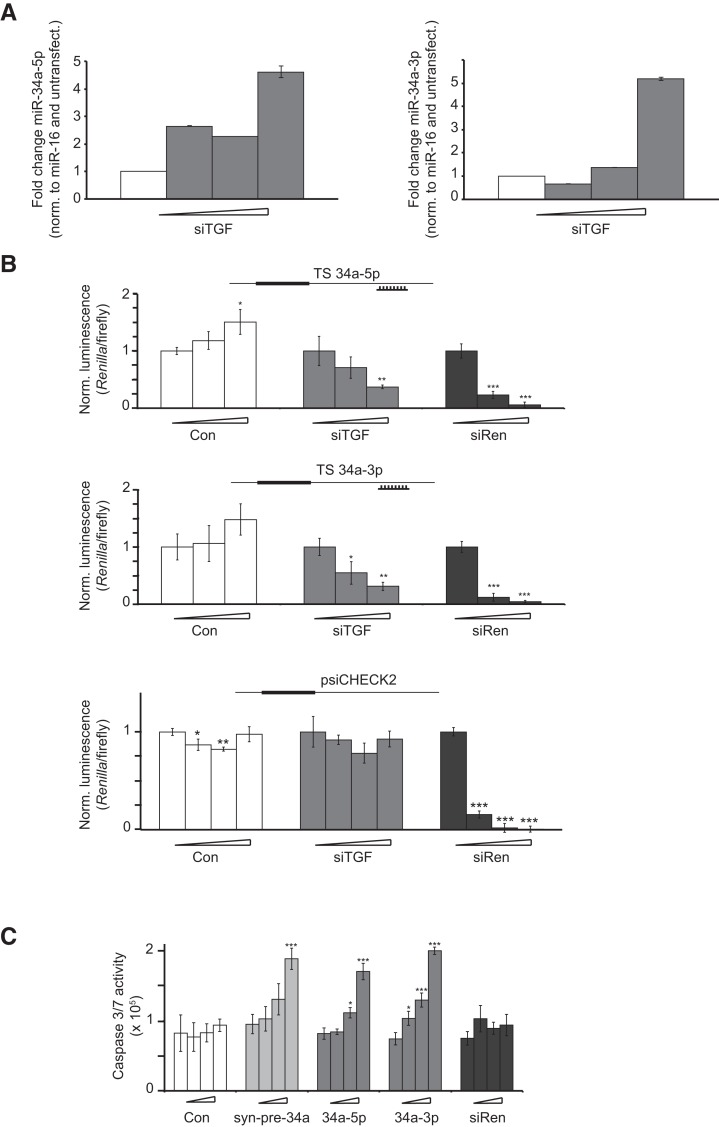
FIGURE 1.
MiR-34a-5p and miR-34a-3p show antiproliferative activity. (A) siTGF was transfected into HeLa cells in increasing doses (0, 2, 9, and 36 nM). RNA was isolated and qPCR was performed for miR-34a-5p and miR-34a-3p. The y-axis shows the relative RNA abundance (normalized against miR-16 and an untransfected control). Error bars, SD of four replicates. (B) Endogenous miR-34a-5p and miR-34a-3p, induced during TGFB RNAi, suppress luciferase reporter genes bearing complementary target sites. Transfections of Con (negative control siRNA), siTGF (siRNA against TGFB), and siRen (siRNA against Renilla luciferase) into HeLa cells were performed at increasing concentrations (0, 2, 9, and 36 nM). Luciferase reporter plasmids bearing no (psiCHECK2) or a single reverse complementary target site (TS) for the miRNA (origin of the TS shown in the inset) were transfected. Renilla luminescence values were normalized against firefly luminescence and then to the value from the corresponding 0-nM transfection for each treatment. Error bars, SD of three transfections. (C) MiR-34a-5p and miR-34a-3p, induced during TGFB RNAi, contribute to apoptosis. HeLa cells were transfected with Con (negative control), syn-pre-34a, miRNA mimics 34a-5p and 34a-3p, and siRen at increasing concentrations (0, 2, 9, and 36 nM). Caspase 3/7 activity was measured from lysates of HeLa cells 72 h post-transfection. Error bars, SD of three transfections (*) P < 0.05; (**) P < 0.01; (***) P < 0.001.
A straightforward way to assay for the activity of a miRNA is by dual-reporter assays (Doench et al. 2003) expressing Renilla and firefly luciferase. We generated two (“sensor”) plasmids, each bearing a single complementary site to miR-34a-5p and miR-34a-3p in the 3′ UTR of Renilla. These were transfected separately into HeLa cells after induction of endogenous miR-34a via TGF-β1 RNAi (siTGF). An especially potent siRNA targeting the Renilla luciferase (siRen) served as a control for transfection efficiency and a commercially available, unrelated double-stranded RNA (Con) served as a control for siRNAs and miRNAs to indicate nonspecific effects or toxicity. We observed a sequence-specific suppression of Renilla activity of both sensors up to 70% (Fig. 1B), suggesting a simultaneous activity of RISC complexes bearing endogenous miR-34a-5p and miR-34a-3p. An empty vector was unaffected by siTGF treatment, confirming a sequence-specific effect (Fig. 1B, bottom panel). To examine the activity of 5p and 3p miRNAs from miR-34a independently, we used commercially available miRNA mimics (34a-5p, 34a-3p). These are typically dsRNA, which are modified so as to allow only guide strand incorporation into RISC. We transfected 34a-5p and 34a-3p separately into cells and observed that both mimics induced caspase 3/7 activity in a concentration-dependent manner to a similar extent (Fig. 1C). Taken together, the experiments suggested that both arms of miR-34a contributed to the apoptotic phenotype resulting from TGF-β1 RNAi. It implied that further investigation of miR-34a in this pathway would be better served using reagents for pre- or pri-mir-34a, either of which could be expected to produce miRISC complexes from both 5p and 3p miRNAs. As we preferred not to use expression vectors because of their inefficient transfection of routinely used cell lines, we turned therefore to synthesis of pre-miRNAs.
Design and use of syn-pre-miRNA mimics
Pre-miRNAs can be prepared by in vitro transcription from DNA templates, although the method suffers several limitations including the promoter-derived constraints at the 5′ end of the sequence, a heterogeneous incorporation of untemplated nucleotides at both termini (Helm et al. 1999) and the inclusion of a 5′ triphosphate (Milligan and Uhlenbeck 1989). One solution to obtain homogeneous 5′ and 3′ ends is the use of dual cis-cleaving ribozymes cotranscribed with the desired RNA (Birikh et al. 1997); however, we experienced irregular cleavage activity by the ribozymes and a difficult purification of the desired hairpin. The expression of complete pre-miRNAs in cells from a DNA plasmid has been described (McManus et al. 2002; Terasawa et al. 2011); however, DNA vectors do not have the convenience of use of an easily transfectable small dsRNA. Given these limitations, we turned to a chemical synthesis of pre-miRNAs (syn-pre-miRNA). The length of human pre-miRNAs (50–90 nt) is challenging for routine laboratory synthesis (Shiba et al. 2007). Consequently, reports of the use of syn-pre-miRNAs as miRNA mimics are rare (see, e.g., Macrae et al. 2008; Koscianska et al. 2011; Starega-Roslan et al. 2011) (see Siolas et al. 2005 for a synthesis of shRNAs). To our surprise, the synthesis of pre-miRNAs using standard reagents (2′-O-tert-butyldimethylsilyl [TBS]-protected phosphoramidites) proved rather straightforward if an appropriate controlled pore glass (CPG) solid support was used (1000 Å universal support). Yields of full-length pre-miRNAs are low (1–8 nmol), but are sufficient for many transfections. Product purification by reverse-phase high-performance liquid chromatography (RP-HPLC) separates failure sequences (see Fig. 2A) and characterization by electrospray mass spectrometry (MS) provides an unambiguous proof of product. To characterize the general properties of syn-pre-miRNAs, ∼40 sequences were prepared, using sequences from miRBase (Griffiths-Jones 2006; Supplemental Table 1).
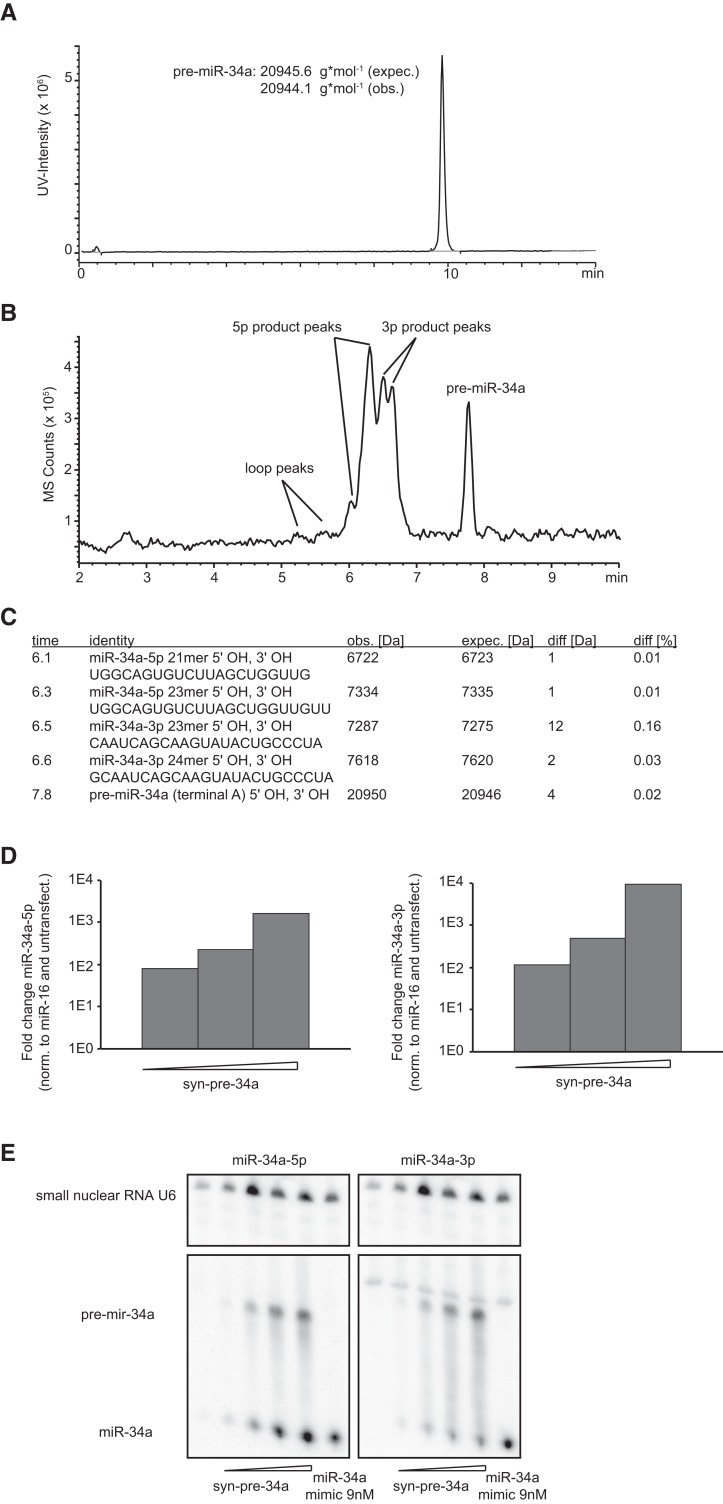
FIGURE 2.
Properties of chemically synthesized pre-miRNA mimics. (A) HPLC-MS chromatogram of syn-pre-34a. The x-axis represents column retention time; the y-axis shows UV-peak intensity. (B) Syn-pre-34a was incubated with recombinant Dicer for 2 h at 37°C in buffer. Analysis by HPLC-MS indicates processing of the precursor (slowest migrating product) into distinct product groups: the pre-miR-34a terminal loop, the miR-34a-5p, and miR-34a-3p strands. (C) Table of predicted products generated by Dicer cleavage of syn-pre-34a, including observed molecular masses and predicted molecular masses. (D) Abundance of miR-34a-5p and miR-34a-3p strands after transfection of syn-pre-miR34a into HeLa cells at increasing doses (2, 9, and 36 nM), followed by RNA isolation and qPCR using primers for miR-34a-5p and miR-34a-3p strands. The y-axis shows the relative RNA abundance (normalized against internal miR-16 and an untransfected control). (E) Syn-pre-34a (0, 0.5, 2, 9, and 36 nM) and ds-34a (9 nM) were transfected into HeLa cells. Total RNA was isolated and probed by Northern blotting for miR-34a-5p and miR-34a-3p, as well as U6 snRNA, with radioactively labeled probes.
To ensure that syn-pre-miR-34a (syn-pre-34a) was recognized by Dicer, we performed in vitro processing assays. Syn-pre-34a was incubated with recombinant Dicer, using a similar protocol as previously described (Davies and Arenz 2008) and analyzed by RP-HPLC-MS (Fig. 2B). Syn-pre-34a was processed into the terminal loop, miR-34a-5p and miR-34a-3p (Fig. 2C). The main 5p 23-nt product was one of the two most abundant miR-34a-5p strands cataloged in miRBase. Two product sequences matched closely the miR-34a-3p sequences, one of which was identical to the second most abundant sequence in miRBase. The second sequence had an additional 5′ guanine. Both 3p strands carried 5′ terminal hydroxyl groups according to MS instead of the expected phosphate group, possibly due to dephosphorylation after processing. The 3p strands were seen in similar amounts, while the 23-nt 5p product was more abundant than the shorter 21-nt product. Taken together the data indicated that syn-pre-34a formed a hairpin that was processed in vitro into miR-34a-5p and miR-34a-3p, even in the absence of auxiliary factors such as TRBP (Koscianska et al. 2011). We then transfected syn-pre-34a into HeLa cells. We have shown recently that a Cy3-labeled pre-miRNA is efficiently delivered into HeLa cells using the same protocol as for miRNA mimics (Pradère et al. 2013). We detected a concentration-dependent production of miR-34a-5p and miR-34a-3p strands by stem–loop quantitative real time PCR (qPCR) and Northern blotting (Fig. 2, D and E, respectively). The qPCR analysis and the Northern blotting suggested a higher abundance for the 5p strand, mirroring strand abundances reported in miRBase and a large collection of libraries from many cell types, except T-cells (Landgraf et al. 2007). The Northern blot showed that at transfection concentrations of 9 nM, syn-pre-34a, and the miR-34a mimic produced comparable levels of miR-34a-5p in cells. The pre-miRNA was also visible on the blot, indicating incomplete processing (Fig. 2E). Taken together, this data shows that syn-pre-34a undergoes processing in cells to bona fide miR-34a miRNAs, and therefore represents a chemical mimic of pre-miR-34a.
Synthetic pre-miRNAs are processed in cells to 5p and 3p siRNAs and miRNAs
In order to characterize the properties of syn-pre-miRNAs, we used dual-luciferase reporter assays. Four plasmids were initially constructed containing experimentally validated miRNA target sites flanked by ∼60 nt of natural sequence from: CDKN1A for miR-17-5p and its family members (Ivanovska et al. 2008); THBS1 for miR-194 (Sundaram et al. 2011), GYS1 for miR-122 (Esau et al. 2006), and SIRT1 for miR-34a-5p (Yamakuchi et al. 2008). Cotransfections of the plasmids and the syn-pre-miRNAs, as well as a commercial miRNA mimic were carried out in HeLa cells. Use of the siRen positive control allows monitoring transfection efficiency, which can vary between experiments. The 5p strands of all syn-pre-miRNAs were active and suppressed their targets to a similar level as the miRNA mimics (Fig. 3A). The level of reporter inhibition varied considerably: For example, syn-pre-34a inhibited its target weakly, whereas syn-pre-194 was as potent as the siRNA-targeting Renilla luciferase. Next, we conducted a comparison of syn-pre-miRNAs of the miR-17-5p family against target sites in the 3′ UTRs from CDKN1A, TGFBR2 (Calin 2004), as well as a fully complementary site to miR-106a-5p. All family members were active as miRNAs, albeit with small variations in their inhibitory power (Fig. 3B). In contrast, only syn-pre-106a and syn-pre-17-5p strongly suppressed the miR-106a-5p sensor, perfectly consistent with the degree of complementarity between the miRNA and the target site (Fig. 3C). This data was strong evidence for direct and sequence-specific effects of the syn-pre-miRNAs. Taken together the experiments showed that syn-pre-miRNAs make active 5p miRNAs, and that the reagents are of comparable biological potency to commercial mimics.
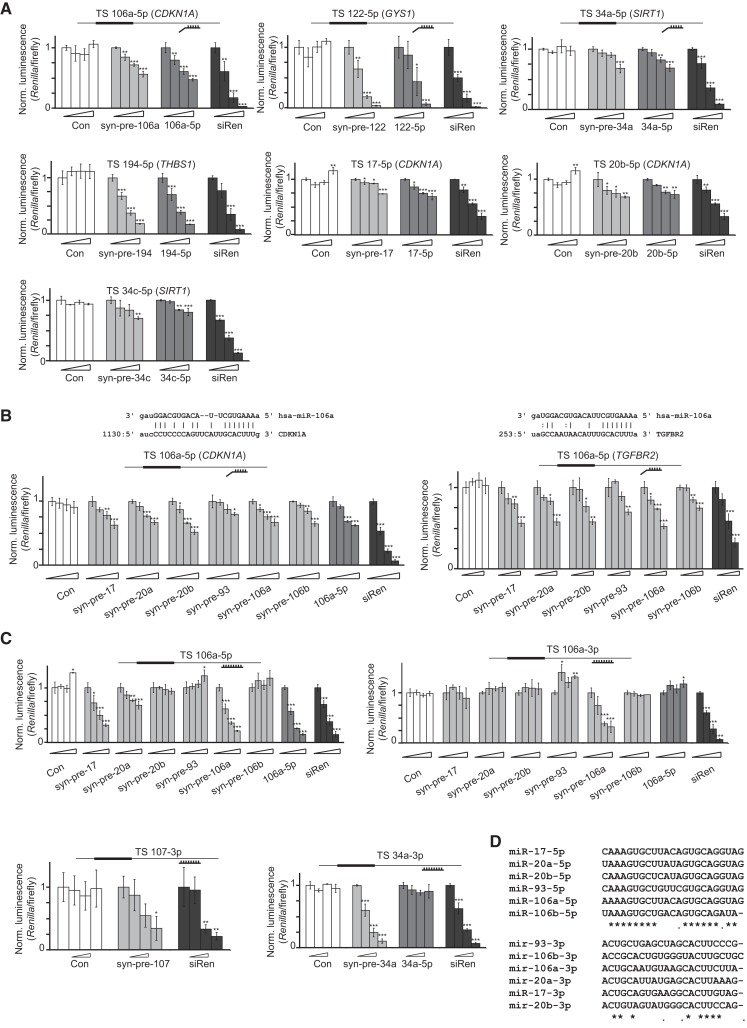
FIGURE 3.
Syn-pre-miRNAs are processed in cells to functional 5p and 3p miRNAs. (A) Syn-pre-miRNAs and their corresponding miRNA mimics were compared for inhibitory activity in dual-luciferase reporter assays. Transfections of Con, syn-pre-miRNAs, corresponding miRNA mimics, and siRen into HeLa cells were performed at increasing concentrations (0, 2, 9, and 36 nM). Luciferase reporter plasmids bearing a single validated target site (TS) for the miRNA (origin of the TS shown in the inset) were transfected. Renilla luminescence values were normalized against firefly luminescence, and then to the value from the corresponding 0 nM transfection. Error bars, SD of three transfections (*) P < 0.05; (**) P < 0.01; (***) P < 0.001. (B) Syn-pre-miRNAs from the miR-106 family show varying levels of inhibition of luciferase reporters containing target sites from CDKN1A and TGFBR2. Transfections and work-up were performed as in A. Error bars, SD of three transfections (*) P < 0.05; (**) P < 0.01; (***) P < 0.001. (C) Syn-pre-miRNAs (miR-106a, miR-107, miR-34a) are processed to functional 5p and 3p miRNAs in HeLa cells and suppress luciferase reporters containing a single complementary target site. Syn-pre-miRNAs and controls were transfected with increasing concentrations (0, 2, 9, and 36 nM) into HeLa cells. Work-up and data analysis were performed as in A. Error bars, SD of three transfections (*) P < 0.05; (**) P < 0.01; (***) P < 0.001. (D) Sequence homology between the 5p and 3p miR-17 family members.
A previous study using reporters showed that 14 from 22 plasmid-expressing pre-miRNAs produced 3p miRNAs active against artificial complementary and bulged targets (Yang et al. 2011). We generated sensor constructs for the 3p strands of pre-miR-106a, pre-miR-34a, pre-miR-107, and pre-miR-122. We cotransfected these plasmids as well as their cognate syn-pre-miRNAs and the controls into cells. For the miR-106a target site plasmid we observed a strong sequence-specific repression from syn-pre-106a, but no effects from the other family members, presumably because of insufficient 3p strand complementarity to the target (Fig. 3C). As expected, we saw no effects from the mimics, presumably due to their inactivated counterstrands. Similarly, the 3p sensors for syn-pre-107 and syn-pre-34a were equally suppressed, as was the reporter for syn-pre-122 (data not shown). These experiments confirmed that the processing of syn-pre-miRNAs produced a 3p miRNA active against artificial target sites, in broad agreement with the previous report (Yang et al. 2011). However, because these targets were fully complementary it did not confirm that Dicer cleaved the syn-pre-miRNAs precisely to natural 3p miRNAs strands with the correct seed sequences. Dicer cleavage is influenced by the overhang of the pre-miRNA (Vermeulen et al. 2005; Park et al. 2011) and miRNAs show considerable heterogeneity at their 3′ termini (Newman et al. 2011), so defining the sequence of the 3′ overhang of syn-pre-miRNAs can be ambiguous, and therefore critical.
Identification of new targets for miR-34a-3p
Many targets of miR-34a-5p involved in cell growth/death have been identified, including BCL2, CD44, CDK6, CCND1, E2F3, MET, MYB, MYC, SIRT1, VEGF, WNT, and SIRT1 (Yamakuchi et al. 2008). We examined the effects of syn-pre-34a on endogenous SIRT1. Both 34a-5p mimic, as well as siTGF suppressed a luciferase reporter containing the validated target site of the SIRT1 3′ UTR, whereas the 34a-3p mimic was inactive (Fig. 4A). Syn-pre-34a showed a particularly strong inhibition of the SIRT1 reporter, and also endogenous SIRT1 protein in HeLa cells (Fig. 4B).
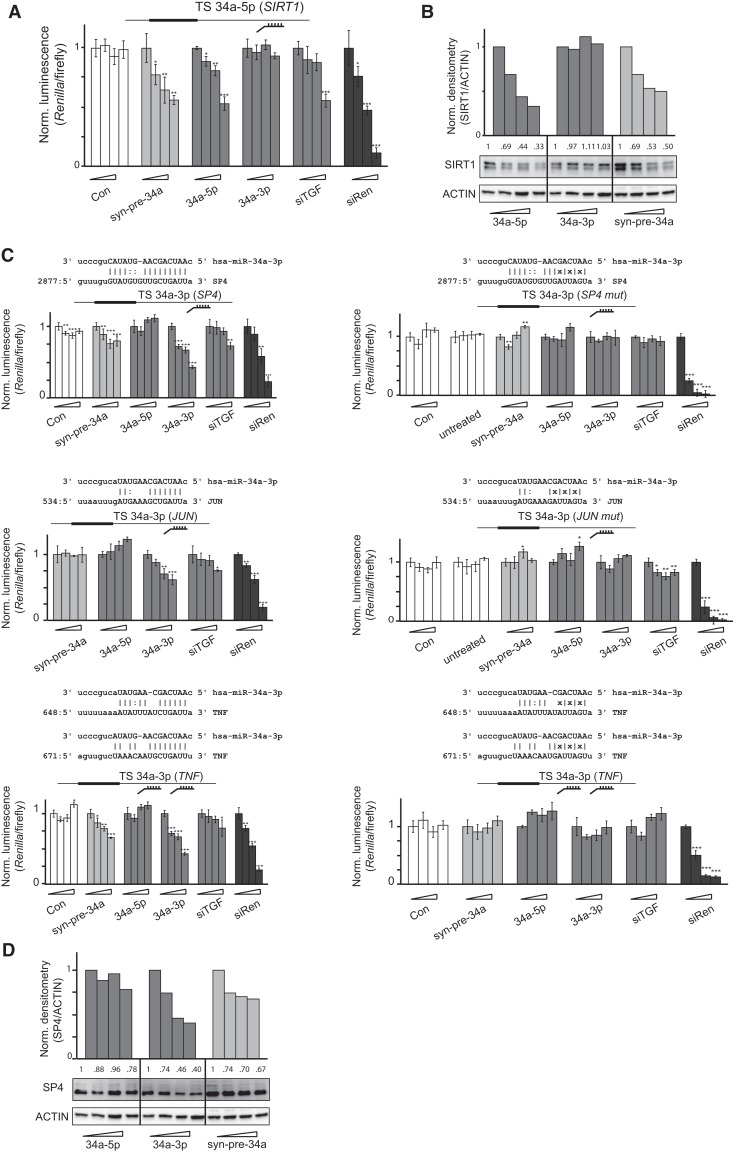
FIGURE 4.
Identification of new targets for miR-34a-3p. (A) SiTGF and miR-34a-5p repress SIRT1 3′ UTR in a dual-luciferase reporter assay. Transfections of dsRNAs into HeLa cells were performed at increasing concentrations (0, 2, 9, and 36 nM). Luciferase reporter plasmid bearing a single validated target site from SIRT1 (Yamakuchi et al. 2008) for miR-34a-5p was transfected. Renilla luminescence values were normalized against firefly luminescence and then to the value from the corresponding 0-nM transfection. Error bars, SD of three transfections (*) P < 0.05; (**) P < 0.01; (***) P < 0.001. (B) Syn-pre-34a and 34a-5p mimic repress SIRT1 protein in HeLa cells. Cells were transfected with increasing concentrations (0, 2, 9, and 36 nM) of dsRNAs. Total protein was isolated 48 h post-transfection. SIRT1 protein expression was analyzed by Western blot using a specific antibody and was quantified by densitometry using ImageJ (rsbweb.nih.gov/ij/). (C) Syn-pre-34a and 34a-3p mimic repress target sites from SP4, JUN, and TNF 3′ UTRs predicted by the mirSVR algorithm (Betel et al. 2010). Transfections of dsRNAs into HeLa cells were performed at increasing concentrations (0, 2, 9, and 36 nM). Luciferase reporter plasmids (identity shown in inset) bearing either a single predicted target site (SP4 and JUN) or two predicted target sites (TNFα) and their corresponding mutated controls were transfected. Renilla luminescence values were normalized against firefly luminescence and then to the value from the corresponding 0-nM transfection. Error bars, SD of three transfections (*) P < 0.05; (**) P < 0.01; (***) P < 0.001. (D) Syn-pre-34a and 34a-3p repress SP4 protein in HeLa cells. Cells were transfected with increasing concentrations (0, 2, 9, and 36 nM) of dsRNAs. Total protein was isolated 48 h post-transfection. SP4 protein expression was analyzed by Western blot using a specific antibody and was quantified by densitometry using ImageJ.
To our knowledge, the only experimentally validated target of miR-34a-3p is XIAP, although the location of the target site in the 3′ UTR was not identified (Niederer et al. 2011a). In a search for candidate miR-34a-3p targets, which may have contributed to cell death during TGF-β1 RNAi, we used the miRanda-mirSVR algorithm (Betel et al. 2010). We identified potential target sites in the 3′ UTRs of SP4, JUN, and TNF (two sites) (Fig. 4C). It has been postulated previously that miR-34a-3p regulates JUN, but no target site for the miRNA was proposed (Guled et al. 2009). We therefore generated reporter constructs bearing these sites, as well as mutated analogs, embedded in ∼120 nt of natural flanking sequence. We cotransfected them into cells together with the 34a-5p and 34a-3p mimics, syn-pre-34a, and siTGF. The SP4 and TNFα reporters were inhibited by 34a-3p mimic, suggesting that their 3′ UTRs are targets of miR-34a-3p, but not of miR-34a-5p (Fig. 4C, left panels). Mutation of the putative binding sites in the reporters abolished the activities of the mimics (Fig. 4C, right panels). Syn-pre-34a also suppressed these reporters, showing for the first time that the synthetic hairpin indeed produced a bona fide 3p miRNA. The potency of the syn-pre-34a was less than that of the individual 3p mimic, but similar to that of siTGF. The JUN target site showed a lower level of suppression by the miR-34a-3p mimic, and mutation of the site also abolished its activity. Here, syn-pre-34a was inactive, possibly because of a reduced sensitivity of the reporter, or from an inherently lower level of activity at this target site. Importantly, cotransfection of all three reporters separately with siTGF also led to their suppression at the highest concentrations and implied that these genes are regulated by induced endogenous miR-34a-3p during apoptosis from TGF-β1 RNAi (Fig. 4C). Finally, a Western blot analysis on protein lysates using an antibody specific for SP4 confirmed its regulation by 34a-3p and syn-pre-34a (Fig. 4D). Taking together the data generated using exogenously delivered commercial mimics and syn-pre-34a, we confirmed that the increased levels of endogenous miR-34a-5p and miR-34a-3p during TGF-β1 RNAi suppress target sites in the 3′ UTRs of SIRT1 (miR-34a-5p) and SP4, JUN, and TNFα (miR-34a-3p). Hence, we conclude that miR-34a demonstrates a dual functional activity from both of its miRNAs in HeLa cells.
The suppression of the TNFα reporter by syn-pre-34a was the most robust of the three (Fig. 4C). A similar trend of this activity extended to the endogenous target, secreted TNFα, in spite of very low levels of TNFα produced by HeLa cells (data not shown). Consequently, we looked for other systems in which miR-34a might use both of its 5p and 3p strands in TNFα signaling. Specifically, we examined whether miR-34a might play a role during the lipopolysaccharide (LPS)-induced secretion of TNFα in macrophages.
The dual activity of miR-34a-5p and miR-34a-3p strands in TNF signaling
TNFα protein is secreted by activated, monocyte-derived macrophages during bacterial infections (Beutler et al. 1985). Increased TNFα derives from transcription as well as post-transcriptional regulation of its 3′ UTR (Raabe et al. 1998). MiR-34 has been linked with the TNFα pathway previously (Jiang et al. 2012). We hypothesized therefore that miR-34a-3p may also play a role in monocyte-derived macrophages, by suppressing TNFα levels directly.
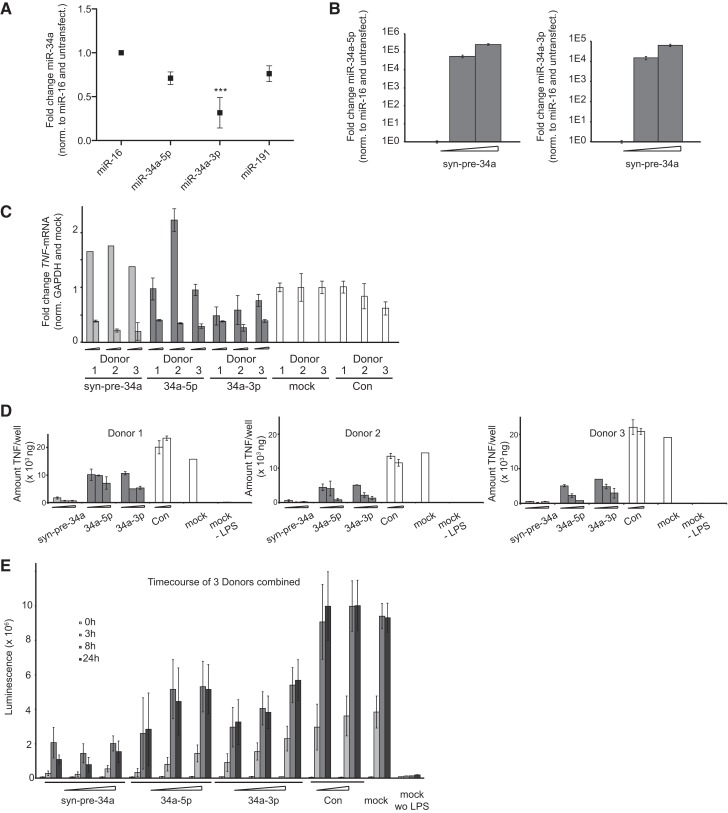
We used primary human monocyte-derived macrophages because of their ease of transfection in comparison to, for example, the THP1 cell line, which requires harsh electroporation methods. In five samples generated from the blood of individual donors we measured baseline expression of miR-34a, miR-16, and miR-191. Several genes have been proposed for the normalization of miRNA expression levels under changing conditions. Stably expressed miRNAs are reportedly superior internal controls to other small RNAs, e.g., snoRNAs (Wotschofsky et al. 2011). We and other groups have found that expression of miR-16 and miR-191 are invariant under a variety of conditions and therefore can be useful as internal controls for normalization (Lindsay 2008; Peltier and Latham 2008; Mestdagh et al. 2009). MiR-191 showed similar expression across all five human samples, whereas miR-16 expression was constant, but distinct in samples 1 and 3, and in 4–6, which were processed in two groups. In the five samples we found expression of miR-34a-5p and miR-34a-3p in ratios from 1:3–3:1 (Supplemental Table 2). In sharp contrast to most other cell types in which it is hardly expressed (data from Landgraf et al. 2007), macrophages have relatively high levels of miR-34a-3p, possibly consistent with an important function in these cells.
We treated the five samples with LPS and measured the response of miRNAs (Fig. 5A). Mir-34a-5p decreased in all cases when normalized to miR-16 (Supplemental Table 2). However, the magnitude of the response was similar to that of the controls, and thus we were unsure that this was a bona fide effect, despite the previously published data obtained in the murine macrophage cell line (Jiang et al. 2012). In contrast, LPS treatment reduced miR-34a-3p several-fold in four from five samples, leading overall to a significant effect using either miR-16 or miR-191 for normalization (Fig. 5A). It confirmed for the first time regulation of miR-34a-3p in monocyte-derived macrophages by LPS. Furthermore, it was consistent with a role for miR-34a-3p in maintaining homeostatic levels of TNFα under nonactivated conditions.
FIGURE 5.
MiR-34a-5p and MiR-34a-3p strands are active in TNFα signaling. (A) Primary monocyte-derived macrophages from five human donors were stimulated with LPS (100 ng/mL). RNA was isolated and qPCR for miR-34a-5p and miR-34a-3p strands and internal controls (miR-16 and miR-191) was performed. The y-axis shows the relative RNA abundance (normalized against internal miR-16 and an unstimulated control). Error bars, SEM of five donors. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001. (B) Syn-pre-34a was transfected at increasing concentrations (0, 2, and 9 nM) into macrophages. RNA was isolated and qPCR was performed for miR-34a-5p and miR-34a-3p. The y-axis shows the relative RNA abundance (normalized against internal miR-16 and an untransfected control). Error bars, SD of four replicates. (C) Syn-pre-34a, 34a-5p, and 34a-3p (2 and 9 nM), Con (9 nM) and transfection reagent alone (mock) were transfected into macrophages derived from three human donors. Total RNA was isolated and SYBR-Green qPCR was used to assay TNFα mRNA. The y-axis shows Ct-values representing the absolute abundance of the mRNA normalized to GAPDH mRNA, and then to mock control. Error bars, the SD of four replicates. (D) Syn-pre-34a, 34a-5p, and 34a-3p (2, 9, and 36 nM), Con (9 and 36 nM), and transfection reagent alone (mock) were transfected into macrophages derived from three human donors. Cells were stimulated with LPS (100 ng/mL) and secreted TNFα protein was measured by ELISA after 12 h. The y-axis shows amounts of secreted TNFα protein (nanogram/well). The transfection reagent without LPS treatment (mock – LPS) shows background levels of TNFα protein in unstimulated cells. Error bars, the SD of two transfections. (E) Transfection of syn-pre-34a, 34a-5p, and 34a-3p (2, 9, and 36 nM), Con (9 and 36 nM), and transfection reagent alone (mock) were performed in macrophages derived from three donors. Secreted TNFα protein was assayed (0, 3, 8, and 24 h) after stimulation with LPS (100 ng/mL) using ELISA. The y-axis shows relative amounts of secreted TNFα protein. The transfection reagent without LPS stimulation (mock – LPS) shows background levels of TNFα protein prior to stimulation. Error bars, the SD of three donors with duplicate transfections.
We examined further the regulation of TNFα mRNA by miR-34a in monocyte-derived macrophages derived from three donors. Treatment of macrophages with syn-pre-34a produced high levels of miR-34a-5p and miR-34a-3p dose dependently, confirming successful transfection and correct processing (Fig. 5B). Next, cells were transfected separately with syn-pre-34a, 34a-5p, and 34a-3p, and were then treated with LPS to induce TNFα mRNA as described (Beutler et al. 1985). Induction of TNFα mRNA was unaffected by the pretreatment with the controls (mock, Con). However, we observed strong suppression of TNFα mRNA after treatment at the 9-nM dose with syn-pre-34a and mir-34a-3p mimic in all three samples, consistent with a direct effect on the mRNA (Fig. 5C). MiR-34a-5p mimic also suppressed TNFα mRNA. Neither miRanda-mirSVR (Betel et al. 2010) nor Targetscan (www.targetscan.org) showed a likely target site for miR-34a-5p in the TNFα UTR. However, miR-34a-5p reportedly regulates TNFα through NOTCH1 (Jiang et al. 2012). We showed that NOTCH1 protein expression is attenuated in both HeLa cells and in LPS-stimulated macrophages by treatment with both 34a-5p and syn-pre-34a (Supplemental Fig. 1).
If miR-34 occupies a key role in the regulation of TNFα by LPS, then addition of miR-34a during activation should rescue LPS-induction of TNFα. We used an ELISA to probe for the combined effects of LPS and miR-34a on secreted TNFα. We treated separately macrophages from three donors with syn-pre-34a, 34a-5p, and 34a-3p. We stimulated cells with LPS (100 ng/mL) after 24 h and measured a dramatic increase in TNFα secretion (from 6 ng/mL to 16–18 μg/mL) (Fig. 5D) in the supernatants from cells treated with controls. In contrast, monocyte-derived macrophages transfected with 34a-5p or 34a-3p suppressed TNFα secretion dose dependently. Levels of secreted TNFα in syn-pre-34a transfected probes were decreased at much higher efficiency (to 0.9–0.3 µg/mL) at all concentrations. The strength of these effects was confirmed in a time-course experiment in which we measured TNFα before LPS stimulation (0 h), and after 3, 8, and 24 h. Secreted TNFα levels reached their highest values after 8 h and were stable for at least 24 h in control-treated cells (Fig. 5E). Supernatants from Con-transfected and mock-treated cells showed similar levels of TNFα. However, syn-pre-34a, 34a-5p, and 34a-3p repressed TNFα protein level in the supernatants from all three samples. Treated cells appeared viable throughout the experiment and amounts of isolated total RNA from wells treated with miRNAs were similar to those from mock-treated cells, suggesting that attenuated TNFα did not derive from reduced cell numbers in this short experiment. Once again, the magnitude of the effect was the greatest from the syn-pre-34a, suggesting an additive or possibly a synergistic effect from the action of both miRNA strands of miR-34a (Fig. 5E).
Taken together, the data from these experiments indicates that in monocyte-derived macrophages the transcriptional and post-transcriptional activation of TNFα by LPS requires that homeostatic levels of miR-34a be lowered. Both strands of miR-34a share important roles in regulating macrophage TNFα: miR-34a-5p targets TNFα, possibly indirectly and the miR-34a-3p strand targets TNFα mRNA directly. The roles of the two miRNA strands are emulated by the syn-pre-34a reagent, which in primary human monocyte-derived macrophages (and in HeLa cells) is processed into functional 5p and 3p miRNAs. These, in combination, show a superior level of inhibitory activity than the miRNA mimics delivered separately in monocyte-derived macrophages.
DISCUSSION
Today we know that both arms of many miRNAs are functional and that their abundance varies in a context-dependent fashion. Consequently, for a complete picture of a miRNA's biology, reagents are required that provide both strands from the miRNA gene. Commercially available miRNA mimics are vital tools for assigning function to single miRNAs, but, typically, they are modified to ensure loading of only one strand into miRISC. Pri- or pre-miRNAs expressed from DNA plasmids are able to provide both strands of a miRNA, but plasmids do not have the convenience of use of an easily transfectable small dsRNA (e.g., siRNA).
Here, we introduce chemically synthesized pre-miRNAs (syn-pre-miRNA) as a general class of transfectable pre-miRNA mimics, which recapitulates the properties of endogenous pre-miRNAs. These hairpins, complete with their terminal loops and any associated regulatory sites, with 5′ and 3′ terminal hydroxyl groups, are synthesized, purified, and characterized using protocols accessible in most oligonucleotide facilities. Alternatively, and contrary to popular belief, syn-pre-miRNAs are available from oligonucleotide vendors at affordable prices. To date, we have shown with more than 40 examples that they are processed to functional miRNAs in cells. We have seen with miR-106a, miR-34a, and miR-122 that syn-pre-miRNAs provide both 5p and 3p strands to the RISC machinery, and that the resultant miRNAs for the most part demonstrate a similar level of inhibitory activity as commercial miRNA mimics.
We used syn-pre-34a to investigate the two miRNAs expressed from hsa-miR-34a. MiR-34a is a tumor suppressor: Accounts of its antiproliferative properties are invariably focused on its 5p strand. We demonstrated here that induction of miR-34a during TGF-β1 RNAi gives comparable levels of endogenous miR-34a-5p and miR-34a-3p in HeLa cells. Both miRNAs were functional, as shown by their repression of luciferase reporters. Furthermore, by transfecting cells with 5p and 3p mimics we saw that caspase 3/7 was induced by both reagents, suggesting that each of the endogenous miRNA strands contributed to the phenotype. Lastly, we showed that miR-34a miRNAs suppress target sites in the 3′ UTRs of SIRT1 (miR-34a-5p), as well as SP4, JUN, and TNFα, three previously unrecognized targets of miR-34a-3p. The activities of both endogenous miRNAs were jointly recapitulated by syn-pre-34a, which, in addition to the aforementioned reporters (except JUN), suppressed endogenous NOTCH1, SP4, and possibly TNFα proteins in HeLa cells. In summary, the data showed that miR-34a induces both its miRNAs during TGF-β1 RNAi in order to suppress expression of proliferative genes, and that this behavior is phenocopied by syn-pre-34a.
Tissue-resident macrophages respond to LPS produced from bacteria with secretion of TNFα, resulting in tissue inflammation, activation of an immune response, or induction of apoptosis. Overproduction of TNFα leads to pathological conditions, and consequently the pharmacological inhibition of TNFα in inflammation is of therapeutic value (Walsh 2010). We found comparable levels of miR-34a-5p and miR-34a-3p in primary human monocyte-derived macrophages. Comparable expression of both arms of a miRNA in a cell has been proposed to indicate targeting of overlapping genes and pathways (Marco et al. 2012; Uchino et al. 2013). We also showed that in macrophages TNFα is targeted by miR-34a-5p and by miR-34a-3p directly, as well as by syn-pre-34a, demonstrating that both strands of the pre-miRNA are functional. We then examined the effects of LPS activation of TNFα on levels of miR-34 in monocyte-derived macrophages from human donors. We were unable to confirm that miR-34a-5p is repressed by LPS as described previously (Jiang et al. 2012). On the other hand, LPS treatment led to a strong attenuation of the miR-34a-3p strand. Taken together, the data suggested that one role of homeostatic levels of miR-34a in macrophages may be to hold TNFα expression in check. If so, then the TNFα response to LPS would be rescued by addition of exogenous miR-34a-3p and possibly also by miR-34a-5p. Indeed, TNFα mRNA levels in LPS-treated macrophages from three donors were suppressed to comparable degrees by transfection of 34a-3p, syn-pre-34a, but also 34a-5p. Possibly, TNFα mRNA is targeted directly by the action of miR-34a-3p on its 3′ UTR, and indirectly by miR-34a-5p. Similar effects were seen on TNFα protein except that its suppression by syn-pre-34a in LPS-treated macrophages was considerably more pronounced than from treatment with either of the mono-functional 34a-5p and 34a-3p mimics alone. This suggests an additive or a synergistic response of the cells to the dual activity of miR-34a.
Here, we have described the preparation/properties of chemically synthesized pre-miRNAs. These reagents are transfected into cells similarly to siRNAs and are processed to functional 5p and 3p miRNAs. With a growing appreciation of interplay between both strands of some miRNAs, these reagents represent practical chemical tools to explore such relationships. We show that miR-34a uses both of its arms in TGF-β-induced apoptosis and LPS-induced secretion of TNFα in human monocyte-derived macrophages.
MATERIALS AND METHODS
Cell culture
HeLa cells (ATCC) were maintained in Dulbecco's Modified Eagle's medium (Gibco, Invitrogen) supplemented with 10% fetal bovine serum (FBS; Sigma-Aldrich).
Peripheral blood mononuclear cells (PBMC) were isolated from healthy blood donors based on Ficoll-Hypaque separation. Monocytes were isolated from PBMC with anti-CD14-coated magnetic beads (Miltenyi Biotech) by positive selection. For the first 7 d, 5% human serum and 0.02 µg/mL human M-CSF (macrophage colony stimulating factor, PeproTech) were added to generate macrophages. Macrophages were cultured in RPMI-1640 (Sigma-Aldrich), supplemented with 10% FBS (Sigma-Aldrich) and 1% penicillin/streptomycin.
Cell transfections
SiRNA against Renilla (siRen) was obtained from Dharmacon, the control siRNA (Con) was obtained from Ambion (No. AM4640), and siTGF (Dogar et al. 2011) was obtained from Dharmacon. RNAs were transfected using Oligofectamine (Invitrogen) according to the manufacturer's instructions.
Macophages were transfected 10 d after isolation from human blood. RNAs were diluted in 0.1 M sodium chloride solution and mixed with jetPRIME (Polyplus-Transfection) transfection reagent according to the manufacturer's instructions. Shortly before transfection, culture media was replaced with serum-free media (OPTI-MEM I Reduced Serum Media, Gibco, Invitrogen) and the transfection mixture was added onto the macrophages. FBS (Sigma-Aldrich) was added to a total amount of 10% after 4 h. After 24 h the media was exchanged with macrophage media (RPMI1640 supplemented with 10% FBS and 1% penicillin/streptomycin) and stimulated with LPS (Sigma-Aldrich) when required.
Luciferase assays
Dual luciferase reporter plasmids containing the indicated target sites were cloned into the psiCHECK2 Vector (Promega). HeLa cells were seeded in white 96-well plates and RNAs were transfected after 8 h. There were no observable differences in cell viability between cells transfected with syn-pre-miRNAs and commercial mimics, Con or siRen. All transfections were performed in triplicates. After 24 h, plasmid DNA (20 ng/well) was transfected using jetPEI (Polyplus) according to the manufacturer's protocol. After 48 h, supernatants were removed and firefly substrate (15 µL including Lysis Buffer; Dual-Glo Luciferase Assay System, Promega) diluted with 15 µL H2O was added. Luminescence was measured on a microtiter plate reader (Mithras LB940, Berthold Technologies). After 30 min, 15 µL of Renilla substrate (including firefly Quencher Solution; Dual-Glo Luciferase Assay System, Promega) per well was added and the measurement was repeated. Values were normalized against firefly luciferase and the corresponding oligofectamine mock control, respectively.
Apoptosis assays
Caspase-3/7 activity was measured in lysates of transfected cells using a chemiluminescent substrate (Caspase-Glo 3/7 substrate, Promega). Cells were grown in 96-well plates, and transfected with various miRNAs. For measurements of cell-associated caspase 3/7 activity cells were lysed in PBS containing 1% Triton X-100 and 5 µL of lysates were mixed with equal volumes of substrate and chemiluminescence was measured in sealed plates after 30 min in a plate reader.
Stimulation of macrophages and ELISA
For activation of cells LPS (Sigma-Aldrich) was added to the medium at a final concentration of 100 ng/mL. Cell supernatants were collected for TNFα analysis and quantified with human TNFα DuoSet ELISA (R&D Systems), according to the manufacturer's instructions.
Dicer assay
Synthetic pre-miR-34a (final concentration 2.5 µM) was incubated with commercial Dicer (1 unit) (Genlantis) in a buffered aqueous solution (30 mM Tris-HCl, 3 mM MgCl2, 50 mM NaCl, 1 mM Dithiotreitol) for 2 h at 37°C. The reaction was stopped by addition of 0.5 µL of 0.5 M EDTA (pH 8) and kept at 4°C prior to analysis by LCMS using an Agilent 1200/6130 apparatus fitted with a C-18 reverse-phase (RP) column (Waters Acquity OST C18, 2.1 × 50 mm, 1.7 μm) equilibrated at 70°C. Elution conditions were 12%–25% MeOH in aqueous HFIP (0.4 M)/TEA (16.2 mM) buffer in 10 min.
Northern blot
Northern blot analyses were used to determine the levels of precursor and mature expression. Total RNA was extracted using TRIzol (Invitrogen) from HeLa cells transfected with precursor or annealed double-stranded miRNAs and loaded onto an 8%–12% gradient denaturing polyacrylamide gel (30 μg/sample) and semi-dry blotted onto a neutral Hybond-N membrane (Amersham, GE Healthcare) using 2 A/cm2 for 30 min in 0.5× TBE. Blotted RNA was chemically cross-linked to the membrane using EDC for 1 h at 60°C. Specific probes that perfectly matched mature miRNAs were generated by T7 in vitro transcription and radiolabeling using α-32-phosphate UTP from suitable DNA templates. The probes were hybridized using ULTRAhyb Ultrasensitive Hybridization Buffer (Ambion) in order to determine the expression of both mature and pre-miRNA species. The hybridized and washed membranes were exposed to a PhosphorScreen for at least 8 h and scanned on a Typhoon 9410 PhosphorImager (GE Healthcare). Band intensity was determined by densitometrical analyses using ImageJ software (Wayne Rasband, NIH). Blots were stripped using 1% SDS for 30 min at 85°C and reprobed using probes complementary to the counterstrand and complementary to U6 snRNA that was used for normalization.
Protein analysis
Cells were lysed with RIPA lysis buffer (Sigma-Aldrich). Protein concentrations were determined using a BCA assay (Thermo Fisher Scientific); 10–20 µg of protein was mixed with equal quantities of SDS loading buffer (100 mM Tris-HCl, 4% SDS, 20% glycerol, 0.2% bromophenol blue). Samples were heated at 95°C for 5 min, separated on SDS gels, and transferred to polyvinylidene difluoride membranes. Nonspecific membrane binding was blocked for 1 h at room temperature with 5% BSA (or milk) in phosphate-buffered saline containing 0.05% Tween 20. Membranes were incubated overnight at 4°C with appropriate primary antibodies from Santa Cruz Biotechnology (SIRT1: SC-74504; SP4: SC-645; NOTCH1: SC-6014-R) and Sigma-Aldrich (β-ACTIN: #A5316). After washing, membranes were incubated with horseradish peroxidase-conjugated secondary antibodies for 1–2 h in blocking buffer. Signals generated by the chemiluminescent substrate (ECL(+); Amersham Biosciences) were captured by a cooled CCD camera (Bio-Rad). Protein bands were quantified by densitometry using the analysis software ImageJ.
RNA extraction and qPCR
Total RNA was extracted using RNeasy Kit (Qiagen), allPrep (Qiagen), or mirVana miRNA Isolation kit (Ambion). For mRNA analysis, 1 µg of total RNA was reverse transcribed using the M-MLV reverse transcriptase kit (Invitrogen) according to manufacturer's instructions. Expression levels were assayed using Power SYBR Green PCR Master Mix (Applied Biosystems). PCR cycling conditions were 95°C/10 min and 40 cycles of (95°C/15 sec; 60°C/1 min). Values were normalized using GAPDH or average of all measured mRNAs. For miRNA analyses, TaqMan probes (Applied Biosystems) were used. Total RNA was reverse transcribed with miRNA-specific RT primers and amplified with TaqMan miRNA-specific primers. cDNA was synthesized from 10 ng (primary human monocyte-derived macrophages) and 20 ng (HeLa) of total RNA in a 15-µL volume using TaqMan MicroRNA Reverse Transcription kit (Applied Biosystems), according to manufacturer's instructions. Normalization prior to qPCR was made on total RNA. Baseline cycle threshold (Ct)-values can vary according to extraction methods: Donors 1–3 were processed with RNeasy Kit and donors 4 and 5 with allPrep). All samples were measured in quadruplicates; the Ct values were transformed to relative quantities. Fold changes shown in the figures are calculated using the Delta Delta CT method (Livak and Schmittgen 2001). All primer sequences are shown in Supplemental Table 3.
Oligonucleotide synthesis
Oligoribonucleotides were prepared under standard conditions on a MerMade 12 synthesizer (Bioautomation Corporation) using UnySupport controlled-pore glass (CPG; Glen Research) and phosphoramidites from Thermo Fisher Scientific. After completion of synthesis cycles, the CPG was treated with gaseous methylamine at 65°C and 1 bar for 2 h. RNAs were eluted with ethanol/water (1:1). Desilylation was carried out on dry RNAs using freshly prepared 1-methyl-2-pyrrolidone, triethylamine (TEA), and TEA.3HF (6:3:4) for 90 min at 70°C. Isopropoxytrimethylsilane was added and samples were lyophilized. The crude RNA was purified by RP-HPLC (Agilent 1200 Series; Agilent Technologies). Dimethoxytrityl groups were cleaved using 40% acetic acid for 30 min at room temperature. RNAs were again purified by HPLC using a C18 column (XBridge OST, particle size 2.5 mm, Waters). Purified samples were analyzed on an Agilent 6130 Series Quadrupole LC/MS (Agilent Technologies) with electron spray ionization. Purity and yields were determined by HPLC and Nanodrop, respectively. Sequences were taken from miRbase using the most abundant 5p and 3p strands from the reported deep-sequencing reads and are shown in the Supplemental Material.
Plasmid construction
PsiCHECK2 (Promega) reporter constructs were cut with NotI and XhoI restriction enzymes and inserts were cloned into the 3′ UTR of the Renilla mRNA to yield reporter constructs having miRNA or fully complementary binding sites. The inserted sequences in the psiCHECK2 vector are reported in Supplemental Table 3. The psiCHECK2 vectors were sequenced and subsequently transfected as described above.
Statistics
All statistical analyses were performed using a one-way analysis of variance (ANOVA) to evaluate treatment effects. If the ANOVA value was significant, comparisons between the 0 nM and the other transfections were performed using ANOVA followed by Dunnett's t-test to localize the significant difference. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001. All statistics were run with GraphPad Prism 6.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank M. Zimmermann and G. Simplicio for help with RNA synthesis and transfections. We are grateful to C. Halin-Winter and H. Towbin for helpful suggestions. This work was supported in part by grants from the Swiss National Science Foundation to J.H. (205321_124720/1) and to K.J.M. (31003A-127317), and the Krebsforschung Schweiz KFS to J.H. (02648-08-2010).
REFERENCES
- Almeida MI, Nicoloso MS, Zeng L, Ivan C, Spizzo R, Gafà R, Xiao L, Zhang X, Vannini I, Fanini F, et al. 2012. Strand-specific miR-28-5p and miR-28-3p have distinct effects in colorectal cancer cells. Gastroenterology 142: 886–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuma-Mukai A, Oguri H, Mituyama T, Qian ZR, Asai K, Siomi H, Siomi MC 2008. Characterization of endogenous human Argonautes and their miRNA partners in RNA silencing. Proc Natl Acad Sci 105: 7964–7969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP 2009. MicroRNAs: Target recognition and regulatory functions. Cell 136: 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betel D, Koppal A, Agius P, Sander C, Leslie C 2010. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol 11: R90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beutler B, Greenwald D, Hulmes JD, Chang M, Pan YC, Mathison J, Ulevitch R, Cerami A 1985. Identity of tumour necrosis factor and the macrophage-secreted factor cachectin. Nature 316: 552–554 [DOI] [PubMed] [Google Scholar]
- Birikh KR, Heaton PA, Eckstein F 1997. The structure, function and application of the hammerhead ribozyme. Eur J Biochem 245: 1–16 [DOI] [PubMed] [Google Scholar]
- Birmingham A, Anderson E, Sullivan K, Reynolds A, Boese Q, Leake D, Karpilow J, Khvorova A 2007. A protocol for designing siRNAs with high functionality and specificity. Nat Protoc 2: 2068–2078 [DOI] [PubMed] [Google Scholar]
- Byrd AE, Aragon IV, Brewer JW 2012. MicroRNA-30c-2* limits expression of proadaptive factor XBP1 in the unfolded protein response. J Cell Biol 196: 689–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA 2004. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci 101: 2999–3004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Hu S-J 2012. Effect of microRNA-34a in cell cycle, differentiation, and apoptosis: A review. J Biochem Mol Toxicol 26: 79–86 [DOI] [PubMed] [Google Scholar]
- Chiang HR, Schoenfeld LW, Ruby JG, Auyeung VC, Spies N, Baek D, Johnston WK, Russ C, Luo S, Babiarz JE, et al. 2010. Mammalian microRNAs: Experimental evaluation of novel and previously annotated genes. Genes Dev 24: 992–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies BP, Arenz C 2008. A fluorescence probe for assaying micro RNA maturation. Bioorg Med Chem 16: 49–55 [DOI] [PubMed] [Google Scholar]
- Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ 2004. Processing of primary microRNAs by the Microprocessor complex. Nature 432: 231–235 [DOI] [PubMed] [Google Scholar]
- Djuranovic S, Nahvi A, Green R 2012. miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science 336: 237–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench JG, Petersen CP, Sharp PA 2003. siRNAs can function as miRNAs. Genes Dev 17: 438–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogar AM, Towbin H, Hall J 2011. Suppression of latent transforming growth factor (TGF)-β1 restores growth inhibitory TGF-β signaling through microRNAs. J Biol Chem 286: 16447–16458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esau C, Davis S, Murray SF, Yu XX, Pandey SK 2006. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab 3: 87–98 [DOI] [PubMed] [Google Scholar]
- Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R 2004. The Microprocessor complex mediates the genesis of microRNAs. Nature 432: 235–240 [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S 2006. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res 34: D140–D144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Hui JHL, Marco A, Ronshaugen M 2011. MicroRNA evolution by arm switching. EMBO Rep 12: 172–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A, Srivastava M, Fahey B, Woodcroft BJ, Chiang HR, King N, Degnan BM, Rokhsar DS, Bartel DP 2008. Early origins and evolution of microRNAs and Piwi-interacting RNAs in animals. Nature 455: 1193–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guled M, Lahti L, Lindholm PM, Salmenkivi K, Bagwan I, Nicholson AG, Knuutila S 2009. CDKN2A, NF2, and JUN are dysregulated among other genes by miRNAs in malignant mesothelioma—a miRNA microarray analysis. Genes Chromosomes Cancer 48: 615–623 [DOI] [PubMed] [Google Scholar]
- Gurtan AM, Lu V, Bhutkar A, Sharp PA 2012. In vivo structure-function analysis of human Dicer reveals directional processing of precursor miRNAs. RNA 8: 1116–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom K-H, Kim Y-K, Jin H, Kim VN 2004. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev 18: 3016–3027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helm M, Brule H, Giege R, Florentz C 1999. More mistakes by T7 RNA polymerase at the 5′ ends of in vitro-transcribed RNAs. RNA 5: 618–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Yan Z, Xu Y, Hu H, Menzel C, Zhou Y, Chen W, Khaitovich P 2009. Sequence features associated with microRNA strand selection in humans and flies. BMC Genomics 10: 413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvágner G, McLachlan J, Pasquinelli AE, Bálint E, Tuschl T, Zamore PD 2001. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293: 834–838 [DOI] [PubMed] [Google Scholar]
- Ivanovska I, Ball AS, Diaz RL, Magnus JF, Kibukawa M, Schelter JM, Kobayashi SV, Lim L, Burchard J, Jackson AL, et al. 2008. MicroRNAs in the miR-106b family regulate p21/CDKN1A and promote cell cycle progression. Mol Cell Biol 28: 2167–2174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Huang Q, Zhang S, Zhang Q, Chang J, Qiu X, Wang E 2010. Hsa-miR-125a-3p and hsa-miR-125a-5p are downregulated in non-small cell lung cancer and have inverse effects on invasion and migration of lung cancer cells. BMC Cancer 10: 318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang P, Liu R, Zheng Y, Liu X, Chang L, Xiong S, Chu Y 2012. MiR-34a inhibits lipopolysaccharide-induced inflammatory response through targeting Notch1 in murine macrophages. Exp Cell Res 318: 1175–1184 [DOI] [PubMed] [Google Scholar]
- Khvorova A, Reynolds A, Jayasena SD 2003. Functional siRNAs and miRNAs exhibit strand bias. Cell 115: 209–216 [DOI] [PubMed] [Google Scholar]
- Kim S, Lee UJ, Kim MN, Lee E-J, Kim JY, Lee MY, Choung S, Kim YJ, Choi Y-C 2008. MicroRNA miR-199a* regulates the MET proto-oncogene and the downstream extracellular signal-regulated kinase 2 (ERK2). J Biol Chem 283: 18158–18166 [DOI] [PubMed] [Google Scholar]
- Koscianska E, Starega-Roslan J, Krzyzosiak WJ 2011. The role of Dicer protein partners in the processing of microRNA precursors. PLoS One 6: e28548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol J, Loedige I, Filipowicz W 2010. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet 11: 597–610 [DOI] [PubMed] [Google Scholar]
- Kuchenbauer F, Mah SM, Heuser M, McPherson A, Rüschmann J, Rouhi A, Berg T, Bullinger L, Argiropoulos B, Morin RD, et al. 2011. Comprehensive analysis of mammalian miRNA* species and their role in myeloid cells. Blood 118: 3350–3358 [DOI] [PubMed] [Google Scholar]
- Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. 2007. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 129: 1401–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau P-W, Guiley KZ, De N, Potter CS, Carragher B, Macrae IJ 2012. The molecular architecture of human Dicer. Nat Struct Mol Biol 19: 436–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HY, Doudna JA 2012. TRBP alters human precursor microRNA processing in vitro. RNA 18: 2012–2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay MA 2008. microRNAs and the immune response. Trends Immunol 29: 343–351 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔC(T) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- López JA, Alvarez-Salas LM 2011. Differential effects of miR-34c-3p and miR-34c-5p on SiHa cells proliferation apoptosis, migration and invasion. Biochem Biophys Res Commun 409: 513–519 [DOI] [PubMed] [Google Scholar]
- Lund E, Güttinger S, Calado A, Dahlberg JE, Kutay U 2004. Nuclear export of microRNA precursors. Science 303: 95–98 [DOI] [PubMed] [Google Scholar]
- Ma E, Zhou K, Kidwell MA, Doudna JA 2012. Coordinated activities of human dicer domains in regulatory RNA processing. J Mol Biol 422: 466–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macrae IJ, Li F, Zhou K, Cande WZ, Doudna JA 2006. Structure of Dicer and mechanistic implications for RNAi. Cold Spring Harb Symp Quant Biol 71: 73–80 [DOI] [PubMed] [Google Scholar]
- Macrae IJ, Ma E, Zhou M, Robinson CV, Doudna JA 2008. In vitro reconstitution of the human RISC-loading complex. Proc Natl Acad Sci 105: 512–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marco A, Macpherson JI, Ronshaugen M, Griffiths-Jones S 2012. MicroRNAs from the same precursor have different targeting properties. Silence 3: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathonnet G, Fabian MR, Svitkin YV, Parsyan A, Huck L, Murata T, Biffo S, Merrick WC, Darzynkiewicz E, Pillai RS, et al. 2007. MicroRNA inhibition of translation initiation in vitro by targeting the cap-binding complex eIF4F. Science 317: 1764–1767 [DOI] [PubMed] [Google Scholar]
- McManus MT, Petersen CP, Haines BB, Chen J, Sharp PA 2002. Gene silencing using micro-RNA designed hairpins. RNA 8: 842–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mestdagh P, Van Vlierberghe P, De Weer A, Muth D, Westermann F, Speleman F, Vandesompele J 2009. A novel and universal method for microRNA RT-qPCR data normalization. Genome Biol 10: R64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan JF, Uhlenbeck OC 1989. Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol 180: 51–62 [DOI] [PubMed] [Google Scholar]
- Montag J, Hitt R, Opitz L, Schulz-Schaeffer WJ, Hunsmann G, Motzkus D 2009. Upregulation of miRNA hsa-miR-342-3p in experimental and idiopathic prion disease. Mol Neurodegener 4: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman MA, Mani V, Hammond SM 2011. Deep sequencing of microRNA precursors reveals extensive 3′ end modification. RNA 17: 1795–1803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederer F, Ospelt C, Brentano F, Hottiger MO, Gay RE, Gay S, Detmar M, Kyburz D 2011a. SIRT1 overexpression in the rheumatoid arthritis synovium contributes to proinflammatory cytokine production and apoptosis resistance. Ann Rheum Dis 70: 1866–1873 [DOI] [PubMed] [Google Scholar]
- Niederer F, Trenkmann M, Ospelt C, Karouzakis E, Neidhart M, Stanczyk J, Kolling C, Gay RE, Detmar M, Gay S, et al. 2011b. Downregulation of microRNA-34a* in rheumatoid arthritis synovial fibroblasts promotes apoptosis resistance. Arthritis Rheum 64: 1771–1779 [DOI] [PubMed] [Google Scholar]
- Okamura K, Balla S, Martin R, Liu N, Lai EC 2008a. Two distinct mechanisms generate endogenous siRNAs from bidirectional transcription in Drosophila melanogaster. Nat Struct Mol Biol 15: 581–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Phillips MD, Tyler DM, Duan H, Chou Y-t, Lai EC 2008b. The regulatory activity of microRNA* species has substantial influence on microRNA and 3′ UTR evolution. Nat Struct Mol Biol 15: 354–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packer AN, Xing Y, Harper SQ, Jones L, Davidson BL 2008. The bifunctional microRNA miR-9/miR-9* regulates REST and CoREST and is downregulated in Huntington's disease. J Neurosci 28: 14341–14346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J-E, Heo I, Tian Y, Simanshu DK, Chang H, Jee D, Patel DJ, Kim VN 2011. Dicer recognizes the 5′ end of RNA for efficient and accurate processing. Nature 475: 201–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltier HJ, Latham GJ 2008. Normalization of microRNA expression levels in quantitative RT-PCR assays: Identification of suitable reference RNA targets in normal and cancerous human solid tissues. RNA 14: 844–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradère U, Brunschweiger A, Gebert LFR, Lucic M, Roos M, Hall J 2013. Chemical synthesis of mono- and bis-labeled pre-microRNAs. Angew Chem Int Ed 10.1002/anie.201304986 [DOI] [PubMed] [Google Scholar]
- Raabe T, Bukrinsky M, Currie RA 1998. Relative contribution of transcription and translation to the induction of tumor necrosis factor-α by lipopolysaccharide. J Biol Chem 273: 974–980 [DOI] [PubMed] [Google Scholar]
- Ro S, Park C, Young D, Sanders KM, Yan W 2007. Tissue-dependent paired expression of miRNAs. Nucleic Acids Res 35: 5944–5953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby JG, Stark A, Johnston WK, Kellis M, Bartel DP, Lai EC 2007. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res 17: 1850–1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai K, Furukawa C, Haraguchi T, Inada K, Shiogama K, Tagawa T, Fujita S, Ueno Y, Ogata A, Ito M, et al. 2011. MicroRNAs miR-199a-5p and -3p target the Brm subunit of SWI/SNF to generate a double-negative feedback loop in a variety of human cancers. Cancer Res 71: 1680–1689 [DOI] [PubMed] [Google Scholar]
- Schwarz DS, Hutvágner G, Du T, Xu Z, Aronin N, Zamore PD 2003. Asymmetry in the assembly of the RNAi enzyme complex. Cell 115: 199–208 [DOI] [PubMed] [Google Scholar]
- Shan SW, Lee DY, Deng Z, Shatseva T, Jeyapalan Z, Du WW, Zhang Y, Xuan JW, Yee S-P, Siragam V, et al. 2009. MicroRNA MiR-17 retards tissue growth and represses fibronectin expression. Nat Cell Biol 11: 1031–1038 [DOI] [PubMed] [Google Scholar]
- Shiba Y, Masuda H, Watanabe N, Ego T, Takagaki K, Ishiyama K, Ohgi T, Yano J 2007. Chemical synthesis of a very long oligoribonucleotide with 2-cyanoethoxymethyl (CEM) as the 2′-O-protecting group: Structural identification and biological activity of a synthetic 110mer precursor-microRNA candidate. Nucleic Acids Res 35: 3287–3296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siolas D, Lerner C, Burchard J, Ge W, Linsley PS, Paddison PJ, Hannon GJ, Cleary MA 2005. Synthetic shRNAs as potent RNAi triggers. Nat Biotechnol 23: 227–231 [DOI] [PubMed] [Google Scholar]
- Starega-Roslan J, Krol J, Koscianska E, Kozlowski P, Szlachcic WJ, Sobczak K, Krzyzosiak WJ 2011. Structural basis of microRNA length variety. Nucleic Acids Res 39: 257–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaram P, Hultine S, Smith LM, Dews M, Fox JL, Biyashev D, Schelter JM, Huang Q, Cleary MA, Volpert OV, et al. 2011. p53-responsive miR-194 inhibits thrombospondin-1 and promotes angiogenesis in colon cancers. Cancer Res 71: 7490–7501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terasawa K, Shimizu K, Tsujimoto G 2011. Synthetic pre-miRNA-based shRNA as potent RNAi triggers. J Nucleic Acids 2011: 131579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang WP, Kwok TT 2009. The miR-18a* microRNA functions as a potential tumor suppressor by targeting on K-Ras. Carcinogenesis 30: 953–959 [DOI] [PubMed] [Google Scholar]
- Uchino K, Takeshita F, Takahashi R-u, Kosaka N, Fujiwara K, Naruoka H, Sonoke S, Yano J, Sasaki H, Nozawa S, et al. 2013. Therapeutic effects of microRNA-582-5p and -3p on the inhibition of bladder cancer progression. Mol Ther 21: 610–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen A, Behlen L, Reynolds A, Wolfson A, Marshall WS, Karpilow J, Khvorova A 2005. The contributions of dsRNA structure to Dicer specificity and efficiency. RNA 11: 674–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh G 2010. Biopharmaceutical benchmarks 2010. Nat Biotechnol 28: 917–924 [DOI] [PubMed] [Google Scholar]
- Wotschofsky Z, Meyer HA, Jung M, Fendler A, Wagner I, Stephan C, Busch J, Erbersdobler A, Disch AC, Mollenkopf HJ, et al. 2011. Reference genes for the relative quantification of microRNAs in renal cell carcinomas and their metastases. Anal Biochem 417: 233–241 [DOI] [PubMed] [Google Scholar]
- Wu H, Ye C, Ramirez D, Manjunath N 2009. Alternative processing of primary microRNA transcripts by Drosha generates 5′ end variation of mature microRNA. PLoS One 4: e7566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakuchi M, Ferlito M, Lowenstein CJ 2008. miR-34a repression of SIRT1 regulates apoptosis. Proc Natl Acad Sci 105: 13421–13426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J-S, Phillips MD, Betel D, Mu P, Ventura A, Siepel AC, Chen KC, Lai EC 2011. Widespread regulatory activity of vertebrate microRNA* species. RNA 17: 312–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Kolb FA, Jaskiewicz L, Westhof E, Filipowicz W 2004. Single processing center models for human Dicer and bacterial RNase III. Cell 118: 57–68 [DOI] [PubMed] [Google Scholar]
- Zhou H, Huang X, Cui H, Luo X, Tang Y, Chen S, Wu L, Shen N 2010. miR-155 and its star-form partner miR-155* cooperatively regulate type I interferon production by human plasmacytoid dendritic cells. Blood 116: 5885–5894 [DOI] [PubMed] [Google Scholar]