The Lsm1-7 ring is required for rapid histone mRNA degradation and binds to the oligo(U) tail added to the 3′ end of histone mRNA. This paper shows that the C-terminal tail of Lsm4 directly interacts with two proteins, SLBP and 3′hExo, bound to the 3′ end of histone mRNA. Mutant Lsm4 proteins that cannot bind the histone mRNP were stably expressed and were incorporated into the Lsm1-7 ring. Cells expressing these proteins do not degrade histone mRNAs rapidly. It is possible that the Lsm1-7 ring may be recruited to a number of mRNA targets by directly interacting with specific mRNPs.
Keywords: Lsm proteins, histone mRNA, mRNA degradation, SLBP, ERI1
Abstract
Metazoan replication-dependent histone mRNAs are the only known eukaryotic mRNAs that lack a poly(A) tail, ending instead in a conserved stem–loop sequence, which is bound to the stem–loop binding protein (SLBP) on the histone mRNP. Histone mRNAs are rapidly degraded when DNA synthesis is inhibited in S phase in mammalian cells. Rapid degradation of histone mRNAs is initiated by oligouridylation of the 3′ end of histone mRNAs and requires the cytoplasmic Lsm1-7 complex, which can bind to the oligo(U) tail. An exonuclease, 3′hExo, forms a ternary complex with SLBP and the stem–loop and is required for the initiation of histone mRNA degradation. The Lsm1-7 complex is also involved in degradation of polyadenylated mRNAs. It binds to the oligo(A) tail remaining after deadenylation, inhibiting translation and recruiting the enzymes required for decapping. Whether the Lsm1-7 complex interacts directly with other components of the mRNP is not known. We report here that the C-terminal extension of Lsm4 interacts directly with the histone mRNP, contacting both SLBP and 3′hExo. Mutants in the C-terminal tail of Lsm4 that prevent SLBP and 3′hExo binding reduce the rate of histone mRNA degradation when DNA synthesis is inhibited.
INTRODUCTION
The final step in mRNA metabolism is degradation of the mRNA, and rapid degradation of mRNA is the most effective way of stopping protein expression. Regulation of mRNA half-life is an important component of regulation of gene expression. The pathways for degrading polyadenylated mRNAs are relatively well understood. The initial step in degradation is deadenylation [shortening the poly(A) tail to ∼10 nt]. The cytoplasmic Lsm complex, Lsm1-7, binds the oligo(A) tail and also recruits the degradation factors, although the molecular details of how Lsm1-7 function are not known. The Lsm1-7 complex is found in a complex with the translation inhibitor Rck/p54 (in yeast dhh1) (Cougot et al. 2004) and also recruits the decapping complex, hDcp1, and hDcp2 (for review, see Parker and Song 2004). Following decapping, the mRNA can be degraded 5′–3′ by the processive exonuclease Xrn1. After deadenylation, the mRNA can also be degraded 3′–5′ by the exosome, a complex of 10–12 polypeptides that contains Rrp44, a processive 3′–5′ exonuclease (Liu et al. 2006; Makino et al. 2013).
For some mRNAs, degradation is initiated by an endonucleolytic cleavage, followed by degradation of the two fragments. In NMD, the Smg6 endonuclease cleaves the mRNA (Huntzinger et al. 2008; Eberle et al. 2009), and Xrn1 degrades the 3′ fragment. The molecular details of degradation of the 5′ fragment are not clear (Franks et al. 2010), but they are dependent on exosome-mediated degradation (Gatfield and Izaurralde 2004; Huntzinger et al. 2008; Eberle et al. 2009). The endonuclease PMR1 has been shown to initiate the degradation of some mRNAs, and the mechanism of degradation of the 5′ fragment is not known (Schoenberg 2011). Finally, in Chlamydomonas reinhardtii and Arabidopsis thaliana, following cleavage of mRNAs by an siRNA/Argonaute protein complex, the 5′ fragment can be oligouridylated or oligoadenylated and could then potentially bind Lsm1-7 (Shen and Goodman 2004; Ibrahim et al. 2006) as part of its degradation pathway. In metazoans, particularly Drosophila, degradation of the 5′ fragment following siRNA-directed cleavage requires the exosome, and depletion of Xrn1 does not alter the half-life of these fragments (Orban and Izaurralde 2005). How the exosome is recruited to the 5′ fragment is not understood.
The Lsm1-7 ring consists of seven polypeptides that form a ring as a result of interactions between their core Sm domains. In addition, there are extensions on both the N and C termini of several Lsm proteins. The Lsm1-7 ring can bind either oligo(A) or oligo(U) tails (Chowdhury et al. 2007; Song and Kiledjian 2007). The specific role(s) of individual Lsm proteins in interacting with factors involved in mRNA degradation and translational inhibition is not known, although mutations in yeast Lsm1 that reduce binding to oligoadenylated mRNA stabilize yeast mRNAs (Chowdhury and Tharun 2008). It is not known whether Lsm1-7 is recruited to specific deadenylated mRNAs by components of the mRNP.
Metazoan replication-dependent histone mRNAs are unique among eukaryotic mRNAs in that they are not spliced and terminate in a conserved 26-nt sequence containing a stem–loop rather than the canonical poly(A) tail (Marzluff et al. 2008). The levels of these mRNAs are tightly coupled with DNA synthesis, as histone proteins are needed to immediately package the newly replicated genome. Precise regulation of new histone protein synthesis is critical for proper chromatin assembly during DNA replication. Following the completion of S phase or the inhibition of DNA synthesis by pharmacological agents, histone mRNAs are rapidly degraded (Heintz et al. 1983; Sittman et al. 1983). The half-life of these messages decreases from 45–60 min to ∼10 min. The stem–loop at the 3′ end of histone mRNA is the cis-element that is responsible for histone mRNA degradation (Pandey and Marzluff 1987), ensuring that all five classes of histone mRNAs are regulated coordinately. The stem–loop is bound by the stem–loop binding protein (SLBP), which participates in all steps in histone mRNA metabolism. It also interacts specifically with a 3′–5′ histone exonuclease, 3′hExo (ERI1), which together with SLBP can form a ternary complex at the 3′ end of histone mRNA (Dominski et al. 2003; Yang et al. 2006; Tan et al. 2013).
During the past several years, we have defined several features of histone mRNA degradation. Histone mRNA degradation requires that the histone mRNA be actively translated and requires the nonsense-mediated decay (NMD) factor Upf1, suggesting that translation termination may become inefficient as part of triggering initiation of histone mRNA degradation (Kaygun and Marzluff 2005a,b). The initial biochemical step in degradation is covalent alteration of the histone mRNA by addition of an oligo(U) tail to the 3′ end (Mullen and Marzluff 2008; Su et al. 2013). Knockdown of Lsm1 inhibits degradation of histone mRNA consistent with Lsm1-7 being recruited to the oligo(U) tail. Knockdown of either component of the exosome or the decapping enzyme each partially stabilized histone mRNA, suggesting that both 5′–3′ and 3′–5′ degradation pathways are involved in degrading histone mRNA. Consistent with this observation, molecules that had been decapped and partially degraded from both the 5′ end and 3′ end were detected by circular RT-PCR (Mullen and Marzluff 2008).
Since histone mRNA degradation requires that the histone mRNA be translated (Graves et al. 1987; Kaygun and Marzluff 2005b), it is likely that SLBP is associated with the histone mRNP when degradation is initiated. Consistent with this possibility, we detected the association of SLBP with both Upf1 and Lsm1 after treatment of cells with inhibitors of DNA replication (Kaygun and Marzluff 2005a; Mullen and Marzluff 2008), suggesting that the oligo(U) tail is added to the histone mRNA while it is still associated with polyribosomes. 3′hExo, which specifically binds the stem–loop and forms a stable ternary complex with the 3′ end of histone mRNA (Yang et al. 2006; Tan et al. 2013), is also required for rapid degradation of histone mRNA (Hoefig et al. 2013).
The molecular details of how histone mRNA is degraded are not known. In particular, we do not understand how the degradation factors are recruited to histone mRNA. The Lsm1-7 ring is required for degradation of both histone mRNA and polyadenylated mRNA. Given the many targets of Lsm 1-7 and the widely varying half-lives of individual mRNAs, it is not known whether Lsm1-7 simply recognizes the oligo(A) or oligo(U) tail on mRNA or whether binding to the tail involves other components of the mRNP. It is likely that the amount of Lsm1-7 may be limiting relative to its potential target mRNAs and that efficient mRNA metabolism may require additional sequence elements other than simply binding to the oligo(A) or oligo(U) homopolymer. In particular, for the rapid degradation of histone mRNAs when DNA replication is inhibited, efficient recruitment of Lsm1-7 to the target message is essential. Here we demonstrate that the C-terminal tail of Lsm4 interacts directly with both SLBP and 3′hExo and show that this interaction is necessary for rapid histone mRNA degradation. We propose that this interaction is an example of a more general mechanism that may function to help target Lsm1-7 to specific sets of mRNAs.
RESULTS
SLBP and Lsm1-7 bind the oligouridylated histone stem–loop
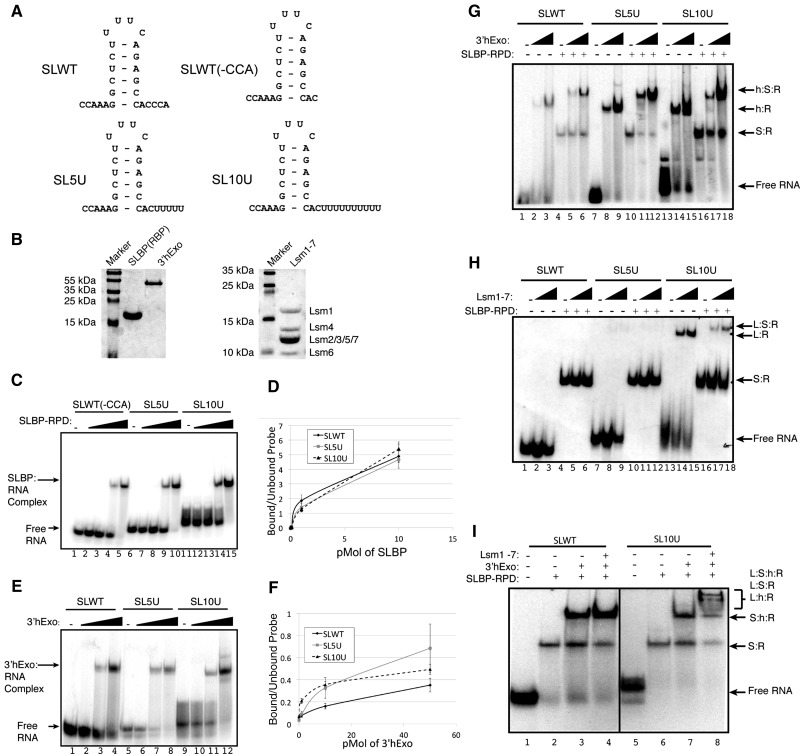
In mammals, the initial cleavage of histone pre-mRNA in the nucleus occurs 5 nt after the stem–loop (Scharl and Steitz 1994), and SLBP remains bound to the processed RNA and participates in export to the cytoplasm (Sullivan et al. 2009). The cytoplasmic histone mRNA ends 2–3 nt after the stem–loop as a result of exonucleolytic trimming by the 3′hExo (Hoefig et al. 2013), which forms a stable ternary complex (SLBP:SL:3′hExo) with SLBP and the stem–loop (Yang et al. 2006; Tan et al. 2013). The initial intermediate in histone mRNA degradation is oligouridylation of the 3′ end of histone mRNA (Su et al. 2013). Much of the histone mRNA is degraded 3′–5, since knockdown of the exosome has a much greater effect on histone mRNA degradation than knockdown of Dcp2 or Xrn1 (Mullen and Marzluff 2008). Removal of SLBP and possibly also 3′hExo is likely required for 3′–5′ degradation of histone mRNA past the stem–loop. Once the integrity of the stem–loop is destroyed, 3′hExo has a greatly reduced affinity for the RNA (Yang et al. 2006). We asked whether SLBP and/or 3′hExo can interact with the oligouridylated RNA and whether Lsm1-7 can bind at the same time as SLBP, 3′hExo, or both. To determine whether oligouridylation affects the ability of SLBP or 3′hExo to bind the histone stem–loop, we prepared stem–loop constructs that mimic the cytoplasmic histone mRNA ending in 2 nt after the stem–loop followed by zero, five, or 10 uridines [SLWT(-CCA), SL5U, SL10U] (Fig. 1A). The SLBP RNA processing domain (RPD, amino acids 127–220) was used in these experiments to facilitate resolution of the SLBP:SL complex from the 3′hExo:SL complex. The proteins used in these experiments are shown in Figure 1B. Mobility shift experiments showed that baculovirus expressed SLBP-RPD bound to all three probes, and the presence of uridyl groups at the 3′ end of the stem–loop did not alter the affinity of SLBP-RPD for the stem–loop structure (Fig. 1C,D). Thus, uridylation does not disrupt or weaken the SLBP:SL interaction. We also tested whether 3′hExo would bind the uridylated stem–loops, since previous experiments suggested that 3′hExo did not bind histone pre-mRNA (Dominski et al. 2003). Surprisingly, uridylation did not decrease the affinity of 3′hExo for the stem–loop (Fig. 1E,F) consistent with the finding that 3′hExo is required for initial degradation of histone mRNAs and may be responsible for degrading the oligo(U) tail (Hoefig et al. 2013).
FIGURE 1.
Binding of SLBP, 3′hExo, and Lsm1-7 to the histone stem–loop. (A) Stem–loop constructs used in mobility shift experiments. (B) The purified proteins used in these experiments were analyzed by SDS-gel electrophoresis. The purified His-SLBP-RPD and His-3′hExo were resolved on a 10% SDS–polyacrylamide gel (left), and the Lsm1-7 proteins were resolved on a 15% SDS–polyacrylamide gel (right). (C) Electrophoretic mobility shift assays (EMSA) were carried out using increasing amounts (0.01, 0.1, 1, and 10 pmol) of recombinant SLBP-RPD with 10 fmol of the uniformly labeled histone stem–loop probes. Lanes labeled (–) had no added protein. Complexes were analyzed on an 8% polyacrylamide gel. (D) The data in panel B were quantified as the percent of probe in complex versus unbound probe: (circle) SLWT(-CCA), (gray square) SL5U, (triangle) SL10U (n = 3). (E) EMSAs using recombinant 3′hExo under the same conditions except tRNA were omitted. Each reaction was done with 10 fmol of the indicated uniformly labeled histone stem–loop probe and increasing amounts (0.1, 1.0, and 10 pmol) of 3′hExo. (F) The data were quantified as in panel D: (circle) SLWT(-CCA), (gray square) SL5U, (triangle) SL10U (n = 3). (G) EMSAs with 3′hExo, SLBP RPD, or mixtures of the two proteins. Increasing amounts (1, 10 pmol) of 3′hExo were incubated with (lanes 5,6,11,12,17,18) or without (lanes 2,3,8,9,15,16) 1 pmol of SLBP-RPD as indicated above each lane. The probes used were SLWT (lanes 1–6), SL5U (lanes 7–12), or SL10U (lanes 13–18). Complexes were analyzed on a 6% polyacrylamide gel. The monomer and heterodimer complexes formed are indicated. Complex components are indicated by h (3′hExo), S (SLBP), or R (RNA). (H) The indicated probes were incubated without (lanes 1–3,7–9,13–15) or with (lanes 4–6,10–12,16–18) 1 pmol of SLBP-RPD and either 24 pmol (lanes 2,5,8,11,14,17) or 48 pmol (lanes 3,6,9,12,15,18) of Lsm1-7. Complexes were analyzed on a 6% polyacrylamide gel. Complexes components are indicated by L (Lsm1-7), S (SLBP), or R (RNA). (I) The SLWT (lanes 1–4) or SL10U probe (lanes 5–8) were incubated with 1 pmol of SLBP-RPD (lanes 2,6); SLBP and 10 pmol of 3′hExo (lanes 3,7); or SLBP, 3′hExo, and 48 pmol of Lsm1-7 (lanes 4,8). Complexes were analyzed on a 6% polyacrylamide gel. Complex components are indicated by L (Lsm1-7), S (SLBP), h (3′hExo), or R (RNA).
To determine whether the triple complex consisting of SLBP, stem–loop RNA, and 3′hExo (SLBP:SL:3′hExo) also forms with the same efficiency regardless of whether or not the 3′ end is uridylated, we incubated the radiolabeled stem–loop (Fig. 1G, lanes 1–6), the stem–loop with five uridines (lanes 7–12), or with 10 uridines added (lanes 13–18) with increasing amounts of 3′hExo. SLBP-RPD was added in a constant amount in lanes 4–6, 10–12, and 16–18. 3′hExo bound to each of the probes with similar affinity, and the addition of the SLBP-RPD resulted in efficient formation of a triple complex with all three probes. Thus, uridylation does not dramatically alter the affinity of SLBP, 3′hExo, or the triple complex for the histone stem–loop.
Lsm1-7 has a similar affinity for both oligo(A) and oligo(U) tails at the 3′ end of RNAs (Chowdhury et al. 2007; Song and Kiledjian 2007). Since the Lsm1-7 complex is required for histone mRNA degradation after inhibition of DNA synthesis (Mullen and Marzluff 2008), presumably by binding the oligouridylated RNA, we tested whether the presence of SLBP and/or 3′hExo affected the binding of Lsm1-7. To determine the conditions under which Lsm1-7 could bind the histone stem–loop, we performed mobility shift experiments with recombinant Lsm1-7 and the SLBP-RPD. Lsm1-7 was not able to bind the SLWT (Fig. 1H, lanes 2, 3) and had weak affinity for SL5U (Fig. 1H, lanes 8, 9), but formed a stable complex with the SL10U RNA (Fig. 1H, lanes 14, 15). When both Lsm1-7 and SLBP were added to the SL10U RNA, formation of a ternary complex was readily detected (Fig. 1H, lanes 17, 18).
We then determined whether a quaternary complex containing SLBP-RPD:SL:3′hExo:Lsm1-7 could form in vitro on uridylated histone mRNA. Using the mobility shift assay, we were able to detect higher mobility complexes forming on the SL10U RNA (Fig. 1I, lane 8), but not on the SLWT RNA (Fig. 1I, lane 4). These complexes result from Lsm1-7 interacting with the ternary SLBP:3′hExo:stem–loop complex; however, an unambiguous determination of components of the individual complexes was not possible. These results show that oligouridylation likely also does not disrupt the SLBP:3′hExo:histone mRNA complex. Note that these experiments are done in the presence of EDTA where 3′hExo is not active.
Lsm4 interacts directly with the RNA binding domain of SLBP
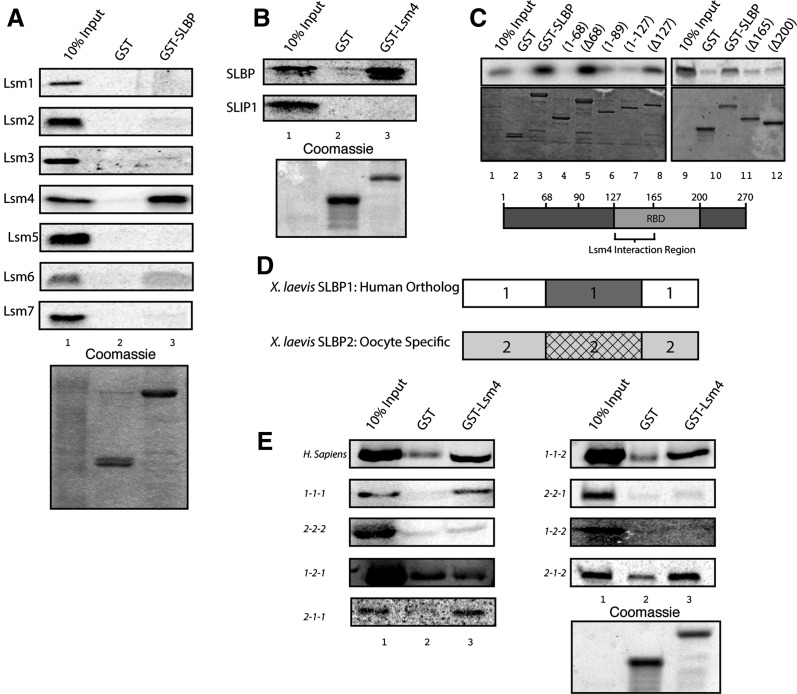
To determine whether any of the individual Lsm1-7 proteins interact directly with SLBP, we cloned each member of the ring, labeled them with 35S-methionine by in vitro translation, and tested their ability to interact with GST-SLBP expressed in baculovirus. Of the seven Lsm proteins, Lsm4 interacted strongly with recombinant full-length GST-SLBP in vitro, while Lsm6 showed low levels of binding (Fig. 2A). As Lsm4 showed the most robust binding, we focused only on it for this study. We also tested the ability of the seven Lsm proteins to interact with GST-SLIP1, a component of the cytoplasmic histone mRNP that is involved in translation (Cakmakci et al. 2008). None of the subunits of the Lsm1-7 complex interacted directly with SLIP1 (Supplemental Data).
FIGURE 2.
Lsm4 interacts with the RNA binding domain of SLBP. (A) Five micrograms of recombinant GST or GST-SLBP was bound to glutathione Sepharose resin for 2 h at 4°C followed by incubation with 8 μL of the indicated 35S-methionine-labeled Lsm protein in reticulocyte lysate in a 100-μL total volume for 2 h at 4°C. Glutathione beads were washed with TEN100 buffer, and the proteins were eluted by boiling in SDS, resolved on a 12% SDS-PAGE gel, dried and visualized on a PhosphorImager. (B) SLBP (top) or SLIP1 (bottom) labeled with 35S-methionine by in vitro translation was incubated with GST or GST-Lsm4 and analyzed as in panel A. (C) 35S-methionine-labeled Lsm4 was incubated with the indicated recombinant GST-SLBP N- and C-terminal truncations, and the bound proteins were analyzed as in panel A. (Top) PhosphorImage of bound Lsm4. (Bottom) Coomassie-stained gel. (D) The diagram of the xSLBP1 and xSLBP2 proteins, with the N-terminal domain, the RNA binding domain, or the C-terminal domains indicated. (E) All possible combinations of these three domains were created (Sanchez and Marzluff 2002), and the six chimeric proteins were tested for their ability to interact GST-Lsm4. The indicated chimeric xSLBP proteins were labeled with 35S-methionine and incubated with GST or GST-Lsm4, and the bound proteins were analyzed as in panel A.
To confirm these interactions, we purified recombinant GST-Lsm4 from Escherichia coli and tested its interaction with SLBP and SLIP1 labeled with 35S-methionine by in vitro translation in rabbit reticulocyte lysate. These experiments confirmed that Lsm4 interacted in vitro with SLBP and not with SLIP1 (Fig. 2B). To determine the region of SLBP that interacts with Lsm4, we purified both N- and C-terminal truncations of SLBP fused to GST and tested the interaction by GST pull-down with full-length Lsm4. N-terminal fragments 1–68, 1–89, and 1–127 did not interact with Lsm4 (Fig. 2C, lanes 4,6,8), while the reciprocal fragments Δ68, Δ91, and Δ127 bound Lsm4 (Fig. 2C, lanes 3,5,7). Deleting either 165 (Δ165) or 200 amino acids (Δ200) from the N terminus abolished Lsm4 binding (Fig. 2C, lanes 10,11). C-terminal truncations extending to amino acid 165 interacted with Lsm4. Since the RNA binding domain (RBD) extends from amino acids 127–199, these data suggest that the RNA binding domain of SLBP interacts with Lsm4 and that the N-terminal portion of the RBD contains amino acids (between amino acids 127 and 165) that are necessary for binding to Lsm4.
To determine whether the RBD is the only essential region for binding and to further identify amino acids in SLBP required for binding to Lsm4, we took advantage of the fact that Xenopus contains two SLBPs: xSLBP1, the ortholog of mammalian SLBP, and xSLBP2, which functions to translationally silence histone mRNA during oogenesis (Wang et al. 1999; Sanchez and Marzluff 2002). We used chimeric SLBP proteins constructed from xSLBP1 and xSLBP2, which contain all possible combinations of the domains of the two SLBPs (Fig. 2D). The chimeric proteins have been described previously and were used to determine the region of xSLBP1 required for histone mRNA translation (Sanchez and Marzluff 2002). xSLBP2 has the same overall organization as xSLBP1, with a central RNA binding domain (domain 2), which is 62% identical (80% similar) to the xSLBP1 binding domain but has no similarity in the N-terminal (domain 1) or C-terminal (domain 2) domains of the protein. Chimeric proteins are referred to by which domains they contain. For example, 1-2-1 contains the N- and C-terminal domains of xSLBP1 and the RNA binding domain of xSLBP2. Using the six constructs, we found that the three constructs that contained the RNA binding domain of xSLBP1 (1-1-2, 2-1-1, 2-1-2) retained binding to Lsm4. In contrast, the three proteins that contained the RNA binding domain of xSLBP2 (1-2-1, 2-2-1, 1-2-2) did not bind Lsm4 (Fig. 2E). This result demonstrates that SLBP binds Lsm4 with high specificity and confirmed that the amino acids required for binding are found within the RNA binding domain and must include the small number of residues that differ in this domain between xSLBP1 and xSLBP2.
Amino acids in the unstructured C-terminal tail of Lsm4 are required for binding to SLBP
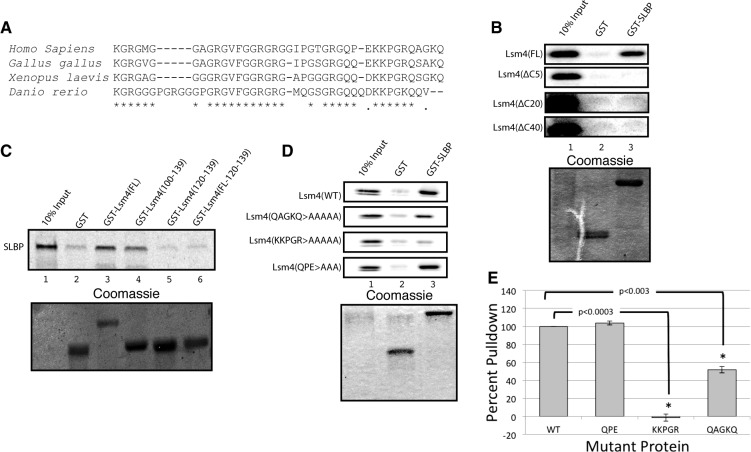
The first 100 amino acids of Lsm4 contain the Sm fold, and the C-terminal 40 amino acids of Lsm4 are predicted to be unstructured. This region contains several of GRG repeats that are arginine methylation sites (Fig. 3A; Brahms et al. 2001). The entire Lsm4 protein, including the C-terminal domain, is highly conserved in vertebrates (Fig. 3A). We expressed full-length Lsm4 and a truncation of Lsm4 with the C-terminal unstructured tail deleted (Lsm4 ΔC40) in vitro, labeled with 35S-methionine, and tested their ability to interact with GST-SLBP. Full-length Lsm4 interacted strongly with GST-SLBP (Fig. 3B, top), but deletions of as few as five amino acids from the C-terminal end of Lsm4 no longer interacted with SLBP, demonstrating that the C-terminal tail is necessary for binding. We then expressed a protein in which just the C-terminal 40-amino-acid tail of Lsm4 was fused to GST, GST-Lsm4 (100–139), and this protein bound SLBP with similar affinity as the full-length GST-Lsm4 (Fig. 3C, lane 4). These data demonstrated that the C-terminal extension of Lsm4 is necessary and sufficient for binding to SLBP.
FIGURE 3.
The C-terminal extension of Lsm4 is required for binding to SLBP. (A) Comparison of the sequence of the C terminus of Lsm4 from several vertebrates. (B) Full-length Lsm4 or Lsm4 with the indicated deletions of the C terminus were labeled with 35S-Met in reticulocyte lysate and tested for binding with recombinant GST-SLBP or GST as in Figure 2A. The stained gel is shown below the autoradiogram. (C) SLBP labeled with 35S-methionine was incubated with GST fused with full-length length Lsm4, the final 40 amino acids of Lsm4 [GST-Lsm4(100–139)], the final 20 amino acids of Lsm4 [GST-Lsm4(120–139)], or GST fused to a 20-amino-acid random flexible sequence replacing amino acids 100–119 of Lsm4 followed by amino acids 120–139 of Lsm4. (D) The indicated point mutations were introduced to the C-terminal tail of Lsm4, the proteins labeled with 35S-Met in rabbit reticulocyte lysate, and then incubated with GST or GST-SLBP. (E) The results from three independent experiments were quantified using ImageQuant. The amount of binding to WT-Lsm4 was normalized to 100, and statistical significance was determined by a Student's t-test.
To further define the binding site for Lsm4, we expressed and purified GST-Lsm4(120–139), which contained only the final 20 amino acids of Lsm4 fused to GST (Fig. 3C, lane 5). This protein did not interact with SLBP. The failure to interact could be attributed to the terminal amino acids not being sufficient for binding or the close juxtaposition of the tail to the GST protein. Therefore, we expressed and purified GST-Lsm4(FL19/120-139), which contained only the last 20 amino acids of Lsm4, but separated them from the globular GST with a 19-amino-acid flexible linker. This protein also did not bind SLBP (Fig. 3C, lane 6).
To more precisely determine which amino acids in the Lsm4 tail are required for binding, we made point mutations in the last 13 amino acids to define which were the critical amino acids for binding. Changing the final five amino acids to alanines (QAGKQ > AAAAA) reduced binding by 50%, while mutation of the penultimate five amino acids completely abolished binding to SLBP (KKPGR > AAAAA) (Fig. 3D,E). However, changing the three amino acids N-terminal of KKPGR (QPE > AAA) had no effect on binding to SLBP. These data show that a distinct set of amino acids located at the most C-terminal region of Lsm4 are required for binding to SLBP, but that amino acids between 100 and 119 were also required for binding. Note that this region of Lsm4 is conserved in all vertebrates (Fig. 3A).
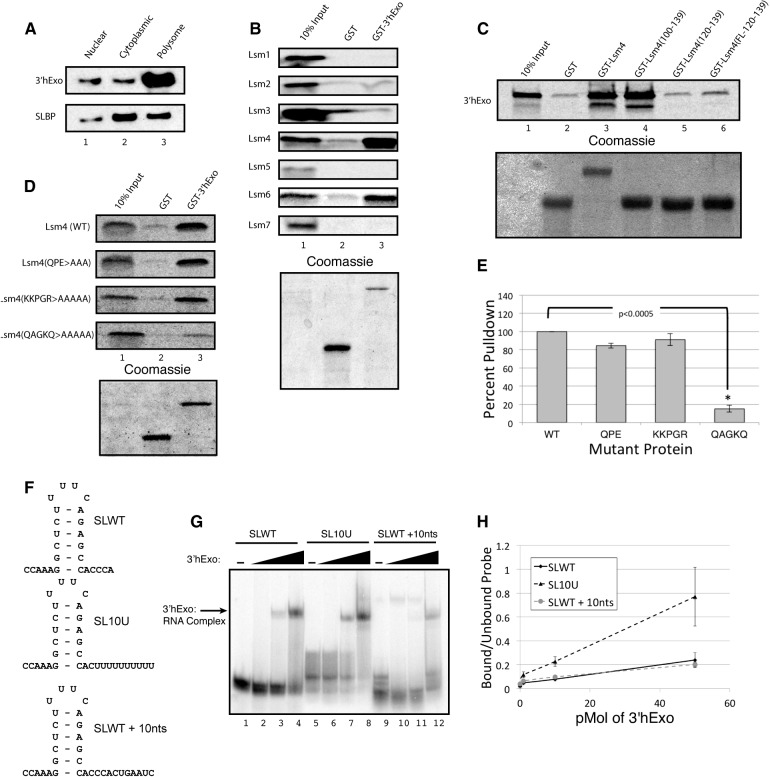
3′hExo interacts with Lsm4 and Lsm6
In mouse cells that have 3′hExo deleted, histone mRNAs are degraded more slowly when DNA replication is inhibited, demonstrating that the 3′hExo also plays an important role in histone mRNA degradation (Hoefig et al. 2013). 3′hExo is predominantly found on polysomes (Fig. 4A; Ansel et al. 2008), consistent with a role in mRNA metabolism. Since 3′hExo is able to bind the uridylated stem–loop (Fig. 1D), we investigated the possibility that the Lsm1-7 ring may also form direct interactions with 3′hExo. We tested the ability of full-length his-tagged GST-3′hExo expressed in E. coli to interact with all seven members of the Lsm1-7 ring that were expressed in rabbit reticulocyte lysate in the presence of 35S-methionine. We found that, similar to SLBP, GST-3′hExo interacts with Lsm4, and in addition, there is also a strong interaction in vitro with Lsm6 (Fig. 4B). Using the same deletions and mutations we used to characterize the SLBP:Lsm4 interaction, we determined the regions of Lsm4 required to bind to 3′hExo. The 3′hExo bound to the last 40 amino acids of Lsm4 as well as it bound the full-length proteins (Fig. 4C, lanes 3, 4). The 3′hExo, like SLBP, did not bind to the last 20 amino acids fused to GST or to the last 20 amino acids fused to GST through a 19-amino-acid flexible linker, indicating that, like SLBP, amino acids 100–119 were also necessary for binding to 3′hExo (Fig. 4C, lane 6).
FIGURE 4.
3′hExo directly interacts with the C-terminal tail of Lsm4. (A) Exponentially growing Jurkat cells were separated into nuclear, free cytoplasmic, and polysomal fractions. Equal cell equivalents of each fraction were analyzed by Western blotting for 3′hExo and SLBP. (B) The Lsm proteins were each labeled with 35S-methionine and incubated with recombinant GST or GST-3′hExo and analyzed as in Figure 2A. (C) The 3′hExo was labeled with 35S-methionine and incubated with the GST proteins fused to the different Lsm4 C-terminal fragments used in Figure 3C. The bound proteins were analyzed as in Figure 2A. (D,E) Full-length Lsm4 and the three mutants in the C-terminal amino acids described in Figure 3D were labeled with 35S-methionine and incubated with GST-3′hExo. The bound proteins were analyzed as in Figure 2A. (E) The results of three independent experiments were quantified. (F) The probes used for mobility shift assays in panels G and H are shown. (G,H) Increasing amounts of 3′hExo (0.1, 1, and 10 pmol) were incubated with 10 fmol of each probe. Complexes were analyzed on a 6% polyacrylamide gel. (H) The results are quantified.
Deletion of the last 10 amino acids of Lsm4 abolished binding of 3′hExo (data not shown) and also abolished binding of SLBP. We tested the three point mutants in the last 13 amino acids we used to define SLBP binding in Figure 3 and found that there are distinct regions required for 3′hExo and SLBP binding (Fig. 4D). Mutation of the last five amino acids, QAGKQ, to alanines abolished 3′hExo binding but only reduced SLBP binding (Fig. 3D), while mutations of the adjacent five amino acids, KKPGR, to alanines had no effect on 3′hExo binding (Fig. 4D) but abolished binding of SLBP (Fig. 3D). The mutation of amino acids 127–129 (QPE) to alanines did not affect either 3′hExo (Fig. 4D) or SLBP binding (Fig. 3D). Although the last 10 amino acids at the 3′ end of Lsm4 were essential for 3′hExo and SLBP binding, the last 20 amino acids of the Lsm4 tail are not sufficient for binding either protein since inserting a different amino acid sequence in the first 20 amino acids after the Sm domain but keeping the C terminus intact also abolished binding of both proteins. Each protein requires a distinct element in the C-terminal 10 amino acids for binding. Thus, it is likely that the C-terminal end of Lsm4 interacts with the ternary complex at the 3′ end of histone mRNA, contacting both SLBP and 3′hExo.
We showed above that 3′hExo interacts with the oligouridylated 3′ end of histone mRNA (Fig. 1D), which also binds Lsm1-7. 3′hExo has weak affinity for the histone pre-mRNA containing 36 nt downstream from the cleavage site (Dominski et al. 2003). We compared the affinity of 3′hExo for a stem–loop containing 10 random nucleotides following the stem–loop to the uridylated stem–loops terminating 10 nt downstream from that canonical 3′ cleavage site (SLWT + 10 nt) (Fig. 1A). The results show that the 3′hExo can interact with either RNA, but that it has a higher affinity for the oligouridylated RNA, suggesting that the 3′hExo might interact directly with the oligo(U) tail, consistent with the data presented in Figure 1, E and F. It is likely that 3′hExo is bound together with SLBP and Lsm1-7 at the 3′ end of oligouridylated histone mRNA when degradation of histone mRNA is initiated.
Mutants in the C-terminal tail of Lsm4 assemble into the Lsm1-7 complex
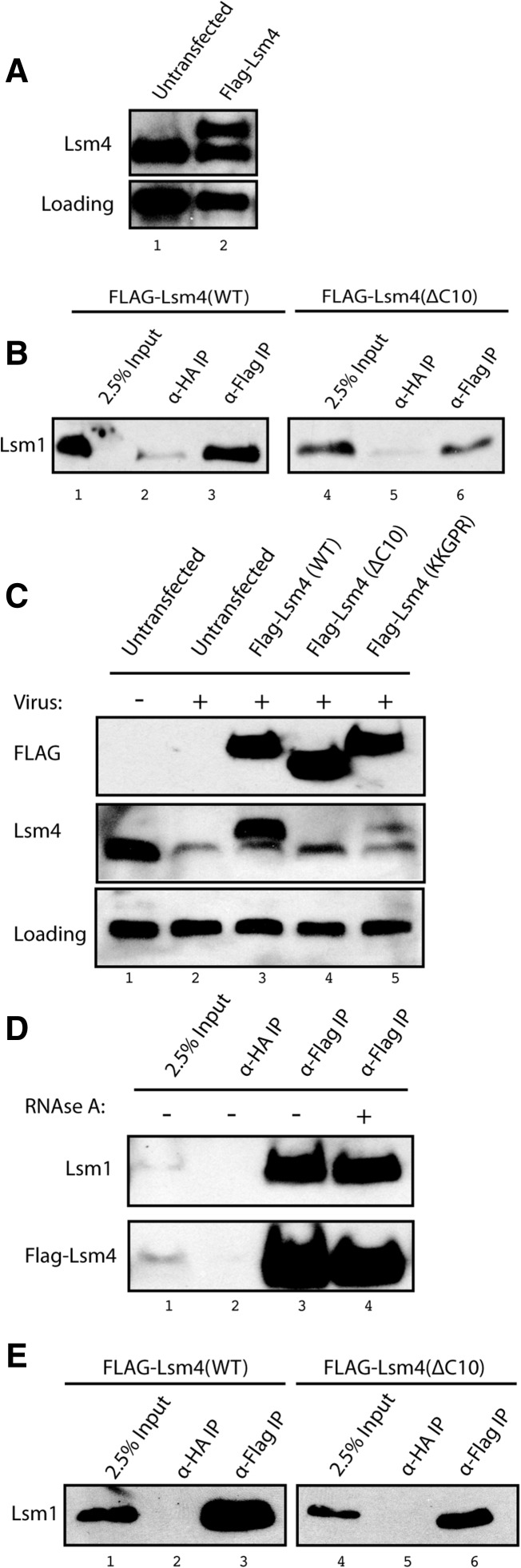
To investigate whether the SLBP:Lsm4 interaction is required for efficient degradation of histone mRNA in vivo, we first had to create a tagged mutant Lsm4 that would be incorporated into the Lsm1-7 complex. Of several tags tested on wild-type Lsm4, only the Lsm4 construct with a single Flag-tag at the N terminus was incorporated into the Lsm1-7 ring. To determine whether the tagged proteins were assembled into the Lsm1-7 ring, we immunoprecipitated the tagged-Lsm4 and assayed the immunoprecipitates for the presence of Lsm1 by Western blotting. The Flag-Lsm4 constructs, which were wild type Flag-Lsm4(WT) or had the final 10 amino acids truncated [Flag-Lsm4(ΔC10)], were incorporated into the Lsm1-7 ring when overexpressed in transient transfections (data not shown).
Based on these results, we constructed stable HeLa cell lines expressing Flag-Lsm4 (Fig. 5A, lanes 1, 2) and Flag-Lsm4(ΔC10) and the Flag-Lsm4(KKGPR) mutant. The stable cell line expressed similar amounts of Flag-tagged Lsm4 as endogenous Lsm4 (Fig. 5A). We determined how well the Flag-Lsm4 was incorporated into the Lsm1-7 rings by immunoprecipitation with anti-Flag and Western blotting for Lsm1. In the stable cell lines, only a small fraction (<3%) of the Lsm1-7 complexes contained Flag-Lsm4 (Fig. 5B, lanes 1–3) or the Flag-Lsm4(ΔC10) mutant (Fig. 5B, lanes 4–6). Thus the Flag-tagged proteins did not compete effectively with the endogenous Lsm4 for incorporation into the Lsm1-7 ring.
FIGURE 5.
Flag-Lsm4 proteins are incorporated into the Lsm1-7 complex. (A) HeLa cells were transfected with Flag-Lsm4 and stable cells lines selected with G418. Proteins were resolved by SDS-gel electrophoresis and analyzed by Western blotting with anti-Lsm4. (Bottom panel) A loading control (cross-reacting band with anti-Lsm4). (B) Equal amounts of lysate from cells stably transfected with Flag-Lsm4 (lanes 1–3) or the Flag-Lsm(ΔC10) (lanes 4–6) were incubated overnight with 1.0 μg of either anti-Flag or anti-HA antibody, and the immunoprecipitates were collected and analyzed by Western blotting for Lsm1. (Lanes 1,4) 2.5% of the input was analyzed. (C) The same cells were treated with viruses encoding an shRNA targeting the Lsm4 3′ UTR and stably selected with puromycin. Ten micrograms of total cell protein from lysates of untransfected cells and the stably transfected cells expressing Flag-Lsm4(WT), Flag-Lsm4(ΔC10), or Flag-Lsm4(KKPGR) was resolved by SDS-gel electrophoresis and analyzed by Western blotting with anti-Flag (top) and anti-Lsm4 (middle). The Flag-tagged Lsm4(ΔC10) protein comigrates with Lsm4 (lane 4) and reacts poorly, if at all, with the Lsm4 antibody. The Lsm4(KKPGR) mutant also reacts weakly with the Lsm4 antibody (lane 5). (D) Lysates from cells transfected with Flag-Lsm4(WT) and treated with shRNA against endogenous Lsm4 were immunoprecipitated with anti-Flag, and the immunoprecipitates were probed with anti-Lsm1 (top) and anti-Lsm4 (bottom). The lysates were analyzed without (lane 2) or with (lane 3) treatment with RNase. (E) Lysates from cells transfected with Flag-wild-type Lsm4 (lanes 1–3) or Flag-Lsm4(ΔC10) (lanes 4–6) and with endogenous Lsm4 knocked down were incubated with anti-HA (lanes 2,5), anti-Flag (lanes 3,6) as in panel B, and the immunoprecipitates were probed with anti-Lsm1. (Lanes 1,4) 2.5% of the input lysate was analyzed.
To increase the percentage of Lsm1-7 complexes containing Flag-Lsm4, we knocked down the endogenous Lsm4 with a lentivirus expressing an shRNA directed against the 3′ UTR of the Lsm4 mRNA (Fig. 5C). The knockdown of the endogenous Lsm4 was efficient (Fig. 5C, lane 2), and these cells grew very slowly compared with cells with Lsm1 knocked down and died between 7 and 14 d post-infection. The cells expressing the Flag-tagged wild-type and mutant Lsm4 proteins survived and continued to proliferate. The shRNA treatment knocks down both Lsm1-7 and Lsm2-8 complexes, and knockdown of the Lsm2-8 complex, required for splicing, has a more deleterious effect on the cell. Thus, the mutant Lsm4 proteins support the essential function of Lsm4, which likely includes formation of a functional Lsm2-8 ring in the nucleus. In the knockdown cells, the Flag-Lsm4 is ∼90% of the total Lsm4 protein in the cell (Fig. 5C, lane 3). We could not detect the tagged mutant proteins effectively using the Lsm4 antibody, which did not detect proteins with deletion or mutation of the last 10 amino acids efficiently (Fig. 5C, lanes 4, 5). Note that the Lsm4(ΔC10) protein migrates identically with the endogenous Lsm4, also preventing unambiguous detection of the Flag-tagged protein with the Lsm4 antibody (Fig. 5C, lane 4). However, the Western signal for the Flag-tagged proteins was similar in the three cell lines (Fig. 5C, top panel), indicating that there were similar expression levels in all three cell lines.
Knockdown of endogenous Lsm4 in the cells expressing Flag-Lsm4 resulted in a dramatic increase in the fraction of Lsm1-7 complexes that contained the Flag-Lsm4 (cf. Fig. 5D and 5E). Flag-Lsm4 was efficiently precipitated with anti-Flag antibody (Fig. 5D, lanes 3, 4), and a large fraction of the Lsm1 protein was coimmunoprecipitated with the Flag-Lsm4; this association was insensitive to RNase A (Fig. 5D, lanes 3, 4) and did not affect the ability to coprecipitate with endogenous Lsm1 (Fig. 5D, top panel, lanes 3, 4). The mutant Flag-Lsm4(ΔC10) protein was also incorporated into a large fraction of the Lsm1-7 complexes after knockdown of endogenous Lsm4 (Fig. 5E). Since the knockdown is not complete, there will be a percentage of wild-type Lsm1-7 rings in the cells. Thus, we were able to create a healthy cell line expressing a mutant Lsm4 that is incorporated into a substantial fraction of the Lsm1-7 (and presumably also Lsm 2-8) rings, allowing us to determine the effect of this mutation on histone mRNA degradation.
Interaction of SLBP and Lsm4 is required for efficient histone mRNA degradation
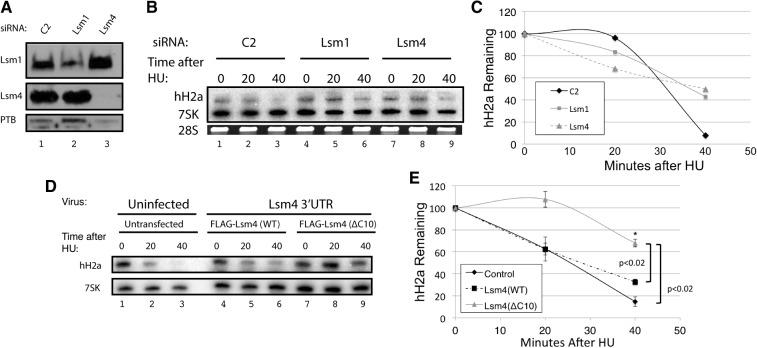
Histone mRNAs are rapidly degraded after treatment of exponentially growing HeLa cells with 5 mM hydroxyurea (HU) to inhibit DNA replication. When factors involved in histone mRNA degradation (e.g., Upf1, Lsm 1, exosome subunits) are knocked down, the degradation of histone mRNA is slowed although the histone mRNA is ultimately degraded (Kaygun and Marzluff 2005a; Mullen and Marzluff 2008), since the knockdowns are never complete and there is bidirectional decay of histone mRNAs. Total cell RNA was prepared 0, 20, and 40 min after addition of HU. We knocked down either Lsm1 or Lsm4 in HeLa cells using siRNAs (Fig. 6A). Each knockdown resulted in a decrease in histone mRNA degradation (Fig. 6B,C). We carried out a similar analysis in cells expressing the Flag-Lsm4(WT) and Flag-Lsm4(ΔC10), which had the endogenous Lsm4 knocked down. In cells depleted of Lsm4 that were expressing the Flag-Lsm4(WT), the histone mRNAs were degraded similarly to histone mRNAs in the untransfected cells, indicating that the Flag-Lsm4(WT) functioned normally in histone mRNA degradation. In contrast, in cells depleted of Lsm4 that were expressing Flag-Lsm4(ΔC10), histone mRNAs were degraded more slowly, similar to the degradation in the cells with Lsm1 or Lsm4 knocked down, indicating that the mutant Lsm4(ΔC10) did not function normally in histone mRNA degradation (Fig. 6D,E). Since Lsm4(ΔC10) does not bind either SLBP or 3′hExo, these data support the conclusion that the interaction between Lsm4 and the complex at the 3′ end of histone mRNA is necessary for efficient histone degradation.
FIGURE 6.
The interaction of Lsm4 with SLBP and 3′hExo is necessary for degradation of histone mRNA after inhibiting DNA replication. (A) HeLa cells were treated with a control siRNA (lane 1), an siRNA targeting Lsm1 (lane 2), and an siRNA targeting Lsm4 (lane 3), as previously described (Mullen and Marzluff 2008). Total cell lysates were prepared and analyzed by Western blotting for Lsm1 (top), Lsm4 (middle), and polypyrimidine track binding protein (PTB, bottom). (B,C) The cells in panel A (exponentially growing) were treated with 5 mM HU, and RNA was harvested before HU treatment and 20 and 40 min after HU treatment (HU). Total cell RNA was purified, and 1 μg of whole cell RNA was resolved on 8 M urea–6% polyacrylamide gels, and histone H2a mRNA and 7SK RNA were detected by Northern blotting and visualized with a PhosphorImager. The RNA samples were also analyzed by gel electrophoresis and stained with ethidium bromide for 28S rRNA (bottom). (C) The PhosphorImager images were analyzed by ImageQuant. (D,E) Exogenous Flag-Lsm4 with a wild-type C-terminal tail or Flag-Lsm4(ΔC10) was stably expressed in HeLa cells. Endogenous Lsm4 was knocked down by an shRNA targeting the 3′ UTR of endogenous Lsm4 (Fig. 5C). Histone mRNA degradation was initiated by treating exponentially growing cells with 5 mM HU, and the total cell RNA was analyzed as in panel B. The results of three independent experiments were quantified in panel E; statistical significance was determined by a Student's t-test.
DISCUSSION
Degradation of histone mRNA requires the assembly of a complex of factors on the 3′ end of the mRNA, resulting in oligouridylation of the 3′ end of the mRNA followed by binding of the Lsm1-7 complex to the oligo(U) tail and subsequent degradation of the histone mRNA via both 5′–3′ and 3′–5′ pathways (Mullen and Marzluff 2008). The 3′hExo is essential for the initial step of degradation of the histone mRNA after oligouridylation (Hoefig et al. 2013). The 3′ end of the cytoplasmic, translationally competent histone mRNA likely is a ternary complex of SLBP and 3′hExo bound to the stem–loop at the 3′ end (Yang et al. 2006; Tan et al. 2013). Since addition of the oligo(U) tail does not evict SLBP or 3′hExo from the histone 3′ end, recruitment of other factors, such as Lsm1-7, likely precedes disassembly of the mRNP.
The Lsm1-7 ring also plays a critical role in degradation of polyadenylated mRNAs (Parker and Song 2004). Following deadenylation, it binds to the short (<15 nt) oligo(A) tail of the mRNA resulting in the recruitment of factors that inhibit translation and activate degradation. The Lsm 2-7 proteins are shared as part of a second complex, Lsm 2-8, which is associated with the oligo(U) tail on the 3′ end of U6 snRNA in the nucleus, where it is essential for splicing (Mayes et al. 1999; Pannone et al. 2001). How the relative amounts of the two different Lsm complexes are controlled is not understood, but the Lsm2-7 proteins are components of two critical complexes, one involved in pre-mRNA splicing in the nucleus and the other in mRNA degradation in the cytoplasm.
Specific roles for individual Lsm proteins in these complexes have not been identified. The complex consists of a ring of proteins that interact through their Sm domains. Each of the members of the ring has additional sequences at the N and C termini that may be available for interacting with specific factors. Some of these tails are likely essential for assembly of the Lsm 1-7 ring. These sequences are highly conserved among vertebrates, suggesting that they have specific functions. The Lsm1-7 ring interacts with Rck/p54, Pat 1, Hedls, and hEdc3, although whether regions of the Lsm1-7 ring directly contact all these factors is not known (Fenger-Gron et al. 2005; Ozgur et al. 2010).
Lsm4 plays a specific role in histone mRNA degradation
Our finding that a specific site on Lsm4 directly binds with both proteins at the 3′ end of the histone mRNP suggests that recruitment of the Lsm1-7 ring to its target mRNAs may involve protein–protein contacts with the mRNP as well as recognition of an unstructured oligo(A) or oligo(U) stretch. Several lines of evidence support this conclusion. The C-terminal tail of Lsm4 specifically interacts with the RNA binding domain of SLBP. This interaction is conserved between frogs and humans, but the RNA binding domain of frog SLBP2, an SLBP that functions in storing histone mRNA in an inactive form in the oocyte, does not interact with Lsm4. Lsm4 also interacts with another component of the histone mRNP, 3′hExo, suggesting that it forms a specific complex with the 3′ end of histone mRNP. A mutant in Lsm4 that disrupts its binding to SLBP and 3′hExo specifically interferes with histone mRNA degradation.
Since the Lsm proteins are small, any alterations in their structure may affect their ability to assemble into the Lsm1-7 ring. We found that many tagged wild-type constructs we made of either Lsm4 or Lsm1 would not assemble into the Lsm1-7 ring even after RNAi knockdown of most of the endogenous proteins. Of several tags we tested, only the singly N-terminally Flag-tagged Lsm4 (and not a triple-tagged Flag Lsm4) was capable of assembly into the Lsm1-7 ring in vivo. Even this tagged protein did not compete effectively with the endogenous Lsm4, and <10% of the protein was incorporated into the Lsm1-7 ring in cells expressing the tagged Lsm4 together in similar amounts to endogenous Lsm4. We were only able to assemble substantial amounts of tagged-Lsm4 into the Lsm1-7 ring by knocking down the endogenous Lsm4.
SLBP, 3′hExo, and Lsm1-7 can bind simultaneously to oligouridylated histone mRNAs
Throughout the life cycle of the histone mRNA, SLBP interacts with the histone mRNA. The SLBP:SL complex provides a scaffold that helps to coordinate different processes. In the nucleus, during pre-mRNA processing, SLBP interacts with the nascent histone mRNA. Following the maturation of histone mRNAs, SLBP is essential for transport of the mRNA to the cytoplasm (Sullivan et al. 2009), likely by interacting with SLIP1 and/or CTIF (Kim et al. 2009; Choe et al. 2013) and Dbp5 (von Moeller et al. 2013) to facilitate TAP-dependent mRNA export (Erkmann et al. 2005a). During S phase in the cytoplasm, SLBP interacts with SLIP1 and/or CTIF (Choe et al. 2013), which, in turn, interact with the translation initiation complex at the 5′ end of the message. At the 3′ end of the histone mRNA, there is likely a ternary complex of SLBP:SL:3′hExo (Yang et al. 2006; Tan et al. 2013).
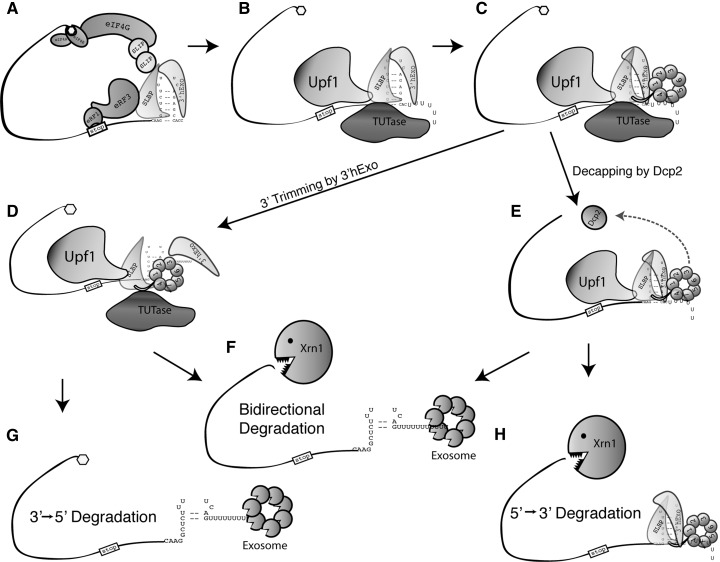
The rapid decrease in the half-life of histone mRNA when DNA synthesis is inhibited is mediated by the 3′ end of histone mRNA, which must be close to the terminating ribosome. We have argued that the initial signal for histone mRNA degradation acts by reducing the efficiency of termination of histone mRNA translation (Kaygun and Marzluff 2005b), which, in turn, results in recruitment of Upf1 as a critical factor required for histone mRNA degradation. Upf1 interacts directly with SLBP (Kaygun and Marzluff 2005a; S Meaux and WF Marzluff, unpubl.). We hypothesize that recruitment of Upf1 results in the subsequent recruitment of a TUTase that adds an oligo(U) tail to the histone mRNA. Subsequent recruitment of Lsm1-7, possibly promoted by the direct interaction of SLBP and 3′hExo with Lsm4, is essential for efficient histone mRNA degradation. In cells lacking 3′hExo, the oligouridylated histone mRNA accumulates (Hoefig et al. 2013), suggesting that one role of 3′hExo may be to remove the oligo(U) tail and initiate degradation of the 3′ end of the histone mRNA. A limiting step in 3′–5′ degradation of the histone mRNA is likely removal of SLBP and any associated proteins from the 3′ end of the histone mRNA to allow 3′–5′ exonucleolytic digestion (Fig. 7).
FIGURE 7.
Model of histone mRNA degradation after HU treatment. Possible pathways for degradation of histone mRNAs are shown. (A–C) The initial step is recruitment of Upf1 and oligouridylation of the 3′ end of histone mRNA. Lsm1-7 binds to the oligo(U) tail, promoted by Lsm4 binding to the SLBP and 3′hExo. (D–F) Subsequently, there is partial degradation of the stem–loop by 3′hExo and/or decapping of the histone mRNA. This intermediate is uridylated and binds the Lsm1-7 ring (Hoefig et al. 2013). (F–H) Subsequently the histone mRNA can be degraded bidirectionally (Mullen and Marzluff 2008) (F), 3′–5′ (G), or 5′–3′ (H). The relative importance of these three pathways is not known.
The precise role of the Lsm4:SLBP interaction is not clear. It may enhance the recruitment of limiting amounts of Lsm1-7 to the oligouridylated histone mRNA, or it may help in remodeling and eventually dissembling the SLBP:SL:3′hExo complex, making the histone 3′ end available for degradation to the exosome, and it could participate in both functions. Since 3′hExo is responsible for the initial step in histone mRNA degradation, including probably the removal of the oligo(U) tail (Hoefig et al. 2013), it is also possible that the C-terminal tail plays a role in promoting this reaction. An intermediate in histone mRNA degradation is partial degradation into the stem–loop, likely catalyzed by the 3′hExo (Hoefig et al. 2013). This intermediate is also uridylated and again binds Lsm1-7 presumably to recruit the exosome to promote degradation and/or recruitment of the decapping complex. For histone mRNAs being degraded 3′–5′, the rate-limiting step in degradation may be the removal of SLBP from the 3′ end of the histone mRNA, allowing complete degradation of the mRNA from the 3′ end.
Function of the Lsm1-7 complex in degradation of polyadenylated mRNAs
A question that remains is whether Lsm1-7 specifically recognizes its proper RNA substrates with high affinity from a broad group of substrates that have similar features. The mechanism by which Lsm1-7 is recruited to any RNA substrate is not well understood. Lsm1-7 has a higher affinity for oligo(A) or oligo(U) RNA at the 3′ end of mRNAs (Chowdhury et al. 2007; Chowdhury and Tharun 2008), and the complex can also bind to oligo(A) stretches at the 5′ end of orthopox viral mRNAs (Bergman et al. 2007), as well as internal oligo(A) sites in other viral mRNAs (Galao et al. 2010). Protein–protein interactions, such as those shown here with SLBP and 3′hExo, may provide additional specificity that allows for association of the Lsm1-7 complex preferentially with RNAs being targeted for degradation. All of the Lsm proteins contain a core Sm fold. However, some of these proteins contain substantial N- or C-terminal extensions, particularly Lsm1, Lsm4, and Lsm8. In yeast, the C-terminal domain of Lsm1 was shown to have an additional function in binding RNA substrates that is distinct from the Sm core of the protein (Chowdhury and Tharun 2008). For mammalian histone mRNAs, the C-terminal domain of Lsm4 may play a similar role, functioning to provide additional specificity through interaction with the protein components of the mRNP in addition to the Lsm1-7 ring binding the oligo(U) tail. In yeast, Lsm4 also contains an unstructured C-terminal extension, although there is no conservation between the vertebrate Lsm4 C-terminal extension, with yeast, Drosophila or Caenorhabditis elegans. However, other groups have presented data that this C-terminal extension may play a role in formation of P-bodies (Decker et al. 2007). Our results suggest that there are likely to also be specific protein–protein interactions between proteins associated with poly(A) mRNPs and the Lsm1-7 complex, which may contribute to the half-life of specific poly(A) mRNAs following removal of the poly(A) tail.
After binding of Lsm1-7 to oligouridylated or oligoadenylated mRNA, one pathway that is activated is decapping of the mRNA. Factors that assist in recruiting the decapping complex interact directly with the Lsm1-7 ring, resulting in decapping followed by degradation by the 5′–3′ exonuclease Xrn1 (Parker and Sheth 2007). In yeast growing in rich media, the major pathway of mRNA degradation is 5′–3′. In mammalian cells, the major pathway is less clear. In cytoplasmic extracts prepared from tissue culture cells, 3′–5′ degradation is active, although it is impossible to extrapolate from the in vitro results what happens in vivo. An attractive possibility is that many mRNAs are degraded both 5′–3′ and 3′–5′ (Murray and Schoenberg 2007), either as individual mRNAs entering different pathways or the same mRNA molecule being degraded simultaneously from the 5′ end and the 3′ end, as we have found for some histone mRNAs (Mullen and Marzluff 2008).
A fraction of the histone mRNAs is certainly degraded by the 5′–3′ degradation pathway (Mullen and Marzluff 2008; Su et al. 2013). However, after inhibition of DNA replication, there is an increase in the fraction of the histone mRNAs that is degraded by the 3′–5′ pathway (Su et al. 2013), requiring the exosome (Mullen and Marzluff 2008). It seems likely that removal of the oligo(U) tail either by 3′hExo or the exosome is initiated with Lsm1-7 bound to the tail, and indeed it is possible that Lsm1-7 plays a direct role in removal of the U-tail rather than simply protecting the tail from degradation. The details of how the Lsm1-7 ring may direct the subsequent pathway of histone mRNA degradation await elucidation in the future.
MATERIALS AND METHODS
Protein purification
Baculovirus-encoded proteins were produced as described for the 3′hExo (Dominski et al. 2003) and GST-SLBP fusion proteins (Erkmann et al. 2005b). Lsm1-7 complexes were expressed in bacteria and assembled into complexes (Zaric and Kambach 2008). DNA encoding the Lsm1-7 proteins was cloned into pxFRM, and the cloned DNA was used as a template for in vitro transcription. Lsm4 recombinant proteins fused to GST were obtained by cloning appropriate DNAs into pET42a, and the His-tagged proteins were purified.
Transcription and purification of stem–loop probes
The radiolabeled stem–loop probes were synthesized as previously described using two oligonucleotides (Milligan et al. 1987), one encoding the T7 promoter and the other containing the reverse complement of the T7 promoter and the desired stem–loop sequence oligo with the coding strand of the stem–loop, as previously described (Pandey et al. 1991). The RNA was purified by electrophoresis on a 15% polyacrylamide–7 M urea gel.
Electrophoretic mobility shift assay
The electrophoretic mobility shift assay (EMSA) was performed essentially as previously described using 10 fmol of uniformly labeled RNA; the RNA was incubated on ice with various amounts of purified recombinant protein in 10 mM HEPES (pH 7.60, 50 mM KCl, 0.1 mM EDTA, 10% glycerol, 1 μg/μL yeast tRNA, 0.1 μg/μL, BSA) (Williams and Marzluff 1995). For shifts with 3′hExo, yeast tRNA was omitted from the binding reactions. Reactions were directly loaded onto a native polyacrylamide gel (acrylamide:bisacrylamide 29:1 in 1× TBE buffer). The radioactive complexes were visualized by autoradiography or using a PhosphorImager. Images were analyzed by ImageQuant.
GST pull-down assay
In vitro translation was carried out using a coupled rabbit reticulocyte or Wheat germ extract kit for the Lsm proteins with similar molecular weight (MW) to globin (Promega). Five micrograms of recombinant GST proteins was incubated at 4°C with pre-equilibrated glutathione-Sepharose resin (GE life sciences) in 100 μL of TEN100 buffer (20 mM Tris at pH 7.5, 0.1 mM EDTA, 100 mM NaCl). Ten microliters of the lysate containing in vitro–translated protein was added to the beads along with 10 μL of 10× TEN100 buffer, 14 μL of GDB buffer (10% glycerol, 10 mM DTT, 0.05 mg/mL BSA), and 76 μL of dH2O. Afterward proteins were allowed to bind for 2 h at 4°C while rotating. Glutathione beads were washed four times with 1 mL of TEN100 buffer. Twenty-five microliters of 2× SDS loading dye (4% SDS, 10% β-mercaptoethanol, 0.125 M Tris at pH 6.8, 20% glycerol, 0.2% Bromophenol Blue) was added to the beads and boiled for 10 min. The bound proteins were eluted from the resin and analyzed on an SDS–polyacrylamide gel. The gel percentage is indicated in each figure legend. Gels were stained with Coomassie blue to confirm pull-down of recombinant GST protein. Gels were dried, and the bound proteins were visualized by autoradiography or on a PhosphorImager.
Cloning of Flag-tagged proteins
Lsm4 cDNA was cloned into pcDNA-Flag between the BamHI and XhoI sites. This vector contains a single N-terminal Flag tag (DYKDDDDK). It is driven by the CMV promoter and contains a β-globin intron and the bGH polyadenylation signal. Mutations were introduced by PCR and recloned between BamHI and XhoI. For all in vitro translation assays, proteins were cloned into pxFRM (Sanchez and Marzluff 2002) between NcoI and XbaI. This vector contains a synthetic poly(A) tail to aid in translation.
Immunoprecipitation
HeLa cells were cultured in DMEM supplemented with 10% FBS and 100 IU/mL penicillin and streptomycin. Cells were trypsinized and washed once with PBS. Cell pellets were lysed in hypotonic lysis buffer (10 mM Tris at pH 7.5, 10 mM NaCl, 2 mM EDTA, 0.1% Triton X) for 10 min on ice and then adjusted to 150 mM NaCl with 5 M NaCl. Cell debris was removed by centrifugation. The cell lysate was quantified by Bradford assay. For each immunoprecipitation, 0.5 mL of 1.0 mg/mL lysates was used. Lysates were precleared with 25 μL of protein G Sepharose (GE Lifesciences) for 1 h at 4°C. For immunoprecipitation, 1 μL of 1 mg/mL antibody was added to each sample and rotated overnight at 4°C. The following day, 50 μL of 1:1 protein G Sepharose slurry was added and rotated for 4 h at 4°C. Following incubation, protein G Sepharose was washed four times with hypotonic lysis buffer supplemented to 150 mM NaCl, and then the beads were resuspended in 25 μL of 2× SDS loading buffer and boiled for 10 min. The resulting supernatant was loaded on an SDS-PAGE gel.
Measurement of histone mRNA half-life
Histone mRNA degradation was measured as previously described (Mullen and Marzluff 2008). Exponentially growing HeLa cells (<50% confluent) were treated with 5 mM HU for the indicated time. Total cell RNA was prepared, resolved by gel electrophoresis on a 6% urea–acrylamide gel, and the histone mRNA and 7SK RNA were detected by Northern blotting as previously described (Mullen and Marzluff 2008).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
This work was supported by NIH grant GM29832 to W.F.M. A.S.R. was supported by a fellowship from the Beckman Foundation. We thank Patrick Lackey and Deirdre Tatomer for critical reading of this manuscript.
REFERENCES
- Ansel KM, Pastor WA, Rath N, Lapan AD, Glasmacher E, Wolf C, Smith LC, Papadopoulou N, Lamperti ED, Tahiliani M, et al. 2008. Mouse Eri1 interacts with the ribosome and catalyzes 5.8S rRNA processing. Nat Struct Mol Biol 15: 523–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman N, Moraes KC, Anderson JR, Zaric B, Kambach C, Schneider RJ, Wilusz CJ, Wilusz J 2007. Lsm proteins bind and stabilize RNAs containing 5′ poly(A) tracts. Nat Struct Mol Biol 14: 824–831 [DOI] [PubMed] [Google Scholar]
- Brahms H, Meheus L, de Brabandere V, Fischer U, Luhrmann R 2001. Symmetrical dimethylation of arginine residues in spliceosomal Sm protein B/B′ and the Sm-like protein LSm4, and their interaction with the SMN protein. RNA 7: 1531–1542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cakmakci NG, Lerner RS, Wagner EJ, Zheng L-X, Marzluff WF 2008. SLIP1, a factor required for activation of histone mRNA translation by the stem-loop binding protein. Mol Cell Biol 28: 1182–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe J, Kim KM, Park S, Lee YK, Song OK, Kim MK, Lee BG, Song HK, Kim YK 2013. Rapid degradation of replication-dependent histone mRNAs largely occurs on mRNAs bound by nuclear cap-binding proteins 80 and 20. Nucleic Acids Res 41: 1307–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury A, Tharun S 2008. lsm1 mutations impairing the ability of the Lsm1p-7p-Pat1p complex to preferentially bind to oligoadenylated RNA affect mRNA decay in vivo. RNA 14: 2149–2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury A, Mukhopadhyay J, Tharun S 2007. The decapping activator Lsm1p-7p-Pat1p complex has the intrinsic ability to distinguish between oligoadenylated and polyadenylated RNAs. RNA 13: 998–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cougot N, Babajko S, Seraphin B 2004. Cytoplasmic foci are sites of mRNA decay in human cells. J Cell Biol 165: 31–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker CJ, Teixeira D, Parker R 2007. Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J Cell Biol 179: 437–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski Z, Yang X, Kaygun H, Marzluff WF 2003. A 3′ exonuclease that specifically interacts with the 3′ end of histone mRNA. Mol Cell 12: 295–305 [DOI] [PubMed] [Google Scholar]
- Eberle AB, Lykke-Andersen S, Muhlemann O, Jensen TH 2009. SMG6 promotes endonucleolytic cleavage of nonsense mRNA in human cells. Nat Struct Mol Biol 16: 49–55 [DOI] [PubMed] [Google Scholar]
- Erkmann JA, Sanchez R, Treichel N, Marzluff WF, Kutay U 2005a. Nuclear export of metazoan replication-dependent histone mRNAs is dependent on RNA length and is mediated by TAP. RNA 11: 45–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erkmann JA, Wagner EJ, Dong J, Zhang YP, Kutay U, Marzluff WF 2005b. Nuclear import of the stem–loop binding protein and localization during the cell cycle. Mol Biol Cell 16: 2960–2971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenger-Gron M, Fillman C, Norrild B, Lykke-Andersen J 2005. Multiple processing body factors and the ARE binding protein TTP activate mRNA decapping. Mol Cell 20: 905–915 [DOI] [PubMed] [Google Scholar]
- Franks TM, Singh G, Lykke-Andersen J 2010. Upf1 ATPase-dependent mRNP disassembly is required for completion of nonsense-mediated mRNA decay. Cell 143: 938–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galao RP, Chari A, Alves-Rodrigues I, Lobao D, Mas A, Kambach C, Fischer U, Diez J 2010. Lsm1-7 complexes bind to specific sites in viral RNA genomes and regulate their translation and replication. RNA 16: 817–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatfield D, Izaurralde E 2004. Nonsense-mediated messenger RNA decay is initiated by endonucleolytic cleavage in Drosophila. Nature 429: 575–578 [DOI] [PubMed] [Google Scholar]
- Graves RA, Pandey NB, Chodchoy N, Marzluff WF 1987. Translation is required for regulation of histone mRNA degradation. Cell 48: 615–626 [DOI] [PubMed] [Google Scholar]
- Heintz N, Sive HL, Roeder RG 1983. Regulation of human histone gene expression: Kinetics of accumulation and changes in the rate of synthesis and in the half-lives of individual histone mRNAs during the HeLa cell cycle. Mol Cell Biol 3: 539–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoefig KP, Rath N, Heinz GA, Wolf C, Dameris J, Schepers A, Kremmer E, Ansel KM, Heissmeyer V 2013. Eri1 degrades the stem-loop of oligouridylated histone mRNAs to induce replication-dependent decay. Nat Struct Mol Biol 20: 73–81 [DOI] [PubMed] [Google Scholar]
- Huntzinger E, Kashima I, Fauser M, Sauliere J, Izaurralde E 2008. SMG6 is the catalytic endonuclease that cleaves mRNAs containing nonsense codons in metazoan. RNA 14: 2609–2617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim F, Rohr J, Jeong WJ, Hesson J, Cerutti H 2006. Untemplated oligoadenylation promotes degradation of RISC-cleaved transcripts. Science 314: 1893. [DOI] [PubMed] [Google Scholar]
- Kaygun H, Marzluff WF 2005a. Regulated degradation of replication-dependent histone mRNAs requires both ATR and Upf1. Nat Struct Mol Biol 12: 794–800 [DOI] [PubMed] [Google Scholar]
- Kaygun H, Marzluff WF 2005b. Translation termination is involved in histone mRNA degradation when DNA replication is inhibited. Mol Cell Biol 25: 6879–6888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KM, Cho H, Choi K, Kim J, Kim BW, Ko YG, Jang SK, Kim YK 2009. A new MIF4G domain-containing protein, CTIF, directs nuclear cap-binding protein CBP80/20-dependent translation. Genes Dev 23: 2033–2045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Greimann JC, Lima CD 2006. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell 127: 1223–1237 [DOI] [PubMed] [Google Scholar]
- Makino DL, Baumgartner M, Conti E 2013. Crystal structure of an RNA-bound 11-subunit eukaryotic exosome complex. Nature 495: 70–75 [DOI] [PubMed] [Google Scholar]
- Marzluff WF, Wagner EJ, Duronio RJ 2008. Metabolism and regulation of canonical histone mRNAs: Life without a poly(A) tail. Nat Rev Genet 9: 843–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayes AE, Verdone L, Legrain P, Beggs JD 1999. Characterization of Sm-like proteins in yeast and their association with U6 snRNA. EMBO J 18: 4321–4331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan JF, Groebe DR, Witherell GW, Uhlenbeck OC 1987. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res 15: 8783–8794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen TE, Marzluff WF 2008. Degradation of histone mRNA requires oligouridylation followed by decapping and simultaneous degradation of the mRNA both 5′ to 3′ and 3′ to 5′. Genes Dev 22: 50–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray EL, Schoenberg DR 2007. A+U-rich instability elements differentially activate 5′–3′ and 3′–5′ mRNA decay. Mol Cell Biol 27: 2791–2799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orban TI, Izaurralde E 2005. Decay of mRNAs targeted by RISC requires XRN1, the Ski complex, and the exosome. RNA 11: 459–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozgur S, Chekulaeva M, Stoecklin G 2010. Human Pat1b connects deadenylation with mRNA decapping and controls the assembly of processing bodies. Mol Cell Biol 30: 4308–4323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey NB, Marzluff WF 1987. The stem-loop structure at the 3′ end of histone mRNA is necessary and sufficient for regulation of histone mRNA stability. Mol Cell Biol 7: 4557–4559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey NB, Sun J-H, Marzluff WF 1991. Different complexes are formed on the 3′ end of histone mRNA in nuclear and polysomal extracts. Nucleic Acids Res 19: 5653–5659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannone BK, Kim SD, Noe DA, Wolin SL 2001. Multiple functional interactions between components of the Lsm2–Lsm8 complex, U6 snRNA, and the yeast La protein. Genetics 158: 187–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R, Sheth U 2007. P bodies and the control of mRNA translation and degradation. Mol Cell 25: 635–646 [DOI] [PubMed] [Google Scholar]
- Parker R, Song H 2004. The enzymes and control of eukaryotic mRNA turnover. Nat Struct Mol Biol 11: 121–127 [DOI] [PubMed] [Google Scholar]
- Sanchez R, Marzluff WF 2002. The stem-loop binding protein is required for efficient translation of histone mRNA in vivo and in vitro. Mol Cell Biol 22: 7093–7104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharl EC, Steitz JA 1994. The site of 3′ end formation of histone messenger RNA is a fixed distance from the downstream element recognized by the U7 snRNP. EMBO J 13: 2432–2440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenberg DR 2011. Mechanisms of endonuclease-mediated mRNA decay. Wiley Interdiscip Rev RNA 2: 582–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B, Goodman HM 2004. Uridine addition after microRNA-directed cleavage. Science 306: 997. [DOI] [PubMed] [Google Scholar]
- Sittman DB, Graves RA, Marzluff WF 1983. Histone mRNA concentrations are regulated at the level of transcription and mRNA degradation. Proc Natl Acad Sci 80: 1849–1853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song MG, Kiledjian M 2007. 3′ Terminal oligo U-tract-mediated stimulation of decapping. RNA 13: 2356–2365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su W, Slepenkov SV, Slevin MK, Lyons SM, Ziemniak M, Kowalska J, Darzynkiewicz E, Jemielity J, Marzluff WF, Rhoads RE 2013. mRNAs containing the histone 3′ stem–loop are degraded primarily by decapping mediated by oligouridylation of the 3′ end. RNA 19: 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan KD, Mullen TE, Marzluff WF, Wagner EJ 2009. Knockdown of SLBP results in nuclear retention of histone mRNA. RNA 15: 459–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan D, Marzluff WF, Dominski Z, Tong L 2013. Structure of histone mRNA stem-loop, human stem-loop binding protein, and 3′hExo ternary complex. Science 339: 318–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Moeller H, Lerner R, Ricciardi A, Basquin C, Marzluff WF, Conti E 2013. Structural and biochemical studies of SLIP1–SLBP identify DBP5 and eIF3g as SLIP1-binding proteins. Nucleic Acids Res 14: 7960–7971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z-F, Ingledue TC, Dominski Z, Sanchez R, Marzluff WF 1999. Two Xenopus proteins that bind the 3′ end of histone mRNA: Implications for translational control of histone synthesis during oogenesis. Mol Cell Biol 19: 835–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams AS, Marzluff WF 1995. The sequence of the stem and flanking sequences at the 3′ end of histone mRNA are critical determinants for the binding of the stem-loop binding protein. Nucleic Acids Res 23: 654–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XC, Purdy M, Marzluff WF, Dominski Z 2006. Characterization of 3′hExo, a 3′ exonuclease specifically interacting with the 3′ end of histone mRNA. J Biol Chem 281: 30447–30454 [DOI] [PubMed] [Google Scholar]
- Zaric BL, Kambach C 2008. Reconstitution of recombinant human LSm complexes for biochemical, biophysical, and cell biological studies. Methods Enzymol 448: 57–74 [DOI] [PubMed] [Google Scholar]