It is well established that protein synthesis occurs with high fidelity. This paper describes a sensitive system to analyze the frequency of all possible missense errors by several tRNAs. The only errors that occur with appreciable frequency are observed when codon–anticodon pairing involves noncanonical base-pairing with geometries similar to canonical Watson–Crick base pairs.
Keywords: Escherichia coli, β-galactosidase, mistranslation, non-Watson–Crick base pairs, protein synthesis
Abstract
Protein synthesis must rapidly and repeatedly discriminate between a single correct and many incorrect aminoacyl-tRNAs. We have attempted to measure the frequencies of all possible missense errors by tRNA , tRNA
, tRNA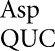 and tRNA
and tRNA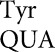 . The most frequent errors involve three types of mismatched nucleotide pairs, U•U, U•C, or U•G, all of which can form a noncanonical base pair with geometry similar to that of the canonical U•A or C•G Watson–Crick pairs. Our system is sensitive enough to measure errors at other potential mismatches that occur at frequencies as low as 1 in 500,000 codons. The ribosome appears to discriminate this efficiently against any pair with non-Watson–Crick geometry. This extreme accuracy may be necessary to allow discrimination against the errors involving near Watson–Crick pairing.
. The most frequent errors involve three types of mismatched nucleotide pairs, U•U, U•C, or U•G, all of which can form a noncanonical base pair with geometry similar to that of the canonical U•A or C•G Watson–Crick pairs. Our system is sensitive enough to measure errors at other potential mismatches that occur at frequencies as low as 1 in 500,000 codons. The ribosome appears to discriminate this efficiently against any pair with non-Watson–Crick geometry. This extreme accuracy may be necessary to allow discrimination against the errors involving near Watson–Crick pairing.
INTRODUCTION
The fidelity of protein synthesis is critical to cellular health. Errors are kept at low levels by a variety of mechanisms at each step in the transfer of information from DNA to protein. Errors during transcription are extremely infrequent, for Escherichia coli RNA polymerase ∼1 × 10−5 per nucleotide (Springgate and Loeb 1975; Erie et al. 1992). Errors are more frequent during protein synthesis, resulting either from misacylation of tRNAs or from tRNA selection errors that cause insertion of an incorrect amino acid (misreading) shifting out of the normal reading frame (frameshifting), or spontaneous release of the peptidyl-tRNA (drop-off) (Kurland et al. 1996). Misreading errors are arguably the most common translational errors (Kramer and Farabaugh 2007; Kramer et al. 2010; Yadavalli and Ibba 2012).
The ribosome limits errors during tRNA selection by discriminating between the structures formed by correct (cognate) and incorrect (near or noncognate) codon–anticodon interactions. An early proposed accuracy mechanism was kinetic proofreading (Hopfield 1974; Ninio 1975), in which two successive 100-fold discrimination steps separated by the irreversible step of GTP hydrolysis by elongation factor Tu (EF-Tu) explained the proposed 10,000-fold preference for cognate tRNA. Recent kinetic analysis suggested that accuracy mainly derives from differences in the rate of activation of the EF-Tu GTPase, which is ∼1000-fold faster for cognate than near-cognate complexes, and more frequent dissociation of near-cognates before GTP cleavage (for review, see Rodnina et al. 2005). The mechanism of this discrimination had been thought to involve an intimate set of interactions between the ribosome and the codon and anticodon base pairs (Ogle et al. 2003), with only the cognate complex stimulating a large conformational change to promote an interaction between the ribosome's sarcin–ricin loop and the GTPase domain of EF-Tu activating GTP hydrolysis (Voorhees et al. 2010). These interactions were thought to require the geometry of Watson–Crick pairs, which is different from all other base pairs (Leontis et al. 2002). This conclusion has been challenged recently (Demeshkina et al. 2012). High-resolution X-ray structures indicate that the A site interacts similarly with both cognate and near-cognate tRNAs, with mismatched bases being forced into Watson–Crick geometry in the A site. Demeshkina et al. (2012) suggest that deformations caused by structural clashes of mispaired nucleotides in the A site might propagate through the structure to increase tRNA dissociation; presumably, the same effect might also reduce the rate of GTPase activation.
Previously, we developed a system allowing comprehensive analysis of the frequency of missense errors by individual tRNAs (Kramer and Farabaugh 2007; Kramer et al. 2010). That system exploited lysine 529 (K529), an essential amino acid in the active site of Photinus pyralis (firefly) luciferase (Fluc). Mutation of this residue reduces activity to as low as 3 × 10−4 times wild type. Mutants with a subset of single-nucleotide mutations of this codon have higher activity, resulting from misreading of these codons by tRNA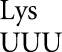 . The spectrum of misreading errors and their frequencies differs between the bacterium Escherichia coli (Kramer and Farabaugh 2007) and the yeast Saccharomyces cerevisiae (Kramer et al. 2010). A major factor identified as determining the observed frequencies is the rate of decoding of the mutant codon by its cognate tRNA; errors by tRNA
. The spectrum of misreading errors and their frequencies differs between the bacterium Escherichia coli (Kramer and Farabaugh 2007) and the yeast Saccharomyces cerevisiae (Kramer et al. 2010). A major factor identified as determining the observed frequencies is the rate of decoding of the mutant codon by its cognate tRNA; errors by tRNA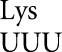 were more frequent at codons decoded by low-abundance competing cognate tRNAs.
were more frequent at codons decoded by low-abundance competing cognate tRNAs.
Here we analyze errors by tRNA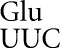 , tRNA
, tRNA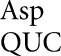 and tRNA
and tRNA showing that they involve only a small subset of all possible non-Watson–Crick base pairs, favoring those with near Watson–Crick geometry (Weixlbaumer et al. 2007). The background frequency of misreading at non-error-prone codons can be remarkably low—approximately 2 × 10–6 per codon—implying that the ribosome is unexpectedly efficient at eliminating most errors involving most base pairs with other than Watson–Crick geometry. These results will be discussed in the context of the structure of codon–anticodon complexes formed during misreading.
showing that they involve only a small subset of all possible non-Watson–Crick base pairs, favoring those with near Watson–Crick geometry (Weixlbaumer et al. 2007). The background frequency of misreading at non-error-prone codons can be remarkably low—approximately 2 × 10–6 per codon—implying that the ribosome is unexpectedly efficient at eliminating most errors involving most base pairs with other than Watson–Crick geometry. These results will be discussed in the context of the structure of codon–anticodon complexes formed during misreading.
RESULTS
Frequent misreading errors by tRNA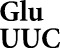 occur at four out of 14 near cognate codons
occur at four out of 14 near cognate codons
The active site of the enzyme β-galactosidase is complex, with many residues making structurally important interactions with the substrate (Juers et al. 2003). The glutamic acid at position 537 (E537) is the principle nucleophile during catalysis (Gebler et al. 1992), and mutants of E537 have very low activity, varying between 10–3 and 3 × 10−6 times wild type (Yuan et al. 1994). These activity differences have been attributed to structural differences among the mutant enzymes; because of our work on misreading by tRNA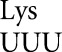 we suspected that the differences might reflect misreading of the mutant codons by the tRNA
we suspected that the differences might reflect misreading of the mutant codons by the tRNA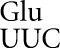 , resulting in a very small proportion of fully active, wild-type proteins. We generated a full set of mutations to near-cognate codons (those that differ from the Glu codons GAA or GAG by a single substitution), as well as all possible synonymous noncognate codons (those synonymous in coding with a near-cognate mutant, but having more than one nucleotide difference from a Glu codon). The variation in the β-galactosidase activity of these mutants could result from the variable activity of the mutant enzymes—the functional replacement model—or from variation in the frequency of misreading of the mutant codons by tRNA
, resulting in a very small proportion of fully active, wild-type proteins. We generated a full set of mutations to near-cognate codons (those that differ from the Glu codons GAA or GAG by a single substitution), as well as all possible synonymous noncognate codons (those synonymous in coding with a near-cognate mutant, but having more than one nucleotide difference from a Glu codon). The variation in the β-galactosidase activity of these mutants could result from the variable activity of the mutant enzymes—the functional replacement model—or from variation in the frequency of misreading of the mutant codons by tRNA —the translational error model (Kramer and Farabaugh 2007). The value of the synonymous noncognate codons is that under the functional replacement model, noncognate mutant enzymes having the same amino acid substitution should have the same activity as the near-cognate mutants, but under the translational error model their activity should be much lower, since misreading of noncognate codons is much less likely.
—the translational error model (Kramer and Farabaugh 2007). The value of the synonymous noncognate codons is that under the functional replacement model, noncognate mutant enzymes having the same amino acid substitution should have the same activity as the near-cognate mutants, but under the translational error model their activity should be much lower, since misreading of noncognate codons is much less likely.
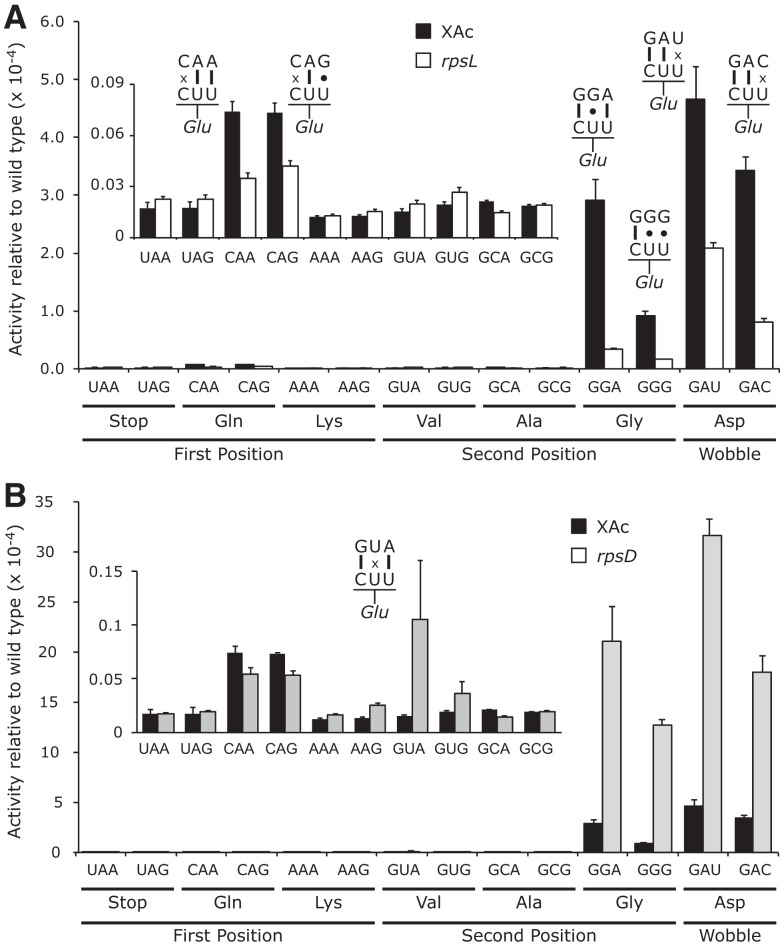
Figure 1 shows that 10 of the 14 E537 near-cognate mutants had extremely low activity averaging 2.3 × 10−6 times the wild-type enzyme; all synonymous noncognate mutants had similar activities (Supplemental Table S1). The activity of these mutants represents the maximum possible frequency of misreading errors by this tRNA, a level ∼200-fold below the previous estimate of random missense errors, ∼5 × 10−4 per codon (Kurland 1992; Parker 1992).
FIGURE 1.
Error frequencies by tRNA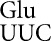 on mutants of codon 537 vary by over two orders of magnitude. The activity of mutants with the indicated codons in place of Glu codon 537 are shown relative to the activity of wild-type β-galactosidase (error bars, SEM). The insets use greatly expanded y-axis to represent the much lower activities of the 10 indicated mutants. (A) Comparison of mutant activities in the wild-type (XAc) and hyperaccurate (rpsL) genetic backgrounds. (B) Comparison of activities in wild-type and error-prone (rpsD) backgrounds.
on mutants of codon 537 vary by over two orders of magnitude. The activity of mutants with the indicated codons in place of Glu codon 537 are shown relative to the activity of wild-type β-galactosidase (error bars, SEM). The insets use greatly expanded y-axis to represent the much lower activities of the 10 indicated mutants. (A) Comparison of mutant activities in the wild-type (XAc) and hyperaccurate (rpsL) genetic backgrounds. (B) Comparison of activities in wild-type and error-prone (rpsD) backgrounds.
Four E537 mutants have activity much higher than this background: The mutants introducing the two Asp codons GAU and GAC and two of the four Gly codons, GGA and GGG (Fig. 1A). Synonymous noncognate Gly mutants (GGU and GGC) have activities ∼90-fold lower than GGA and GGC, implying that the higher activities represent translational error (Supplemental Table S1). To test this conclusion, we assessed the effect on enzyme activity of the hyperaccurate mutation rpsL141, which introduces a K42N mutation in ribosomal protein S12 (Andersson et al. 1982). We previously demonstrated that this mutation strongly reduces misreading by tRNA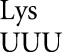 (Kramer and Farabaugh 2007). The rpsL141 mutation had no significant effect on background activity (Supplemental Table S1), but it significantly reduces activity of the four higher-activity mutants (Fig. 1A). An error-prone mutation of ribosomal protein S4, rpsD12 (Dahlgren and Ryden-Aulin 2000), increased errors by tRNA
(Kramer and Farabaugh 2007). The rpsL141 mutation had no significant effect on background activity (Supplemental Table S1), but it significantly reduces activity of the four higher-activity mutants (Fig. 1A). An error-prone mutation of ribosomal protein S4, rpsD12 (Dahlgren and Ryden-Aulin 2000), increased errors by tRNA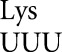 at all error-prone codons (Kramer and Farabaugh 2007). In the rpsD12 the background activity of the four higher-activity mutants of E537 increased as well as one lower-activity mutant that introduces the Val GUA codon; there was no significant effect on the other mutants (Fig. 1B; Supplemental Table S1). To exclude effects of plasmid instability on differences in activity we performed assays of a second protein encoded by the reporter plasmid, chloramphenicol acetyltransferase (CAT). We found no significant differences in activity for any tested transformant (data not shown). Second, to insure that there was no variation in protein expression we performed Western blotting with an anti-β-galactosidase antibody and again found no significant differences (data not shown).
at all error-prone codons (Kramer and Farabaugh 2007). In the rpsD12 the background activity of the four higher-activity mutants of E537 increased as well as one lower-activity mutant that introduces the Val GUA codon; there was no significant effect on the other mutants (Fig. 1B; Supplemental Table S1). To exclude effects of plasmid instability on differences in activity we performed assays of a second protein encoded by the reporter plasmid, chloramphenicol acetyltransferase (CAT). We found no significant differences in activity for any tested transformant (data not shown). Second, to insure that there was no variation in protein expression we performed Western blotting with an anti-β-galactosidase antibody and again found no significant differences (data not shown).
The conclusion of this experiment is that errors by tRNA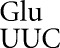 occur with strikingly different frequencies. Most codons are misread at rates as low as an average of 2.3 × 10−6 per codon, whereas four codons are misread with frequencies ∼100-fold higher. At the error-prone codons, misreading errors by tRNA
occur with strikingly different frequencies. Most codons are misread at rates as low as an average of 2.3 × 10−6 per codon, whereas four codons are misread with frequencies ∼100-fold higher. At the error-prone codons, misreading errors by tRNA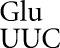 would involve either U34•U3 or U34•C3 wobble or U35•G2 second-position mispairing. All other predicted mispairing produced much less-frequent errors.
would involve either U34•U3 or U34•C3 wobble or U35•G2 second-position mispairing. All other predicted mispairing produced much less-frequent errors.
Frequent misreading errors by tRNA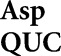 and tRNA
and tRNA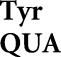 also occur at a small minority of near-cognate codons
also occur at a small minority of near-cognate codons
To expand the range of our analysis, we constructed two more reporter systems based on active site mutations of β-galactosidase, aspartate 201 (D201) and tyrosine 503 (Y503). D201 is one of many amino acids that orient the substrate in the active site, coordinating and acting as a counter ion for bound monovalent cations to indirectly interact with the substrate (Xu et al. 2004). Y503 participates in orienting the transition state of the substrate in the active site (Juers et al. 2012). We used these reporter systems to explore the frequency of missense errors by tRNA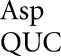 and tRNA
and tRNA at codons D201 and Y503.
at codons D201 and Y503.
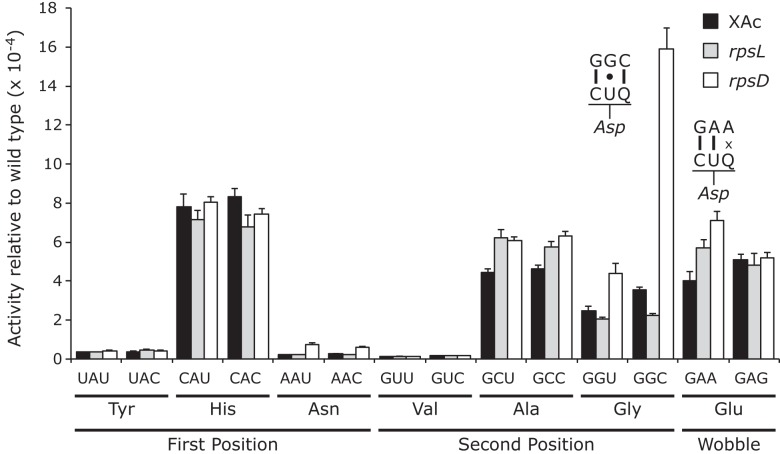
As shown in Figure 2, the D201 mutants that replace Asp by Tyr, Asn, or Val have extremely low activity, averaging 2.1 × 10−5 times wild type. This value establishes a maximum error frequency for tRNA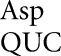 on these codons. The other mutants replacing Asp by His, Ala, Gly, or Glu have substantially more activity, averaging 3.7 × 10−4 times wild type. For these mutants, each synonymous pair has indistinguishable activity, consistent with the functional replacement model rather than translational error (Supplemental Table S2). Consistent with this conclusion, the activity of the mutants in the rpsL141 background is the same as in the wild type, averaging 3.9 × 10−4 times wild type. In the error-prone rpsD12 background, however, some of the D201 mutants have measurable higher activity. There was a large increase in activity only for the Gly GGC mutant, which could be read by tRNA
on these codons. The other mutants replacing Asp by His, Ala, Gly, or Glu have substantially more activity, averaging 3.7 × 10−4 times wild type. For these mutants, each synonymous pair has indistinguishable activity, consistent with the functional replacement model rather than translational error (Supplemental Table S2). Consistent with this conclusion, the activity of the mutants in the rpsL141 background is the same as in the wild type, averaging 3.9 × 10−4 times wild type. In the error-prone rpsD12 background, however, some of the D201 mutants have measurable higher activity. There was a large increase in activity only for the Gly GGC mutant, which could be read by tRNA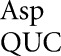 with a middle-position U35•G2 mismatch; the activity suggested an error frequency of 1.6 × 10−3 per codon, equivalent to the frequency of middle-position errors for tRNAGlu (1.3 × 10−3 and 2.1 × 10−3). This implies that for this type of error tRNA
with a middle-position U35•G2 mismatch; the activity suggested an error frequency of 1.6 × 10−3 per codon, equivalent to the frequency of middle-position errors for tRNAGlu (1.3 × 10−3 and 2.1 × 10−3). This implies that for this type of error tRNA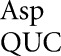 might be as error prone as tRNA
might be as error prone as tRNA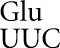 . There were much weaker codon-specific effects on activity of the Gly GGU, Glu GAA, and the Asn AAU/AAC mutants, which would require a second (U35•G2), wobble (Q34•A1) or first-position mismatch (C36•A1), respectively. These data show that the residual activity of many of the D201 mutants is high enough to obscure putative misreading activities, reducing the value of these mutants for analyzing misreading. This fact suggests that great caution should be used in interpreting residual activities of active-site mutants as evidence of translational error, as we have suggested before (Kramer et al. 2010).
. There were much weaker codon-specific effects on activity of the Gly GGU, Glu GAA, and the Asn AAU/AAC mutants, which would require a second (U35•G2), wobble (Q34•A1) or first-position mismatch (C36•A1), respectively. These data show that the residual activity of many of the D201 mutants is high enough to obscure putative misreading activities, reducing the value of these mutants for analyzing misreading. This fact suggests that great caution should be used in interpreting residual activities of active-site mutants as evidence of translational error, as we have suggested before (Kramer et al. 2010).
FIGURE 2.
The activities of mutants of Asp codon 201 show that many codons exhibit high levels of functional replacement. The activities of the D201 mutants relative to wild type are shown as in Figure 1.
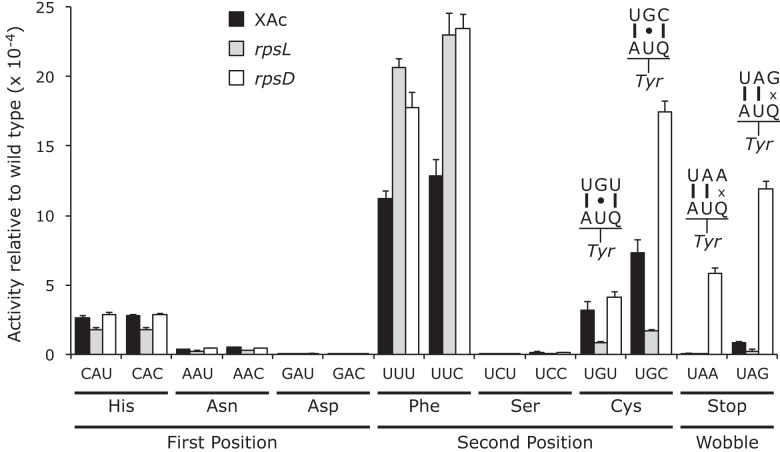
Most of the Y503 mutants showed indistinguishable activities for synonymous codon mutants, including those changing Tyr to His, Asn, Asp, Phe, and Ser (Fig. 3; Supplemental Table S3). The absolute values of the activities in this class varied widely; the Asn, Asp, and Ser mutants had very low activity, averaging 1.9 × 10−5 times wild type, the upper bound of the estimate for the frequency of missense errors by tRNA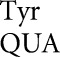 on these codons. The His and Phe mutants had much higher activities, averaging 2.7 × 10−4 and 1.2 × 10−3 times wild type, but the similarity in activity and the failure of the hyperactive rpsL141 mutant to reduce the activity of these mutants again supported the functional replacement model, implying that the activities do not reflect error. The second class, the Cys and termination codon replacements, showed codon-specific variation in activity that was strongly reduced by rpsL141, suggesting that their activities result from translational error. The activities of the Cys and termination codon mutants increased in the error-prone rpsD12 background. The errors at Cys codons UGU and UGC would require a middle-position U35•G2 mismatch, and those at the stop codons UAA and UAG would require wobble-position Q34•A3 or Q34•G3 mismatches. The increased errors at the His and Phe codons in the rpsL141 and rpsD12 mutant backgrounds compared with the wild type are unexpected. The fact that errors are the same in these backgrounds and the synonymous mutants have equal activity is inconsistent with these increases resulting from translational error.
on these codons. The His and Phe mutants had much higher activities, averaging 2.7 × 10−4 and 1.2 × 10−3 times wild type, but the similarity in activity and the failure of the hyperactive rpsL141 mutant to reduce the activity of these mutants again supported the functional replacement model, implying that the activities do not reflect error. The second class, the Cys and termination codon replacements, showed codon-specific variation in activity that was strongly reduced by rpsL141, suggesting that their activities result from translational error. The activities of the Cys and termination codon mutants increased in the error-prone rpsD12 background. The errors at Cys codons UGU and UGC would require a middle-position U35•G2 mismatch, and those at the stop codons UAA and UAG would require wobble-position Q34•A3 or Q34•G3 mismatches. The increased errors at the His and Phe codons in the rpsL141 and rpsD12 mutant backgrounds compared with the wild type are unexpected. The fact that errors are the same in these backgrounds and the synonymous mutants have equal activity is inconsistent with these increases resulting from translational error.
FIGURE 3.
The activities of mutants of Tyr codon 503 indicate significant second- and wobble-position errors. The activities of the Y503 mutants relative to wild type are shown as in Figure 1.
DISCUSSION
Frequent missense errors do not require weak cognate tRNA competition
Our previous work on translational errors by tRNA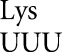 in E. coli came to three general conclusions (Kramer and Farabaugh 2007). First, the frequency of errors varied over 10-fold, with an apparent background frequency of no more than 3.1 × 10–4 per codon. Second, a major factor causing this variation was competition between the correct (cognate) and incorrect (near-cognate) aminoacyl–tRNAs; significantly higher error frequencies were found at codons decoded by limiting cognate tRNAs. Third, wobble mismatches did not depend on weak cognate competition; they occurred at significant frequencies despite the high abundance of the competing cognate tRNA. These conclusions, however, were based on a single tRNA. The primary purpose of the work presented here was to test the generality of these conclusions, and the data show that the second and third conclusions are not general.
in E. coli came to three general conclusions (Kramer and Farabaugh 2007). First, the frequency of errors varied over 10-fold, with an apparent background frequency of no more than 3.1 × 10–4 per codon. Second, a major factor causing this variation was competition between the correct (cognate) and incorrect (near-cognate) aminoacyl–tRNAs; significantly higher error frequencies were found at codons decoded by limiting cognate tRNAs. Third, wobble mismatches did not depend on weak cognate competition; they occurred at significant frequencies despite the high abundance of the competing cognate tRNA. These conclusions, however, were based on a single tRNA. The primary purpose of the work presented here was to test the generality of these conclusions, and the data show that the second and third conclusions are not general.
The data presented here challenge our previous conclusion that significant errors require poor availability of the competing cognate tRNA. Using the comprehensive data on tRNA abundance of Dong et al. (1996), we find no association of high error frequency with low abundance of the cognate tRNA decoding the mutant codon (data not shown). All competing cognate tRNAs are present at concentrations above the median and some are highly abundant. A low availability of the competing cognate tRNA is therefore not necessary for missense errors, although an extremely limiting competing cognate may stimulate higher errors frequencies. The extremely low abundance of the competing cognate tRNAs for AGA and AGG may explain why the apparent error frequency by tRNA on those codons is ∼10-fold greater than for any of the errors in this study.
on those codons is ∼10-fold greater than for any of the errors in this study.
Minimum errors during translation is surprisingly low
The most unexpected conclusion of this work is that the random background frequency of misreading is surprisingly low. Background activities vary among the mutants tested here, but were all substantially lower than for tRNA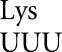 . For the E537 mutants, the lowest activities averaged 2.3 × 10–6 times wild type; the levels for the D201 and Y503 were ∼10-fold higher at 2.1 × 10–5 and 1.9 × 10–5 times wild type. The fact that the ribosome can discriminate to as low as 1 error in 430,000 incorporations is unexpected. Why would the protein synthetic system be configured so that most mistakes are so rare? We can conceive of no process of selection that could have attained that level of accuracy. We argue that this extreme level of accuracy evolved to deal with the more frequent errors resulting from near Watson–Crick base-pairing; these occur at frequencies that are over two orders of magnitude higher. The fact that we have a few similarly high-frequency errors by all tRNAs tested to date suggests that reducing the accuracy of protein synthesis might have affected all tRNAs; we note that mutants with high inaccuracy have an obvious slow growth phenotype (Ehrenberg and Kurland 1984; Kurland 1992), which could result from the burden of increased translational error.
. For the E537 mutants, the lowest activities averaged 2.3 × 10–6 times wild type; the levels for the D201 and Y503 were ∼10-fold higher at 2.1 × 10–5 and 1.9 × 10–5 times wild type. The fact that the ribosome can discriminate to as low as 1 error in 430,000 incorporations is unexpected. Why would the protein synthetic system be configured so that most mistakes are so rare? We can conceive of no process of selection that could have attained that level of accuracy. We argue that this extreme level of accuracy evolved to deal with the more frequent errors resulting from near Watson–Crick base-pairing; these occur at frequencies that are over two orders of magnitude higher. The fact that we have a few similarly high-frequency errors by all tRNAs tested to date suggests that reducing the accuracy of protein synthesis might have affected all tRNAs; we note that mutants with high inaccuracy have an obvious slow growth phenotype (Ehrenberg and Kurland 1984; Kurland 1992), which could result from the burden of increased translational error.
Wobble-position errors requiring a purine–purine clash are less frequent than those requiring a pyrimidine–pyrimidine mismatch
In our original study with tRNA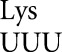 we found that among the most frequent errors were those requiring a wobble-position mismatch. Errors at the Asn codon AAU occurred at a rate of 1.6 × 10–3 per codon, only twofold less than the most error-prone codons, and in the error-prone rpsD12 background they were the most frequent at an astonishing 1.4 × 10–2 per codon (Kramer and Farabaugh 2007). The fact that the cognate tRNA
we found that among the most frequent errors were those requiring a wobble-position mismatch. Errors at the Asn codon AAU occurred at a rate of 1.6 × 10–3 per codon, only twofold less than the most error-prone codons, and in the error-prone rpsD12 background they were the most frequent at an astonishing 1.4 × 10–2 per codon (Kramer and Farabaugh 2007). The fact that the cognate tRNA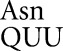 is among the most abundant tRNAs in E. coli (Dong et al. 1996) suggested that these errors did not depend on poor cognate competition. This result is consistent with previous data suggesting that wobble errors are frequent (for example, see Bouadloun et al. 1983; Parker and Precup 1986). There is, however, limited information on wobble misreading errors and we were interested in determining whether frequent wobble errors was a general phenomenon. Our results show a striking difference in the frequencies of wobble errors between those by tRNA
is among the most abundant tRNAs in E. coli (Dong et al. 1996) suggested that these errors did not depend on poor cognate competition. This result is consistent with previous data suggesting that wobble errors are frequent (for example, see Bouadloun et al. 1983; Parker and Precup 1986). There is, however, limited information on wobble misreading errors and we were interested in determining whether frequent wobble errors was a general phenomenon. Our results show a striking difference in the frequencies of wobble errors between those by tRNA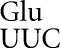 and by tRNA
and by tRNA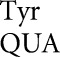 . Wobble-position errors in the E537 system by tRNA
. Wobble-position errors in the E537 system by tRNA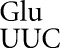 in wild type were as high as 4.7 × 10–4 per codon on GAU and rose to 3.2 × 10–3 in rpsD12, whereas in the Y503 system wobble errors by tRNA
in wild type were as high as 4.7 × 10–4 per codon on GAU and rose to 3.2 × 10–3 in rpsD12, whereas in the Y503 system wobble errors by tRNA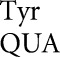 were no higher than 8.7 × 10–6 on UAA and rose only to 5.9 × 10–4 in rpsD12. Functional replacement in the D201 system blocked our measuring errors below 3.4 × 10–4, but in rpsD12 wobble errors were no higher than 7.1 × 10–4 on GAA. The result for tRNA
were no higher than 8.7 × 10–6 on UAA and rose only to 5.9 × 10–4 in rpsD12. Functional replacement in the D201 system blocked our measuring errors below 3.4 × 10–4, but in rpsD12 wobble errors were no higher than 7.1 × 10–4 on GAA. The result for tRNA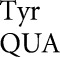 shows that high-frequency wobble errors are not a general rule. We note that the high wobble errors for tRNA
shows that high-frequency wobble errors are not a general rule. We note that the high wobble errors for tRNA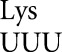 on AAU and tRNA
on AAU and tRNA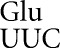 on GAU would both involve a U34•U3 mismatch, and that lower frequency errors by tRNA
on GAU would both involve a U34•U3 mismatch, and that lower frequency errors by tRNA on UAA and perhaps by tRNA
on UAA and perhaps by tRNA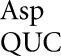 on GAA would require a Q34•A3 clash.
on GAA would require a Q34•A3 clash.
High-frequency errors involve a small subset of possible base-pair mismatches
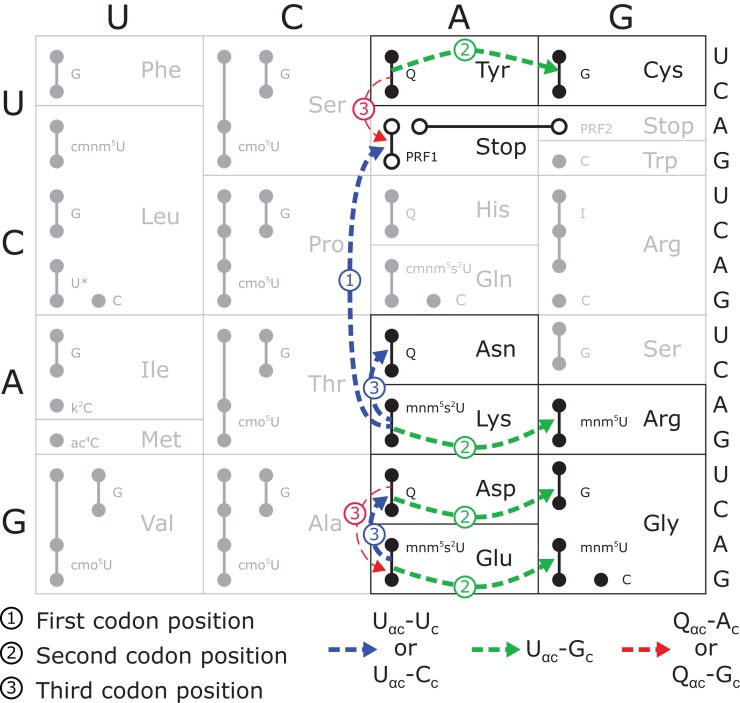
As in our previous studies (Kramer and Farabaugh 2007; Kramer et al. 2010), frequent errors occur at only a small set of codons. Errors by tRNA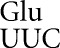 and tRNA
and tRNA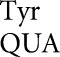 occurred at only four codons each and for tRNA
occurred at only four codons each and for tRNA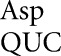 at only three codons, and then only in error-inducing conditions. These errors involve a very restricted set of mismatched bases. As shown in Figure 4, the most frequent errors by these three tRNAs and tRNA
at only three codons, and then only in error-inducing conditions. These errors involve a very restricted set of mismatched bases. As shown in Figure 4, the most frequent errors by these three tRNAs and tRNA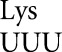 (Kramer and Farabaugh 2007) involved U34•U3 or U34•C3 (to Asn codons), U35•G2 (to Arg codons) or U36•U1 mismatches (to stop codons). We found errors above background involving a few more mismatches in an error-prone background; we found two other types of errors involving U35•U2 pairs (by tRNA
(Kramer and Farabaugh 2007) involved U34•U3 or U34•C3 (to Asn codons), U35•G2 (to Arg codons) or U36•U1 mismatches (to stop codons). We found errors above background involving a few more mismatches in an error-prone background; we found two other types of errors involving U35•U2 pairs (by tRNA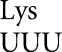 to Ile AUA), and Q34•A3 or Q34•G3 pairs (by tRNA
to Ile AUA), and Q34•A3 or Q34•G3 pairs (by tRNA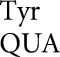 on UAA/UAG) (Supplemental Table S3). There were no significant errors involving eight first-position mismatches: C•U, C•C, C•A, A•C, A•A, A•G, U•C, or U•G. Our present error reporters allow us to test 16 possible mismatches; testing the 20 other possible mismatches would require generating more misreading reporters.
on UAA/UAG) (Supplemental Table S3). There were no significant errors involving eight first-position mismatches: C•U, C•C, C•A, A•C, A•A, A•G, U•C, or U•G. Our present error reporters allow us to test 16 possible mismatches; testing the 20 other possible mismatches would require generating more misreading reporters.
FIGURE 4.
The pattern of tRNA misreading involves two types of base mismatches. Each codon is represented by a small circle; filled circles connected by lines are all recognized by a single tRNA, with the anticodon wobble nucleotide of each tRNA identified immediately to the right using the standard convention (Limbach et al. 1994). The termination codons are represented by unfilled circles and lines connect those recognized by each of the peptide termination factors (PRF1 or PRF2). Misreading events in the wild-type background identified here or previously by Kramer and Farabaugh (2007) are shown as dashed arrows. The thickness of the arrow indicates relative frequency of the events (thick >10–4 per codon; thin <<10–4 per codon); for misreading of Glu codons GAA/GAG by tRNA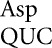 , the frequency of misreading could not be more than 4 × 10–4 per codon. The arrows are labeled to indicate the position of the mismatch and color coded for the nature of the mismatched base pair, as shown.
, the frequency of misreading could not be more than 4 × 10–4 per codon. The arrows are labeled to indicate the position of the mismatch and color coded for the nature of the mismatched base pair, as shown.
What could explain our finding errors at involving only three of 16 possible mismatches? Since errors do not depend on weak cognate tRNA competition, the selection of mismatches does not result from competition restricting the opportunity to generate significant errors. One possibility is that only a restricted set of mismatches can be as efficiently accepted in the A site. The mechanism of acceptance remains controversial. Ogle et al. (2003) suggested that three nucleotides in the ribosomal decoding center—G530, A1492, and A1493—monitor the codon–anticodon complex, discriminating between cognate and near-cognate complexes based on the structure of the base pairs formed. The recognition of cognate complexes was thought to induce a large-scale structural rearrangement of the ribosome, activating the intrinsic GTPase of elongation factor Tu (EF-Tu) (Voorhees et al. 2010). Demeshkina et al. (2012) recently found that the decoding center nucleotides interact with both cognate and near-cognate complexes constraining mismatched bases to adopt Watson–Crick geometry and induce the structural change in both cases. They suggest that discrimination against near-cognate depends on more subtle effects, either the energy cost of tautomeric shifts required for the mismatched pairs to form Watson–Crick geometry or deformations of the tRNA and decoding center elements caused by forcing this pairing.
Demeshkina et al. (2012) tested near-cognate decoding requiring first-position U36•G1 and second-position U35•G2 mismatches. The U35•G2 mismatch is from misreading of the Cys codon UGC by tRNA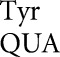 , which we have shown is the most frequent error by this tRNA. These second-position errors are frequent for all tRNAs tested (Fig. 4). U36•G1 errors have been demonstrated for misreading of the Gly GGC codon by tRNA
, which we have shown is the most frequent error by this tRNA. These second-position errors are frequent for all tRNAs tested (Fig. 4). U36•G1 errors have been demonstrated for misreading of the Gly GGC codon by tRNA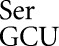 (Toth et al. 1988). Given our observation of large differences in error frequencies among near-cognate tRNAs and the lack of data on the structure formed by other near-cognates, it is premature to conclude that other base mismatches can be constrained to interact in Watson–Crick geometry. We propose that the three mismatches we have identified may do so and that the other eight may not, or constraining them to interact may more radically disturb the structure of the tRNA or decoding center. It is striking that the U•U, U•C, and U•G pairs were recently shown to be capable of pairing with near Watson–Crick geometry (Weixlbaumer et al. 2007). Formation of these pairs is stabilized in the wobble position by modification of uridine to uridine 5-oxyacetic acid (cmo5U) (Nasvall et al. 2004), but this modification is missing from the three tRNAs studied here. Even in the absence of this modification, these pairs may inefficiently form Watson–Crick geometry, so the cost of constraining them into that geometry during misreading may incur a smaller energetic cost than other mispairs, leading to greater misreading. Data on the effect of other tRNA modifications on misreading error frequency will be presented elsewhere (N Manickam and PJ Farabaugh, in prep.).
(Toth et al. 1988). Given our observation of large differences in error frequencies among near-cognate tRNAs and the lack of data on the structure formed by other near-cognates, it is premature to conclude that other base mismatches can be constrained to interact in Watson–Crick geometry. We propose that the three mismatches we have identified may do so and that the other eight may not, or constraining them to interact may more radically disturb the structure of the tRNA or decoding center. It is striking that the U•U, U•C, and U•G pairs were recently shown to be capable of pairing with near Watson–Crick geometry (Weixlbaumer et al. 2007). Formation of these pairs is stabilized in the wobble position by modification of uridine to uridine 5-oxyacetic acid (cmo5U) (Nasvall et al. 2004), but this modification is missing from the three tRNAs studied here. Even in the absence of this modification, these pairs may inefficiently form Watson–Crick geometry, so the cost of constraining them into that geometry during misreading may incur a smaller energetic cost than other mispairs, leading to greater misreading. Data on the effect of other tRNA modifications on misreading error frequency will be presented elsewhere (N Manickam and PJ Farabaugh, in prep.).
In general, then, we conclude that most near-cognate misreading errors by the tRNAs studied here and previously (Kramer and Farabaugh 2007) are discriminated against. The rate of errors at these non-error-prone codons can be extremely low, perhaps near the frequency of errors during transcription, suggesting that screening against some errors can be nearly absolute. Errors at other codons can be orders of magnitude higher, suggesting that the structures of a minority of mismatched codon–anticodon pairs may much more closely resemble correct Watson–Crick pairing, thus eluding the ribosome's discrimination mechanism.
MATERIALS AND METHODS
Bacterial strains
The Escherichia coli strains used in this study were XAc, araΔ[lac-proAB] gyrA(nalR) rpoB(rifR) argE[amber] (Andersson et al. 1982); B210, XAc rpsL141 zcg-174::Tn10; B212, XAc rpsD12 (Andersson et al. 1982); B322, XAc Δtgt770::Kan; B281 XAc mnmE::tn983 and B368, XAc ΔmiaA721::Kan. Strain XAc, B210 and B212 were gifts of Monika Ryden-Aulin (Stockholm University), B281 was a gift of Glenn R. Björk (Umeå University), and B322 and 368 were obtained from the Coli Genetic Stock Center (Yale University). Mutations not in the XAc genetic background were transferred to that background by P1 transduction, selecting for linked drug-resistance markers. Bacterial cultures were grown at 37°C in Luria-Bertani (LB) liquid medium (10 g NaCl, 10 g tryptone, and 5 g yeast extract per liter of distilled water) supplemented with relevant antibiotics, where required.
In vitro mutagenesis to generate β-galactosidase mutants
Mutants of codons 201, 503, and 537 of β-galactosidase were generated using QuikChange mutagenesis (Stratagene) according to the manufacturer's specifications using commercially available DNA oligonucleotides (Integrated DNA Technologies). Sequences of all oligonucleotides are given in Supplemental Table S4. To facilitate mutagenesis, some oligonucleotides were modified with the addition of extended nonoverlapping 3′ ends (Liu and Naismith 2008). The mutations were generated using the plasmid pJC27 as template and with the high-fidelity DNA polymerase PfuTurbo AD (Stratagene) with the addition of 3% (v/v) of DMSO to reduce the formation of primer dimer. The β-galactosidase activity expressed from mutant plasmids was extremely low, producing white colonies when grown on medium with 5-bromo-4-chloro-3-indolyl β-D-galactopyranoside (XGal), a chromogenic substrate for β-galactosidase. The appropriate regions of candidate plasmids were sequenced on an ABI Prism Sequencer according to manufacturer's instructions with the BigDye Terminator v3.1 Sequencing kit (Applied Biosystems), or were sequenced commercially (GENEWIZ).
β-galactosidase assay
Plasmids were introduced into bacterial strains using Z-competent cells (Zymo Research) or by the calcium chloride method (Hanahan 1983). Transformants were grown in 1 mL of liquid LB media in 96-well culture blocks at 37°C overnight to stationary phase. Cultures were inoculated to a dilution of 1:33 in 1 mL of liquid LB media with chloramphenicol and grown at 37°C to an OD600 of 0.5–0.8. Triplicate assays of β-galactosidase activity on each of three independent transformants were performed using the Beta-Glo system (Promega) according to manufacturer's specifications using Microfluor 96-well microtiter plates (ThermoFisher Scientific) in a Turner Biosystems MicroPlate II luminometer (Promega) using the Beta-Glo shake program. The relative light units (RLUs) measured by the luminometer are proportional to the activity of β-galactosidase in the cells. This method was used because of its much greater sensitivity compared with the conventional system using ortho-nitrophenyl-β-galactopyranoside. Misreading error frequency was calculated as a ratio of mutant to wild-type RLU. The significance of the results was tested using the Analysis of Variance (ANOVA) with Tukey's post-hoc test.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
This work was supported by a grant from the National Institutes of General Medical Sciences (GM029480).
REFERENCES
- Andersson DI, Bohman K, Isaksson LA, Kurland CG 1982. Translation rates and misreading characteristics of rpsD mutants in Escherichia coli. Mol Gen Genet 187: 467–472 [DOI] [PubMed] [Google Scholar]
- Bouadloun F, Donner D, Kurland CG 1983. Codon-specific missense errors in vivo. EMBO J 2: 1351–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlgren A, Ryden-Aulin M 2000. A novel mutation in ribosomal protein S4 that affects the function of a mutated RF1. Biochimie 82: 683–691 [DOI] [PubMed] [Google Scholar]
- Demeshkina N, Jenner L, Westhof E, Yusupov M, Yusupova G 2012. A new understanding of the decoding principle on the ribosome. Nature 484: 256–259 [DOI] [PubMed] [Google Scholar]
- Dong H, Nilsson L, Kurland CG 1996. Co-variation of tRNA abundance and codon usage in Escherichia coli at different growth rates. J Mol Biol 260: 649–663 [DOI] [PubMed] [Google Scholar]
- Ehrenberg M, Kurland CG 1984. Costs of accuracy determined by a maximal growth rate constraint. Q Rev Biophys 17: 45–82 [DOI] [PubMed] [Google Scholar]
- Erie DA, Yager TD, von Hippel PH 1992. The single-nucleotide addition cycle in transcription: A biophysical and biochemical perspective. Annu Rev Biophys Biomol Struct 21: 379–415 [DOI] [PubMed] [Google Scholar]
- Gebler JC, Aebersold R, Withers SG 1992. Glu-537, not Glu-461, is the nucleophile in the active site of (lac Z) β-galactosidase from Escherichia coli. J Biol Chem 267: 11126–11130 [PubMed] [Google Scholar]
- Hanahan D 1983. Studies on transformation of Escherichia coli with plasmids. J Mol Biol 166: 557–580 [DOI] [PubMed] [Google Scholar]
- Hopfield J 1974. Kinetic proofreading: A new mechanism for reducing errors in biosynthetic processes requiring high specificity. Proc Natl Acad Sci 71: 4135–4139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juers DH, Hakda S, Matthews BW, Huber RE 2003. Structural basis for the altered activity of Gly794 variants of Escherichia coli β-galactosidase. Biochemistry 42: 13505–13511 [DOI] [PubMed] [Google Scholar]
- Juers DH, Matthews BW, Huber RE 2012. LacZβ-galactosidase: Structure and function of an enzyme of historical and molecular biological importance. Protein Sci 21: 1792–1807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer E, Farabaugh P 2007. The frequency of translational misreading errors in E. coli is largely determined by tRNA competition. RNA 13: 87–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer EB, Vallabhaneni H, Mayer LM, Farabaugh PJ 2010. A comprehensive analysis of translational missense errors in the yeast Saccharomyces cerevisiae. RNA 16: 1797–1808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurland CG 1992. Translational accuracy and the fitness of bacteria. Annu Rev Genet 26: 29–50 [DOI] [PubMed] [Google Scholar]
- Kurland C, Hughes D, Ehrenberg M 1996. Limitations of translational accuracy. In Escherichia coli and Salmonella: Cellular and molecular biology, Vol. 1 (ed. Neidhardt FC, et al. ), pp. 979–1004 ASM Press, Washington, DC [Google Scholar]
- Leontis NB, Stombaugh J, Westhof E 2002. The non-Watson–Crick base pairs and their associated isostericity matrices. Nucleic Acids Res 30: 3497–3531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limbach PA, Crain PF, McCloskey JA 1994. Summary: The modified nucleosides of RNA. Nucleic Acids Res 22: 2183–2196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Naismith JH 2008. An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnol 8: 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasvall SJ, Chen P, Bjork GR 2004. The modified wobble nucleoside uridine-5-oxyacetic acid in tRNAPro(cmo5UGG) promotes reading of all four proline codons in vivo. RNA 10: 1662–1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninio J 1975. Kinetic amplification of enzyme discrimination. Biochimie 57: 587–595 [DOI] [PubMed] [Google Scholar]
- Ogle JM, Carter AP, Ramakrishnan V 2003. Insights into the decoding mechanism from recent ribosome structures. Trends Biochem Sci 28: 259–266 [DOI] [PubMed] [Google Scholar]
- Parker J 1992. Variations in reading the genetic code. In Transfer RNA in protein synthesis (ed. Hatfield DL, et al. ), pp. 191–267 CRC Press, London, UK [Google Scholar]
- Parker J, Precup J 1986. Mistranslation during phenylalanine starvation. Mol Gen Genet 204: 70–74 [DOI] [PubMed] [Google Scholar]
- Rodnina MV, Gromadski KB, Kothe U, Wieden HJ 2005. Recognition and selection of tRNA in translation. FEBS Lett 579: 938–942 [DOI] [PubMed] [Google Scholar]
- Springgate CF, Loeb LA 1975. On the fidelity of transcription by Escherichia coli ribonucleic acid polymerase. J Mol Biol 97: 577–591 [DOI] [PubMed] [Google Scholar]
- Toth MJ, Murgola EJ, Schimmel P 1988. Evidence for a unique first position codon-anticodon mismatch in vivo. J Mol Biol 201: 451–454 [DOI] [PubMed] [Google Scholar]
- Voorhees RM, Schmeing TM, Kelley AC, Ramakrishnan V 2010. The mechanism for activation of GTP hydrolysis on the ribosome. Science 330: 835–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weixlbaumer A, Murphy FV, Dziergowska A, Malkiewicz A, Vendeix FA, Agris PF, Ramakrishnan V 2007. Mechanism for expanding the decoding capacity of transfer RNAs by modification of uridines. Nat Struct Mol Biol 14: 498–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, McRae MA, Harron S, Rob B, Huber RE 2004. A study of the relationships of interactions between Asp-201, Na+ or K+, and galactosyl C6 hydroxyl and their effects on binding and reactivity of β-galactosidase. Biochem Cell Biol 82: 275–284 [DOI] [PubMed] [Google Scholar]
- Yadavalli SS, Ibba M 2012. Quality control in aminoacyl-tRNA synthesis: Its role in translational fidelity. Adv Protein Chem Struct Biol 86: 1–43 [DOI] [PubMed] [Google Scholar]
- Yuan J, Martinez-Bilbao M, Huber RE 1994. Substitutions for Glu-537 of β-galactosidase from Escherichia coli cause large decreases in catalytic activity. Biochem J 299(Pt 2): 527–531 [DOI] [PMC free article] [PubMed] [Google Scholar]