Abstract
The use of genetic information to guide medication decisions holds great promise to improve therapeutic outcomes through increased efficacy and reduced adverse events. As in many areas of medicine, pediatric research and clinical implementation in pharmacogenetics lag behind corresponding adult discovery and clinical applications. In adults, genotype-guided clinical decision support for medications such as clopidogrel, warfarin and simvastatin are in use in some medical centers. However, research conducted in pediatric populations demonstrates that the models and practices developed in adults may be inaccurate in children, and some applications lack any pediatric research to guide clinical decisions. To account for additional factors introduced by developmental considerations in pediatric populations and provide pediatric patients with maximal benefit from genotype-guided therapy, the field will need to develop and employ creative solutions. In this article, we detail some concerns about research and clinical implementation of pharmacogenetics in pediatrics, and present potential mechanisms for addressing them.
Keywords: clinical pharmacology, genomics, pediatrics, pharmacogenetics, pharmacogenomics
For many patients, personalized medication therapy based on individual genotype can be realized through pharmacogenetic testing. Genetic signatures of tumors are now guiding oncology therapy in both adult and pediatric populations. At many institutions, genome-guided therapies are reaching beyond oncology to influence medication prescribing of target medications in specialty and primary care settings, including anticoagulants, statins and psychiatric medications.
Amid these successes of personalized medicine, the role of pharmacogenetic testing for children is less defined. Despite the growing exposure of children to prescription medications [1,101] and the recognition of marked variability in medication response [2–6], evidence-based recommendations for genotype-guided dosing in pediatric patients have not been well established. As described in Table 1, medications with well-characterized and clinically implemented pharmacogenetic testing lack information about how to interpret and apply genotype information for children. Indeed, several of these medications are not even US FDA approved for use in children, leading to widespread ‘off label’ use in pediatric practice.
Table 1.
Select medications with pharmacogenetic associations.
| Drug | Gene(s) | US FDA pharmacogenetic information on drug label | Pediatric data on FDA label |
|---|---|---|---|
| Codeine | CYP2D6 | Summary of Pharmacogenetic Statement: ultrarapid metabolizers who have multiple copies of the CYP2D6 genotype may experience overdose symptoms at labeled doses. Guidance: “When physicians prescribe codeine-containing drugs, they should choose the lowest effective dose for the shortest period of time and inform their patients about these risks and the signs of morphine overdose” |
“The safety, effectiveness and the pharmacokinetics of codeine sulfate in pediatric patients below the age of 18 have not been established” |
| Mercaptopurine | TPMT | Summary of Pharmacogenetic Statement: individuals homozygous for nonfunctional TPMT alleles are unusually sensitive to the myelosuppressive effects of mercaptopurine and are prone to developing rapid bone marrow suppression following the initiation of treatment. Genotype and phenotype tests are available to determine the TPMT status. Guidance: “If a patient has clinical or laboratory evidence of severe toxicity, particularly myelosuppression, TPMT testing should be considered. In patients who exhibit excessive myelosuppression due to 6-mercaptopurine, it may be possible to adjust the mercaptopurine dose and administer the usual dosage of other myelosuppressive chemotherapy as required for treatment” |
Approved for “maintenance therapy of acute lymphatic (lymphocytic, lymphoblastic) leukemia as part of a combination regimen” |
| Warfarin |
CYP2C9 VKORC1 CYP4F2 |
Summary of Pharmacogenetic Statement: CYP2C9 and VKORC1 genotype information, when available, can assist in selection of the initial dose of warfarin Guidance: “If the patient’s CYP2C9 and/or VKORC1 genotype are known, consider these ranges (table provided) in choosing the initial dose” Also, “consult the latest evidence-based clinical practice guidelines from the American College of Chest Physicians…” |
“Adequate and well-controlled studies … have not been conducted in any pediatric population, and the optimum dosing, safety and efficacy in pediatric patients is unknown” |
| Simvastatin | SLCO1B1 | None | “Simvastatin orally disintegrating tablets are indicated as an adjunct to diet to reduce total-C, LDL-C and apoB levels in adolescent boys and girls who are at least 1 year postmenarche, 10–17 years of age, with heterozygous familial hypercholesterolemia, if after an adequate trial of diet therapy the following findings are present: LDL cholesterol remains ≥190 mg/dl; or LDL cholesterol remains ≥160 mg/dl and there is a positive family history of premature CVD or two or more other CVD risk factors are present in the adolescent patient” |
C: Cholesterol; CVD: Cardiovascular disease; LDL: Low-density lipoprotein; TPMT: Thiopurine methyltransferase.
Data taken from [104].
Both the lay public and healthcare providers are increasingly recognizing that “children are not small adults,” owing in large part to successful efforts of the FDA and the Best Pharmaceuticals for Children Act to highlight the special considerations for medication use in children [102]. However, the practice of treating genetic testing as special or exceptional and overemphasizing the differences between children and adults is creating barriers against establishing adequate evidence for introducing changes into clinical practice in the realm of pediatric personalized medicine. This will certainly be the case if randomized controlled trials (RCTs) are the required benchmark before implementing changes in practice. In order to leverage data obtained from adult cohorts, the differences between adults and children need to be described and quantified, followed by an approach in which clinical practice and outcomes are studied as state-of-the-art knowledge is incrementally introduced into clinical care. While some may be uncomfortable with this approach, an honest survey of our current pediatric practices reveals that expert consensus, standard of care and practice evolution, rather than evidence provided by controlled research trials, form the basis for the majority of our current practices as pediatricians and pediatric subspecialists.
In this article, we use contemporary examples to illustrate unique barriers in pediatric pharmacogenetics. We then pose potential solutions to overcome these barriers and accelerate clinical application of research findings to an appropriate pace.
Issue 1: pediatric patients grow & develop over time
A hallmark of pediatric medicine is the emphasis on the growth and developmental maturation of the patient. Along with physical growth and cognitive development, the expression patterns of genes in drug response pathways evolve and change over time [7,8]. Sentinel studies have emphasized the developmental ontogeny of pharmacogenetic pathways, which include drug metabolism enzymes, drug transporters and drug targets [9–12]. The progressive maturation of these pathways as well as the ongoing physical development of pediatric patients may affect the application of adult-derived pharmacogenetic data for the care of children. An example is provided by the drug–gene interaction of simvastatin and SLCO1B1 [13]. Simvastatin is an HMG-CoA reductase inhibitor used to treat hyperlipidemia and is one of the most commonly prescribed medications in the USA. The gene SLCO1B1 encodes the organic anion transporter protein OATP1B1, which has been shown to play a role in drug distribution. In adults, individuals with a variant (specifically rs4149056) in SLCO1B1 have higher plasma drug concentrations and are at increased risk for myopathy and rhabdomyolysis. This has led to the screening of many patients for this SLCO1B1 risk allele prior to initiation or dose escalation of simvastatin therapy. Published guidelines developed by the Clinical Pharmacogenetics Implementation Consortium recommend lowering simvastatin dose, monitoring creatine kinase and/or using an alternative statin for individuals with the SLCO1B1 risk allele(s) [13]. In addition to genotype, increasing age and higher dose are known to increase risk for simvastatin toxicity. The FDA now recommends against starting at the 80-mg simvastatin dose in any individual. Although SLCO1B1 variants have been associated with increased plasma concentrations for related medications, such as pravastatin, a clear relationship of genotype to myopathy risk owing to HMG-CoA reductase inhibitors other than simvastatin has not been demonstrated.
Currently, only a small number of children are prescribed simvastatin, typically for familial hypercholesterolemia or after an organ transplant. However, pediatric exposures can be expected to rise with the adoption of universal cholesterol screening, as advised by the National Heart, Lung and Blood Institute (MD, USA) and the American Academy of Pediatrics (CA, USA) [14]. Simvastatin is approved for use in children aged 10 years and older with heterozygous familial hypercholesterolemia, starting with a dose of 10 mg daily and increasing to a maximum daily dose of 40 mg. Wide variations in height, weight and body surface area are expected in the age range covered by this recommendation, but no guidance is given with respect to dose titration based on patient size. There are no specific guidelines on whether simvastatin prescribing in children should be informed by genotype, as there are no reported studies of the effect of the rs4149056 variant in SLCO1B1 on simvastatin pharmacokinetics or drug effects in pediatric patients. The Pharmacogenomics Knowledgebase (PharmGKB), a curated resource for genetic variation and drug response, states with respect to pediatric patients, “At the time of the development of this recommendation, there are no data available on the possible role of SLCO1B1 in simvastatin-induced myopathies in pediatric patient populations; however, there is no reason to suspect that SLCO1B1 variant alleles would affect simvastatin hepatic uptake differently in children compared with adults” [103].
Studies of simvastatin in pediatrics are lacking, but a report has been published of genotype–phenotype correlation between pravastatin and SLCO1B1 genotype [15]. In this study of 32 children (20 with heterozygous familial hypercholesterolemia and 12 with cardiac transplantation), SLCO1B1 variants were associated with significantly decreased plasma concentrations of pravastatin. Although based on a small number of individuals, these data suggest the influence of SLCO1B1 variants may be inconsequential or even reversed at young ages. In addition to this observation, unanswered practical questions stand as barriers to implementing SLCO1B1-guided clinical decision support for simvastatin dosing in children when other developmental issues are considered. These questions include:
Given the impact of increasing age and dose on toxicity, are children of any genotype at risk for muscle-related complications?
If younger individuals with variant alleles are not at increased risk, then at what age does a patient’s risk for myopathy and rhabdomyolysis increase toward adult levels?
Are there other clinical or genetic variants that are important to consider in children?
How can we best assess myopathy in the youngest patients who are unable to report specific symptoms?
In addition to the unclear impact of SLCO1B1 variants on risk for myopathy in children, the optimal minimum age for use of HMG-CoA reductase inhibitors is not well established. Previous guidelines supported use of these medications after the age of 10 years in boys, and after the onset of menses in girls, owing to theoretical concerns that early exposure may affect sexual maturation, especially in female patients [16]. More recent guidelines indicate both girls and boys over 10 years of age are candidates for drug therapy regardless of the level of sexual maturity, as trials to date have not identified adverse effects of these medications on sexual maturation or growth [14]. However, studies to date have limited follow-up data for this end point. Further studies to determine late effects and those due to chronic exposure, specifically during the critical maturational phase of adolescence, are required. Interactions of age, gender and genotypes may elucidate the best candidates for treatment with this drug class versus other lipid-lowering strategies.
Issue 2: therapeutic goals are often pediatric-specific & influenced by unique extrinsic factors
In addition to the developmental changes intrinsic to pediatric patients, some medications are used for pediatric-specific indications and are given in the setting of environmental influences that do not exist in adults. These pediatric-specific indications and unique extrinsic factors are important to consider in performing or evaluating pharmacogenetic research for application in children. Extrinsic factors that may be pediatric-specific include the duration of medication exposures (longer or shorter than adults), specific adverse drug effects that may not be seen in adults, specific drug–drug interactions due to different medication exposure patterns from adults and unique drug–environment interactions. Given these factors, extrapolation of target drug concentrations or determination of environmental impact from adult populations is not appropriate. The required concentration in the target organ may be substantially different, and there may simply be no data from adults that is even relevant to the pediatric population.
NSAIDs provide an example case. In adult patients, NSAIDs are primarily used for anti-inflammatory and analgesic effects. However, in neonates NSAIDs, such as indomethacin and ibuprofen, are used to pharmacologically induce closure of a patent ductus arteriosus (PDA). A review of the literature reveals that the first report of using indomethacin for PDA closure was over 35 years ago, and yet pharmacokinetic (drug distribution and elimination) and pharmacodynamic (drug effect) studies continue to attempt to determine the ideal dosing for these agents in the neonatal population [17–20]. Difficulties in determining accurate pharmacodynamic models to predict drug effect may be due to failure to include all the relevant covariates; although gestational age, measures of renal function and patient weight are nearly universally included in these studies, other factors, such as concomitant gentamicin, H2 receptor antagonists and heparin (all commonly used in critically ill neonates), may have vasodilatory effects that counteract NSAID effect [21]. Pharmacogenetic associations to indomethacin efficacy may therefore be influenced by these age-specific factors. The risk profiles of these medications in neonates include dangers of nephrotoxicity and gastrointestinal bleeding, as they do for adults. Neonates treated with NSAIDs for persistent PDA have an additional risk of spontaneous intestinal perforation, an adverse event of extreme rarity in adults [22–24]. Studies of pharmacogenetic predictors of NSAID efficacy must also determine the incidence of adverse events, including pediatric-specific adverse events, as variants increasing the levels or duration of exposure may also increase these risks. Finally, recent studies have explored the effect of enteral intake of breastmilk or formula during NSAID administration, another clinical factor in which genetic variants may contribute to adverse events, including the pediatric-specific adverse event of necrotizing enterocolitis [25].
Issue 3: ethical issues regarding clinical pharmacogenetic testing in children
Recent statements from professional organizations including the American Academy of Pediatrics and the American College of Medical Geneticists state that the use of established pharmacogenetic tests to improve the use of drugs in minors is ethically appropriate [26,27]. However, with the development and increased use of multiplex platforms for pharmacogenetic tests and the advent of clinical whole-exome sequencing, the likelihood of identifying pharmacogenetic variants unrelated to any current or planned therapies increases. Current guidance from professional societies does not directly address this scenario, and a consensus on how to balance the risks and benefits of disclosing these results is yet to be determined.
The perspective of performing genetic testing within the context of a family is rarely taken when performing clinical pharmacogenetic testing in adults, but is brought into focus when the patient is accompanied by a parent or other family members. This family context elicits several important considerations. First, the genetic test results can have real pharmacogenetic implications for other family members. For example, if a child is found to be a poor metabolizer for TPMT, which encodes the enzyme responsible for the breakdown of thiopurine medications, this indicates that each parent is either a poor or intermediate metabolizer, and each full sibling has increased risk of being a poor or intermediate metabolizer (at least 25 and 50% risk, respectively). Discussion of this risk with family members to foster preprescription genotyping for relatives may be warranted, but the burden of sharing information with relatives will rest with the parents. Second, reducing risk for the pediatric patient may require testing in other family members. Codeine, a well-characterized and frequently used prodrug, requires metabolism by CYP2D6 to generate its active metabolite, morphine. CYP2D6 is a highly polymorphic gene with variations in gene sequence, copy number and pseudogene arrangement. Given typical doses, individuals who are poor metabolizers are unlikely to achieve adequate analgesia, while ultrarapid metabolizers who have multiple copies of the gene encoding active enzyme can accumulate high concentrations of morphine and experience symptoms of overdose. Mothers who have active CYP2D6 secrete morphine in their breastmilk during codeine therapy. In order to protect newborn and infant patients from dangerous and sometimes lethal doses of morphine transmitted through breastmilk, detection of ultrarapid metabolizer status with the mother’s genotype, rather than the infant’s, is required [28,29]. Third, as with all genetic testing, the risk of detecting misattributed paternity should be considered and discussed prior to testing, including the possibility of false positive misattributed paternity. An added complexity with pharmacogenetic testing results from the conventional ‘star-allele’ nomenclature, where each allele is named based upon a specific combination of variants in the gene. Depending on the variants represented on any given assay and the algorithms used to associate variants to star-allele ‘calls,’ different platforms may assign different star-alleles to the same underlying set of genetic variations, resulting in apparent nonpaternity.
Issue 4: undefined threshold of evidence for implementation
An additional barrier to clinical implementation of pharmacogenetics for the pediatric patient is the unclear minimum threshold of evidence required prior to introducing pharmacogenetic testing into clinical practice. In an ideal world, generation of data from modeling and observational studies would be followed by prospective RCTs to determine whether genotype-guided therapy improved outcomes. Broad clinical implementation would only follow in pediatric patients if evidence from these well designed and executed RCTs proved efficacy and demonstrated a favorable cost–benefit ratio. In personalized medicine, and specifically in personalized pediatrics, the gold-standard RCT may be unfeasible for several reasons including sample size limitations, lack of measureable meaningful outcomes (e.g., mortality or significant adverse drug reaction), cost and perceived lack of equipoise based on adult studies and limited pediatric data.
An example case may be provided by warfarin. In adult studies, variants in genes including VKORC1, CYP2C9 and CYP4F2 have been shown to influence therapeutic dose, and genotype-guided dose selection results in earlier therapeutic drug levels [30,31] and reduced early complications [32,33]. In some centers, this has prompted a move toward genotype-guided therapy for patients beginning warfarin therapy. In pediatric patients, increasing evidence demonstrates that genetic variations play an important role, similar to adults [28]. Several studies have shown that CYP2C9 and VKORC1 polymorphisms influence warfarin dose in children, but have also highlighted important differences between adults and children, such as the important influence of age in pediatric dosing [34–40]. At what point should genotype data be used to clinically guide dosing in children? In adults, two ongoing prospective RCTs to evaluate the value of genotype-guided warfarin dosing are each enrolling over 1200 patients [41,42], a number three-times greater than the total number of children reported in the seven pediatric studies reported to date (n = 415) [34–40]. To appropriately conduct pediatric studies, children in different age ranges would need to be stratified, increasing the number of participants required. Owing to maturational changes in the thrombotic cascade, specific study of adolescents including Tanner staging and stratification by gender may also be necessary. More importantly, randomization to the nongenotyped arm may be unethical given the weight of the current evidence. Global application of the RCT standard would be inappropriately restrictive and would prevent pediatric patients from the benefits of this evolving field. However, a balance must be achieved between minimizing risk in this vulnerable patient population and optimizing delivery of the benefits of progress.
Solution 1: obtaining ontogeny & modeling data
Several strategies may be employed to efficiently bridge knowledge gaps in pediatric pharmacogenetics, pharmacokinetics and pharmacodynamics to enable evidence-based advice for genotype-guided medication dosing in children. First, studies of the maturational patterns of known drug-metabolizing enzymes must be completed across the developmental spectrum, with sufficient sampling across ages, gender, disease states and ethnicities. These studies may be facilitated by new technologies including RNAseq and metabolomics, which enable characterization of a single sample across a spectrum of transcripts and products. Sufficient diversity of sample collection may be facilitated by collaborations across institutions and by leveraging the power of biobank structures. This approach will determine typical trajectories, define at what age children mature to adult patterns and also refine the extent of interindividual variability. Resulting data will facilitate the interpretation and application of adult-derived data and determine the generalizability of pediatric data within children of various ages and clinical presentations.
Our efforts must not stop at profiling for known drug-metabolizing enzymes, transporters and targets because young patients may express unique patterns at specific developmental stages. These novel pathways will shed light on the biology of development, give insight for personalized pediatrics and provide novel targets for intervention in both pediatric and adult patients. These developmentally dynamic enzymes and transporters, known and novel, can be incorporated into modeling programs, such as SimCYP (Simcyp Ltd; Sheffield, UK), with the goal of predicting pharmacokinetics in children in silico [43]. With this knowledge of the developmental ontogeny and predicted pharmacokinetics, clinical study designs can be optimized prior to enrolling any patients so that models can be validated in clinical settings with samples from treated children, greatly increasing the efficiency of such studies [44]. The compilation, publication and use of this ontologic data are critical in advancing the field of pediatric pharmacogenetics.
Solution 2: alternative approaches to sample collection & analysis methodologies
Studies of ontogeny, pharmacokinetics and pharmacogenetics will all rely upon analyses of biospecimens from a diverse range of children. Studies in pediatric pharmacogenetics must consider methods of specimen collection that overcome obstacles frequently encountered in this population. For studies in which repeated or scheduled blood collections are not feasible, either due to parental concerns or limited volume availability (e.g., in neonates), expertise in analysis of sparse, random samples can guide study design and data interpretation. Additionally, investigation of the utility of more readily available biospecimens, including urine and saliva, will allow for more frequent sampling with methods that are more acceptable to patients and families. Novel methods of noninvasive sampling are ideal for pediatric applications and should be incorporated into ongoing collections and studies. For example, microfluidics technology may allow for repeated and accurate sampling of even our smallest patients in the neonatal intensive care unit [45–47]. Combining these collection methods with high-sensitivity analyses, such as high performance liquid chromatography, already shown to enable determination of concentrations of multiple drugs from a single dried blood spot card [48], creates a feasible way to perform suitably large pharmacogenetic studies in the pediatric population.
Opportunistic studies in which patients who are receiving a medication of interest who, as part of their clinical treatment course, are recruited and consented will be useful for pediatric pharmacogenetic research, as additional risks due to medication exposure would not be an issue. Genotypes may be determined from DNA obtained from remnant blood samples, saliva or possibly even urine [49]. Additionally, the drug concentration data measured from these samples of convenience can be incorporated into pharmacokinetic modeling, enabling study of variable drug absorption, distribution, metabolism and elimination. Patients receiving medications as part of their clinical care are monitored for treatment response; thus, pharmacodynamic studies can be also completed using data gathered during medical care. Although this departs from the idealized timed assessments after drug administration, useful data can be gleaned while minimizing risks [50,51].
Solution 3: development of a pediatric pharmacogenetic repository infrastructure
An important mechanism for advancing personalized pediatrics through pharmacogenetics is the development of an annotated biorepository focused on this area of research. Ideally, this would include multiple biospecimen types, complete annotation of exposures, outcomes and clinical data, and a wide spectrum of ages and diseases. The ability to select participants by their genotype or clinical features and recontact them for future studies would markedly enhance the utility of the resource. Although this endeavor could be undertaken by a single institution, its full potential would not be realized unless participants are enrolled from multiple institutions. In a collaborative consortium model, each participating institution can maintain ownership of one or more phenotypes for which all member institutions would contribute systematic data and samples. Existing networks such as the Electronic Medical Records and Genomics (eMERGE) Network, which includes two sites specifically focused on pediatrics, provide a model for this type of collaboration across institutions [52].
One of the most cumbersome yet vital aspects of repository development is the collection of phenotypic data, including data regarding medication exposures, clinical comorbidities and outcomes. Even if lifetime medical records are available, documentation may not be accurate for some exposures (e.g., over-the-counter medications or supplements) or responses (e.g., sleep disturbance due to stimulant mediations). Thus, engaging participants and planning for adequate study personnel will be necessary to accurately assess relevant phenotypes for each submission and to supplement medical records data with prospectively and retrospectively collected information relevant to each exposure. Critical information includes comprehensive medication administration data including all dosages, time of administration and timing of collection for each sample. For specific medication exposures, outcome data not documented as part of routine clinical care may be essential. For example, study of red man syndrome due to vancomycin exposure would include infusion rate as well as a detailed description of the appearance and resolution of the rash; these data are best documented at the time of the event, while long-term outcomes (e.g., cognitive changes due to neonatal exposure to sedatives) require follow-up in subsequent years.
To facilitate complementary avenues of research, the repository should store multiple sample types – including urine, saliva, plasma, whole blood, samples for RNA analysis and extracted DNA. Not every sample would be necessary for each individual, but for specific events or exposures different samples may be targeted. The importance of collecting drug-exposed control samples cannot be overemphasized to enable study of adverse drug events. Additionally, in the setting of polypharmacy, having samples from patients on various combinations of medications may enable discrimination of drug–drug events from interindividual variability. Lastly, the phenotyping done for each patient and each targeted exposure or reaction must be standardized across institutions to allow for the ability to combine cases and controls when performing adequately powered studies of rare events.
The inclusion of samples from healthy volunteers and family members would also strengthen the resource. Genetic cataloging of these samples will accelerate the identification of individuals representing the spectrum of genetic variability. Individuals may then be selected for enrollment in pharmacogenetic studies, circumventing the need for such large enrollment numbers when uncommon or rare variants are of specific interest. Depending on the risks involved, study designs can be optimized by inclusion of healthy individuals. For example, to evaluate CYP2D6 activity, dextromethorphan, a common ingredient in over-the-counter cough syrups, can be administered to assess an individual’s pharmacokinetic profile instead of codeine. While beyond the scope of this manuscript, addressing the ethical concerns involved in the development of such a repository would be mandatory. The inclusion of children and their relatives [53,54], genotype-guided substudies [55] and the management of pharmacogenomic data [56] must be addressed from an ethics perspective.
Solution 4: reaching consensus on evidence
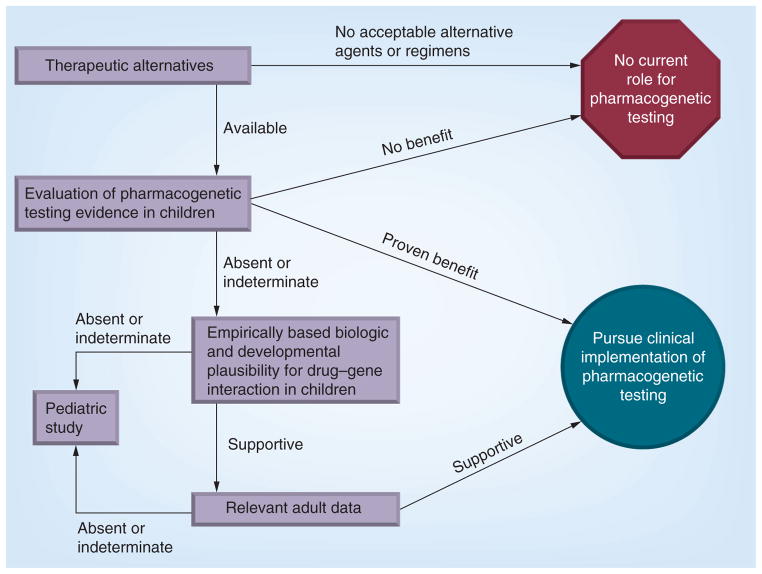
Determining the threshold of evidence required for clinical application of pharmacogenetics in pediatrics has the potential to aid in appropriate and timely clinical implementation, with the understanding that such a definition would serve only as a guideline. Individual institutions, providers or families may present with specific circumstances where reason and judgment guide appropriate clinical care away from a consensus guideline. To further the discussion in this arena, we propose several factors for consideration as depicted in Figure 1. The initial step of evaluation should include a thoughtful analysis of the therapeutic alternatives for the medication under consideration. Is there a viable alternative (a different drug, a different dose or a change to monitoring) if an ‘actionable’ genetic test result is found? Through this process, the determination may be that pharmacogenetic testing is not warranted, as there is no known effective alternative, or because an equally effective alternate agent is available without pharmacogenetic variability; this logic has led some to prescribe alternate agents, such as oxycodone instead of codeine, rather than perform CYP2D6 genotyping. In other cases such as thiopurine drugs, where TPMT genotyping and dose reduction for individuals with reduced or absent functional enzymatic activity can avoid life-threatening bone marrow suppression without a reduction in efficacy, genotype-guided therapy has the potential to play more of a role.
Figure 1. Evaluating pharmacogenetic applications.
This schematic demonstrates multiple factors that need to be considered when evaluating the existing evidence for introducing a pharmacogenetic test and guidance for a specific medication into clinical practice. The relative influence of evidence types and the weight of evidence required may also take into account: cost of pharmacogenetic testing; cost of standard and alternate therapies; severity of the event to be avoided, whether adverse drug event or therapeutic failure; and frequency of the event.
If an effective therapeutic alternative is available, an assessment of any available pediatric-based data supporting pharmacogenetic-guided therapy should be evaluated for strength and relevance. Study type, study design, cohort populations and effect size for the genetic variant should all be considered. In the absence of studies specifically evaluating genotype-guided therapy for the target medication, indirect evidence supporting the drug–gene interaction in children should be examined, including studies of ontogeny of drug metabolism and response pathways in children, and observational or retrospective studies finding associations between the relevant genotypes and outcomes in children. If data exist for the drug–gene interaction in adults, those studies should also be closely evaluated, with additional consideration given to determination of whether the drug and gene of interest have comparable pharmacology, physiology, indication for use, therapeutic goals and adverse event profiles in children as in the reported adult populations.
In an ideal case with available therapeutic alternatives and pediatric-derived, prospectively obtained data supporting use of genetic data to improve medication outcomes, the decision to move forward with clinical implementation of genotype-guided therapeutics is straightforward. However, given the difficulty of ascertaining prospective data with clinically meaningful outcomes for any pediatric cohort, this scenario will be the exception. More commonly, the scenario will include minimal (if any) pediatric-derived data but consistent adult-derived data. In this setting, we propose that pharmacogenetic testing moves forward into clinical use if/when the following conditions are met (Figure 1):
An evidence-based therapeutic alternative (e.g., a different drug, dose or therapeutic monitoring plan) is available for pediatric patients;
Strong, adult-derived data relevant to pediatric therapeutic indications, risks and pharmacology support genotype-guided therapy;
Evidence supporting the same association is present in pediatric-derived data (which may come from retrospective data, pharmacokinetic studies, well-established ontogeny or interventional studies).
In situations where there are pediatric-specific therapeutic indications and/or risks, the threshold for pediatric-specific evidence will necessarily be higher, to include prospective data collection. Depending on the drug and the nature of the genetic association, this prospective trial may include analyses of pharmacokinetic data (e.g., for drug metabolism variants) and/or pharmacodynamic data (e.g., for drug receptor variants).
Conclusion
Moving pediatric pharmacogenetics toward clinical implementation will require collaboration between many providers, researchers and institutions. A consortium with the requisite expertise, patient volume and diversity, and stable funding should be established with the collective goal of optimizing the pharmaceutical care of children, including neonates. Formalizing such a consortium will facilitate the development of multidisciplinary teams bringing together specific skill sets required to address these topics with the samples and phenotypic data available in the consortium’s repository. In addition to fostering research endeavors in this arena, this consortium will prioritize the development of guidelines defining consensus thresholds for clinical application of pharmacogenetics in the pediatric space to guide research efforts and enable more rapid clinical implementation as indicated.
Future perspective
The establishment of a creative and collaborative research consortium will address many current challenges in the field of pediatric pharmacogenetics. Ontologic studies will result in more complete understanding of changes in drug disposition, metabolism and effects across the full pediatric age spectrum. Models developed from these data will be confirmed and refined by employing state-of-the-art, high-throughput, highly sensitive technologies to analyze noninvasively obtained samples, enabling development of age-specific medication regimens and novel therapies specifically designed for pediatric patients. Coupling these data with advances in genome science will lead to a further step: personalized medication choices based on age and genetic variation, thereby maximizing therapeutic benefits and minimizing medication risks.
Executive summary.
Developmental issues in pediatric pharmacogenetics
As children grow and develop, the pathways affecting drug distribution and response also change.
Children have different therapeutic goals and adverse event profiles than adults, as well as unique drug–drug, drug–food and drug–environment exposures.
Special issues for pediatric pharmacogenetic research
Ethical factors and familial context should be considered when genetic testing is performed in children, but should not paralyze progress.
To date, there is no clear consensus on the level of evidence required to determine when pharmacogenetic testing may be clinically implemented for pediatric patients.
Potential solutions
Studies of genetic ontogeny and subsequent modeling can be used to predict pharmacokinetics and drug effects in children even for medications not directly studied.
Use of remnant samples and noninvasive sampling techniques can confirm modeling results despite restricted sample access.
Collaboration across institutions can lead to the collection of annotated samples from adequately exposed pediatric patients facilitating convincing pharmacogenomic research, as well as to the development of guidance on evidence thresholds for clinical implementation.
Acknowledgments
The authors would like to thank EW Clayton, D Roden and L Muglia for their thoughtful review of this manuscript.
Footnotes
For reprint orders, please contact: reprints@futuremedicine.com
Financial & competing interests disclosure
SL Van Driest has been supported by the NIH/NIGMS Clinical Pharmacology Training Program (5T32 GM007569-33). TL McGregor is supported by NIH/NICHD grant 5K23HD000001. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
▪ of interest
▪▪ of considerable interest
- 1.Lasky T, Ernst FR, Greenspan J. Use of analgesic, anesthetic, and sedative medications during pediatric hospitalizations in the United States 2008. Anesth Analg. 2012;115(5):1155–1161. doi: 10.1213/ANE.0b013e31825b6fb2. [DOI] [PubMed] [Google Scholar]
- 2.Bartelink IH, Wolfs T, Jonker M, et al. Highly variable plasma concentrations of voriconazole in pediatric hematopoietic stem cell transplantation patients. Antimicrob Agents Chemother. 2013;57(1):235–240. doi: 10.1128/AAC.01540-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guy-Viterbo V, Scohy A, Verbeeck RK, Reding R, Wallemacq P, Musuamba FT. Population pharmacokinetic analysis of tacrolimus in the first year after pediatric liver transplantation. Eur J Clin Pharmacol. 2013;69(8):1533–1542. doi: 10.1007/s00228-013-1501-0. [DOI] [PubMed] [Google Scholar]
- 4.Ince I, de Wildt SN, Peeters MY, et al. Critical illness is a major determinant of midazolam clearance in children aged 1 month to 17 years. Ther Drug Monit. 2012;34(4):381–389. doi: 10.1097/FTD.0b013e31825a4c3a. [DOI] [PubMed] [Google Scholar]
- 5.Lee JW, Kang HJ, Lee SH, et al. Highly variable pharmacokinetics of once-daily intravenous busulfan when combined with fludarabine in pediatric patients: Phase I clinical study for determination of optimal once-daily busulfan dose using pharmacokinetic modeling. Biol Blood Marrow Transplant. 2012;18(6):944–950. doi: 10.1016/j.bbmt.2011.11.025. [DOI] [PubMed] [Google Scholar]
- 6.Williams JH, Jayaraman B, Swoboda KJ, Barrett JS. Population pharmacokinetics of valproic acid in pediatric patients with epilepsy: considerations for dosing spinal muscular atrophy patients. J Clin Pharmacol. 2012;52(11):1676–1688. doi: 10.1177/0091270011428138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leeder JS. Pharmacogenetics and pharmacogenomics. Pediatr Clin North Am. 2001;48(3):765–781. doi: 10.1016/s0031-3955(05)70338-2. [DOI] [PubMed] [Google Scholar]
- 8▪▪.Wagner J, Leeder JS. Pediatric pharmacogenomics: a systematic assessment of ontogeny and genetic variation to guide the design of statin studies in children. Pediatr Clin North Am. 2012;59(5):1017–1037. doi: 10.1016/j.pcl.2012.07.008. Provides a thorough and systematic discussion of the genetic and developmental influences on HMG-CoA reductase effects in children. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alcorn J, McNamara PJ. Ontogeny of hepatic and renal systemic clearance pathways in infants: part I. Clin Pharmacokinet. 2002;41(12):959–998. doi: 10.2165/00003088-200241120-00003. [DOI] [PubMed] [Google Scholar]
- 10.Blake MJ, Castro L, Leeder JS, Kearns GL. Ontogeny of drug-metabolizing enzymes in the neonate. Semin Fetal Neonatal Med. 2005;10(2):123–138. doi: 10.1016/j.siny.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 11.Bouwmeester NJ, van den Anker JN, Hop WC, Anand KJ, Tibboel D. Age- and therapy-related effects on morphine requirements and plasma concentrations of morphine and its metabolites in postoperative infants. Br J Anaesth. 2003;90(5):642–652. doi: 10.1093/bja/aeg121. [DOI] [PubMed] [Google Scholar]
- 12.de Wildt SN, Kearns GL, Leeder JS, van den Anker JN. Glucuronidation in humans. Pharmacogenetic and developmental aspects. Clin Pharmacokinet. 1999;36(6):439–452. doi: 10.2165/00003088-199936060-00005. [DOI] [PubMed] [Google Scholar]
- 13.Wilke RA, Ramsey LB, Johnson SG, et al. The Clinical Pharmacogenomics Implementation Consortium: CPIC guideline for SLCO1B1 and simvastatin-induced myopathy. Clin Pharmacol Ther. 2012;92(1):112–117. doi: 10.1038/clpt.2012.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Expert Panel on Integrated Guidelines for Cardiovascular Health and Risk Reduction in Children and Adolescents; National Heart, Lung, and Blood Institute. Expert panel on integrated guidelines for cardiovascular health and risk reduction in children and adolescents: summary report. Pediatrics. 2011;128(Suppl 5):S213–S256. doi: 10.1542/peds.2009-2107C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hedman M, Antikainen M, Holmberg C, et al. Pharmacokinetics and response to pravastatin in paediatric patients with familial hypercholesterolaemia and in paediatric cardiac transplant recipients in relation to polymorphisms of the SLCO1B1 and ABCB1 genes. Br J Clin Pharmacol. 2006;61(6):706–715. doi: 10.1111/j.1365-2125.2006.02643.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McCrindle BW, Urbina EM, Dennison BA, et al. Drug therapy of high-risk lipid abnormalities in children and adolescents: a scientific statement from the American Heart Association Atherosclerosis, Hypertension, and Obesity in Youth Committee, Council of Cardiovascular Disease in the Young, with the Council on Cardiovascular Nursing. Circulation. 2007;115(14):1948–1967. doi: 10.1161/CIRCULATIONAHA.107.181946. [DOI] [PubMed] [Google Scholar]
- 17.Friedman WF, Hirschklau MJ, Printz MP, Pitlick PT, Kirkpatrick SE. Pharmacologic closure of patent ductus arteriosus in the premature infant. N Engl J Med. 1976;295(10):526–529. doi: 10.1056/NEJM197609022951003. [DOI] [PubMed] [Google Scholar]
- 18.Heymann MA, Rudolph AM, Silverman NH. Closure of the ductus arteriosus in premature infants by inhibition of prostaglandin synthesis. N Engl J Med. 1976;295(10):530–533. doi: 10.1056/NEJM197609022951004. [DOI] [PubMed] [Google Scholar]
- 19.Hirt D, Van Overmeire B, Treluyer J-M, et al. An optimized ibuprofen dosing scheme for preterm neonates with patent ductus arteriosus, based on a population pharmacokinetic and pharmacodynamic study. Br J Clin Pharmacol. 2008;65(5):629–636. doi: 10.1111/j.1365-2125.2008.03118.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van Overmeire B, Touw D, Schepens PJ, Kearns GL, van den Anker JN. Ibuprofen pharmacokinetics in preterm infants with patent ductus arteriosus. Clin Pharmacol Ther. 2001;70(4):336–343. [PubMed] [Google Scholar]
- 21▪.Reese J, Veldman A, Shah L, Vucovich M, Cotton RB. Inadvertent relaxation of the ductus arteriosus by pharmacological agents that are commonly used in the neonatal period. Semin Perinatol. 2010;34(3):222–230. doi: 10.1053/j.semperi.2010.02.007. Highlights novel considerations with respect to drug–drug interactions in evaluating patent ductus arteriosus and response to attempts at medical closure. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ervens J, Schiffmann L, Berger G, Hoffmeister B. Colon perforation with acute peritonitis after taking clindamycin and diclofenac following wisdom tooth removal. J Craniomaxillofac Surg. 2004;32(5):330–334. doi: 10.1016/j.jcms.2004.05.007. [DOI] [PubMed] [Google Scholar]
- 23.Kara C, Derici H, Nazli O, Tansug T, Bozdag AD. Colonic perforation after short-term use of nonsteroidal antiinflammatory drugs: report of two cases. Tech Coloproctol. 2009;13(1):75–78. doi: 10.1007/s10151-008-0434-6. [DOI] [PubMed] [Google Scholar]
- 24.Wadhawan R, Oh W, Vohr BR, et al. Spontaneous intestinal perforation in extremely low birth weight infants: association with indometacin therapy and effects on neurodevelopmental outcomes at 18–22 months corrected age. Arch Dis Child Fetal Neonatal Ed. 2013;98(2):F127–F132. doi: 10.1136/archdischild-2011-300659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clyman R, Wickremasinghe A, Jhaveri N, et al. Enteral feeding during indomethacin and ibuprofen treatment of a patent ductus arteriosus. J Pediatr. 2013;163(2):406–411.e4. doi: 10.1016/j.jpeds.2013.01.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fallat ME, Katz AL, Mercurio MR, et al. Ethical and policy issues in genetic testing and screening of children. Pediatrics. 2013;131(3):620–622. doi: 10.1542/peds.2012-3680. [DOI] [PubMed] [Google Scholar]
- 27.Ross LF, Saal HM, David KL, Anderson RR American Academy of Pediatrics; American College of Medical Genetics and Genomics. Technical report: ethical and policy issues in genetic testing and screening of children. Genet Med. 2013;15(3):234–245. doi: 10.1038/gim.2012.176. [DOI] [PubMed] [Google Scholar]
- 28▪.Madadi P, Ross CJ, Hayden MR, et al. Pharmacogenetics of neonatal opioid toxicity following maternal use of codeine during breastfeeding: a case-control study. Clin Pharmacol Ther. 2009;85(1):31–35. doi: 10.1038/clpt.2008.157. Illustrates the importance of maternal genotype and drug exposure on neonatal effects. [DOI] [PubMed] [Google Scholar]
- 29.Vandervaart S, Berger H, Sistonen J, et al. CYP2D6 Polymorphisms and codeine analgesia in postpartum pain management: a pilot study. Ther Drug Monit. 2011;33(4):425–432. doi: 10.1097/FTD.0b013e3182272b10. [DOI] [PubMed] [Google Scholar]
- 30.Klein TE, Altman RB, Eriksson N, et al. International Warfarin Pharmacogenetics Consortium, The International Warfarin Pharmacogenetics Consortium. Estimation of the warfarin dose with clinical and pharmacogenetic data. N Engl J Med. 2009;360(8):753–764. doi: 10.1056/NEJMoa0809329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wadelius M, Chen LY, Lindh JD, et al. The largest prospective warfarin-treated cohort supports genetic forecasting. Blood. 2009;113(4):784–792. doi: 10.1182/blood-2008-04-149070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Epstein RS, Moyer TP, Aubert RE, et al. Warfarin genotyping reduces hospitalization rates: results from the MM-WES (Medco-Mayo Warfarin Effectiveness Study) JAm Coll Cardiol. 2010;55(25):2804–2812. doi: 10.1016/j.jacc.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 33.Hirai K, Hayashi H, Ono Y, et al. Influence of CYP4F2 polymorphisms and plasma vitamin K levels on warfarin sensitivity in Japanese pediatric patients. Drug Metab Pharmacokinet. 2013;28(2):132–137. doi: 10.2133/dmpk.dmpk-12-rg-078. [DOI] [PubMed] [Google Scholar]
- 34.Biss TT, Avery PJ, Brandão LR, et al. VKORC1 and CYP2C9 genotype and patient characteristics explain a large proportion of the variability in warfarin dose requirement among children. Blood. 2012;119(3):868–873. doi: 10.1182/blood-2011-08-372722. [DOI] [PubMed] [Google Scholar]
- 35▪.Johnson JA, Gong L, Whirl-Carrillo M, et al. Clinical Pharmacogenetics Implementation Consortium Guidelines for CYP2C9 and VKORC1 genotypes and warfarin dosing. Clin Pharmacol Ther. 2011;90(4):625–629. doi: 10.1038/clpt.2011.185. Highlights the utility of physiologically based pharmacokinetic modeling to predict complex drug–drug interactions and illustrate differences between children and adults. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kato Y, Ichida F, Saito K, et al. Effect of the VKORC1 genotype on warfarin dose requirements in Japanese pediatric patients. Drug Metab Pharmacokinet. 2011;26(3):295–299. doi: 10.2133/dmpk.DMPK-10-NT-082. [DOI] [PubMed] [Google Scholar]
- 37.Kosaki K, Yamaghishi C, Sato R, et al. 1173C>T polymorphism in VKORC1 modulates the required warfarin dose. Pediatr Cardiol. 2006;27(6):685–688. doi: 10.1007/s00246-005-1150-x. [DOI] [PubMed] [Google Scholar]
- 38.Moreau C, Bajolle F, Siguret V, et al. Vitamin K antagonists in children with heart disease: height and VKORC1 genotype are the main determinants of the warfarin dose requirement. Blood. 2012;119(3):861–867. doi: 10.1182/blood-2011-07-365502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nguyen N, Anley P, Yu MY, Zhang G, Thompson AA, Jennings LJ. Genetic and clinical determinants influencing warfarin dosing in children with heart disease. Pediatr Cardiol. 2013;34(4):984–990. doi: 10.1007/s00246-012-0592-1. [DOI] [PubMed] [Google Scholar]
- 40.Nowak-Göttl U, Dietrich K, Schaffranek D, et al. In pediatric patients, age has more impact on dosing of vitamin K antagonists than VKORC1 or CYP2C9 genotypes. Blood. 2010;116(26):6101–6105. doi: 10.1182/blood-2010-05-283861. [DOI] [PubMed] [Google Scholar]
- 41.Do EJ, Lenzini P, Eby CS, et al. Genetics informatics trial (GIFT) of warfarin to prevent deep vein thrombosis (DVT): rationale and study design. Pharmacogenomics J. 2012;12(5):417–424. doi: 10.1038/tpj.2011.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.French B, Joo J, Geller NL, et al. Statistical design of personalized medicine interventions: the Clarification of Optimal Anticoagulation through Genetics (COAG) trial. Trials. 2010;11:108. doi: 10.1186/1745-6215-11-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johnson TN, Rostami-Hodjegan A, Tucker GT. Prediction of the clearance of eleven drugs and associated variability in neonates, infants and children. Clin Pharmacokinet. 2006;45(9):931–956. doi: 10.2165/00003088-200645090-00005. [DOI] [PubMed] [Google Scholar]
- 44.Johnson TN, Rostami-Hodjegan A. Resurgence in the use of physiologically based pharmacokinetic models in pediatric clinical pharmacology: parallel shift in incorporating the knowledge of biological elements and increased applicability to drug development and clinical practice. Paediatr Anaesth. 2011;21(3):291–301. doi: 10.1111/j.1460-9592.2010.03323.x. [DOI] [PubMed] [Google Scholar]
- 45.Harris LF, Rainey P, Castro-López V, O’Donnell JS, Killard AJ. A microfluidic anti-Factor Xa assay device for point of care monitoring of anticoagulation therapy. Analyst. 2013;138(17):4769–4776. doi: 10.1039/c3an00401e. [DOI] [PubMed] [Google Scholar]
- 46▪.Li CG, Lee CY, Lee K, Jung H. An optimized hollow microneedle for minimally invasive blood extraction. Biomed Microdevices. 2013;15(1):17–25. doi: 10.1007/s10544-012-9683-2. Development of a microneedle for taking a minimally invasive blood sample has the potential to facilitate pediatric pharmacologic and pharmacogenetic research. [DOI] [PubMed] [Google Scholar]
- 47.Li T, Barnett A, Rogers KL, Gianchandani YB. A blood sampling microsystem for pharmacokinetic applications: design, fabrication, and initial results. Lab Chip. 2009;9(24):3495–3503. doi: 10.1039/b910508e. [DOI] [PubMed] [Google Scholar]
- 48.Shah NM, Hawwa AF, Millership JS, Collier PS, McElnay JC. A simple bioanalytical method for the quantification of antiepileptic drugs in dried blood spots. J Chromatogr B Analyt Technol Biomed Life Sci. 2013;923–924:65–73. doi: 10.1016/j.jchromb.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 49.Shan Z, Zhou Z, Chen H, et al. PCR-ready human DNA extraction from urine samples using magnetic nanoparticles. J Chromatogr B Analyt Technol Biomed Life Sci. 2012;881–882:63–68. doi: 10.1016/j.jchromb.2011.11.042. [DOI] [PubMed] [Google Scholar]
- 50.Hawwa AF, Collier PS, Millership JS, et al. Population pharmacokinetic and pharmacogenetic analysis of 6-mercaptopurine in paediatric patients with acute lymphoblastic leukaemia. Br J Clin Pharmacol. 2008;66(6):826–837. doi: 10.1111/j.1365-2125.2008.03281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Suyagh M, Collier PS, Millership JS, et al. Metronidazole population pharmacokinetics in preterm neonates using dried blood-spot sampling. Pediatrics. 2011;127(2):e367–e374. doi: 10.1542/peds.2010-0807. [DOI] [PubMed] [Google Scholar]
- 52.McCarty CA, Chisholm RL, Chute CG, et al. The eMERGE Network: a consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med Genomics. 2011;4:13. doi: 10.1186/1755-8794-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Freund CL, Clayton EW. Pharmacogenomics and children: meeting the ethical challenges. Am J Pharmacogenomics. 2003;3(6):399–404. doi: 10.2165/00129785-200303060-00007. [DOI] [PubMed] [Google Scholar]
- 54.Moran C, Thornburg CD, Barfield RC. Ethical considerations for pharmacogenomic testing in pediatric clinical care and research. Pharmacogenomics. 2011;12(6):889–895. doi: 10.2217/pgs.10.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tabor HK, Brazg T, Crouch J, et al. Parent perspectives on pediatric genetic research and implications for genotype-driven research recruitment. J Empir Res Hum Res Ethics. 2011;6(4):41–52. doi: 10.1525/jer.2011.6.4.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Harris ED, Ziniel SI, Amatruda JG, et al. The beliefs, motivations, and expectations of parents who have enrolled their children in a genetic biorepository. Genet Med. 2012;14(3):330–337. doi: 10.1038/gim.2011.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Websites
- 101.Products – Data Briefs – Number 42. 2010 Sep; www.cdc.gov/nchs/data/databriefs/db42.htm#fig3.
- 102.Best Pharmaceuticals for Children Act. Public Law No 107–109, 115 Stat 1408. 2002 www.fda.gov/RegulatoryInformation/Legislation/FederalFoodDrugandCosmeticActFDCAct/SignificantAmendmentstotheFDCAct/ucm148011.htm.
- 103.PharmGBK. CPIC Dosing Guideline for simvastatin and SLCO1B1. www.pharmgkb.org/gene/PA134865839.
- 104.Drugs@FDA. www.accessdata.fda.gov/scripts/cder/drugsatfda.