Abstract
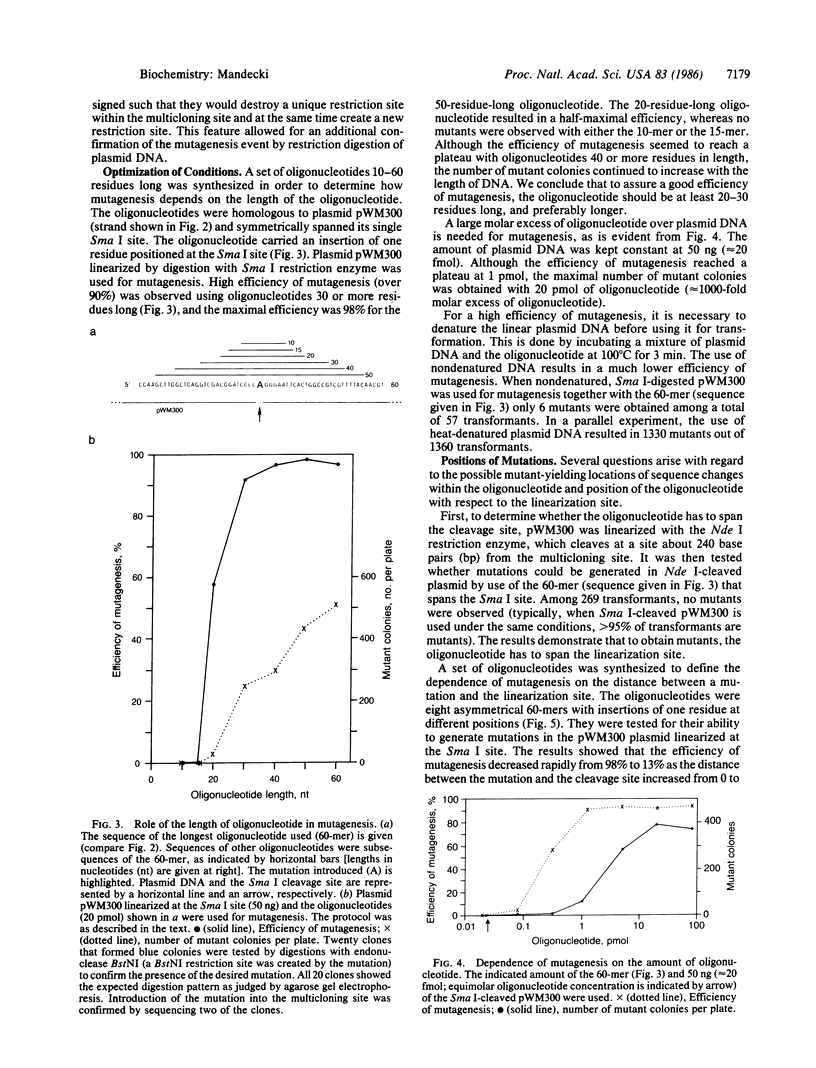
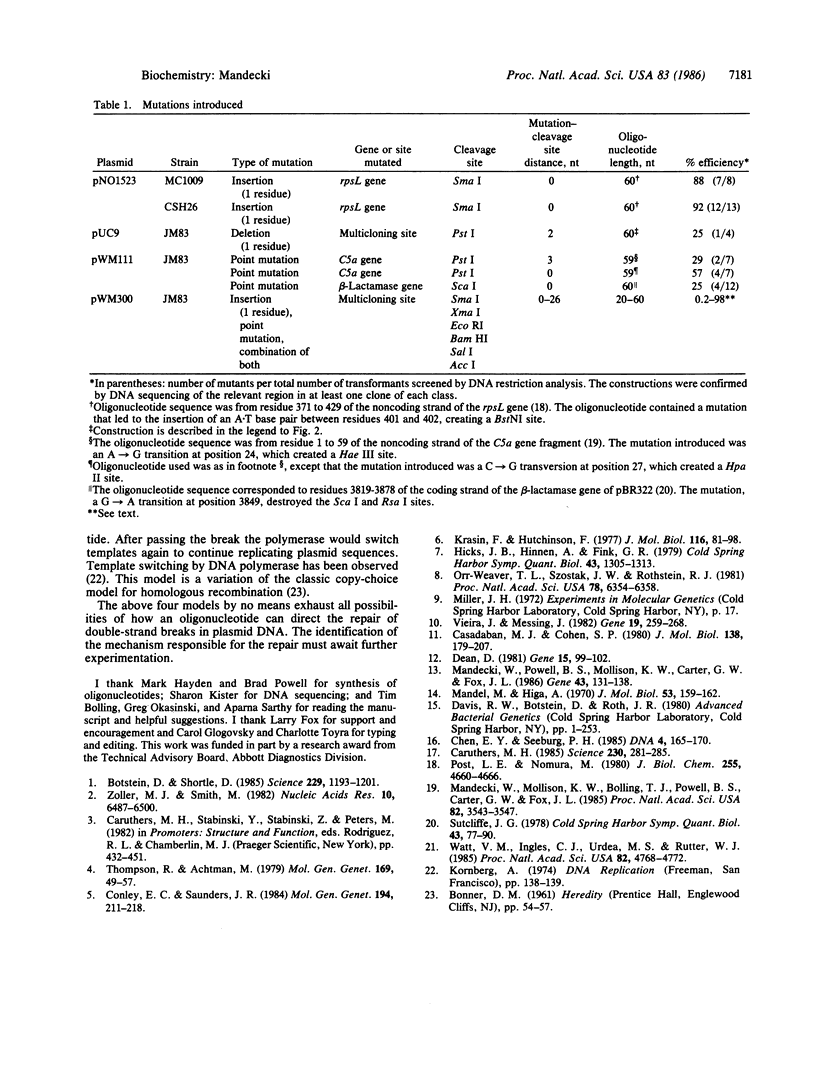
A DNA double-strand break can be efficiently repaired in Escherichia coli if an oligodeoxyribonucleotide is provided to direct the repair. The oligonucleotide must be at least 20 residues long and have a sequence identical to sequences flanking the break. The phenomenon can be used to introduce defined mutations into DNA in the area of a double-strand break. To obtain mutants, the oligonucleotide that carries a mutation and the denatured linearized plasmid DNA are introduced into E. coli by transformation. No enzymatic manipulation in vitro is required. The mutants can constitute up to 98% of the total number of transformants obtained. The efficiency of mutagenesis decreases as the distance between the mutation and the plasmid cleavage site increases. The universality of the method was tested by introducing mutations into four genes, using four plasmids and three E. coli strains, as well as eight restriction enzymes to linearize DNA. Several models of the oligonucleotide-directed DNA double-strand break repair are discussed.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Botstein D., Shortle D. Strategies and applications of in vitro mutagenesis. Science. 1985 Sep 20;229(4719):1193–1201. doi: 10.1126/science.2994214. [DOI] [PubMed] [Google Scholar]
- Caruthers M. H. Gene synthesis machines: DNA chemistry and its uses. Science. 1985 Oct 18;230(4723):281–285. doi: 10.1126/science.3863253. [DOI] [PubMed] [Google Scholar]
- Casadaban M. J., Cohen S. N. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980 Apr;138(2):179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- Chen E. Y., Seeburg P. H. Supercoil sequencing: a fast and simple method for sequencing plasmid DNA. DNA. 1985 Apr;4(2):165–170. doi: 10.1089/dna.1985.4.165. [DOI] [PubMed] [Google Scholar]
- Conley E. C., Saunders J. R. Recombination-dependent recircularization of linearized pBR322 plasmid DNA following transformation of Escherichia coli. Mol Gen Genet. 1984;194(1-2):211–218. doi: 10.1007/BF00383519. [DOI] [PubMed] [Google Scholar]
- Dean D. A plasmid cloning vector for the direct selection of strains carrying recombinant plasmids. Gene. 1981 Oct;15(1):99–102. doi: 10.1016/0378-1119(81)90108-6. [DOI] [PubMed] [Google Scholar]
- Hicks J. B., Hinnen A., Fink G. R. Properties of yeast transformation. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 2):1305–1313. doi: 10.1101/sqb.1979.043.01.149. [DOI] [PubMed] [Google Scholar]
- Krasin F., Hutchinson F. Repair of DNA double-strand breaks in Escherichia coli, which requires recA function and the presence of a duplicate genome. J Mol Biol. 1977 Oct 15;116(1):81–98. doi: 10.1016/0022-2836(77)90120-6. [DOI] [PubMed] [Google Scholar]
- Mandecki W., Mollison K. W., Bolling T. J., Powell B. S., Carter G. W., Fox J. L. Chemical synthesis of a gene encoding the human complement fragment C5a and its expression in Escherichia coli. Proc Natl Acad Sci U S A. 1985 Jun;82(11):3543–3547. doi: 10.1073/pnas.82.11.3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandecki W., Powell B. S., Mollison K. W., Carter G. W., Fox J. L. High-level expression of a gene encoding the human complement factor C5a in Escherichia coli. Gene. 1986;43(1-2):131–138. doi: 10.1016/0378-1119(86)90016-8. [DOI] [PubMed] [Google Scholar]
- Mandel M., Higa A. Calcium-dependent bacteriophage DNA infection. J Mol Biol. 1970 Oct 14;53(1):159–162. doi: 10.1016/0022-2836(70)90051-3. [DOI] [PubMed] [Google Scholar]
- Orr-Weaver T. L., Szostak J. W., Rothstein R. J. Yeast transformation: a model system for the study of recombination. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6354–6358. doi: 10.1073/pnas.78.10.6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post L. E., Nomura M. DNA sequences from the str operon of Escherichia coli. J Biol Chem. 1980 May 25;255(10):4660–4666. [PubMed] [Google Scholar]
- Sutcliffe J. G. Complete nucleotide sequence of the Escherichia coli plasmid pBR322. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):77–90. doi: 10.1101/sqb.1979.043.01.013. [DOI] [PubMed] [Google Scholar]
- Thompson R., Achtman M. The control region of the F sex factor DNA transfer cistrons: physical mapping by deletion analysis. Mol Gen Genet. 1979 Jan 16;169(1):49–57. doi: 10.1007/BF00267544. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Watt V. M., Ingles C. J., Urdea M. S., Rutter W. J. Homology requirements for recombination in Escherichia coli. Proc Natl Acad Sci U S A. 1985 Jul;82(14):4768–4772. doi: 10.1073/pnas.82.14.4768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoller M. J., Smith M. Oligonucleotide-directed mutagenesis using M13-derived vectors: an efficient and general procedure for the production of point mutations in any fragment of DNA. Nucleic Acids Res. 1982 Oct 25;10(20):6487–6500. doi: 10.1093/nar/10.20.6487. [DOI] [PMC free article] [PubMed] [Google Scholar]


