Abstract
Previously, we examined 20 non-HLA SNPs for association with islet autoimmunity (IA) and/or progression to type 1 diabetes (T1D). Our objective was to investigate fourteen additional non-HLA T1D candidate SNPs for stage- and age-related heterogeneity in the etiology of T1D. Of 1634 non-Hispanic white DAISY children genotyped, 132 developed IA (positive for GAD, insulin, or IA-2 autoantibodies at two or more consecutive visits); 50 IA positive children progressed to T1D. Cox regression was used to analyze risk of IA and progression to T1D in IA positive children. Restricted cubic splines were used to model SNPs when there was evidence that risk was not constant with age. C1QTNF6 (rs229541) predicted increased IA risk (HR: 1.57, CI: 1.20–2.05) but not progression to T1D (HR: 1.13, CI: 0.75–1.71). SNP (rs10517086) appears to exhibit an age-related effect on risk of IA, with increased risk before age 2 years (age 2 HR: 1.67, CI: 1.08–2.56) but not older ages (age 4 HR: 0.84, CI: 0.43–1.62). C1QTNF6 (rs229541), SNP (rs10517086), and UBASH3A (rs3788013) were associated with development of T1D. This prospective investigation of non-HLA T1D candidate loci shows that some SNPs may exhibit stage- and age-related heterogeneity in the etiology of T1D.
1. Introduction
Type 1 diabetes (T1D) is a chronic autoimmune disease in which the insulin-producing beta cells of the pancreas are destroyed. There is typically a preclinical phase of circulating autoantibodies, called islet autoimmunity (IA), which precedes the clinical diagnosis of T1D. T1D is widely believed to be caused by an environmental factor on a susceptible genetic background. The major susceptibility locus for T1D maps to the HLA class II genes at chromosome 6p21. These HLA class II alleles account for 30–50% of the familial clustering of T1D [1].
More than 50 non-HLA T1D susceptibility gene markers have been confirmed. The major non-HLA loci include INS [2], CTLA4 [3], PTPN22 [4], IL2RA [5], and IFIH1 [6]. The DAISY study has previously investigated 20 non-HLA SNPs and found SNPs in PTPN22, UBASH3A, INS, and IFIH1 associated with IA and/or progression to T1D [7–11].
Prospective birth cohorts have the unique ability to study two stages in the natural history of T1D: development of IA and progression to T1D in IA positive children. Different exposures have been associated with one or both stages. For instance, DAISY recently identified an association between a gene-gene interaction involving the vitamin D receptor gene (VDR) and protein tyrosine phosphatase, nonreceptor type 2 gene (PTPN2) with progression to T1D in IA positive children, but not with development of IA [12]. This would be an example of stage-related heterogeneity in the natural history of T1D. There is also evidence of age-related heterogeneity in the etiology of T1D when a gene or exposure is associated with the disease at certain ages, but not others. One example is a recent study that found differences in metabolite profiles relative to age, in which there was an association between lower methionine levels and presence of diabetes autoantibodies in younger onset (≤2 years), but not older onset (≥8 years) autoimmunity [13]. The purpose of this analysis was to investigate stage- and age-related heterogeneity of fourteen non-HLA T1D candidate SNPs for their association with development of IA and progression to T1D in a prospective birth cohort of non-Hispanic white (NHW) children at increased genetic risk of T1D. Additionally, we investigated whether the fourteen T1D candidate SNPs that were originally detected by GWAS using a case-control study design would be detected in time-to-event analyses of T1D risk in a prospective birth cohort.
2. Materials and Methods
2.1. Subjects
The Diabetes Autoimmunity Study in the Young (DAISY) is a prospective study composed of two groups of children at increased risk for T1D who were recruited between 1993 and 2004 and are being followed prospectively for the development of IA and T1D. One group is made up of first degree relatives of patients with T1D, identified and recruited between birth and eight years of age, mainly through the Barbara Davis Center for Childhood Diabetes (n = 815). The second group consists of infants born at St. Joseph's Hospital in Denver, CO, whose umbilical cord blood was screened for diabetes-susceptibility HLA-DR, DQ genotypes (n = 819). Details of the newborn screening, sibling and offspring recruitment, and followup of both cohorts have been published previously [14, 15]. Cord blood or the first available blood sample (depending on enrollment group) was sent to Roche Molecular Systems, Inc., Alameda, CA, for PCR-based HLA-DR, DQ typing. All study protocols were approved by the Colorado Multiple Institutional Review Board, and informed consent was given by parents of all participating children.
2.2. Measurement of Autoantibodies
Autoantibodies were tested at 9, 15, and 24 months, and annually thereafter, or at their first visit and annually thereafter if the child enrolled after birth. Radioimmunoassays were used to measure serum autoantibodies to insulin, GAD-65, and IA-2 (BDC512), as previously described [16–19], with rigorous confirmation of all positive and a subset of negative results. The cut-off for positivity was established as the 99th percentile of healthy controls. Children who tested autoantibody positive were put on an accelerated testing schedule of every 3–6 months.
Cases of IA were defined as those children positive for at least one islet autoantibody (IAA, GAD-65, IA-2) on two or more consecutive visits. T1D was diagnosed by a physician and defined as random blood glucose >200 mg/dL and/or HbA1c (A1C) >6.5% with clinical symptoms of T1D.
2.3. Non-HLA SNP Genotyping
DAISY children were genotyped for fourteen non-HLA T1D candidate SNPs: C1QTNF6 (rs229541), C6orf173 (rs9388489), C14orf181 (rs1465788), IL2 (rs2069762), IL2 (rs4505848), IL2RA (rs12722563), IL2RA (rs2104286), IL7R (rs6897932), PRKCQ (rs947474), SKAP2 (rs7804356), SMARCE1 (rs7221109), TLR8 (rs5979785), UBASH3A (rs3788013), and SNP (rs10517086). Thirteen of the fourteen SNPs were chosen from three GWAS meta-analyses [20–22]. UBASH3A (rs3788013) was chosen based on its strong LD with UBASH3A (rs876498), which was discovered for its association with T1D by Concannon et al. [23].
The SNPs were genotyped utilizing the Taqman SNP genotype based OpenArray platform (Applied Biosystems, CA, USA). Custom designed 48-sample arrays and normalized genomic DNA were loaded using the OpenArray AccuFill system and cycling was performed on a GeneAmp 9700 PCR system (Applied Biosystems, CA, USA), all according to manufacturer protocol. Alleles were analyzed using the OpenArray SNP genotyping analysis software v.1.0.3 and Taqman Genotyper Software 2.0 (Applied Biosystems, CA, USA). All fourteen SNPs had a 95% call rate or higher.
Each SNP was tested for consistency with Hardy-Weinberg proportions using a 1-degree of freedom χ 2 goodness-of-fit test with a P value of 0.05 considered as evidence of a departure from Hardy-Weinberg equilibrium; all fourteen SNPs were in Hardy-Weinberg equilibrium. Linkage disequilibrium (LD) was tested in our population using Haploview version 4.2 as measured by r 2 and D', with r 2 = 0.257 and D' = 0.862 for the two IL2RA SNPs and r 2 = 0.222 and D' = 1.0 for the two IL2 SNPs.
2.4. Analysis Population
We obtained genetic data on at least one of the fourteen non-HLA T1D candidate SNPs for 1634 non-Hispanic white children in the DAISY cohort. This included 132 children who developed IA, of whom 50 went on to develop T1D. Fifteen IA cases were positive for autoantibodies on their first clinic visits; these left-censored cases were removed from the development of IA analysis cohort but retained in the progression from IA to T1D cohort. The same 50 IA positive children who went on to develop T1D are the same 50 T1D cases in the development of T1D analyses. However, not all IA positive children went on to develop T1D. All statistical analyses were limited to non-Hispanic whites in the DAISY cohort.
2.5. Statistical Analyses
SAS version 9.3 (SAS Institute Inc., Cary, NC, USA) statistical software package was used for all statistical analyses. SNPs were tested for violation of the proportional hazards assumption using a supremum test, with a P value < 0.20 indicating possible departure from proportional hazards [24]. If a SNP appeared not to meet this assumption, restricted cubic splines were used to evaluate the nature and extent of the violation. SNPs were analyzed for their association both with development of IA and with progression from IA to T1D. For each model, hazard ratios (HR) and 95% confidence intervals (CI) were estimated using Cox regression analyses. There were 302 sibling pairs in the analysis cohort, so a clustered time to event analysis was performed treating siblings from the same family as a cluster, and robust sandwich variance estimates were used for statistical inference [25]. Analyses of time to development of IA were adjusted for the HLA-DR genotype (HLA-DR3/4, DQB1*0302 versus other genotypes) and presence of a first degree relative with T1D. Analyses of time to progression to T1D were adjusted, in addition, for age at first positive autoantibody visit. TLR8 (rs5979785) was additionally adjusted for sex because it is on the X chromosome. The significance threshold was defined as α = 0.05. Because our analyses were based on a priori hypotheses with SNPs previously found to be associated with T1D, P values were not corrected for multiple testing. We analyzed each non-HLA SNP in separate, covariate adjusted models. In the analyses examining development of IA, all of the SNPs were treated additively, except IL2RA (rs12722563) and PRKCQ (rs947474), which were dichotomized on the minor allele due to small sample sizes. Additionally, in the progression to T1D analyses, SNPs were treated additively, except C14orf181 (rs1465788), IL2RA (rs12722563), IL2RA (rs2104286), IL7R (rs6897932), PRKCQ (rs947474), SKAP2 (rs7804356), and SNP (rs10517086), which were dichotomized on the minor allele due to small sample sizes.
3. Results
3.1. Development of IA
We first examined whether non-HLA variants were associated with development of IA. The mean age at first IA positive visit was 6.2 years, and the mean age at last follow-up visit in children who did not develop IA was 9.9 years (Table 1). IA positive children were more likely to have the HLA-DR3/4, DQB1*0302 genotype compared to DAISY children who did not develop IA (HR: 2.97, 95% CI: 2.01, 4.39).
Table 1.
Demographic characteristics of DAISY non-Hispanic white population.
| Characteristic | Development of islet autoimmunity (IA) (n = 1619) | Progression from IA to type 1 diabetes (T1D) (n = 132) | ||||||
|---|---|---|---|---|---|---|---|---|
| Children positive for IA (n = 117) | Children negative for IA (n = 1502) | Univariate HR and 95% CI | P value | IA positive children who progressed to T1D (n = 50) | IA positive children who have not progressed to T1D (n = 82) | Univariate HR and 95% CI | P value | |
| Mean age (years) | 6.2 ± 4.2a | 9.9 ± 5.7b | N/A | N/A | 8.7 ± 3.9c | 14.1 ± 4.2b | N/A | N/A |
| Mean age at first IA positive visit (years) | N/A | N/A | N/A | N/A | 3.9 ± 2.9 | 7.2 ± 4.2 | 0.86 (0.77, 0.95) | 0.003 |
| HLA-DR3/4, DQB1*0302 |
43 (36.8%) | 248 (16.5%) | 2.97 (2.01, 4.39) | <0.0001 | 28 (56.0%) | 19 (23.2%) | 2.79 (1.65, 4.72) | 0.0001 |
| First degree relative with T1D | 75 (64.1%) | 737 (49.1%) | 1.41 (0.96, 2.07) | 0.08 | 36 (72.0%) | 54 (65.9%) | 1.02 (0.56, 1.85) | 0.96 |
| Sex (female) | 62 (53.0%) | 712 (47.4%) | 1.22 (0.85, 1.74) | 0.28 | 24 (48.0%) | 44 (53.7%) | 1.03 (0.59, 1.83) | 0.91 |
CI: confidence interval; DAISY: Diabetes Autoimmunity Study in the Young; HLA: human leukocyte antigen; HR: hazard ratio; IA: islet autoimmunity; T1D: type 1 diabetes.
aAge at first IA positive visit.
bAge at last followup.
cAge at T1D diagnosis.
Unadjusted SNP association analyses are presented in (see Supplemental Table 1 in Supplemetary Material available online at http://dx.doi.org/10.1155/2013/417657). Adjusting for HLA-DR3/4, DQB1*0302 and first degree relative with T1D, C1QTNF6 (rs229541) was associated with development of IA (HR: 1.57, 95% CI: 1.20, 2.05 (for each additional minor allele)) (Table 2). SNP (rs10517086) did not appear to meet the assumptions of proportional hazards in the development of IA analysis and therefore was modeled using a restricted cubic spline. The restricted cubic spline shows that SNP (rs10517086) is associated with an increased risk of developing IA before the age of two or in younger ages but is not associated with developing IA in older ages (Figure 1). Children with a minor allele developed IA significantly earlier than children with no minor alleles (mean age at onset of IA: 7.2, 5.2, and 3.2 for 0, 1, and 2 minor alleles, resp., P = 0.003).
Table 2.
Association between non-HLA T1D candidate SNPs and development of IA, progression from IA to T1D, and development of T1D adjusted for HLA-DR3/4, DQB1*0302 genotype and first degree relative with T1D.
| SNP | Minor allele | Development of IA (n = 1619) |
Progression from IA to T1D (n = 132) |
Development of T1D (n = 1619) |
||||
|---|---|---|---|---|---|---|---|---|
| MAFa | Adjusted HRb and 95% CI | P value | Adjusted HRc and 95% CI | P value | Adjusted HRb and 95% CI | P value | ||
| C1QTNF6 (rs229541) | A | 0.44 | 1.57 (1.20, 2.05)d | 0.001 | 1.13 (0.75, 1.71)d | 0.56 | 1.95 (1.33, 2.87)d | 0.001 |
| C6orf173 (rs9388489) | G | 0.46 | 1.15 (0.88, 1.51)d | 0.31 | 0.87 (0.61, 1.24)d | 0.44 | 1.03 (0.69, 1.54)d | 0.87 |
| C14orf181 (rs1465788) | T | 0.28 | 0.97 (0.73, 1.27)d | 0.80 | 1.09 (0.62, 1.92)e | 0.77 | 0.70 (0.40, 1.23)e | 0.21 |
| IL2 (rs2069762) | C | 0.29 | 1.08 (0.82, 1.43)d | 0.58 | 1.22 (0.79, 1.87)d | 0.38 | 1.20 (0.79, 1.83)d | 0.40 |
| IL2 (rs4505848) | G | 0.35 | 1.04 (0.80, 1.34)d | 0.78 | 0.93 (0.67, 1.28)d | 0.65 | 1.18 (0.82, 1.70)d | 0.37 |
| IL2RA (rs12722563) | A | 0.12 | 0.90 (0.58, 1.41)e | 0.65 | 0.52 (0.17, 1.57)e | 0.25 | 0.43 (0.17, 1.08)e | 0.07 |
| IL2RA (rs2104286) | C | 0.27 | 0.85 (0.63, 1.15)d | 0.28 | 1.26 (0.70, 2.27)e | 0.44 | 0.77 (0.43, 1.38)e | 0.38 |
| IL7R (rs6897932) | T | 0.28 | 0.93 (0.68, 1.28)d | 0.66 | 0.86 (0.49, 1.53)e | 0.61 | 0.88 (0.50, 1.55)e | 0.65 |
| PRKCQ (rs947474) | G | 0.17 | 1.19 (0.80, 1.77)e | 0.40 | 0.76 (0.41, 1.42)e | 0.39 | 0.87 (0.47, 1.64)e | 0.67 |
| SKAP2 (rs7804356) | C | 0.25 | 0.90 (0.66, 1.22)d | 0.49 | 1.54 (0.83, 2.85)e | 0.17 | 1.01 (0.57, 1.78)e | 0.99 |
| SMARCE1 (rs7221109) | T | 0.35 | 0.94 (0.72, 1.22)d | 0.63 | 0.94 (0.57, 1.57)d | 0.82 | 0.73 (0.45, 1.20)d | 0.21 |
| TLR8 (rs5979785) | C | 0.20 | 0.82 (0.64, 1.05)d | 0.11 | 0.84 (0.61, 1.16)d | 0.29 | 0.86 (0.60, 1.24)d | 0.43 |
| UBASH3A (rs3788013) | A | 0.44 | 1.19 (0.89, 1.59)d | 0.25 | 1.03 (0.72, 1.48)d | 0.86 | 1.63 (1.04, 2.54)d | 0.03 |
| rs10517086 | A | 0.30 | ∗ | ∗ | 1.36 (0.77, 2.41)e | 0.30 | 2.03 (1.35, 3.03)d | 0.001 |
CI: confidence interval; DAISY: Diabetes Autoimmunity Study in the Young; HLA: human leukocyte antigen; HR: hazard ratio; IA: islet autoimmunity; MAF: minor allele frequency; T1D: type 1 diabetes.
aMinor allele frequency (MAF) calculated for children negative for IA.
bAdjusted for HLA-DR3/4, DQB1*0302 genotype and first degree relative with T1D. TLR8 (rs5979785) is additionally adjusted for sex because it is on the X chromosome.
cAdjusted for HLA-DR3/4, DQB1*0302 genotype, first degree relative with T1D, and age at first antibody positive visit. TLR8 (rs5979785) is additionally adjusted for sex because it is on the X chromosome.
dSNP analyzed additively with HR representing increase in risk for each additional minor allele.
eSNP analyzed dichotomously with HR representing increase in risk for at least one minor allele.
*SNP rs10517086 did not meet the assumptions of proportional hazards in the development of IA analysis and therefore was modeled using a restricted cubic spline (Figure 1).
Figure 1.
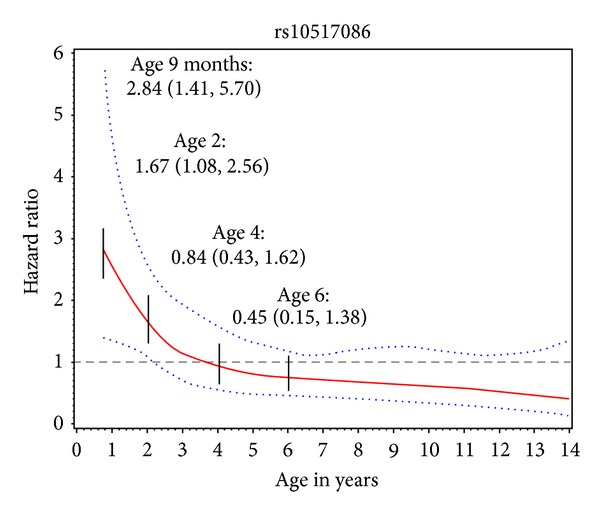
Association between SNP rs10517086 and development of IA modeled using a restricted cubic spline. The hazard ratio at different ages is represented by the solid line and the 95% confidence intervals are represented by the dotted lines. Hazard ratios and 95% confidence intervals are shown at 9 months, 2 years, 4 years, and 6 years (denoted with short vertical lines) illustrating the increased risk of developing IA in the younger ages, but not in older ages.
3.2. Progression to T1D in Children with IA
We then examined whether non-HLA variants were associated with progression to T1D in IA positive children. Of the 132 IA positive children in DAISY, 50 developed T1D; the mean age at T1D diagnosis was 8.7 years (Table 1). The mean age at last followup visit in nondiabetic children with IA was 14.1 years. Children who developed T1D were younger when they first tested positive for an autoantibody than IA positive children who have not progressed to T1D, 3.9 and 7.2 years, respectively (P = 0.003). Children with IA who developed T1D were more likely to have the HLA-DR3/4, DQB1*0302 genotype compared to children with IA who did not progress to T1D (HR: 2.79, 95% CI: 1.65, 4.72). Adjusting for HLA-DR3/4, DQB1*0302, first degree relative with T1D, and age at first IA positive visit, none of the fourteen non-HLA T1D candidate SNPs was associated with progression to T1D in IA positive children (Table 2). SNP association analyses adjusted only for age at first IA positive visit are presented in Supplemental Table 1.
3.3. Development of T1D
In order to evaluate the same outcome as previous GWAS to see if similar associations could be seen using a time-to-event analysis in a prospective birth cohort and to better understand the role these SNPs play in the natural history of T1D, we examined whether these non-HLA variants were associated with T1D in our population of 1619 children. All 50 children who developed T1D had developed IA previously, so the same 50 T1D cases were included in both the progression from IA to T1D (presented in Section 3.2) and these development of T1D analyses. However, not all IA positive children went on to develop T1D during followup. Unadjusted SNP association analyses are presented in Supplemental Table 1. Adjusting for HLA-DR3/4, DQB1*0302 and first degree relative with T1D, three of the fourteen non-HLA T1D candidate SNPs were associated with development of T1D (Table 2). Two of the SNPs associated with development of T1D, C1QTNF6 (rs229541) and SNP (rs10517086), were also associated with development of IA, but not with progression to T1D in IA positive children. The other SNP associated with development of T1D, UBASH3A (rs3788013), was not associated with development of IA nor progression to T1D in IA positive children.
4. Discussion
In exploring associations between fourteen previously discovered non-HLA T1D candidate SNPs and the development of IA and progression to T1D in the prospective DAISY cohort, we found that C1QTNF6 (rs229541) predicts IA but not progression to T1D, demonstrating stage-related heterogeneity. Moreover, SNP (rs10517086) demonstrates age-related heterogeneity with predicting IA only in the youngest ages. These two SNPs were also associated with development of T1D in our cohort, as well as UBASH3A (rs3788013). It is possible that the observed associations between both C1QTNF6 (rs229541) and SNP (rs10517086) and development of T1D were driven by their association with development of IA. Given that all of our T1D cases developed IA prior to clinical diagnosis, it was not possible to determine whether a gene was associated with T1D via a pathway other than IA.
C1QTNF6 (rs229541), which was first identified through a meta-analysis of data from three genome-wide association studies (GWAS) (combined P value = 1.98 × 10−8) [20], is an intronic SNP located on chromosome 22q13 between two genes, C1QTNF6 (C1q and tumor necrosis factor related protein 6) and SSTR3 (somatostatin receptor 3). We found that C1QTNF6 (rs229541) is associated with development of IA but not associated with progression to T1D in IA positive children, suggesting that this gene may play a role early in the development of T1D related to the initial appearance of autoimmunity.
SNP (rs10517086) exhibits an age-related effect with development of IA, with an increased risk of developing IA before the age of two or in younger ages, and a null effect in older ages. Children carrying a risk allele for SNP (rs10517086) developed IA significantly earlier than children without a risk allele. Based on these epidemiologic analyses, future studies should investigate mechanisms as to how this SNP influences risk of early autoimmunity. SNP (rs10517086), which was first discovered through another GWAS and meta-analysis of T1D, is located within a gene desert on chromosome 4. Loci in or near genes without a known function or in regions not containing annotated genes may indicate involvement of long-range gene expression regulatory elements and/or nonprotein-coding RNA genes [26].
UBASH3A (rs3788013) was associated with development of T1D, but not with either of the stages preceding this (development of IA and progression from IA to T1D). UBASH3A (rs3788013) is an intronic SNP located on chromosome 21q22 in the UBASH3A (ubiquitin associated and SH3 domain containing A) gene. UBASH3A is expressed predominantly in T cells suppressing T-cell receptor signaling [27]. UBASH3A has also been associated with other autoimmune diseases, such as celiac disease and rheumatoid arthritis [28]. Another UBASH3A SNP, UBASH3A (rs11203203), was found to be associated with both development of IA and development of T1D in a previous DAISY analysis [11]. The LD between UBASH3A (rs3788013) and UBASH3A (rs11203203) is r 2 = 0.491 and D' = 0.801. DAISY uses two definitions of IA, one that defines IA as the presence of at least one islet autoantibody on two consecutive visits (which is the definition used in the present analysis) and the other that further requires that the children still be autoantibody positive or diabetic on their most recent visit (which is the definition used in the previous analysis). The definition used in the previous study is closer to T1D, which makes our UBASH3A (rs3788013) association with T1D consistent with what was previously found with UBASH3A (rs11203203).
In combination with those presented in this paper, DAISY has now investigated 34 non-HLA T1D candidate SNPs for association with development of IA, progression from IA to T1D, and/or development of T1D with multiple examples of stage-related heterogeneity, which are presented in Table 3. Investigating 20 non-HLA SNPs for development of IA, progression from IA to T1D, and/or development of T1D, Steck et al. found PTPN22 (rs2476601) associated with development of IA, but not progression from IA to T1D, and CTLA4 (rs231775) associated with progression from IA to T1D, but not development of IA [9]. PTPN2 (rs1893217) was only associated with development of IA, while UBASH3A (rs11203203) was associated with development of IA and development of T1D [11]. INS (rs689) was not associated with development of IA nor progression from IA to T1D but was associated with development of T1D [9, 11]. PTPN22 (rs2476601) was also associated with development of T1D [11]. Here we investigated fourteen additional non-HLA T1D candidate SNPs and found C1QTNF6 (rs229541) associated with development of IA and development of T1D, but not progression from IA to T1D. UBASH3A (rs3788013) was associated with development of T1D, but not with either of the stages preceding this (development of IA and progression from IA to T1D) and SNP (rs10517086) was associated with development of T1D, while exhibiting an age-related effect with IA risk but was not associated with progression from IA to T1D. The SNPs investigated in these three studies are summarized in Table 3. The distinction between the risk factors for islet autoimmunity versus progression to type 1 diabetes in IA positive children is important because it may allow us to explore potentially different mechanisms of triggering islet autoimmunity versus epitope spreading and progressive loss of beta-cell mass leading to overt diabetes.
Table 3.
Non-HLA T1D candidate SNPs associated with development of IA, progression from IA to T1D, and/or development of T1D in DAISY.
| SNP | Development of IA | Progression from IA to T1D | Development of T1D | |||
|---|---|---|---|---|---|---|
| Adjusted HR and 95% CI | P value | Adjusted HR and 95% CI | P value | Adjusted HR and 95% CI | P value | |
| C1QTNF6 (rs229541) | 1.57 (1.20, 2.05)abc | 0.001 | 1.13 (0.75, 1.71)abd | 0.56 | 1.95 (1.33, 2.87)abc | 0.001 |
| CTLA4 (rs231775) | 1.12 (0.86, 1.46)aef | 0.42 | 0.54 (0.33, 0.88)aeg | 0.01 | 1.00 (0.70, 1.43)ahc | 1.00 |
| INS (rs689) | 1.39 (0.99, 1.95)aef | 0.05 | 1.34 (0.72, 2.52)aeg | 0.35 | 1.75 (1.08, 2.83)ahc | 0.02 |
| PTPN2 (rs1893217) | 1.42 (1.02, 1.99)ach | 0.04 | 0.65 (0.27, 1.60)ijk | 0.35 | 0.99 (0.60, 1.66)ach | 0.98 |
| PTPN22 (rs2476601) | 1.83 (1.27, 2.63)aef | 0.001 | 0.98 (0.50, 1.93)aeg | 0.96 | 1.74 (1.04, 2.90)ahc | 0.03 |
| UBASH3A (rs11203203) | 1.46 (1.11, 1.91)ach | 0.01 | ∗∗ | ∗∗ | 1.83 (1.28, 2.64)ach | 0.001 |
| UBASH3A (rs3788013) | 1.19 (0.89, 1.59)abc | 0.25 | 1.03 (0.72, 1.48)abd | 0.86 | 1.63 (1.04, 2.54)abc | 0.03 |
| rs10517086 | ∗ | ∗ | 1.36 (0.77, 2.41)bdj | 0.30 | 2.03 (1.35, 3.03)abc | 0.001 |
DAISY: Diabetes Autoimmunity Study in the Young; HLA: human leukocyte antigen; HR: hazard ratio; CI: confidence interval; IA: islet autoimmunity; T1D: type 1 diabetes.
aSNP analyzed additively with HR representing increase in risk for each additional minor allele.
bListed in Table 2 of the current paper.
cAdjusted for HLA-DR3/4, DQB1*0302 genotype and first degree relative with T1D.
dAdjusted for HLA-DR3/4, DQB1*0302 genotype, first degree relative with T1D, and age at first antibody positive visit.
eFrom Steck et al. (2009) [9].
fAdjusted for HLA-DR3/4, DQB1*0302 genotype, ethnicity, sex, and first degree relative with type 1 diabetes.
gAdjusted for HLA-DR3/4, DQB1*0302 genotype, ethnicity, sex, first degree relative with type 1 diabetes, and age at first antibody positive visit.
hFrom Steck et al.(2012) [11].
iFrom Frederiksen et al. (2013) [12]. Frederiksen et al. (2013) [12] did not find an association between PTPN2 (rs1893217) and development of IA, which can be attributed to the use of different IA case definitions used in the manuscripts by Steck et al. (2009) [9] and Frederiksen et al. (2013) [12].
jSNP analyzed dichotomously with HR representing increase in risk for at least one minor allele.
kAdjusted for PTPN2 (rs478582), HLA-DR3/4, DQB1*0302 genotype, first degree relative with type 1 diabetes, ethnicity, and age at first IA positive visit.
**Analysis not conducted.
*SNP rs10517086 did not meet the assumptions of proportional hazards in the development of IA analysis and therefore was modeled using a restricted cubic spline (Figure 1 of the current paper).
GWAS are important for identifying new candidate regions associated with a clinical outcome, such as T1D, in a large number of cases and controls. Prospective birth cohort studies, like DAISY, are then able to take these newly identified candidate regions and look for associations with different stages in the disease process and at different ages. As a prospective birth cohort study following children at increased risk for developing T1D from birth, we are able to capture the preclinical phase of T1D, islet autoimmunity. This allows us to study two separate stages in the natural history of T1D: development of IA and progression to T1D in IA positive children. We are also able to study whether certain exposures are important at one age, but not another.
Due to the cost associated with following a large group of children from birth into adulthood, our sample sizes are much smaller than those obtained for GWAS. Our lack of association for many of these SNPs is not evidence against their association with T1D but may result from limited power, especially in the progression from IA to T1D stage. We also have a very unique population comprised of children with high risk HLA genotypes and it is possible that the effect of these SNPs differs based on one's HLA risk status. The risk for non-HLA loci appears to be lower in individuals carrying high-risk HLA genotypes, as has been seen with PTPN22 (rs2476601) [21, 29] and TCF7 (rs5742913) [30, 31].
We believe that taking these GWAS identified candidate regions and studying them in the context of the natural history of T1D are central to better understanding the disease process and where in the disease process genetic loci may be important. This will allow us to create more accurate risk prediction models for both stages in the natural history of the disease, as well as inform the design of targeted interventions to prevent or slow the progression of IA and subsequent development of T1D.
5. Conclusions
The effect of a SNP may act nonlinearly, with an effect at early ages but not later ages or vice versa. Our results provide evidence that SNP (rs10517086) is acting on early risk of IA, with the age at onset of IA occurring significantly earlier in children with a minor allele compared to children with no minor alleles. By ignoring heterogeneity in the etiology of disease, valuable associations may be missed that could aid in better understanding complex diseases, such as T1D.
Supplementary Material
Unadjusted SNP association analyses are presented in Supplemental Table 1. In the unadjusted SNP analyses, C1QTNF6 (rs229541) was associated with development of IA. None of the SNPs were associated with progression to T1D in IA positive children. Four SNPs, C1QTNF6 (rs229541), IL2RA (rs12722563), UBASH3A (rs3788013), and SNP (rs10517086) were associated with development of T1D.
Conflict of Interests
The authors declare no conflict of interests.
Acknowledgments
This research was supported by NIH Grants R01-DK49654, R01-DK32493, and R01-DK32083, DERC Molecular Biology Core NIH P30 DK57516, and the NIH/NCRR Colorado CTSI Grant no. UL1 RR025780.
Abbreviations
- CI:
Confidence interval
- C1QTNF6:
C1q and tumor necrosis factor related protein 6
- C6orf173:
Chromosome 6 open reading frame 173
- C14orf181:
Chromosome 14 open reading frame 181
- CTLA4:
Cytotoxic T-lymphocyte-associated protein 4
- HLA:
Human leukocyte antigen
- HR:
Hazard ratio
- IA:
Islet autoimmunity
- IFIH1:
Interferon induced with helicase C domain 1
- IL2:
Interleukin 2
- IL2RA:
Interleukin 2 receptor, alpha
- IL7R:
Interleukin 7 receptor
- INS:
Insulin
- PRKCQ:
Protein kinase C, theta
- PTPN22:
Protein tyrosine phosphatase, nonreceptor type 22
- SKAP2:
Src kinase associated phosphoprotein
- SMARCE1:
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily E, member 1
- SNP:
Single nucleotide polymorphism
- T1D:
Type 1 diabetes
- TLR8:
Toll-like receptor 8
- UBASH3A:
Ubiquitin associated and SH3 domain containing A.
References
- 1.Noble JA, Valdes AM, Cook M, Klitz W, Thomson G, Erlich HA. The role of HLA class II genes in insulin-dependent diabetes mellitus: molecular analysis of 180 Caucasian, multiplex families. American Journal of Human Genetics. 1996;59(5):1134–1148. [PMC free article] [PubMed] [Google Scholar]
- 2.Bell GI, Horita S, Karam JH. A polymorphic locus near the human insulin gene is associated with insulin-dependent diabetes mellitus. Diabetes. 1984;33(2):176–183. doi: 10.2337/diab.33.2.176. [DOI] [PubMed] [Google Scholar]
- 3.Nisticò L, Buzzetti R, Pritchard LE, et al. The CTLA-4 gene region of chromosome 2q33 is linked to, and associated with, type 1 diabetes. Human Molecular Genetics. 1996;5(7):1075–1080. doi: 10.1093/hmg/5.7.1075. [DOI] [PubMed] [Google Scholar]
- 4.Bottini N, Musumeci L, Alonso A, et al. A functional variant of lymphoid tyrosine phosphatase is associated with type I diabetes. Nature Genetics. 2004;36(4):337–338. doi: 10.1038/ng1323. [DOI] [PubMed] [Google Scholar]
- 5.Vella A, Cooper JD, Lowe CE, et al. Localization of a type 1 diabetes locus in the IL2RA/CD25 region by use of tag single-nucleotide polymorphisms. American Journal of Human Genetics. 2005;76(5):773–779. doi: 10.1086/429843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nejentsev S, Walker N, Riches D, Egholm M, Todd JA. Rare variants of IFIH1, a gene implicated in antiviral responses, protect against type 1 diabetes. Science. 2009;324(5925):387–389. doi: 10.1126/science.1167728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Steck AK, Bugawan TL, Valdes AM, et al. Association of non-HLA genes with type 1 diabetes autoimmunity. Diabetes. 2005;54(8):2482–2486. doi: 10.2337/diabetes.54.8.2482. [DOI] [PubMed] [Google Scholar]
- 8.Steck AK, Liu S, McFann K, et al. Association of the PTPN22/LYP gene with type 1 diabetes. Pediatric Diabetes. 2006;7(5):274–278. doi: 10.1111/j.1399-5448.2006.00202.x. [DOI] [PubMed] [Google Scholar]
- 9.Steck AK, Zhang W, Bugawan TL, et al. Do non-HLA genes influence development of persistent islet autoimmunity and type 1 diabetes in children with high-risk HLA-DR,DQ genotypes? Diabetes. 2009;58(4):1028–1033. doi: 10.2337/db08-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Steck AK, Rewers MJ. Genetics of type 1 diabetes. Clinical Chemistry. 2011;57(2):176–185. doi: 10.1373/clinchem.2010.148221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Steck AK, Wong R, Wagner B, et al. Effects of non-HLA gene polymorphisms on development of islet autoimmunity and type 1 diabetes in a population with high-risk HLA-DR,DQ genotypes. Diabetes. 2012;61(3):753–758. doi: 10.2337/db11-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frederiksen B, Liu E, Romanos J, et al. Investigation of the vitamin D receptor gene (VDR) and its interaction with protein tyrosine phosphatase, non-receptor type 2 gene (PTPN2) on risk of islet autoimmunity and type 1 diabetes: the Diabetes Autoimmunity Study in the Young (DAISY) The Journal of Steroid Biochemistry and Molecular Biology. 2013;133:51–57. doi: 10.1016/j.jsbmb.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pflueger M, Seppänen-Laakso T, Suortti T, et al. Age- and islet autoimmunity-associated differences in amino acid and lipid metabolites in children at risk for type 1 diabetes. Diabetes. 2011;60(11):2740–2747. doi: 10.2337/db10-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rewers M, Bugawan TL, Norris JM, et al. Newborn screening for HLA markers associated with IDDM: diabetes autoimmunity study in the young (DAISY) Diabetologia. 1996;39(7):807–812. doi: 10.1007/s001250050514. [DOI] [PubMed] [Google Scholar]
- 15.Norris JM, Yin X, Lamb MM, et al. Omega-3 polyunsaturated fatty acid intake and islet autoimmunity in children at increased risk for type 1 diabetes. Journal of the American Medical Association. 2007;298(12):1420–1428. doi: 10.1001/jama.298.12.1420. [DOI] [PubMed] [Google Scholar]
- 16.Yu L, Rewers M, Gianani R, et al. Antiislet autoantibodies usually develop sequentially rather than simultaneously. Journal of Clinical Endocrinology and Metabolism. 1996;81(12):4264–4267. doi: 10.1210/jcem.81.12.8954025. [DOI] [PubMed] [Google Scholar]
- 17.Yu L, Robles DT, Abiru N, et al. Early expression of antiinsulin autoantibodies of humans and the NOD mouse: evidence for early determination of subsequent diabetes. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(4):1701–1706. doi: 10.1073/pnas.040556697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grubin CE, Daniels T, Toivola B, et al. A novel radioligand binding assay to determine diagnostic accuracy of isoform-specific glutamic acid decarboxylase antibodies in childhood IDDM. Diabetologia. 1994;37(4):344–350. doi: 10.1007/BF00408469. [DOI] [PubMed] [Google Scholar]
- 19.Gianani R, Rabin DU, Verge CF, et al. ICA512 autoantibody radioassay. Diabetes. 1995;44(11):1340–1344. doi: 10.2337/diab.44.11.1340. [DOI] [PubMed] [Google Scholar]
- 20.Cooper JD, Smyth DJ, Smiles AM, et al. Meta-analysis of genome-wide association study data identifies additional type 1 diabetes risk loci. Nature Genetics. 2008;40(12):1399–1401. doi: 10.1038/ng.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barrett JC, Clayton DG, Concannon P, et al. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nature Genetics. 2009;41(6):703–707. doi: 10.1038/ng.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bradfield JP, Qu H, Wang K, et al. A genome-wide meta-analysis of six type 1 diabetes cohorts identifies multiple associated loci. PLoS Genetics. 2011;7(9) doi: 10.1371/journal.pgen.1002293.e1002293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Concannon P, Onengut-Gumuscu S, Todd JA, et al. A human type 1 diabetes susceptibility locus maps to chromosome 21q22.3. Diabetes. 2008;57(10):2858–2861. doi: 10.2337/db08-0753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin DY, Wei LJ, Ying Z. Checking the Cox model with cumulative sums of martingale-based residuals. Biometrika. 1993;80(3):557–572. [Google Scholar]
- 25.Wei LJ, Lin DY, Weissfeld L. Regression analysis of multivariate incomplete failure time data by modeling marginal distributions. Journal of the American Statistical Association. 1989;84:1065–1073. [Google Scholar]
- 26.Pociot F, Akolkar B, Concannon P, et al. Genetics of type 1 diabetes: what’s next? Diabetes. 2010;59(7):1561–1571. doi: 10.2337/db10-0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsygankov AY. Multidomain STS/TULA proteins are novel cellular regulators. IUBMB Life. 2008;60(4):224–231. doi: 10.1002/iub.36. [DOI] [PubMed] [Google Scholar]
- 28.Zhernakova A, Stahl EA, Trynka G, et al. Meta-analysis of genome-wide association studies in celiac disease and rheumatoid arthritis identifies fourteen non-HLA shared loci. PLoS Genetics. 2011;7(2) doi: 10.1371/journal.pgen.1002004.e1002004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smyth DJ, Cooper JD, Howson JMM, et al. PTPN22 Trp620 explains the association of chromosome 1p13 with type 1 diabetes and shows a statistical interaction with HLA class II genotypes. Diabetes. 2008;57(6):1730–1737. doi: 10.2337/db07-1131. [DOI] [PubMed] [Google Scholar]
- 30.Noble JA, White AM, Lazzeroni LC, et al. A polymorphism in the TCF7 gene, C883A, is associated with type 1 diabetes. Diabetes. 2003;52(6):1579–1582. doi: 10.2337/diabetes.52.6.1579. [DOI] [PubMed] [Google Scholar]
- 31.Erlich HA, Valdes AM, Julier C, Mirel D, Noble JA. Evidence for association of the TCF7 locus with type I diabetes. Genes and Immunity. 2009;10(supplement 1):S54–S59. doi: 10.1038/gene.2009.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Unadjusted SNP association analyses are presented in Supplemental Table 1. In the unadjusted SNP analyses, C1QTNF6 (rs229541) was associated with development of IA. None of the SNPs were associated with progression to T1D in IA positive children. Four SNPs, C1QTNF6 (rs229541), IL2RA (rs12722563), UBASH3A (rs3788013), and SNP (rs10517086) were associated with development of T1D.


