SUMMARY
E2F/DP transcription factors regulate cell proliferation and apoptosis. Here, we investigated the mechanism of the resistance of Drosophilad DP mutants to irradiation-induced apoptosis. Contrary to the prevailing view, this is not due to an inability to induce the apoptotic transcriptional program, since we show that this program is induced, but rather due to a mitochondrial dysfunction of dDP mutants. We attribute this defect to E2F/DP-dependent control of expression of mitochondria associated genes. Genetic attenuation of several of these E2F/DP targets mimics the dDP mutant mitochondrial phenotype and protects from irradiation-induced apoptosis. Significantly, the role of E2F/DP in the regulation of mitochondrial function is conserved between flies and humans. Thus, our results uncovered a role of E2F/DP in the regulation of mitochondrial function and demonstrate that this aspect of E2F regulation is critical for the normal induction of apoptosis in response to irradiation.
Keywords: dE2f1 transcription factor, Drosophila, apoptosis
INTRODUCTION
The E2F/DP heterodimeric transcription factor (referred to as E2F) plays a critical role in promoting cell cycle progression and apoptosis. While net E2F activity results from the contributions from amongst all of the E2F transcription factors and dictates cell cycle regulation, induction of apoptosis is primarily associated with E2f1 (Biswas and Johnson, 2012; Polager and Ginsberg, 2009). Experiments in which E2f1 is overexpressed or inappropriately activated, as in the case of mutation of the E2F negative regulator, Retinoblastoma protein (pRB), have demonstrated the pro-apoptotic function of E2f1 (DeGregori et al., 1997; Tsai et al., 1998). E2f1 has also been shown to promote apoptosis in both p53-dependent and p53-independent manners. Mechanistically, E2f1 functions by directly regulating the transcription of apoptotic genes such as Apaf1, caspase 7, caspase 3, p73, p19ARF (Aslanian et al., 2004; Irwin et al., 2000; Müller et al., 2001; Pediconi et al., 2003).
The apoptotic function of E2f1 is conserved in Drosophila and has been extensively studied in the context of DNA damage response, where dE2f1 acts in parallel to dp53. In this setting, apoptosis is triggered as a result of the transcriptional induction of the key Drosophila apoptotic genes hid and rpr. Both of these genes are direct transcriptional targets of dE2f1 and their regulation by dE2f1 is an important event in apoptosis induction in other contexts as well (Davidson and Duronio, 2012; Moon et al., 2005; Tanaka-Matakatsu et al., 2009). Therefore, it is thought that in an analogous way dE2f1 directly contributes to the induction of apoptotic genes in irradiated cells. This interpretation appears to fit with the genetic data and is best exemplified by the failure of dDP mutants to undergo cell death in response to irradiation. dDP is an obligatory heterodimeric binding partner of dE2f1, and dE2f1 was shown to be functionally inactivated in dDP mutants (Frolov et al., 2005). Surprisingly, in spite of a complete block in apoptotic response, a dDP mutation does not prevent an irradiation-dependent induction of rpr and hid (Moon et al., 2008). This is in contrast to the effect of inactivation of dp53, another key player in the cellular response to irradiation. Loss of dp53 blocks apoptosis and induction of rpr and hid in irradiated cells (Brodsky et al., 2000; Moon et al., 2008). One explanation for the puzzling dDP mutant phenotype is that there are other, unidentified E2F-regulated apoptotic genes responsible for irradiation-induced apoptosis. Alternatively, dE2f1 might have a separate role distinct from its ability to regulate expression of apoptotic genes, which is needed for cell death to occur.
Here, we investigated the molecular mechanism that defines protection of dDP mutants from irradiation-induced apoptosis. Unexpectedly, we found that an apoptotic transcriptional program is properly executed in irradiated dDP mutants. This suggests that contrary to the prevailing view, dE2f1 does not normally contribute to the induction of the apoptotic genes in irradiated cells. However, we show that dE2F/dDP directly regulates mitochondria associated genes, while a dDP mutation results in downregulation of their expression and concomitant severe mitochondrial defects. Significantly, the dDP mutant mitochondrial defects and protection from irradiation-induced apoptosis can be mimicked by downregulation of mitochondria associated dE2F/dDP targets. We suggest that mitochondrial dysfunction due to the reduced expression of mitochondria associated genes makes dDP mutants refractory to irradiation-induced apoptosis. Importantly, the reduction in mitochondrial activity, direct binding of E2F/DP to mitochondria associated genes, and resistance to DNA damage-induced apoptosis is conserved between flies and humans.
RESULTS
dDP mutants properly induce the apoptotic gene expression signature in response to irradiation
Exposing Drosophila to 40Gy of γ-irradiation induces a G2/M cell cycle arrest and causes a high level of apoptosis. Both phenotypes can be easily visualized in larval eye imaginal discs by immunofluorescence using antibodies against phosphorylated histone H3 (phospho-H3, mitotic marker) and active caspase (C3, apoptotic marker). In untreated eye discs, there are dozens of phospho-H3 positive cells and very few C3-positive cells (Figure 1A and B). This pattern is reversed in irradiated eye discs: the phospho-H3 staining is completely absent due to a cell cycle arrest while the appearance of a large number of C3 positive cells reflects the induction of a DNA damage-dependent apoptosis (Figure 1A and B). The C3 staining accurately reflects the pattern of irradiation-induced apoptosis since a similar pattern is observed when apoptotic cells are labeled by a TUNEL assay (Moon et al., 2008).
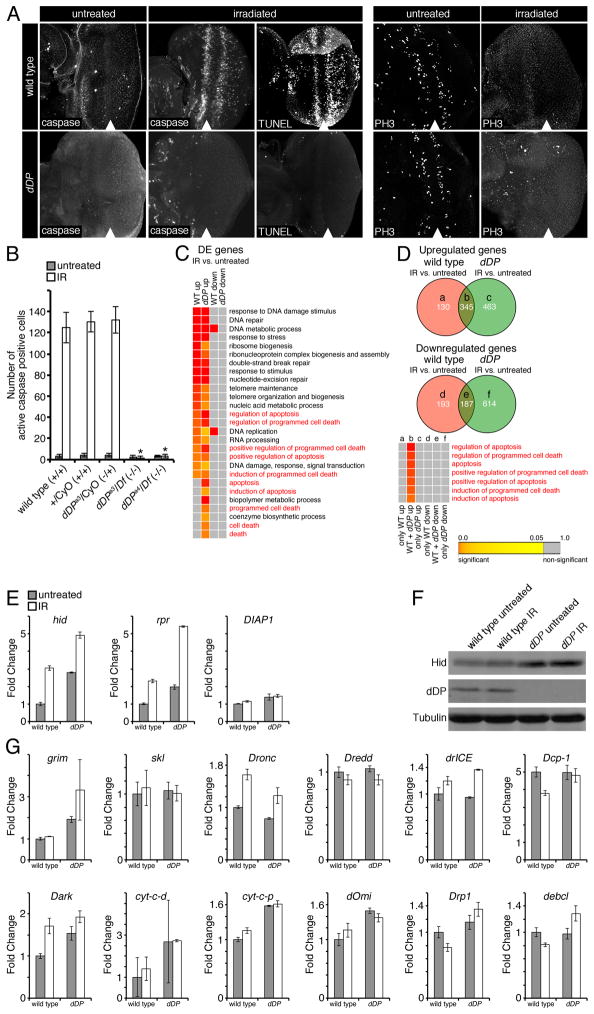
Figure 1. dDP mutant eye discs fail to undergo DNA damage-induced apoptosis despite a normal apoptotic transcriptional response.
Wild type and dDP mutant third instar larval eye discs were untreated or irradiated (IR) with 40 Gy of ionizing radiation. (A) Active caspase immunostaining or TUNEL was used to detect apoptotic cells 4h after IR. Phosphorylated histone H3 (PH3) immunostaining was used to detect mitotic cells 1h after IR. All eye discs are oriented with the posterior to the right. An arrowhead indicates the morphogenetic furrow (MF). (B) Quantification of active caspase cells in the posterior compartment of eye discs either untreated (gray bars) or 4h after IR (white bars) (± SD, with ten discs quantified for each genotype and treatment.) Asterisks (*) indicate that the number of active caspase cells in IR dDP mutant eye discs was statistically significantly different compared to IR control discs for all three control genotypes labeled. P< 0.05 in paired t-tests. (C and D) RNA was isolated from wild type and dDP mutant third instar larval eye discs either untreated or 4h after IR and transcriptional profiles were determined by microarrays. Differentially expressed (DE) genes comparing IR to untreated eye discs were determined for wild type and dDP mutants. Heat maps show enrichment of gene ontology biological process (GOBP) categories for wild type and dDP mutants with a false discovery rate cutoff of 0.05. The heat map colors represent the value of statistical significance of the enrichment. DE genes were separated into either upregulated (up) or downregulated (down) genes. (D) The Venn diagrams display the number of genes either uniquely or commonly differentially expressed following IR in either wild type or dDP mutant backgrounds. Letters denote gene lists corresponding to the respective compartments on the Venn diagrams. The heat map displays the most highly enriched apoptosis-related GOBP categories. (E and G) RNA was isolated from wild type and dDP mutant larvae which were either untreated or 4h after IR. Quantitative real-time PCR was used to measure the expression of the indicated genes. Expression levels of indicated genes for untreated (gray bars) or IR (white bars), for wild type and dDP mutant eye discs are normalized to wild type untreated (± SD, using 3 replicates for each genotype and treatment). (F) Western blot of Hid protein. See also Supplemental Figure S1 and Supplemental Table S1.
To characterize the irradiation-induced apoptotic transcriptional program, we performed gene expression microarrays on untreated and irradiated wild type eye discs and identified 855 genes that are differentially expressed (DE) in response to irradiation. Gene ontology of biological processes (GOBP) enrichment analysis revealed that irradiation led to an upregulation of a statistically significant number of genes involved in DNA damage response, DNA repair and apoptosis (Figure 1C, Supplemental Table S1). Notably, apoptotic genes, rpr and hid, which have been shown to be induced in response to irradiation (Brodsky et al., 2000; Moon et al., 2008; Truscott et al., 2011), were present among upregulated genes.
As mentioned above, the lack of apoptosis in irradiated dDP mutants (Moon et al., 2005) (Figure 1A and B) is thought to be due to the failure to induce the apoptotic gene expression program after DNA damage. Surprisingly, this was not the case. In irradiated dDP mutants, as in wild type, there were a statistically significant number of upregulated genes involved in the DNA damage response, DNA repair and apoptosis (Figure 1C, Supplemental Table S1). Importantly, apoptosis related GO terms remained enriched among upregulated genes that are common to both irradiated wild type and dDP mutants, but not among genes that are DE exclusively in wild type or in dDP mutants (Figure 1D, Supplemental Table S1). Thus, dDP mutants induce the same set of apoptotic genes as wild type animals.
To quantitatively measure the level of expression of apoptotic genes in irradiated dDP mutants, we employed real-time qPCR. Following irradiation, the levels of hid and rpr, that have been previously shown to be necessary for irradiation-induced apoptosis, were elevated in wild type discs (Figure 1E). Remarkably, their expression was higher in dDP mutants even prior to irradiation and there was even further upregulation in irradiated dDP mutant eye discs compared to irradiated wild type eye discs (Figure 1E). Importantly, Hid protein was also expressed at a higher level in dDP mutants as revealed by western blot (Figure 1F). Conversely, the level of Drosophila Inhibitor of Apoptosis 1 (DIAP1), the critical negative regulator of apoptosis in Drosophila (Wang et al., 1999), was indistinguishable between dDP mutants and wild type which were either untreated or irradiated (Figure 1E).
Analysis was expanded to include known canonical pro-apoptotic genes. In almost every case, the level of expression in irradiated dDP mutant eye discs was the same or higher than in irradiated wild type eye discs (Figure 1G). We note, that the level of Dronc in irradiated dDP mutants was approximately 20% lower than in the irradiated control. However, Dronc heterozygous mutant eye imaginal discs undergo apoptosis normally in response to irradiation even though Dronc is expressed at a lower level in Dronc heterozygotes than in irradiated dDP mutants (Supplemental Figure S1). Thus, it seems unlikely that an incomplete induction of Dronc could account for the lack of apoptosis in irradiated dDP mutants.
In summary, these results demonstrate that a dDP mutation does not prevent induction of apoptotic genes in response to irradiation. Moreover, the apoptotic genes are generally expressed at a higher level in irradiated dDP mutants than in irradiated wild type eye discs. We concluded therefore, that the failure of irradiated dDP mutants to undergo apoptosis is not a consequence of an inability to induce the apoptotic transcriptional program.
dE2f/dDP directly regulates expression of mitochondria associated genes
Since dDP mutants properly induce apoptotic genes following irradiation, we focused on the group of genes that are differentially expressed in the irradiated mutant, but not in irradiated wild type eye discs. Kyoto Encyclopedia of Genes and Genomes pathway (KEGG) enrichment analysis showed that while no pathways were enriched using the list of upregulated genes, there was a highly statistically significant enrichment for metabolic and oxidative phosphorylation pathways for the downregulated genes (Figure 2A, Supplemental Table S2). Oxidative phosphorylation and mitochondrial electron transport related GOBP terms were found to be at the top among the most highly enriched categories (Figure 2A, Supplemental Table S2). In agreement, the gene ontology of cellular compartments (GOCC) analysis showed the highest enrichment for mitochondrial terms (Figure 2A, Supplemental Table S2). Intriguingly, while there are 87 genes with a mitochondrion GOCC term that were downregulated following irradiation in dDP mutants, there are 72 mitochondria associated genes which were already downregulated in dDP mutants even prior to irradiation. To validate the results of the gene expression microarrays, mRNA levels of these mitochondria associated genes was determined by qPCR. As shown in Figure 2B and C, a dDP mutation results in downregulation of expression either prior to (Figure 2C) or in response to irradiation (Figure 2B). These results suggest that the mitochondria related transcriptional program could be intrinsically perturbed in dDP mutants.
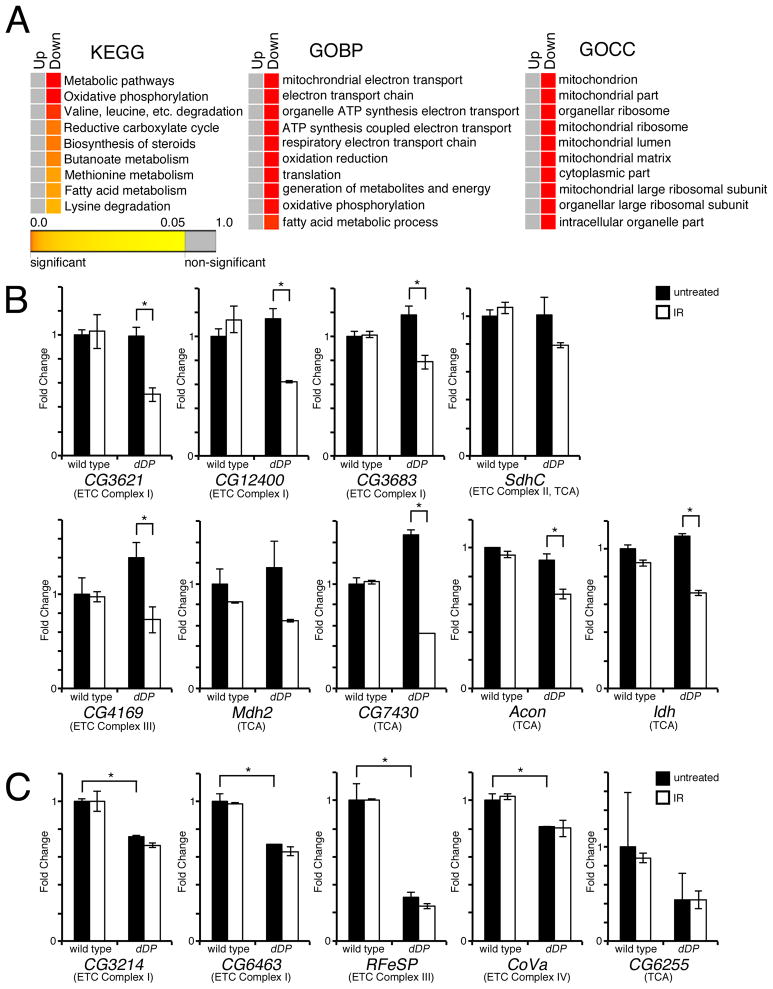
Figure 2. Oxidative phosphorylation genes are downregulated in dDP mutant eye discs following DNA damage-induced irradiation.
(A) Differentially expressed (DE) genes comparing IR to untreated eye discs were determined for wild type and dDP mutants. The heat map colors represent the value of statistical significance of the enrichment. The enriched categories for heat maps of Kyoto encyclopedia of genes and genomes pathway analysis (KEGG), gene ontology biological process (GOBP), and gene ontology cellular compartment (GOCC) are shown. (B and C) qPCR shows expression levels of indicated genes for untreated (black bars) or IR (white bars), for wild type and dDP mutant eye discs (± SD, using 3 replicates for each genotype and treatment). (*) P < 0.05 in paired t-tests. See also Supplemental Table S2.
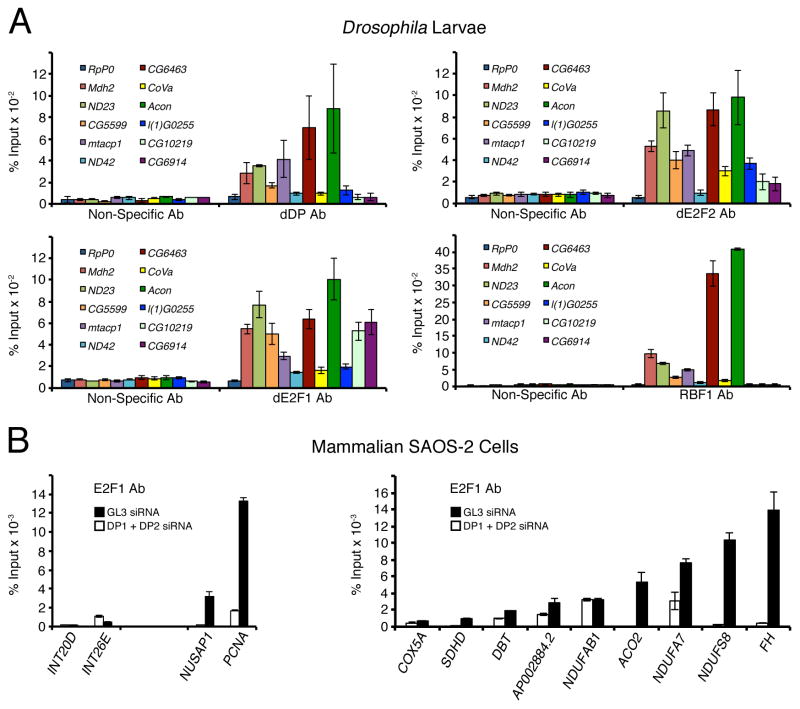
To determine how many of the mitochondria associated genes are direct targets of dE2F/dDP, we performed Chromatin Immunoprecipitation (ChIP) followed by Solexa sequencing (ChIP-Seq) using Drosophila S2R+ cells. Reads were used to assign the binding sites to the closest gene and led to the identification of 2,630 genes bound by dDP. To complement these results, publicly available genome-wide location data of the dE2f2/dDP complexes obtained in Drosophila Kc cells (Georlette et al., 2007) were added. The GOCC enrichment analysis revealed that dE2F/dDP directly targets 61% of the genes with a mitochondrion GOCC term, however, given the large number of dDP targets such enrichment is not statistically significant. Next, eleven mitochondria associated dE2F/dDP target genes were selected to confirm dDP binding and to examine the occupancy of other components of the Rb pathway: dE2f1, dE2f2, and RBF on their promoters by a conventional ChIP-qPCR assay in Drosophila larvae. Importantly, these genes were all down regulated in dDP muntants either prior to or following irradiation. As shown in Figure 3A, the promoter regions of nine out of these eleven genes (Mdh2 (malate dehydrogenase 2), ND23 (NADH:ubiquinone reductase 23kD), CG5599 (lipoamide acyltransferase), mtacp1 (mitochondrial acyl carrier protein of electron transport chain complex I), ND42 (NADH:ubiquinone reductase 42kD), CG6463 (NADH:ubiquinone oxidoreductase 13kDa-βsubunit), CoVa (cytochrome c oxidase subunit Va), Acon (aconitate hydratase), and l(1)G0255 (fumarate hydratase)) were specifically enriched in immunoprecipitates with a dDP antibody while no enrichment was seen for a negative control gene, RpP0. Conversely, a non-specific antibody failed to immunoprecipitate any of these targets. dE2f1 was detected on the promoters of these same nine genes and two other genes (CG10219 (succinate dehydrogenase), and CG6914 (NADH dehydrogenase)), which did not display significant enrichment with a dDP antibody (Figure 3A). dE2f2, the second E2F protein in flies, shows a similar profile of binding. This is consistent with other studies demonstrating that many of dE2f1 targets are bound by dE2f2 (Dimova et al., 2003; Korenjak et al., 2012). Additionally, RBF1, a negative regulator of dE2f/dDP, closely parallels occupancy of dE2f1, dE2f2 and dDP (Figure 3A), which suggests that these genes are under canonical regulation by the RBF/E2F pathway.
Figure 3. Mitochondria associated genes are direct dE2F/dDP targets.
(A) ChIP of Drosophila larvae using dDP, dE2f1, dE2f2 or RBF1 antibodies to detect binding to promoters of oxidative phosphorylation genes (± SD, using 3 replicates for each genotype). A Myc (Non-specific) antibody was used as a negative control. Primers for each experimental gene were designed to flank predicted dE2F binding sites up to 2 kb upstream of the TSS. RpP0 was used as a negative control gene. (B) ChIP of SAOS-2 cells using an E2F1 antibody to detect binding to promoters of oxidative phosphorylation genes (± SD, using 3 replicates for each treatment). White bars indicate SAOS-2 cells treated with DP1 and DP2 siRNA. Primers targeting two intergenic regions, INT20D and INT26E, which lack E2F1 binding, were used as negative controls, while known E2F1 targets NUSAP1 and PCNA were used as positive controls.
To ask whether E2F targets mitochondria associated genes in human cells, we selected eight Drosophila genes ND23, CG5599, mtacp1, CoVa, Acon, l(1)G0255, CG6914, and CG10219 analyzed by ChIP-qPCR and identified human NDUFS8, DBT, NDUFAB1, COX5A, ACO2, FH, NDUFA7, AP002884.2 and SDHD genes as their respective orthologs. (The latter two being orthologs of CG10219, while the other six genes had single human orthologs.) Occupancy of mammalian E2f1 on promoters of these genes was examined by ChIP using chromatin prepared from asynchronously dividing SAOS-2 cells. As positive controls for E2f1 binding, two cell cycle genes, NUSAP1 and PCNA, were included while two intergenic regions, INT20D and INT26E, that lack E2f1 binding were used as negative controls (Figure 3B) (Beshiri et al., 2012). The promoters of NDUFS8, NDUFAB1, ACO2, FH, NDUFA7 and AP002884.2 were enriched in E2f1 immunopreciptates to the same level as for the PCNA and NUSAP1 promoters (Figure 3B). Importantly, E2f1 binding to these promoters was highly specific because it was blocked when DP1 and DP2 were depleted by transfection of siRNA. E2f1 was less abundant on three other targets DBT, COX5A, and SDHD, however, E2f1 enrichment was still higher than in negative controls (INT20D and INT26E) or when DP1 and DP2 were depleted. We concluded that dE2f/dDP directly regulates expression of mitochondria associated genes and that this function of E2F is conserved in mammalian cells.
Mitochondrial function is compromised in dDP mutants
Changes in expression of oxidative phosphorylation genes prompted us to investigate mitochondrial activity in dDP mutants. In wild type cells, functional electron transport chains maintain mitochondrial membrane polarization, which is used to drive ATP production by oxidative phosphorylation. This mitochondrial activity can be measured by MitoTracker dye since its binding to mitochondria is dependent on mitochondrial membrane potential. Compared to the wild type discs, the intensity of MitoTracker staining was significantly reduced in dDP mutant eye discs (Figure 4A). Consistently, dDP trans-heterozygous mutants had a significantly lower level of ATP than matching controls, dDP heterozygous mutants (Figure 4B). Thus, mitochondrial activity is reduced in dDP mutants.
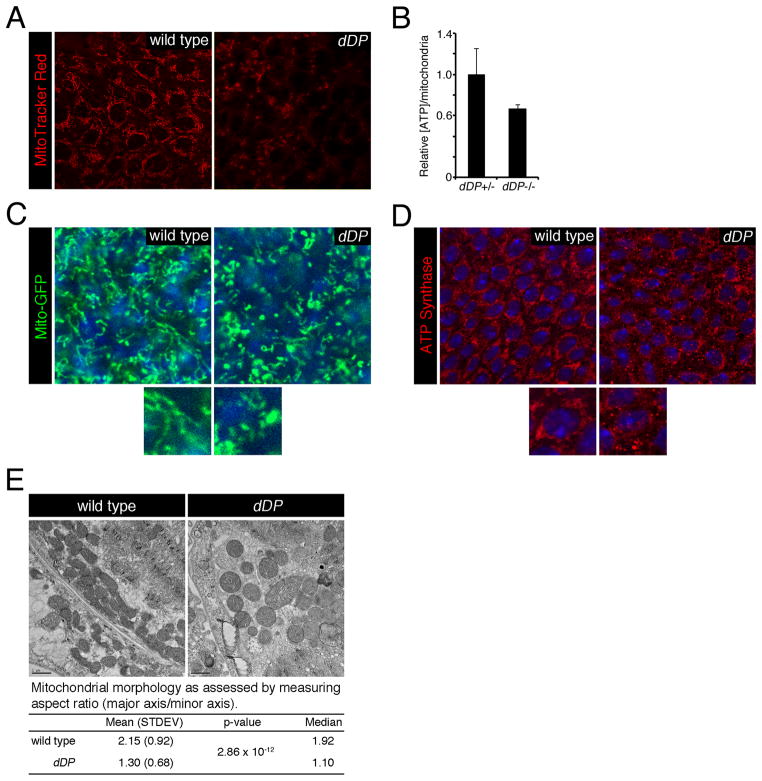
Figure 4. Reduced mitochondria activity and abnormal mitochondrial morphology in dDP mutant eye discs.
(A) MitoTracker (red) was used to examine the mitochondria membrane potential. (B) ATP levels were measured in the eye-antenna discs and brain of control and dDP mutant larvae (± SD, using 3 replicates for each genotype). The ATP levels were normalized to the relative number of mitochondria per animal. The relative number of mitochondria was determined by the ratio of mitochondria genome to nuclear genome content. (C and D) Mitochondria tagged GFP (mito-GFP, green), or immunostaining against ATP Synthase (red) was used to visualize mitochondrial morphology. DNA (blue) is stained with either DAPI (C) or PicoGreen (D). (E) Transmitting electron microscopy. For each genotype 100 mitochondria were measured (± SD) to determine mitochondrial aspect ratios.
Next, the mitochondrial morphology of dDP mutant cells in eye discs was examined by immunofluorescence. Mitochondria were visualized with a green fluorescence protein tagged with a mitochondrial localization signal (mito-GFP). In wild type eye discs, mito-GFP reveals an extensive network of branched mitochondria (Figure 4C). In contrast, the mitochondrial network appeared abnormal in dDP mutants. Particularly, the extent of the mitochondrial network was reduced and mitochondria were more fragmented in dDP mutants than in wild type. This conclusion was confirmed with another mitochondrial marker, ATP-synthase α (Figure 4D).
Transmitting electron microscopy was employed to examine dDP mutant mitochondria at the ultrastructural level. In wild type eye discs, mitochondria were largely elongated. In dDP mutants, mitochondria appeared to be more globular and swollen (Figure 4E). This morphological difference of mitochondria shape was quantified by measuring the aspect ratio (AR), which is the ratio between the major and minor axes of the ellipse equivalent to the mitochondrion. Thus, an AR has a minimal value of 1 for perfectly circular mitochondria and the value increases for more elongated mitochondria. Indeed, the AR of dDP mutant mitochondria was statistically significantly lower than in wild type indicating a larger number of swollen mitochondria in dDP mutants than in wild type (Figure 4E). From these results we concluded that a dDP mutation leads to mitochondrial defects including abnormal mitochondrial shape, reduced mitochondrial membrane potential and lowered ATP generation.
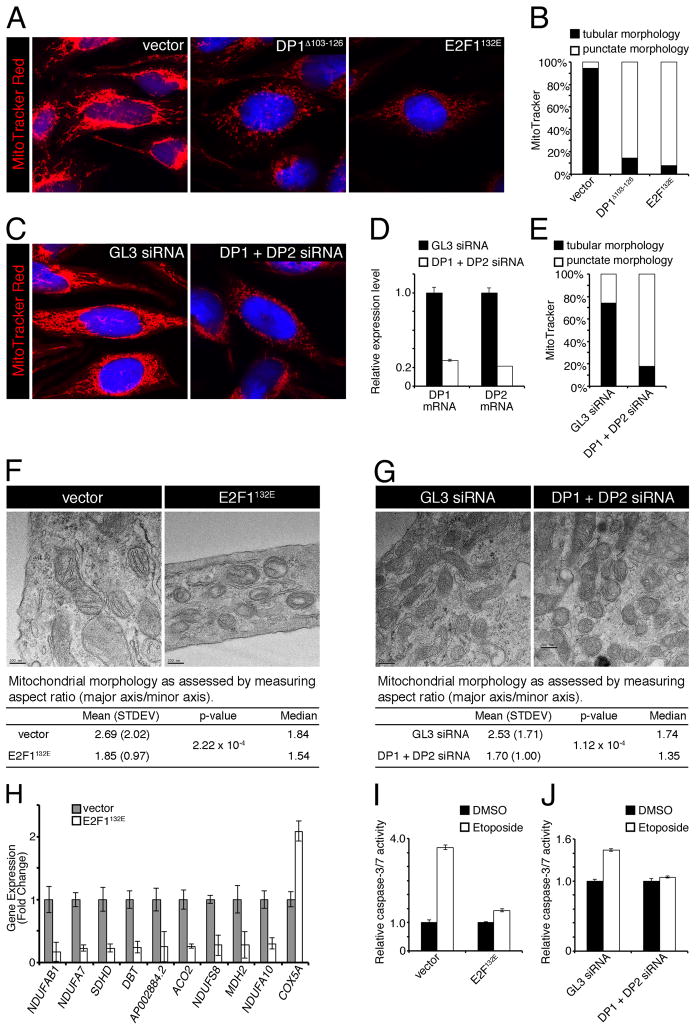
To extend these findings beyond the Drosophila model system, we examined the impact of inactivation of E2F on mitochondrial morphology in mammalian cells by MitoTracker staining. To inactivate E2F, a dominant negative form of DP1Δ103-126, which retains a dimerization domain but lacks a DNA-binding domain (Wu et al., 1996), was used. DP1Δ103-126 was transiently expressed in SAOS-2 cells together with a neomycin resistance gene and a GFP-expressing plasmid to identify transfected cells. In control cells transfected with an empty vector, mitochondria had an elongated, tubular shape in the majority of cells (Figure 5A and B). In contrast, overexpression of DP1Δ103-126 resulted in a larger population of cells which had a punctate and reduced MitoTracker staining indicating that mitochondria structures and membrane potential were compromised. Similar results were observed with the E2F132E dominant negative mutant form of E2F, which contains a point mutation that prevents DNA binding, but does not interfere with DP1 binding (Cress et al., 1993) (Figure 5A and B). Importantly, the abnormal mitochondrial phenotype is not an indirect cell cycle effect caused by E2F or DP mutants since overexpression of a cdk inhibitor p21 in SAOS-2 cells does not elicit such a mitochondrial phenotype (Lopez-Bigas et al., 2008).
Figure 5. Mitochondria activity and structure are compromised in SAOS-2 cells upon E2F inactivation.
(A and C) MitoTracker (red) staining of SAOS-2 cells that were either transfected with empty vector, dominant negative DP1 (DP1Δ103-126), dominant negative E2F1 (E2F1132E), unrelated GL3 siRNA, or DP1 and DP2 siRNAs. (B and E) Cells represented by images from A and C were scored for displaying a tubular or punctate mitochondria phenotype. (D) DP1 and DP2 mRNA levels (± SD, using 3 replicates for each treatment) are efficiently depleted by siRNAs. (F and G) TEM of control or E2F/DP inactivated SAOS-2 cells. For each genotype 100 mitochondria were measured (± SD) to determine mitochondrial aspect ratios. (H) qPCR of mitochondria associated genes in SAOS-2 cells that were either transfected with empty vector (gray bars) or dominant negative E2F1 (E2F1132E) (white bars) (± SD, using 3 replicates for each treatment). (I and J) SAOS-2 cells transfected with the indicated plasmid or siRNAs, as described above, were treated with DMSO (black bars) or 25μM etoposide (white bars) for 24h. Apoptosis was quantified by luminescence emission by cleavage of a proluminescent caspase-3/7 DEVD-aminoluciferin substrate (± SD, using 3 replicates for each treatment). See also Supplemental Figure S2.
To complement the results obtained with overexpression of dominant negative forms of E2F and DP, siRNA was used to inactivate endogenous DP1 and DP2. As shown in Figure 5C–E, the number of cells with abnormal mitochondrial staining was greatly increased following DP1/DP2 siRNA treatment in comparison with cells that were treated with an unrelated GL3 siRNA. These results made using MitoTracker were confirmed by immunofluorescent staining against a mitochondrial marker, Cytochrome c Oxidase Subunit IV (COXIV) (Supplemental Figure S2). Next, mitochondria ultrastructure was examined by transmitting electron microscopy. Compared to control cells which exhibited more elongated mitochondria (mean AR = 2.69), the mitochondria of cells overexpressing the E2F132E mutant construct were found to be more circular (mean AR = 1.85) (Figure 5F). Similar results were obtained when siRNA were used to inactivate endogenous DP1 and DP2 (Figure 5G). As an additional characterization of the mitochondrial defects, we examined expression of mitochondria associated E2F target genes that were identified by ChIP (Figure 3). Notably, nine out of ten genes were downregulated following overexpression of the E2F132E mutant (Figure 5H).
Finally, the effect of E2F inactivation on DNA damage-induced apoptosis was examined. Caspase-3 and -7 activity was measured to assess apoptosis in SAOS-2 cells following treatment with a DNA damage-inducing drug, etoposide. In control cells, etoposide treatment resulted in a strong induction of caspase-3/7 activity, while in cells overexpressing the E2F132E mutant construct or when DP1 and DP2 were depleted by siRNAs, the apoptotic response was muted (Figure 5I and J). We note that we cannot rule out that cell cycle effects induced by E2F inactivation may contribute to the reduced sensitivity to DNA damage-induced apoptosis. Nevertheless, similar to Drosophila eye imaginal discs, inactivation of E2F/DP prevents induction of DNA damage-induced apoptosis in mammalian cells. Taken together, these data suggest that the loss of E2F/DP leads to strikingly similar phenotypes in flies and in mammalian cells indicating that the role of E2F/DP in the regulation of mitochondrial function is conserved.
Downregulation of mitochondria associated dE2f/dDP target genes mimics mitochondrial defects and protection from irradiation-induced apoptosis of dDP mutant eye discs
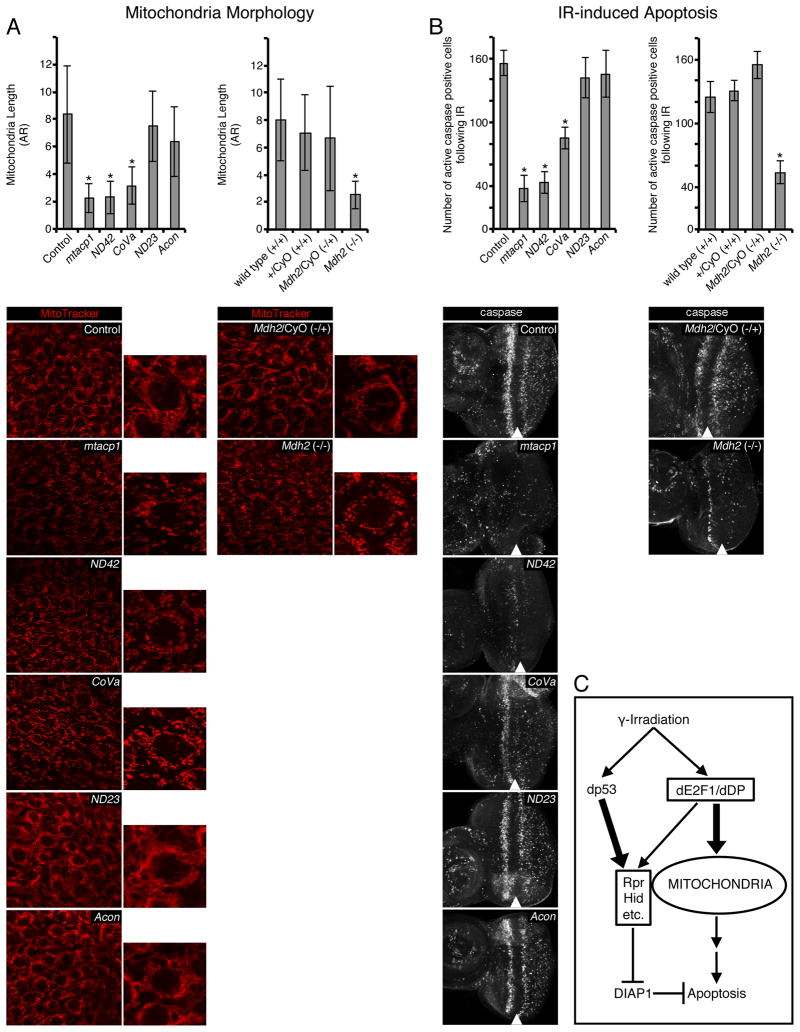
Reduced expression of dE2F/dDP mitochondria associated targets may explain abnormal mitochondrial morphology in dDP mutants. To test this idea directly, we examined the impact of downregulation of these genes by RNAi on the mitochondrial network in the eye imaginal discs. Five candidates, mtacp1, ND42, CoVa, ND23, and Acon, were selected. Mitochondrial activity and morphology was visualized by MitoTracker dye and the phenotype was quantified by measuring aspect ratio (AR). Downregulation of mtacp1, ND42 and CoVa resulted in altered mitochondrial morphology and a reduced mitochondrial network, with mtacp1 and ND42 knockdown giving the strongest effect (AR = 2.24 and 2.31 respectively, compared to AR = 8.36 for control eye discs) (Figure 6A). We note that the mitochondrial phenotype was not as severe as in dDP mutants (Figure 4). In contrast, expression of ND23 and Acon dsRNAs had no effect, which could be due to inefficient protein depletion by RNAi. Additionally, mutation of Mdh2, another dE2F/dDP target gene, also resulted in an abnormal mitochondrial network (AR = 2.54, compared to AR = 7.99 for wild type control eye discs) (Figure 6A). Thus, downregulation of mitochondria associated dE2F/dDP targets is sufficient to, at least partially, phenocopy the mitochondrial defects of dDP mutants.
Figure 6. RNAi mediated knockdown of mitochondria associated dE2f/dDP targets mimics the mitochondrial and apoptosis defects indDP mutant eye discs.
For each gene indicated, the corresponding dsRNA under Gal4-UAS control was expressed in the eye disc with ey-FLP; Act≫Gal4 except for the Mdh2 gene for which a mutant allele was used. (A) MitoTracker (red) was used to label mitochondria and mitochondrial aspect ratios were quantified (± SD, with 30 mitochondria from at least 5 discs quantified for each genotype). (B) Active caspase immunostaining was used to detect and quantify apoptotic cells 4h after irradiation (± SD, with at least 5 discs quantified for each genotype). (*) P < 0.05 in paired t-tests comparing the indicated line to an ey-FLP; Act≫Gal4 control except for Mdh2 which was compared to all three of the control genotypes indicated. We note that many controls were used and none of them were statistically significantly different from each other (Supplemental Figure S3A). (C) In irradiated cells, dp53, but not dE2f1, is primarily responsible for induction of the apoptotic gene expression program. In dDP mutants, mitochondrial activity is reduced, likely due to the reduction in expression of mitochondria associated E2F targets. This mitochondrial defect reduces the overall readiness of dDP mutants to undergo cell death and therefore, the properly induced irradiation transcriptional program is insufficient to trigger apoptosis. See also Supplemental Figure S3.
Recent studies have pointed to the emerging role of mitochondria in the regulation of apoptosis in Drosophila (Abdelwahid et al., 2011; 2007; Goyal et al., 2007; Sandu et al., 2010). This raises the possibility that mitochondrial defects in dDP mutants could alter the apoptotic response of these cells to DNA damage. Having established that knockdown of mitochondria associated dE2F/dDP targets induces a range of mitochondrial defects in eye imaginal discs, we asked whether the apoptotic response to irradiation is altered.
Eye discs of various genotypes were irradiated and four hours after irradiation apoptotic cells were detected by Caspase 3 staining (Figure 6B). Almost no apoptotic cells were detected prior to irradiation in knockdowns and in matching controls (Supplemental Figure S3A). In control eye discs, there were approximately 156 apoptotic cells per disc. Downregulation of mtacp1 and ND42, which had the most severe mitochondrial phenotypes, strongly suppressed the apoptotic response as the number of caspase positive cells were reduced approximately four fold (38 and 43 apoptotic cells per disc respectively). Knockdown of CoVa provided an intermediate effect (85 apoptotic cells). In contrast, ND23 and Acon RNAi which had no effect on the mitochondrial morphology (Figure 6A), did not suppress apoptosis (142 and 145 apoptotic cells respectively). The reduction in the number of apoptotic cells reflects a muted response to irradiation rather than a delay in the induction of apoptosis since the number of caspase positive cells remained low ten hours post irradiation (Supplemental Figure S3B). Furthermore, apoptosis was strongly reduced in irradiated Mdh2 mutant discs (54 apoptotic cells per disc) to the extent seen in the case of mtacp1 and ND42 knockdown, which exhibited similar AR values. The suppression of apoptosis is not an indirect effect of the cell cycle since the pattern of mitoses in Mdh2 mutants remained unaffected (Supplemental Figure S3C).
We concluded that downregulation of mitochondria associated dE2F/dDP target genes is sufficient to induce both the mitochondrial phenotype and protection from irradiation-induced apoptosis similar to what is observed in dDP mutants. Significantly, the genes that elicited the strongest mitochondrial defects, such as mtacp1, ND42 or Mdh2, provided the strongest reduction of irradiation-induced apoptosis. Conversely, knockdown of ND23 and Acon exerted no effect on mitochondrial morphology and apoptosis. Additionally, CoVa elicited both an intermediate effect on mitochondrial morphology as well as irradiation-induced apoptosis. Thus, the severity of mitochondrial defects induced by downregulation of dE2F/dDP regulated mitochondria associated genes correlates well with the degree of protection from irradiation-induced apoptosis.
DISCUSSION
E2F transcription factors are best understood for their role in the control of the cell cycle, apoptosis and differentiation. In this report, we present evidence that E2F is also involved in the regulation of mitochondrial function and identify a specific biological context, DNA damage-induced apoptosis, when such aspect of E2F control becomes critical. We suggest that mitochondrial dysfunction, and not the failure to induce the apoptotic gene expression program makes E2F deficient cells refractory to apoptosis.
In flies and mammals, the conserved mechanism by which E2F triggers apoptosis is transcriptional control of apoptotic targets (Asano et al., 1996; Aslanian et al., 2004; Irwin et al., 2000; Moon et al., 2005; Pediconi et al., 2003). Therefore, it is believed that in irradiated cells, dE2f1, like dp53, contributes to the normal transcriptional induction of apoptotic genes (Moon et al., 2008). However, our data do not support such a model since the apoptotic gene expression program was induced properly in irradiated dDP mutants. Thus, in the context of DNA damage response, the contribution of dE2f1 to the normal transcriptional induction of apoptotic genes is negligible. We emphasize that our data do not imply that dE2f1 is unimportant. For example, unrestrained dE2f1 activity in rbf mutants has been shown to markedly increase the induction of hid and rpr in response to DNA damage and this increase determines the elevated sensitivity of rbf mutants to irradiation-induced apoptosis (Moon et al., 2008). Since ablation of dp53 completely blocks irradiation-induced apoptosis and induction of apoptotic genes (Brodsky et al., 2000; Moon et al., 2008) the irradiation-induced apoptotic program is primarily governed by dp53, while hyperactive dE2f1 can provide additional assistance to dp53 in activating apoptotic genes (Figure 6C). This contribution of dE2f1 becomes evident in certain settings such as in rbf mutants.
Given that the apoptotic gene expression program is properly induced, in irradiated dDP mutants, to undergo apoptosis, their failure to do so is puzzling. We suggest that the resistance of dDP mutants to apoptosis is the consequence of a mitochondrial dysfunction. In dDP mutants, mitochondria exhibit an abnormal morphology, reduced mitochondrial membrane potential and ATP levels. The lower level of expression of dE2f/dDP mitochondria associated target genes is a critical event in determining the response of dDP mutants to irradiation since genetic attenuation of their expression mimics the dDP mutant mitochondrial phenotype and protection from irradiation-induced apoptosis. Significantly, the strongest protection was observed with the genes that exerted the most severe mitochondrial defects upon downregulation. Thus, the response to irradiation-induced apoptosis correlates with the extent of the mitochondrial defects. One possibility is that the mitochondrial dysfunction of dDP deficient cells lowers their mitochondrial readiness to apoptosis and therefore, the irradiation-induced apoptotic transcriptional program is insufficient to trigger cell death. Intriguingly, a concept of mitochondrial readiness to apoptosis is thought to be the molecular basis of a differential response to chemotherapy in cancer patients with acute myelogenous leukemia (Vo et al., 2012).
Among mitochondria associated dE2F/dDP target genes investigated in this work, Mdh2 is particularly interesting. It has been previously shown that an Mdh2 mutation prevents apoptosis in another context during ecdysone-induced cell death in salivary glands (Wang et al., 2010). Destruction of salivary glands is normally triggered by the induction of hid and rpr and both genes were induced in Mdh2 mutants to the level observed in wild type. In addition, Mdh2 mutants display a defect in energy production and exhibit reduced ATP levels which is thought to compromise their ability to undergo apoptosis. This setting is highly reminiscent of dDP mutants, which are also remarkably resistant to cell death even in the face of a high level of induction of a DNA damage-dependent apoptotic transcription program.
The idea that mitochondrial defects could impact execution of apoptosis is consistent with the recently uncovered importance of mitochondria in cell death in Drosophila. Several studies demonstrated that Rpr, Grim, and Hid, the key apoptotic proteins in flies (Hay and Guo, 2006; Steller, 2008), are localized to mitochondria and that this localization is required for efficient activation of apoptosis (Abdelwahid et al., 2007; Clavería et al., 2002; Goyal et al., 2007; Sandu et al., 2010). Thus, one possibility is that pro-apoptotic proteins are not efficiently localized to mitochondria in dDP mutants. It is also possible that the dysfunctional mitochondria of dDP mutants fail to remodel in response to irradiation which has been shown to be necessary for execution of stress induced apoptosis (Abdelwahid et al., 2007; Goyal et al., 2007). We note that our findings do not imply that dE2F/dDP normally triggers apoptosis by modulation of mitochondrial function but rather that mitochondrial function is compromised in E2F deficient cells which would, in turn, result in less efficient apoptosis.
Another important conclusion of our study is that a mechanistic link between the Rb pathway and mitochondria is conserved in mammalian cells. We show that several Drosophila dE2F/dDP-regulated mitochondria associated genes are also E2F targets in mammalian cells and their expression is similarly reduced in cells when E2F is inactivated. Significantly, this leads to strong mitochondrial defects that are highly reminiscent of the mitochondrial phenotype in Drosophila dDP mutant eye discs. Our data are consistent with the recent finding that mammalian E2f1 and pRB regulate expression of oxidative metabolism genes during the adaptive metabolic response in mice (Blanchet et al., 2011). The Rb pathway has been also implicated in the regulation of the mitochondrial biogenesis transcriptional program in erythropoiesis (Sankaran et al., 2008). Intriguingly, a recent study demonstrated that a fraction of endogenous pRB is present at mitochondria where it directly participates in mitochondrial apoptosis (Hilgendorf et al., 2013). Given the prominent role of mitochondrial pathways in apoptosis in mammalian cells it is conceivable that the loss of E2F could impact the efficacy of apoptosis in mammals by an analogous mechanism that operates in Drosophila. Such an idea is consistent with our finding that inactivation of E2F reduces DNA damage-induced apoptosis in mammalian cells.
Interestingly, Nicolay and colleagues recently demonstrated that an rbf mutation alters cellular metabolism and that an abnormal metabolism sensitizes Drosophila to various types of stress (Nicolay et al., 2013). In this work, we found that inactivation of E2F results in a severe mitochondrial dysfunction, which is the basis for the failure of dDP mutants to undergo DNA damage-induced apoptosis. Thus, a general emerging theme is that perturbation of the Rb pathway may exert profound metabolic changes within the cell that can have a major impact on cell survival.
EXPERIMENTAL PROCEDURES
Fly stocks
Canton S (wild type), dDPa3/Df(2R)Exel7124 (dDP), dDPa4/Df(2R)Exel7124 and Dronc51/TM3 (Chew et al., 2004), Mdh22 (Wang et al., 2010) were used. Df(2R)Exel7124 deletes the entire dDP gene. dDPa3 and dDPa4 are null alleles of dDP (Frolov et al., 2005; Royzman et al., 1997). dsRNA of mitochondria associated genes was expressed by Act≫Gal4/P{TRiP}. As a control, ey-FLP; Act≫Gal4/CyOGFP was used as well as ey-FLP; +/CyO GFP; UAS-mtacp1RNAi, ey-FLP; +/CyOGFP; UAS-ND42RNAi, and ey-FLP; +/CyOGFP; UAS-CoVaRNAi. Mitochondria were visualized in CyOGFP/+; sqh-mito-GFP (control) and dDPa4/Df(2R)Exel7124; sqh-mito-GFP (dDP mutant with mito-GFP).
Immunofluorescence
Larval eye disc dissection, fixation, antibody incubation and incorporation of bromodeoxyuridine (BrdU) were done as previously described (Ambrus et al., 2007). For MitoTracker assays, eye discs were incubated with MitoTracker Red dye (500 nM) for 20 min. For TUNEL assay, eye discs were labeled with In Situ Cell Death Detection Kit, TMR red (Roche).
SAOS-2 cells were plated out onto cover slips 24 hours after transfection. Transfected cells were placed under puromycin selection 24 hours afterwards. siRNA and plasmid transfected cells were fixed 72 hours and 96 hours after transfection, respectively.
The primary antibodies were mouse anti-ATP Synthase (1:1000) (MitoSciences), mouse anti-BrdU (1:50) (BD Bioscience), rabbit anti-C3 (Cleaved Caspase3) lot 26, (1:100) (Cell Signaling), rabbit anti-phosphorylated histone H3 (1:150) (Upstate), rabbit anti-COXIV (1:200) (ab16056, Abcam). The secondary conjugated antibodies used in this work were anti-mouse Cy3 (1:100), anti-rabbit Cy3 (1:100), and anti-rabbit Cy5 (1:100) from Jackson Immunolaboratories.
Real Time qPCR
Total RNA was isolated from 50–80 eye discs with TRIzol (Invitrogen). iScript kit (Bio-Rad) and LightCycler 480 SYBR Green I Master (Roche) kits were used for qPCR. Primer sequences are in Supplemental Experimental Procedures.
ATP determination
Eye discs and brains from 25 third instar larvae were homogenized in PBS, and boiled for 5 minutes. ATP was determined with ATP determination kit (Molecular Probes). In parallel, total DNA was isolated from an equal number of eye discs and brains. Samples were subjected to qPCR with primers for both the nuclear genome and the mitochondrial genome. Ratios between the relative quantities of the nuclear to mitochondrial DNA content were determined and used as a normalization factor to determine the relative amount of ATP per mitochondrion. All samples were assayed in triplicate.
Mammalian Cell Culture and Transfection
SAOS-2 cells were grown in DMEM (Mediatech) supplemented with 10% fetal bovine serum (FBS). SAOS-2 cells were transfected by Lipofectamine 2000 (Invitrogen) for siRNA and Fugene 6 (Roche) for plasmid DNA. GL3 siRNA was used as a control (Beshiri et al., 2012). Single siRNA transfections were performed at 50 nM; for double transfection, 25 nM of each siRNA was used. The plasmids used for transfections were pCMV-neo empty vector, pCMV-E2F1-E132, and pCMV-HA-DP1Δ103-126 in 1:1:1 ratio with pmax-GFP (Lonza) and pBabePuro.
ChIP
Drosophila ChIP was performed with either rabbit anti-dDP, mouse anti-RBF1, rabbit anti-dE2f1, rabbit anti-dE2f2, or mouse anti-Myc (9E10) as described (Korenjak et al., 2012). The DNA was analyzed by qPCR and the enrichment for each antibody was calculated relative to the quantity of input DNA. Primers were selected based on their proximity to gene regions within 2 kb upstream of the translation start site that had the highest density of predicted dE2F binding sites which were determined using Regulatory Sequence Analysis Tools.
Mammalian ChIP was performed as described (Lopez-Bigas et al., 2008) using mouse anti-E2F1 (KH20). For probe design see Supplemental Experimental Procedures, sequences from 2 kb upstream of the TSS were retrieved from EnsEMBL database (v69) and analyzed for E2F1 transcription binding sites using TFSEARCH program in the TRANSFAC software (Heinemeyer et al., 1998).
ChIP-seq
ChIP-seq was performed with 7.5×108 S2R+ cells per sample and dDP1 212 antiserum as described (Beshiri et al., 2010). The genomic libraries were generated using AMPure XP beads (Agencourt) following instructions in the TruSeq DNA sample preparation v2 guide (Illumina, Inc.). Library fragments of ~470 bp (insert plus adaptor and PCR primer sequences) were isolated from an agarose gel. The purified DNA was captured on an Illumina flow cell for cluster generation. A total of 9,099,790 and 18,529,623 reads were generated for two biological replicate ChIP samples, respectively and 18,406,223 reads were generated for the input sample.
Illumina/Solexa sequencer output images were processed using Solexa image extraction pipeline [v 1.6 (Cassava)]. Identified 36 bp short reads were uniquely aligned allowing at best two mismatches to the UCSC Drosophila melanogaster reference genome (April 2006, BDGP R5/dm3) using BOWTIE. Peak caller algorithm MACS (v 1.4) was used to determine the enriched peak region against total genomic input as background. Peak shifts were calculated using the strand cross correlation method implemented in Pyicos (Althammer et al., 2011). Enriched peaks were annotated to the closest EnsEMBL (version 62) transcript start site and thus to genes using BEDTools. ChIP-seq data have been deposited in Gene Expression Omnibus with the accession number GSE39393.
Expression data analysis
RNA was harvested from third instar larval eye discs in biological triplicates of Canton S and dDPa3/Df(2R)Exel7124. Samples were subjected to Affymetrix GeneChip microarray. Microarray data were analyzed as previously described in (Nicolay et al., 2011). Gitools was used for enrichment analysis and heatmap generation (Perez-Llamas and Lopez-Bigas, 2011). Resulting p-values were adjusted for multiple testing using the Benjamin and Hochberg’s method of False Discovery Rate (FDR). Microarray data have been deposited in Gene Expression Omnibus with the accession number GSE39389.
Supplementary Material
HIGHLIGHTS.
E2F deficient animals have severe mitochondrial defects;
E2F directly regulates expression of mitochondria associated genes;
Mitochondrial defects explain the resistance of dDP mutants to IR induced apoptosis
An important function of E2F/DP transcription factors is in regulating apoptosis. Ambrus et al. show that, in flies and in mammalian cells, E2F/DP regulate mitochondrial function which in turn is critical for the normal induction of apoptosis in DNA damage response.
Acknowledgments
We thank A. Katzen, G. Ramsey and K. White for discussions, N. Dyson and F. Dick for reagents, and B. Nicolay, N. Dyson and J. Lees for sharing data prior to publication. This work was supported by the National Institutes of Health grants CA138631 (E.V.B.) and GM93827 (M.V.F.), by a Scholar Award from the Leukemia and Lymphoma Society (M.V.F.), by Natural Science and Engineering Research Council of Canada grant 355760-2008 to (N.S.M.), and by grants SAF2009-06954 and SAF2012-36199 of Spanish Ministry of Economy and Competitivity (N.L.-B.). A.I. is supported by a fellowship from AGAUR of the Catalonian government.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abdelwahid E, Rolland S, Teng X, Conradt B, Hardwick JM, White K. Mitochondrial involvement in cell death of non-mammalian eukaryotes. Biochim Biophys Acta. 2011;1813:597–607. doi: 10.1016/j.bbamcr.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelwahid E, Yokokura T, Krieser RJ, Balasundaram S, Fowle WH, White K. Mitochondrial disruption in Drosophila apoptosis. Dev Cell. 2007;12:793–806. doi: 10.1016/j.devcel.2007.04.004. [DOI] [PubMed] [Google Scholar]
- Althammer S, González-Vallinas J, Ballaré C, Beato M, Eyras E. Pyicos: a versatile toolkit for the analysis of high-throughput sequencing data. Bioinformatics. 2011;27:3333–3340. doi: 10.1093/bioinformatics/btr570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrus AM, Nicolay BN, Rasheva VI, Suckling RJ, Frolov MV. dE2F2-independent rescue of proliferation in cells lacking an activator dE2F1. Mol Cell Biol. 2007;27:8561–8570. doi: 10.1128/MCB.01068-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano M, Nevins JR, Wharton RP. Ectopic E2F expression induces S phase and apoptosis in Drosophila imaginal discs. Genes Dev. 1996;10:1422–1432. doi: 10.1101/gad.10.11.1422. [DOI] [PubMed] [Google Scholar]
- Aslanian A, Iaquinta PJ, Verona R, Lees JA. Repression of the Arf tumor suppressor by E2F3 is required for normal cell cycle kinetics. Genes Dev. 2004;18:1413–1422. doi: 10.1101/gad.1196704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beshiri ML, Holmes KB, Richter WF, Hess S, Islam ABMMK, Yan Q, Plante L, Litovchick L, Gévry N, Lopez-Bigas N, et al. Coordinated repression of cell cycle genes by KDM5A and E2F4 during differentiation. Proceedings of the National Academy of Sciences. 2012;109:18499–18504. doi: 10.1073/pnas.1216724109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beshiri ML, Islam A, DeWaal DC, Richter WF, Love J, Lopez-Bigas N, Benevolenskaya EV. Genome-wide analysis using ChIP to identify isoform-specific gene targets. J Vis Exp. 2010 doi: 10.3791/2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas AK, Johnson DG. Transcriptional and nontranscriptional functions of E2F1 in response to DNA damage. Cancer Res. 2012;72:13–17. doi: 10.1158/0008-5472.CAN-11-2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchet E, Annicotte JS, Lagarrigue S, Aguilar V, Clapé C, Chavey C, Fritz V, Casas F, Apparailly F, Auwerx J, et al. E2F transcription factor-1 regulates oxidative metabolism. Nat Cell Biol. 2011;13:1146–1152. doi: 10.1038/ncb2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky MH, Nordstrom W, Tsang G, Kwan E, Rubin GM, Abrams JM. Drosophila p53 binds a damage response element at the reaper locus. Cell. 2000;101:103–113. doi: 10.1016/S0092-8674(00)80627-3. [DOI] [PubMed] [Google Scholar]
- Chew SK, Akdemir F, Chen P, Lu WJ, Mills K, Daish T, Kumar S, Rodriguez A, Abrams JM. The apical caspase dronc governs programmed and unprogrammed cell death in Drosophila. Dev Cell. 2004;7:897–907. doi: 10.1016/j.devcel.2004.09.016. [DOI] [PubMed] [Google Scholar]
- Clavería C, Caminero E, Martínez-A C, Campuzano S, Torres M. GH3, a novel proapoptotic domain in Drosophila Grim, promotes a mitochondrial death pathway. Embo J. 2002;21:3327–3336. doi: 10.1093/emboj/cdf354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cress WD, Johnson DG, Nevins JR. A genetic analysis of the E2F1 gene distinguishes regulation by Rb, p107, and adenovirus E4. Mol Cell Biol. 1993;13:6314–6325. doi: 10.1128/mcb.13.10.6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson JM, Duronio RJ. S phase-coupled e2f1 destruction ensures homeostasis in proliferating tissues. PLoS Genet. 2012;8:e1002831. doi: 10.1371/journal.pgen.1002831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGregori J, Leone G, Miron A, Jakoi L, Nevins JR. Distinct roles for E2F proteins in cell growth control and apoptosis. Proc Natl Acad Sci USA. 1997;94:7245–7250. doi: 10.1073/pnas.94.14.7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimova DK, Stevaux O, Frolov MV, Dyson NJ. Cell cycle-dependent and cell cycle-independent control of transcription by the Drosophila E2F/RB pathway. Genes Dev. 2003;17:2308–2320. doi: 10.1101/gad.1116703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolov MV, Moon NS, Dyson NJ. dDP is needed for normal cell proliferation. Mol Cell Biol. 2005;25:3027–3039. doi: 10.1128/MCB.25.8.3027-3039.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georlette D, Ahn S, MacAlpine DM, Cheung E, Lewis PW, Beall EL, Bell SP, Speed T, Manak JR, Botchan MR. Genomic profiling and expression studies reveal both positive and negative activities for the Drosophila Myb MuvB/dREAM complex in proliferating cells. Genes Dev. 2007;21:2880–2896. doi: 10.1101/gad.1600107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyal G, Fell B, Sarin A, Youle RJ, Sriram V. Role of mitochondrial remodeling in programmed cell death in Drosophila melanogaster. Dev Cell. 2007;12:807–816. doi: 10.1016/j.devcel.2007.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay BA, Guo M. Caspase-dependent cell death in Drosophila. Annu Rev Cell Dev Biol. 2006;22:623–650. doi: 10.1146/annurev.cellbio.21.012804.093845. [DOI] [PubMed] [Google Scholar]
- Heinemeyer T, Wingender E, Reuter I, Hermjakob H, Kel AE, Kel OV, Ignatieva EV, Ananko EA, Podkolodnaya OA, Kolpakov FA, et al. Databases on transcriptional regulation: TRANSFAC, TRRD and COMPEL. Nucleic Acids Res. 1998;26:362–367. doi: 10.1093/nar/26.1.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgendorf KI, Leshchiner ES, Nedelcu S, Maynard MA, Calo E, Ianari A, Walensky LD, Lees JA. The retinoblastoma protein induces apoptosis directly at the mitochondria. Genes Dev. 2013;27:1003–1015. doi: 10.1101/gad.211326.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin M, Marin MC, Phillips AC, Seelan RS, Smith DI, Liu W, Flores ER, Tsai KY, Jacks T, Vousden KH, et al. Role for the p53 homologue p73 in E2F-1-induced apoptosis. Nature. 2000;407:645–648. doi: 10.1038/35036614. [DOI] [PubMed] [Google Scholar]
- Korenjak M, Anderssen E, Ramaswamy S, Whetstine JR, Dyson NJ. RBF binding to both canonical E2F targets and noncanonical targets depends on functional dE2F/dDP complexes. Mol Cell Biol. 2012;32:4375–4387. doi: 10.1128/MCB.00536-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Bigas N, Kisiel TA, DeWaal DC, Holmes KB, Volkert TL, Gupta S, Love J, Murray HL, Young RA, Benevolenskaya EV. Genome-wide analysis of the H3K4 histone demethylase RBP2 reveals a transcriptional program controlling differentiation. Mol Cell. 2008;31:520–530. doi: 10.1016/j.molcel.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon NS, Di Stefano L, Morris EJ, Patel R, White K, Dyson NJ. E2F and p53 induce apoptosis independently during Drosophila development but intersect in the context of DNA damage. PLoS Genet. 2008;4:e1000153. doi: 10.1371/journal.pgen.1000153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon NS, Frolov MV, Kwon EJ, Di Stefano L, Dimova DK, Morris EJ, Taylor-Harding B, White K, Dyson NJ. Drosophila E2F1 has context-specific pro- and antiapoptotic properties during development. Dev Cell. 2005;9:463–475. doi: 10.1016/j.devcel.2005.08.015. [DOI] [PubMed] [Google Scholar]
- Müller H, Bracken AP, Vernell R, Moroni MC, Christians F, Grassilli E, Prosperini E, Vigo E, Oliner JD, Helin K. E2Fs regulate the expression of genes involved in differentiation, development, proliferation, and apoptosis. Genes Dev. 2001;15:267–285. doi: 10.1101/gad.864201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolay BN, Bayarmagnai B, Islam ABMMK, Lopez-Bigas N, Frolov MV. Cooperation between dE2F1 and Yki/Sd defines a distinct transcriptional program necessary to bypass cell cycle exit. Genes Dev. 2011;25:323–335. doi: 10.1101/gad.1999211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolay BN, Gameiro PA, Tschöp K, Korenjak M, Heilmann AM, Asara JM, Stephanopoulos G, Iliopoulos O, Dyson NJ. Loss of RBF1 changes glutamine catabolism. Genes Dev. 2013;27:182–196. doi: 10.1101/gad.206227.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pediconi N, Ianari A, Costanzo A, Belloni L, Gallo R, Cimino L, Porcellini A, Screpanti I, Balsano C, Alesse E, et al. Differential regulation of E2F1 apoptotic target genes in response to DNA damage. Nat Cell Biol. 2003;5:552–558. doi: 10.1038/ncb998. [DOI] [PubMed] [Google Scholar]
- Perez-Llamas C, Lopez-Bigas N. Gitools: analysis and visualisation of genomic data using interactive heat-maps. PLoS ONE. 2011;6:e19541. doi: 10.1371/journal.pone.0019541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polager S, Ginsberg D. p53 and E2f: partners in life and death. Nat Rev Cancer. 2009;9:738–748. doi: 10.1038/nrc2718. [DOI] [PubMed] [Google Scholar]
- Royzman I, Whittaker AJ, Orr-Weaver TL. Mutations in Drosophila DP and E2F distinguish G1-S progression from an associated transcriptional program. Genes Dev. 1997;11:1999–2011. doi: 10.1101/gad.11.15.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandu C, Ryoo HD, Steller H. Drosophila IAP antagonists form multimeric complexes to promote cell death. J Cell Biol. 2010;190:1039–1052. doi: 10.1083/jcb.201004086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankaran VG, Orkin SH, Walkley CR. Rb intrinsically promotes erythropoiesis by coupling cell cycle exit with mitochondrial biogenesis. Genes Dev. 2008;22:463–475. doi: 10.1101/gad.1627208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steller H. Regulation of apoptosis in Drosophila. Cell Death Differ. 2008;15:1132–1138. doi: 10.1038/cdd.2008.50. [DOI] [PubMed] [Google Scholar]
- Tanaka-Matakatsu M, Xu J, Cheng L, Du W. Regulation of apoptosis of rbf mutant cells during Drosophila development. Dev Biol. 2009;326:347–356. doi: 10.1016/j.ydbio.2008.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truscott M, Islam ABMMK, Lopez-Bigas N, Frolov MV. mir-11 limits the proapoptotic function of its host gene, dE2f1. Genes Dev. 2011;25:1820–1834. doi: 10.1101/gad.16947411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai KY, Hu Y, Macleod KF, Crowley D, Yamasaki L, Jacks T. Mutation of E2f-1 suppresses apoptosis and inappropriate S phase entry and extends survival of Rb-deficient mouse embryos. Mol Cell. 1998;2:293–304. doi: 10.1016/s1097-2765(00)80274-9. [DOI] [PubMed] [Google Scholar]
- Vo TT, Ryan J, Carrasco R, Neuberg D, Rossi DJ, Stone RM, DeAngelo DJ, Frattini MG, Letai A. Relative Mitochondrial Priming of Myeloblasts and Normal HSCs Determines Chemotherapeutic Success in AML. Cell. 2012;151:344–355. doi: 10.1016/j.cell.2012.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Lam G, Thummel CS. Med24 and Mdh2 are required for Drosophila larval salivary gland cell death. Dev Dyn. 2010;239:954–964. doi: 10.1002/dvdy.22213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SL, Hawkins CJ, Yoo SJ, Müller HA, Hay BA. The Drosophila caspase inhibitor DIAP1 is essential for cell survival and is negatively regulated by HID. Cell. 1999;98:453–463. doi: 10.1016/s0092-8674(00)81974-1. [DOI] [PubMed] [Google Scholar]
- Wu CL, Classon M, Dyson N, Harlow E. Expression of dominant-negative mutant DP-1 blocks cell cycle progression in G1. Mol Cell Biol. 1996;16:3698–3706. doi: 10.1128/mcb.16.7.3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.