Abstract
Stress responses play an important role in shaping species distributions and robustness to climate change. We investigated how stress responses alter the contribution of additive genetic variation to gene expression during development of the purple sea urchin, Strongylocentrotus purpuratus, under increased temperatures that model realistic climate change scenarios. We first measured gene expression responses in the embryos by RNA-seq to characterize molecular signatures of mild, chronic temperature stress in an unbiased manner. We found that an increase from 12 to 18 °C caused widespread alterations in gene expression including in genes involved in protein folding, RNA processing and development. To understand the quantitative genetic architecture of this response, we then focused on a well-characterized gene network involved in endomesoderm and ectoderm specification. Using a breeding design with wild-caught individuals, we measured genetic and gene–environment interaction effects on 72 genes within this network. We found genetic or maternal effects in 33 of these genes and that the genetic effects were correlated in the network. Fourteen network genes also responded to higher temperatures, but we found no significant genotype–environment interactions in any of the genes. This absence may be owing to an effective buffering of the temperature perturbations within the network. In support of this hypothesis, perturbations to regulatory genes did not affect the expression of the genes that they regulate. Together, these results provide novel insights into the relationship between environmental change and developmental evolution and suggest that climate change may not expose large amounts of cryptic genetic variation to selection in this species.
Keywords: gene expression, gene network, gene–environment interaction, stress response, Strongylocentrotus purpuratus
Introduction
Global climate change is exposing many species to stressful, and often, novel environments (Parmesan & Yohe 2003; Gienapp et al. 2008). Many species have shown changes in their phenology or distribution over the past century, and these changes are predominantly in the direction predicted by increasing temperatures (Parmesan & Yohe 2003; Root et al. 2003). However, organisms possess mechanisms to buffer their development and physiology against the effects of deleterious temperatures (Hochachka & Somero 2002). These include the heat shock response (Lindquist 1986), developmental arrest (Lindsley & Poodry 1977) and behavioural accommodation (Huey 1974). The robustness of buffering mechanisms to increased temperatures may be a key determinant of a species’ future health and persistence (Huey et al. 2009; Somero 2010; Tomanek 2010).
While many buffering mechanisms (such as heat shock gene regulation) are highly conserved over long evolutionary periods (Lindquist 1986; Feder & Hofmann 1999), closely related species can differ drastically in their tolerance of higher temperatures (Huey et al. 2009; Kellermann et al. 2009). This observation suggests that temperature tolerance can evolve rapidly. The mechanisms and evolution of stress responses can be directly linked: the way that a species responds to stress determines not only its immediate tolerance level, but can affect its potential to evolve increased tolerance over the long term. Stressful conditions can alter the expression of standing genetic variation, or expose novel genetic variants, by activating different genes and pathways and by lifting internal buffers on noise and disturbing homoeostasis. This ‘cryptic’ genetic variation then becomes visible to selection (Hoffmann & Hercus 2000; Gibson & Dworkin 2004; Schlichting 2008; Pfennig et al. 2010). These gene–environment interactions (GEIs) modify the genetic architecture of traits and can accelerate or hinder adaptations (Masel 2006). Near-term climate changes are likely to be too rapid for new mutations to provide material for adaptation (Bürger & Lynch 1997), so evolutionary adaptations may rely on standing (but potentially hidden or cryptic) genetic variation within species.
The purple sea urchin, Strongylocentrotus purpuratus, is a keystone species along the Pacific coast of North America, found in intertidal and subtidal waters from Alaska to Cedros Island, Mexico (Cochran & Engelmann 1975; Pearse 2006). It has an enormous population size and high genetic diversity, averaging at least one mismatch between two individuals every 50 bases along the genome (Sodergren et al. 2006). S. purpuratus has little large-scale genetic structure along its range (Palumbi & Wilson 1990; Kenner 1992; Pespeni et al. 2011), although there may be local genetic differentiation (Edmands et al. 1996). Seawater temperatures in this region vary greatly with season, latitude and depth, reaching 4 °C in the winter at the northern extreme, and >20 °C during the summer in the south (Osovitz & Hofmann 2005). However, not all temperatures are equally permissive of growth and development. Seasonality in temperature likely plays a role in determining reproductive timing: temperatures above 17 °C are detrimental to gametogenesis (Basch & Tegner 2007) and may prevent spawning (Cochran & Engelmann 1975). Early embryonic stages are generally thought to be the most sensitive to temperature stress (Andronikov 1975; Fujisawa 1989; Fujisawa & Shigei 1990), and thus may determine tolerance ranges for normal development (Sewell & Young 1999). Climate changes this century are predicted to warm global surface temperatures by 1.1–6.4 °C (IPCC 2007). While such an increase is unlikely to prevent development of embryos from all but perhaps the most southern populations, it will expose more populations to conditions that cause abnormal development (Farmanfarmaian & Giese 1963; Strathmann 1987; Azad et al. 2011) and create stressful conditions that affect dispersal and reproductive success (Hammond & Hofmann 2010; Byrne et al. 2011). Here, we investigated the response of S. purpuratus embryos to a stressful, but realistic temperature range (12–18 °C), and asked if such temperature variation exposed evolutionary relevant GEIs by perturbing developmental gene regulatory networks.
In doing so, we took advantage of both a key feature of S. purpuratus biology and the rich history of prior work on this system. S. purpuratus is a broadcast spawner that produces large numbers of synchronous embryos (Strathmann 1987), making it amenable to studies of functional genetic variation in wild outbred individuals using controlled breeding designs (Lynch & Walsh 1998). In part because of their high fecundity, they have also become an important model system for animal development (http://www.SpBase.org, Sodergren et al. 2006), and for responses to natural and unnatural environmental stresses (Giese & Farmanfarmaian 1963; Fujisawa 1993; Roepke et al. 2005; Todgham & Hofmann 2009). Most importantly for this work is the availability of an extremely well-characterized gene regulatory network for endomesodermal and ectodermal specification in embryos—probably among the best-characterized developmental networks currently available in any animal system (Davidson et al. 2002; Angerer & Angerer 2003; Oliveri et al. 2008; Peter & Davidson 2010). This network consists of transcription factors, signalling molecules and terminal differentiation (structural) genes that are expressed in distinct spatial and temporal patterns during development. The known regulatory relationships among these genes constitute a large proportion of the regulatory interactions necessary for specifying gene expression patterns and determining cell fates throughout embryogenesis. We used this network to test how genetic variation and environmental perturbations during development may be buffered or transmitted through known network relationships.
Specifically, we took a novel systems-genetics approach to study how environmental stress influences gene–gene interactions and GEIs, and how this may be important for future adaptation in this species. At the molecular level, stress responses require the activation or repression of specific genes and pathways to buffer other critical cellular systems (Richter et al. 2010). Therefore, measuring the activity and success of a stress response requires characterizing both the responses of genes involved in the buffering, and the stability of the buffered systems. To do so, we measured transcriptome-wide plasticity to identify components of the S. purpuratus stress response. Next, in a more in-depth analysis, we focused on the critically important and well-characterized endomesodermal developmental gene regulatory network to study how gene regulatory interactions are buffered in the face of such perturbations. Lastly, we tested for GEIs in this network across genetic backgrounds from a natural population, and asked if the response of the network to environmental stress was genetically variable. These combined approaches provide a powerful lens through which we can assess the functional and evolutionary implications of environmental change in an important developmental model species.
Methods
Animal rearing
Adult sea urchins were collected by professional divers at the Monterey Abalone Company (Monterey, CA), and shipped overnight to Durham, NC, where they were kept in artificial seawater (ASW; Coral Life®) basins at 15 °C. Individuals that spawned in transit were removed and the remaining individuals were kept for <1 month before spawning. As fertilization is external in this species, there are no constraints on the number or order of crossings among male or female parents. 32 crosses in a North Carolina II breeding design (Comstock & Robinson 1948; Lynch & Walsh 1998; see Appendix S1, supporting information) were performed to create pools of full-sib and half-sib embryos appropriate for genetic analysis. Standard procedures were used for spawning and raising urchin embryos (Strathmann 1987; Wray et al. 2004). For each of two experimental replicates, eggs from four females were split among 16 beakers and fertilized by the addition of a dilute concentration of activated sperm from one of four males in a factorial design. Fertilization rates were all above 90%.
After fertilization at 12 °C, embryos were poured into rearing dishes with 250 mL ASW preconditioned to one of three temperatures (12, 15 or 18 °C). Each condition was maintained in a separate environmental growth chamber in the Duke Phytotron. Two replicate dishes of each family were cultured at each temperature in a randomized design. 15 and 18 °C cultures were allowed to warm slowly in 50 mL of water for 30 min before being poured into their experimental dishes. All steps from fertilization to final culture dish were accomplished within 15 min for cultures maintained at a given temperature. In total, the entire process required ~1 h from the time of fertilization until all cultures were growing at the appropriate temperature. Embryos were grown at moderate densities (c. 5–10/mL) to reduce effects of crowding.
Sample collection
Cultures were raised until the embryos reached the early gastrula stage (Fig. 1a,b). We chose the point when the archenterons extended 1/3 through the blastocoel as this provided a reliable, time-independent morphological indicator of stage across temperature treatments. Once several indicator cultures had reached this stage within a given temperature condition, we collected all cultures at that temperature for morphological analysis and RNA extraction. For morphological analyses, at least 20 embryos were fixed with 4% paraformaldehyde in ASW at pH 8.3 and then washed into 100% ice-cold methanol for storage at −20 °C. For RNA collection, samples were centrifuged briefly, the ASW removed by aspiration, 600 mL of RLT buffer (Qiagen, Valencia, CA) with 1% β-mercaptoethanol added and then vortexed vigorously for 15 s to lyse the cells. Samples were stored in RLT at −80 °C.
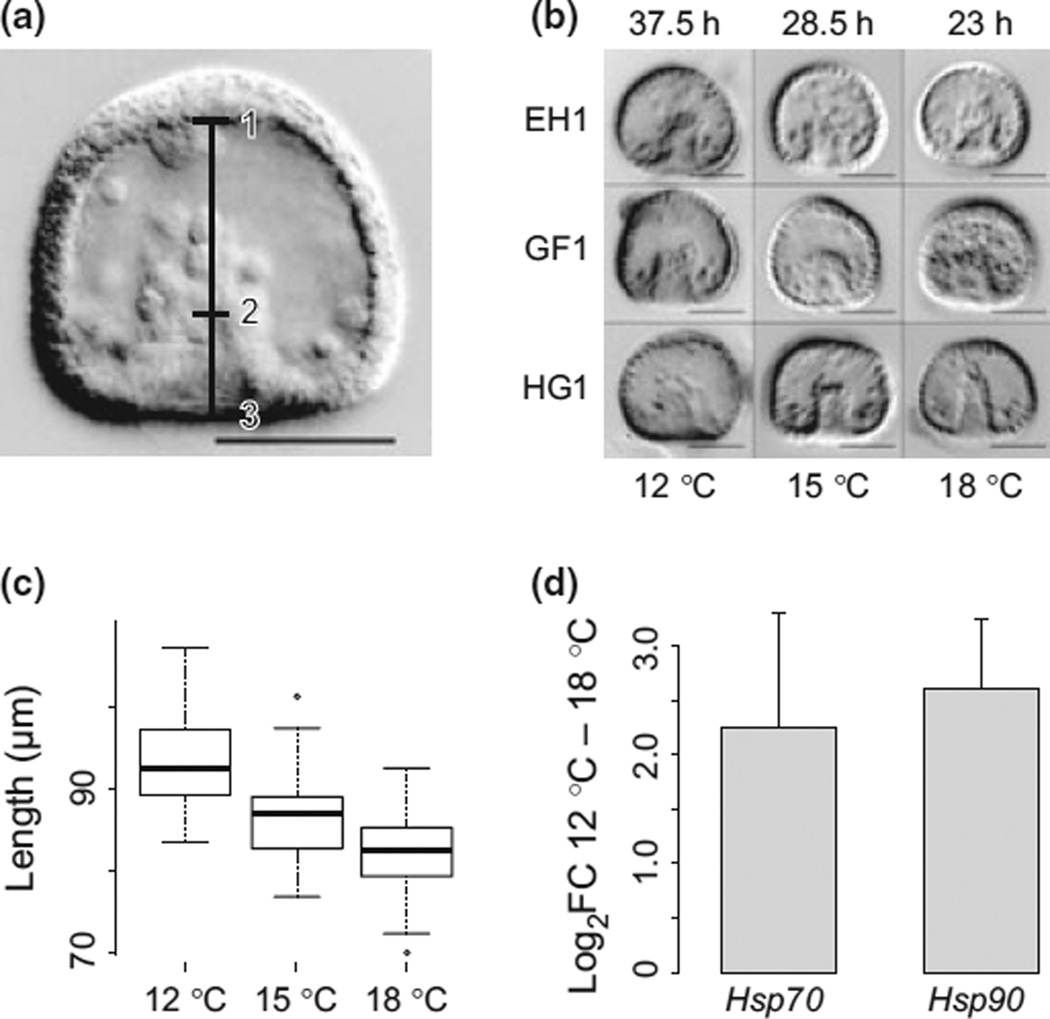
Fig. 1.
Growth temperature subtly alters embryonic development. (a) Gastrula-stage Strongylocentrotus purpuratus embryo. Cultures were sampled when the invaginating gut (archenteron) reached ~1/3 through the central region (blastocoel). Bars show measurements taken on voucher embryos: embryo length 1–3; embryo stage: ratio of 1–2:1–3. (b) Embryonic developmental rates increased greatly at higher temperatures, and caused subtle changes in embryonic morphology between families. The top row of numbers shows the time to gastrulation at each temperature. Despite these changes, embryos appeared generally healthy at all three temperatures. Example embryos sampled from each of three cultures at each of the three experimental temperatures are shown. (a, b) All scale bars are 50 lm. (c) Boxplots of embryo length. 18 and 15 °C embryos were shorter along the animal-vegetal axis than 12 °C embryos. (d) 18 °C cultures increased in both Hsp70 and Hsp90 expression, relative to 12 °C cultures. Expression is log2FC from 12 to 18 °C relative to RBM8A. Error bars show two standard errors estimated from the model: log2(exp)ij = Tempi + Femalej + eij.
Morphological analysis
We photographed 12–23 embryos for morphological analysis from each culture to assay developmental rate and size differences among cultures. Embryos were washed into phosphate-buffered saline and photographed in lateral view using a Zeiss Axioskop 2 with Axiovision v4.6 so that the extent of archenteron invagination could be assessed. Two measures were collected from each picture using ImageJ v1.41 (Abramoff et al. 2004, Fig. 1a): (i) the distance from the vegetal pole to the exterior tip of the archenteron and (ii) the distance from the vegetal pole to the interior edge of the ectoderm at the animal pole. The ratio of these two lengths was used as a measure of the stage of the embryo (0 prior to gastrulation and 1 when gastrulation is complete). The length of the embryo along the animal-vegetal axis was used as an estimate of the total size of the embryo. We accounted for slight differences in developmental stage among cultures in all downstream analyses using a linear model as described below.
RNA preparation
RNA was extracted using the RNeasy 96 kit (Qiagen), following the manufacturer’s instructions. RNA quantity was measured using a NanoDrop (Thermo Scientific, Wilmington, DE) and concentrations were adjusted to be 50–200 ng/lL with RNase-free water (the range appropriate for a DASL (cDNA-mediated Annealing, Selection, extension, and Ligation) assay; see below).
Gene expression measurements
We used three different platforms to measure gene expression differences among cultures. Details of the experimental procedures and analytical methods are presented in the Appendix S1 (Supporting Information). All analyses were performed in R (v1.13.1, R Development Core Team 2010).
Quantitative real-time PCR (qPCR) was used to measure the expression of two chaperone genes involved in environmental stress responses (Hsp70 and Hsp90) in all 12 and 18 °C cultures from the first experimental replicate. Expression was quantified relative to a control gene (RBM8A) using the methods of Pfaffl (2001) and Hellemans et al. (2007). Tests for differential expression were based on three-way anova on log2 relative expression values (Appendix S1, Supporting information).
Strand-specific RNA-seq on the SOLiD 3 plus platform (Life Technologies) was used to measure the transcriptome-wide expression of seven samples representing three female and two male parents at 12 and 18 °C (Table S3, Supporting information). An average of 37 million reads per sample were mapped to the S. purpuratus genome v3.1 (http://www.SpBase.org, Sodergren et al. 2006) using Bowtie (v.0.12.7, Langmead et al. 2009). Uniquely mapped reads were counted in 26 428 nonoverlapping gene models with HTseq (http://www-huber.embl.de/users/anders/HTSeq). As counting variance is much higher for low-expressed transcripts, we excluded all gene models with fewer than an average of 10 reads per sample. We assessed differential expression by culture temperature condition among the remaining 14 454 genes using the R package edgeR (v.2.2.6, Robinson et al. 2010, McCarthy et al. 2012). False discovery rates (FDR) were controlled using the method of Benjamini & Hochberg (1995). Genes were assigned to functional categories based on the mappings of S. purpuratus genes to ENSEMBL proteins of Oliver et al. (2010). Tests for significant enrichments of differentially expressed genes in functional categories were performed using the wilcoxon.py script of the python package pyEnrichment (http://www.duke.edu/~ofedrigo/Olivier_Fedrigo/PythonScripts.html).
The microarray-like Illumina DASL Assay (Kuhn et al. 2004) was used to measure the expression of genes within the sea urchin endomesodermal and ectodermal gene regulatory network in all 192 cultures. We designed 384 probes to target 72 important developmental genes, and then curated these probes to identify sets that were consistent across each transcript (Table S4, Appendix S1, supporting information). Samples were first normalized to the a priori chosen control gene, RBM8A. However, as the RNA-seq data suggested RBM8A gene expression slightly declined at 18 °C (log2FC = −0.19), we performed a second normalization step so that the mean expression of four genes with the smallest estimated temperature effects in the RNA-seq data (Chordin, Not, Fmo2 and FoxN-2/3) was identical across temperatures. Little is known about splicing variation among transcripts or conditions in this species. We attempted to avoid differential splicing events by excluding probes from the analysis if they were not correlated with other probes targeting the same gene.
Quantitative genetics analysis
We used a Bayesian hierarchical mixed effects model to quantify the effects of the temperature treatment, genetic background and other parental differences on the expression of each gene measured in the DASL assay. We fit models for each gene by Markov Chain Monte Carlo simulation using a Gibbs sampler coded in R. Details and derivation of this model are given in the Appendix S1 (Supporting information), and code will be posted on Dryad. Briefly, we fit the transcript expression level of each gene as an unobserved latent variable, using the 2–6 probe measurements of this transcript per sample as independent estimates of this expression. We modelled the variation in transcript expression using a linear mixed effect model with fixed effect predictors of temperature and developmental stage, and random effects corresponding to the male and female parents of each culture. We also fit all pair-wise interactions between male, female and temperature as additional random effects. We used a heavy-tailed t-distribution for the residual error structure to account for the apparent non-normal error distribution of individual probe measurements in the DASL assay. Posterior distributions were summarized as a mean and credible interval spanning the central 95% of posterior samples. We assessed the significance of temperature or parental effects on gene expression levels by asking whether this credible interval overlapped zero.
Network-level analyses
We collected the set of known interactions among our genes in the BioTapestry representations of the endomesodermal network (http://www.biotapestry.org, Longabaugh et al. 2005) and the ectodermal network from the Davidson lab website (http://www.its.caltech.edu/~mirsky/) as of 5 September 2011. In total, 52 of the 73 assayed genes contribute to 93 transcriptional regulatory events (interactions) among genes between 28 and 30 h postfertilization (hpf), at 15 °C, equivalent to the timing of sampling in the current study (Table S5, Supporting information). When two genes were known to form a complex that then regulates a downstream gene, changes in the concentration of either upstream gene could regulate the activity of the complex, so we counted two interactions: one linking each upstream gene to the downstream target.
We then explored the influence of network connections by measuring the correlation in gene expression owing to either temperature or parental effects between pairs of genes known to interact. Because some regulatory genes encode activators and others repressors, we tested for positive correlations between activators and their targets, and negative correlations between repressors and their targets.
For temperature effects, we tested the Pearson correlation of temperature effects of upstream and downstream genes. We also explored other metrics that may be less sensitive to the distribution of temperature effects, such as a binary sign test in which we counted the number of upstream–downstream pairs that either both increased or both decreased more than an a threshold of |log2FC| = 0.05. Results were similar for all methods.
For male parent effects, we calculated the Pearson correlation between the eight male parent effects for each upstream gene–downstream gene pair, multiplied this correlation by −1 if the upstream gene was a repressor and averaged these correlations across the network. We used the posterior means of male parent effects from the quantitative genetics models described above for each gene. Female effects were tested similarly.
To assess the significance of network effects, we used randomization tests to account for the fact that each interaction is not independent (for example, one upstream gene can regulate several downstream genes). For each of 10 000 randomizations, we permuted the labels of all 52 genes involved in interactions and recalculated the test statistic. This preserved both the distribution of responses among genes and the network topology, but randomized the interaction partners of each gene.
To test for an influence of spatial expression patterns on gene expression variation, we extracted the annotated expression domains for each gene at 27 hpf from BioTapestry (Table S4, Supporting information). There are 18 regions specified in BioTapestry, and most of the 72 target genes were expressed in one or more of these regions. Three regions (skeletal micromeres, the oral ectoderm and the veg 1 region) each had five or more genes expressed throughout the region and nowhere else. We tested if gene expression patterns were correlated among genes if they shared entirely overlapping expression domains. For temperature effects, we tested the effect of expression domain using an F-test from a one-way anova with the three above regions (plus ‘other’) as predictors. To assess significance, we permuted the region labels as above and compared the actual P-value to the distribution of randomized P-values. For male and female effects, we calculated the pair-wise correlations of breeding values among all genes, and tested if the mean correlation among genes with entirely overlapping expression domains was higher than for randomly selected genes.
Results
Higher temperatures cause mild developmental stress
Embryos raised at each temperature developed successfully through the gastrula stage. Even those at the highest temperature (18 °C) showed no obvious increase in mortality or developmental abnormalities at gastrulation (Fig. 1b) or through the pluteus stage (~5 days at 12 °C). Developmental rates were strongly dependent on temperature (Loeb 1916; Fujisawa 1993): cultures reached the mid-point of gastrulation after 23 h postfertilization (hpf) at 18 °C, 28.5 hpf at 15 °C and 37.5 hpf at 12 °C (Fig. 1b). Embryo morphology was also affected by temperature: embryos grown at 18 °C were significantly shorter along the animal-vegetal axis (Fig. 1c). Decreased larval size has been observed previously in larvae grown at higher temperatures, possibly owing to increased metabolic demands of development (Chen & Chen 1992). Here, embryos cultured at higher temperatures appeared to have more irregular-shaped blastocoels, and slightly less ordered cells in the ectoderm. Additionally, the expression of two chaperone genes, Hsp70 and Hsp90, was increased at 18 °C (Hsp70: log2FC = 2.24, t39 = 4.22, P = 0.0001, Hsp90: log2FC = 2.60, t37 = 8.03, P = 1e-9. Both models were log2 (exp)ij = Tempi + Femalej + eij), providing additional evidence that this temperature was stressful for development (Fig. 1d).
Several important cellular processes are affected by temperature
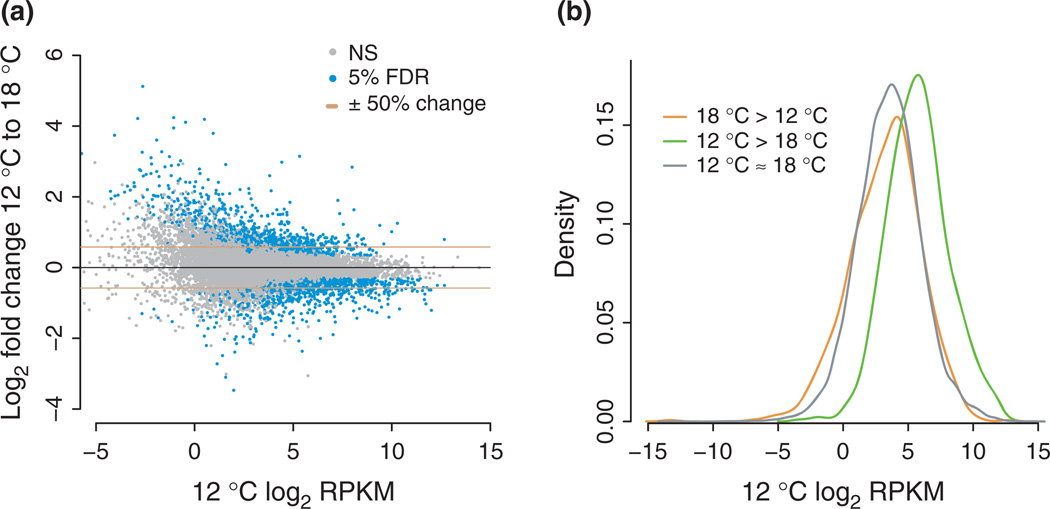
The higher heat shock gene expression but uncompromised gastrulation at 18 °C suggests that the embryos successfully buffered their development against temperature stress. Before focusing in more detail on how our focal network of developmental regulatory genes responded to this stress, we used RNA-seq to perform a relatively unbiased assay of temperature plasticity throughout the sea urchin embryonic transcriptome. This analysis allowed us to characterize the range of developmental changes that occurred between cultures grown at 12 °C or 18 °C and provided context for interpreting the observations of genetic and GEI variation within the focal network. At a FDR of 5%, seawater temperature affected 2036 of the 14 420 assayed genes (~14%, Fig. 2a). Of the differentially expressed genes, 1207 changed more than 50% (absolute value of log2 fold change (log2FC) > 0.58) and 478 changed more than twofold (|log2FC| > 1), with more genes in each category increasing at 18 °C relative to 12 °C (50%: 696 up, 511 down, twofold: 325 up, 153 down). The genes that decreased at 18 °C relative to 12 °C tended to be moderate-to-highly expressed, while genes that increased were similar in distribution to temperature-insensitive genes (Fig. 2b). The lack of down-regulated low-expressed genes was not due either to a floor effect, which would occur if low-expressed genes could not decrease further, or to reduced statistical power. Indeed, some low-expressed genes did exhibit further decreases in expression at 18 °C (Fig. 2a), and we observed a similar lack of down-regulated low-expressed genes if genes were ordered by their average expression over both temperatures, instead of their 12 °C expression. By ordering genes in this way, we removed the discrepancy in power between detecting up- and down-regulated genes.
Fig. 2.
The expression of many genes was affected by higher temperatures. (a) M-A plot showing the average 12 °C expression level (in Reads per Kilobase per Million mapped reads, RPKM) of all 14 420 genes with an average of 10 RNA-seq counts per sample, and the log2 fold change of each gene from 12 to 18 °C. Genes with a significant response at a False discovery rates (FDR) of 0.05 are coloured red. The horizontal blue lines show a ± 50% change in expression (log2FC = ±0.58). The figure is truncated at RPKM −5.8 and log2FC 6 for clarity, hiding 26 genes. (b) Genes that decreased at 18 °C relative to 12 °C tended to be high expressed at 12 °C. Kernel density curves of the expression (RPKM) those genes that increased (orange) decreased (green), or did not change (grey) at 18 °C relative to 12 °C at a FDR of 0.05.
To explore cellular processes affected by temperature stress, we tested for enrichment of specific functionally related genes sets among the genes either up-regulated or down-regulated at 18 °C. Each S. purpuratus gene model was assigned functional classifications in the PANTHER Molecular Function ontology (v6.1) as previously described (Oliver et al. 2010). Among genes up-regulated at 18 °C, we found 14 Molecular Function categories with a significant enrichment at a FDR of 5% (Table 1A, Fig. S1A, supporting information). These categories were consistent with induction of cellular responses to unfolded proteins and reactive oxygen species, and alterations in membrane properties (ion channels), cell–cell signalling (G-protein coupled receptors) and protein or energy metabolism (transferases). At the same FDR threshold, 26 Molecular Function categories were enriched for down-regulated genes (Table 1B, Fig. S1B, Supporting information). These categories included genes involved in energy-expensive processes such as transcription and RNA processing, as well as cell structure and skeletal elements, cellular defence components and developmental processes.
Table 1.
PANTHER categories enriched for up- and down-regulated genes based on RNA-seq analysis
| PANTHER molecular function |
Adjusted P |
No. of genes in category |
|---|---|---|
| A) | ||
| Chaperone | 8.23E−06 | 108 |
| G-protein coupled receptor | 8.23E−06 | 143 |
| Glycosyltransferase | 1.13E−05 | 196 |
| Hsp 70 family chaperone | 0.000470567 | 11 |
| Other ligase | 0.000470567 | 122 |
| Chaperonin | 0.000470567 | 18 |
| Other ion channel | 0.002488457 | 47 |
| Ligase | 0.0039995 | 343 |
| Other chaperones | 0.008856111 | 49 |
| Vesicle coat protein | 0.01024964 | 28 |
| Transferase | 0.01024964 | 761 |
| Oxidoreductase | 0.01684667 | 489 |
| Kinase activator | 0.02111923 | 37 |
| Dehydrogenase | 0.03509571 | 216 |
| B) | ||
| Nucleic acid binding | 0.00E+00 | 1583 |
| Ribosomal protein | 1.57E−09 | 124 |
| Ribonucleoprotein | 1.27E−08 | 49 |
| Tubulin | 2.90E−08 | 30 |
| mRNA processing factor | 0.000194877 | 132 |
| mRNA splicing factor | 0.000194877 | 93 |
| Other RNA-binding protein | 0.000226697 | 136 |
| Membrane-bound signalling molecule | 0.000247238 | 122 |
| Transcription factor | 0.003690222 | 803 |
| Other zinc finger transcription factor | 0.0060876 | 56 |
| Amino acid transporter | 0.006321818 | 40 |
| Homoeobox transcription factor | 0.007221357 | 76 |
| RNA helicase | 0.007221357 | 82 |
| Cytoskeletal protein | 0.007221357 | 555 |
| Actin binding cytoskeletal protein | 0.01309733 | 244 |
| Cadherin | 0.01668833 | 23 |
| Signalling molecule | 0.01668833 | 350 |
| Defence/immunity protein | 0.01668833 | 133 |
| Translation initiation factor | 0.01756 | 57 |
| Serine protease inhibitor | 0.018069 | 15 |
| Cell adhesion molecule | 0.01904318 | 231 |
| Protein kinase | 0.01904318 | 369 |
| Exoribonuclease | 0.02502217 | 26 |
| Other transcription factor | 0.0341525 | 144 |
| Kinase | 0.0356668 | 483 |
| Translation factor | 0.04440885 | 84 |
Genes were assigned to PANTHER Molecular Function categories as in Oliver et al. (2010). Categorical enrichments were performed using scripts written by O. Fedrigo (See Methods), with significance assigned using a Wilcoxon signed rank test. Adjusted P-values were assigned based on the Benjamini & Hochberg (1995) multiple comparisons method using the R function p.adjust. A) PANTHER Molecular Function categories enriched for up-regulated genes with adjusted P < 0.05. B) PANTHER Molecular Function categories enriched for down-regulated genes with adjusted P < 0.05.
Gene network analysis reveals a well-buffered temperature response
While the RNA-seq data were useful for identifying developmental and physiological processes altered at high temperatures, with such a modest sample size, they provide limited insight into the genetic architecture of these responses. Genetic variation in responses to the environment (gene–environment interactions, or GEIs) may be important for a species to evolve under changing climates. Gene expression traits offer an opportunity to understand the mechanisms that create GEIs by studying the effects of the environment on the control of gene expression. GEIs can arise because of environmental effects on the upstream (trans-acting) regulatory genes, interactions of these regulatory genes with promoter regions or from changes in post-transcriptional regulation. As such, transcriptional gene regulatory networks that control gene expression may be important for understanding GEIs in gene expression.
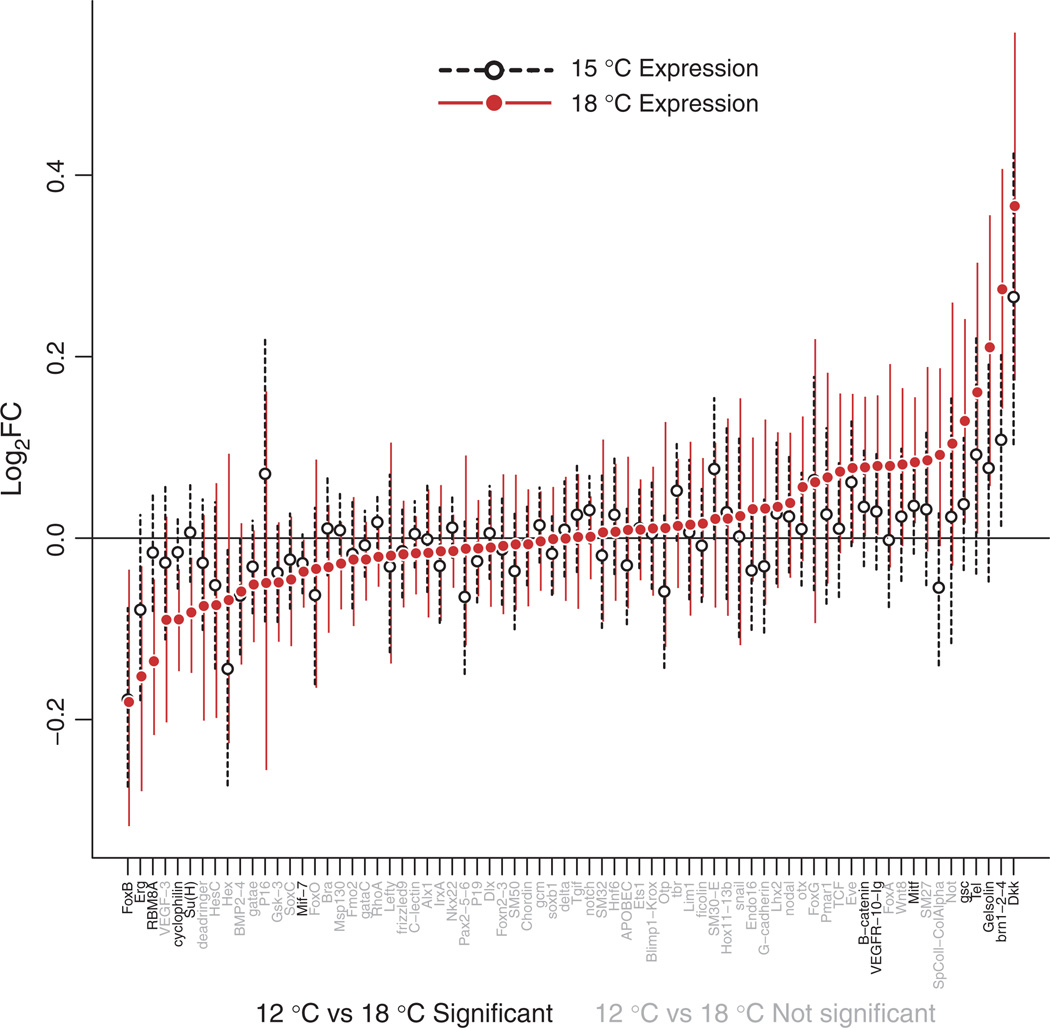
To explore how transcriptional regulatory networks influence GEIs, we investigated the transmission of temperature effects (an environmental perturbation) and genetic effects (a genetic perturbation) through the well-characterized endomesodermal and ectodermal gene regulatory network that controls cell type specification and development in sea urchin embryos. To do so, we used a DASL assay to greatly expand our sample size relative to the RNA-seq data set to capture these responses both across a larger set of genetic backgrounds, and over three temperatures (12, 15 and 18 °C) instead of two. Higher temperatures caused considerable changes in the expression of genes in the network (Fig. 3). Overall, 14 of the 72 assayed genes exhibited changes in gene expression between the 12 and 18 °C temperature conditions, with similar numbers increasing and decreasing with temperature (r = 0.71 between the two independent replicates of the experiment: Fig. S2A, Supporting information). Temperature effects were generally gradual: in most cases, the expression at 15 °C was intermediate between the expressions at 12 and 18 °C (Fig. 3, r = 0.77 between 15 and 18 °C responses), and all but six were smaller in absolute value than 0.15(log2)-fold. We observed no relationship between the presence of temperature effects and embryonic territory (Fig. 4) or functional classes of genes. Temperature accounted for 0.27–17% (median 3.7%, Fig. S3, Supporting information) of the total expression variation among pooled samples of embryos (residual biological variation among individuals and cultures was confounded with technical variation in the DASL assay). Slight variations in developmental stage among cultures had minimal effects on expression overall (Figs S3 and S4, Supporting information).
Fig. 3.
Estimates and credible intervals for the gene expression reaction norms to temperature. The magnitude in log2 fold change (log2FC) of the expression change from 12 to 15 °C (dotted lines) or 18 °C (red) for each of the 72 target genes. Bars cover the central 95% of posterior samples for each gene. Estimates are averaged across the two experimental replicates. The 15 °C response was generally similar to, but smaller than, the 18 °C response. 18 °C responses for gene names coloured black were significantly nonzero.
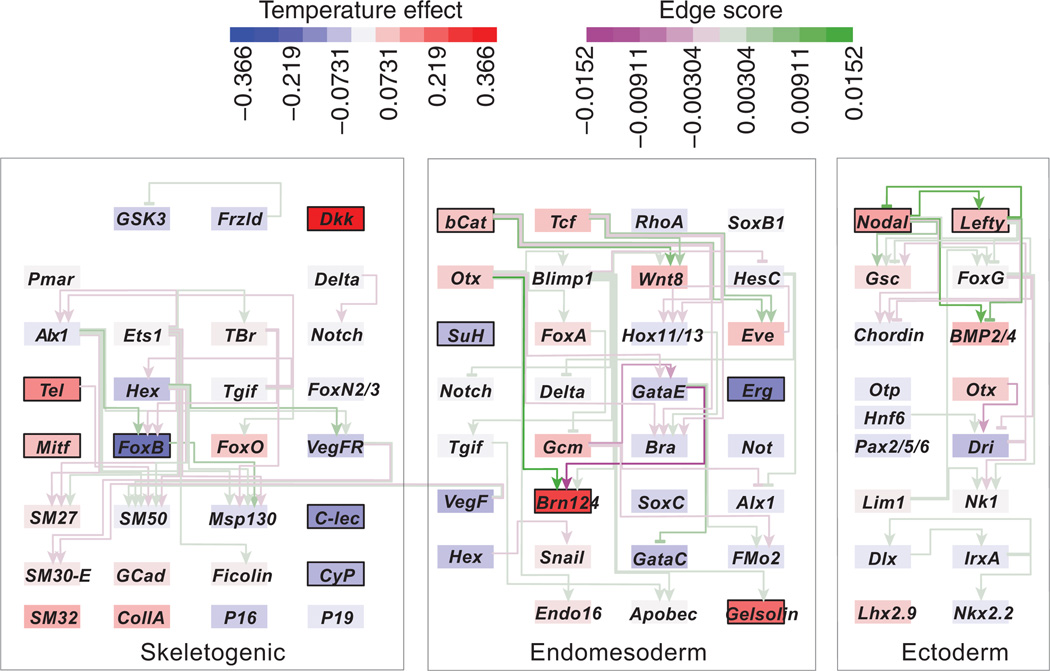
Fig. 4.
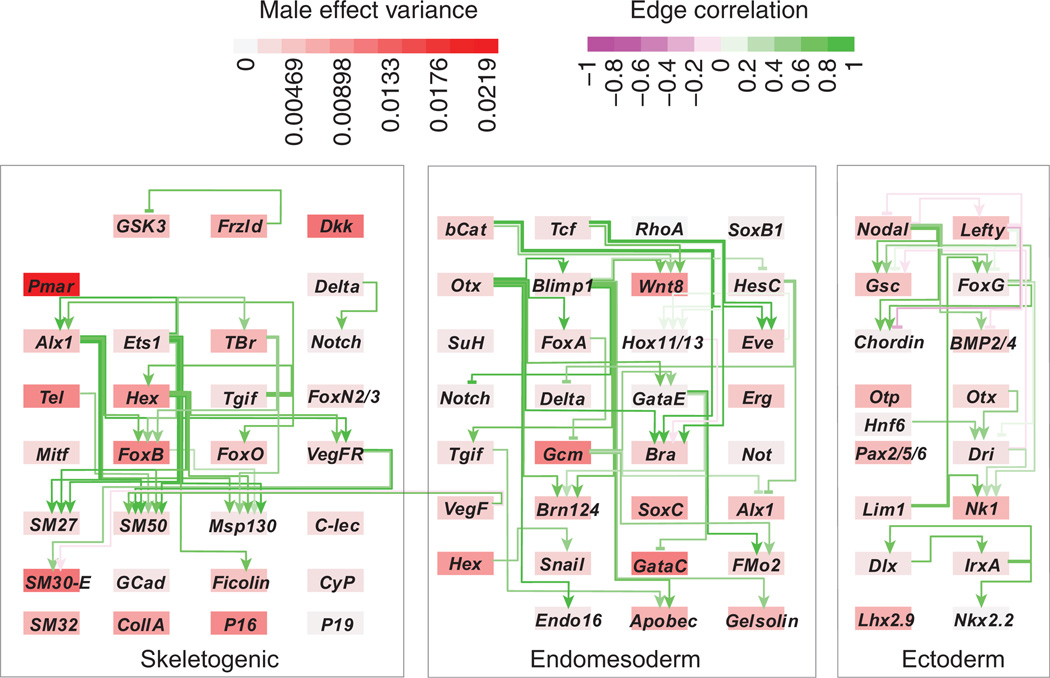
Endomesoderm and ectoderm gene network response to higher temperatures. The sea urchin endomesoderm gene regulatory network controls early cell fate specification and morphological patterning in embryogenesis. This representation of the network is adapted from the BioTapestry model from the Davidson lab website: (http://www.its.caltech.edu/~mirsky/). The network becomes subdivided during embryogenesis into three major territories—the skeletogenic, the endomesodermal and ectodermal cells. All 72 network genes assayed are displayed, including the known regulatory relationships at gastrulation. Genes are placed in a territory where they are expressed at 27 h postfertilization (hpf) at 15 °C, corresponding to the sampling time in this study according to Bio-Tapestry. Arrows show positive regulatory events that are active at 27 hpf, and lines ending in bars show repressive regulatory events. All ectoderm regulatory events shown here (http://www.its.caltech.edu/~mirsky/) are assumed active. Genes are coloured according to the magnitude of response to 18 °C temperatures (more red = larger increase, more blue = larger decrease). Lines are coloured according to the product of the temperature responses of the two connected genes. Higher scores (more green for positive regulation and more purple for repression) indicate a greater similarity between the temperature response of a regulatory gene and the response of the gene it regulates. The mean correlation of interacting genes (r = 0.04, corrected for repressive regulators by multiplying by −1) was well within the range based on randomizing the temperature responses among genes, but preserving the topology of the network (P = 0.30, by randomization).
Importantly, the modest effects of temperature in the DASL assay did not reflect a lack of accuracy of the DASL assay relative to the RNA-seq analysis. Sixty-six genes in the network were successfully measured by RNA-seq, and the estimated change in gene expression (in log2FC) between 12 and 18 °C was correlated across the two platforms (r = 0.68, Fig. S2B, Supporting information). However, the log2FC estimates from the RNA-seq data were considerably larger (~8×, Fig. S2B, Supporting information) than the DASL estimates, likely reflecting the greater dynamic range of the RNA-seq platform. This difference does not affect comparisons among genes or between environmental and genetic perturbations within the network because all genes were subject to the same compression in DASL. The range of temperature effects observed in this target set of developmental genes was not unusual across the transcriptome (median |log2FC| for network genes: 0.22 vs. 0.25 for randomly selected genes in RNA-seq, 95% interval 0.18–0.34 based on 10 000 samples).
To test if the regulatory interactions among the genes in the network influenced the pattern of responses to temperature, we considered all pairs of genes that interact during early development and tested whether they were more correlated in their expression responses than expected by chance. We found no evidence that the temperature responses of upstream genes and their downstream targets (n = 93 transcriptional interactions among 52 genes) were more similar than expected (r = 0.04, P = 0.30, Fig. 4). Additionally, genes with completely overlapping expression domains at 27 hpf did not have similar responses to temperature (anova F3,48 = 1.13, permutation-based P = 0.34). Therefore, perturbations to genes in this network caused by temperature appear to be buffered sufficiently by the developmental system to the extent that the observed differences in the expression of upstream regulators did not systematically result in similar responses in the downstream genes they regulate.
Common genetic and maternal effects, but few gene–environment interactions in gene expression
If significant GEIs contribute to the variation in gene expression among embryos, then selection could potentially lead to more optimal gene expression responses to temperature. Our full-factorial crossing design allowed us to test for, and estimate the magnitude of, maternal, genetic and GEI effects on the gene expression traits. Each of the 32 cohorts of embryos was started with eggs from one of eight wild-caught, out-bred female urchins, and sperm from one of eight similarly out-bred male urchins. Thus, the embryos within each culture were full-sibs, embryos in cultures that shared either a male parent or a female parent were half-sibs and all other embryos were unrelated, permitting estimates of genetic parameters with mixed effect models (Lynch & Walsh 1998).
We found little evidence for GEIs between the sampled genetic backgrounds and temperature in the genes of the endomesodermal and ectodermal gene regulatory network. Five genes had significant female parent-by-temperature (FxT) effects, but no genes had significant male parent-by-temperature effects (MxT). Both FxT and MxT effects contributed little to the overall variation (maximum variance explained by FxT effects was 4%; maximum by MxT effects was 0.5%, Fig. S4, Supporting information). In urchins, male parents only contribute to their offspring through their sperm, which carry little more than DNA. Thus, male parent effects in the embryos are likely due to genetic variation. In contrast, female parents contribute nutritional yolk as well as packaged proteins and mRNAs in eggs (Bertram & Strathmann 1998). Thus, female parent-associated variation is owing to a combination of genetic variation and maternal effects (some, but not all, of which may be genetically based as well). Therefore, female parent-by-temperature effects are not by themselves convincing evidence of GEIs. We found a similar result for the two chaperone genes, Hsp70 and Hsp90. Both of these genes had significant female parent effects at 18 °C, but not at 12 °C, and no significant male parent effects at either temperature (Table S6, Supporting information).
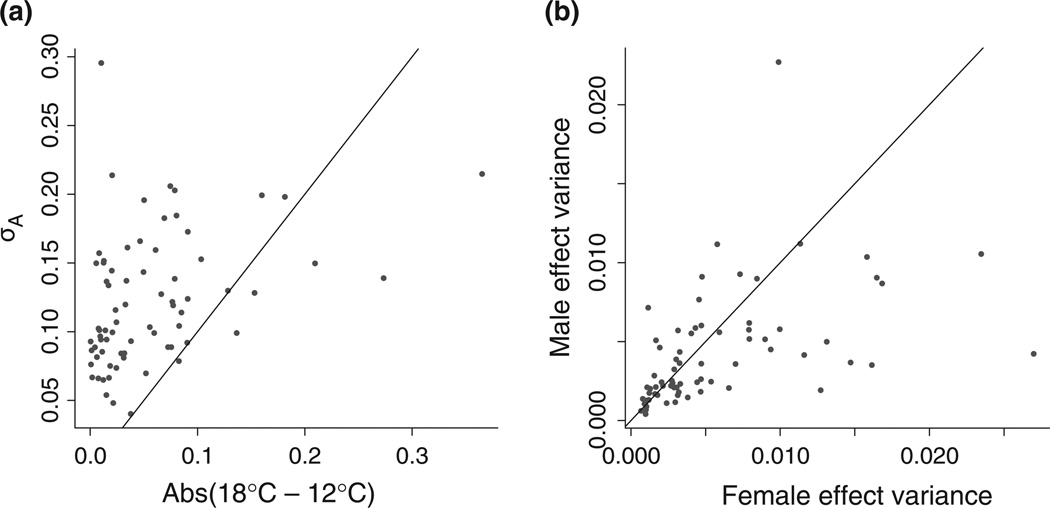
In contrast, parental main effects, which average genetic and maternal effects over the three temperatures, were common among the developmental genes. We detected significant female parent effects in 24 of the 72 genes, and significant male parent effects in nine of the same 24 genes as well as nine others. These parental main effects on gene expression tended to be larger than the effects of temperature. We estimated the additive genetic variance in each gene expression trait as four times its fitted male effect variance (Lynch & Walsh 1998). For 82% of the genes (all but 13), the square root of the estimated additive genetic variance was greater than the difference in expression between 12 and 18 °C (Fig. 5a). Female effects tended to be larger than male effects (Wilcoxon signed rank test of fitted female and male variances: V = 1686, P = 0.037, Fig. 5b), as expected, suggesting that maternal effects were also important for several genes.
Fig. 5.
Genetic effects tended to be larger than temperature effects, and female effects were larger than male effects among network genes. (a) The square root of additive genetic variation (estimated as four times the fitted male variance) and the absolute value of the difference in expression between 12 and 18 °C are plotted for each of the 72 genes. (b) The fitted female effect and male effect variance for each gene. (a, b) The diagonal line in each plot shows y = x.
We tested if the maternal and genetic effects on developmental genes were also independent of the regulatory interactions in the network as the effects of temperature had been. For male effects, the mean correlation of breeding values across interacting gene pairs was 0.45, considerably higher than expected (0.34, P = 0.0045 compared to 10 000 permuted data sets: Fig. 6). This result suggests that, in contrast to temperature perturbations, perturbations to gene expression owing to genetic variants were transmitted through the network. Surprisingly, female effects were not similarly transmitted. The mean correlation of the female effects among interacting genes was lower than for male effects, 0.28, and was not significantly greater than expected by chance (0.23, P = 0.20). As with temperature effects, we observed no significant relationship between spatial expression domain and either male effect correlations or female effect correlations (P = 0.51 and 0.98, respectively).
Fig. 6.
Male effect correlations were greater among directly interacting genes. Network diagram is as in Fig. 4. Here, genes are coloured according to the magnitude of the male effect variance, and edges are coloured according to the correlation between the male breeding values for the upstream and downstream genes. Overall, male effect correlations (corrected for repressive regulators by multiplying by −1) among interacting genes were higher than expected by chance (P = 0.0045).
Discussion
In Monterey Bay, on the central coast of California, warming temperatures over the past century have been linked to widespread changes in species distributions and abundances (Barry et al. 1995). We collected adult S. purpuratus from this region and tested the robustness of their embryos to further increases in seawater temperature. Current seasonal variation in sea surface temperature in the bay ranges from about 12 °C in the winter to about 15 °C in the summer (NOAA NVODS ferret.pmel.noaa.gov/NVODS). The typical spawning season of S. purpuratus across its range is December to March (Strathmann 1987), although ripe females can be found in June (Pearse et al. 1986). Thus, early embryos in Monterey Bay are unlikely to experience temperatures above 13–14 °C. However, climate models such as the A1B scenario of the International Panel on Climate Change predict that by the year 2010, temperatures in the bay will increase 1–3 °C (CM2.1U-H2_SresA1B_X1 model from NOAA GFDL, Delworth et al. 2006), potentially bringing June temperatures as high as 17 °C. Even today, in the southern-most portion of the species’ range, June sea surface temperatures are 17.5 °C (Cedros Island, Mexico. NOAA NVODS), suggesting that our experimental treatment of 18 °C, while high, is not unreasonable for this species, and may even be a common temperature for early embryos from southern populations within a century.
In embryos cultured at 18 °C, we observed both an up-regulation of stress response pathways and changes in embryonic morphology at the gastrula stage, suggesting that this environment was stressful for embryogenesis. Specifically, we observed a decrease in size along the embryonic animal-vegetal axis in embryos raised at 18 °C, potentially signifying a more rapid depletion of energy stores during development (Koehn & Bayne 1989; Chen & Chen 1992) or increased asynchrony between cell division and morphogenesis at a greatly increased (60% faster) growth rate. On the molecular level, these effects were reflected by signatures of temperature-induced stress throughout the embryonic transcriptome: a dramatic up-regulation of chaperones; consistent changes in the regulation of genes involved in development, RNA processing and metabolism reflective of developmental stress (Gasch et al. 2000); and subtle, but potentially important alterations in the expression of key developmental regulatory genes involved in the sea urchin endomesodermal and ectodermal gene regulatory network. Together, these results provide insight into the molecular mechanisms by which sea urchin embryos use global stress responses to buffer critical developmental pathways, and how developmental systems can affect evolutionary responses to climate change.
An induction of heat shock gene expression was expected given similar observations in a wide array of species (reviewed in Lindquist 1986; Feder & Hofmann 1999). However, such an induction of Hsp70 (or any other Hsp) had not been previously demonstrated in S. purpuratus embryos cultured at 18 °C, a relatively subtle but environmentally relevant thermal stress. All previous studies on Hsp70 expression in S. purpuratus embryos had tested the gene expression response to a sudden, transient and larger increase in temperature (e.g. Bédard & Brandhorst 1986; Sanders & Martin 1994; Hammond & Hofmann 2010). In these prior experiments, increased Hsp70 expression was not observed until temperature shocks reached 21–25 °C. Here, we have shown that chronic exposure to much lower intensities of thermal stress can induce clear and consistent heat shock gene expression responses. This is significant, given that global climate change will probably involve continual exposure to more moderate temperature increases. A detailed comparison of gene expression responses to chronic vs. transient high temperatures is an important avenue for future study.
While transcriptome-wide measures of stress responses are useful for discovering important cellular, developmental and physiological processes, we were also interested in how developmental mechanisms of gene regulation would be affected by environmental stress. By themselves, changes in the expression of individual genes provide little insight into the impact of environmental change on developmental processes. It is difficult to know whether a given change in the expression of a particular gene might have functional consequences within the organism, as the threshold for downstream effects is likely to be different for every gene (Oleksiak et al. 2005).
We therefore focused attention on the regulation of a smaller set of genes that compose an exceptionally well-studied gene regulatory network (Davidson et al. 2002; Angerer & Angerer 2003; Oliveri et al. 2008; Peter & Davidson 2010), namely the endomesodermal and ectodermal developmental network. Within the context of this network, we hypothesized that the importance of a perturbation to a particular gene could be measured, at least in part, by assessing the correlation between that perturbation and perturbations to the downstream genes that the focal gene is known to regulate. In this way, an explicit regulatory network provides a partial solution to the problem of classifying ‘important’ vs. ‘unimportant’ changes in gene expression. Certainly, many developmental genes have functions beyond their role within known networks. However, within a network, specific predictions can be made about the outcome of perturbations to particular genes, and these predictions can be tested by comparison with the observed data. Here, we tested if either temperature perturbations or functional genetic variants segregating in wild sea urchins are transmitted through known gene regulatory relationships.
Overall, we found that environmental perturbations to the expression of the network genes were well buffered, at least at the 15 and 18 °C temperatures we tested. We did observe effects of higher temperature on several genes in the network, but there was no evidence that perturbations to upstream genes were transmitted through the network to their downstream regulatory targets. For example, the two most up- and down-regulated genes with known upstream regulators in the network were Brn1/2/4 and FoxB, respectively (Figs 3 and 4). Brn1/2/4 is a midgut-specific transcription factor with a later role in the regulation of Endo16 (Yuh et al. 2005). FoxB is a forkhead transcription factor involved in skeletal differentiation in the embryos (Tu et al. 2006). At the developmental stage we measured, Brn1/2/4 has no known downstream targets but is itself positively regulated by GataE, Blimp1-Krox and Otx (Fig. 4). Brn1/2/4 was significantly up-regulated at 18 °C, but GataE appeared slightly down-regulated, Otx slightly up-regulated and Blimp1-Krox was unchanged at 18 °C (Fig. 3, although none of these latter effects were significant). FoxB is activated by three other skeletogenic transcription factors: Alx1, Ets1 and Tbr (Fig. 4, Oliveri et al. 2008). While FoxB was down-regulated at 18 °C, Alx1, Ets1 and Tbr were all virtually unchanged (Figs 3 and 4). In both of these cases, the expression plasticity of the downstream genes (Brn1/2/4 or FoxB) could not be explained by similar plasticity in their regulators. We observed similar patterns of low correlation between the expression levels of regulatory genes and their targets throughout the network (Fig. 4).
Furthermore, temperature effects on individual genes increased gradually with higher temperatures, suggesting that the overall network state did not change dramatically at 18 °C. One explanation for the observed robustness of this network to temperature may be that the parameters we measured—slight variations in steady-state transcript levels—were not sufficient to perturb its dynamics. Using a dynamic model of a developmental network, Bolouri & Davidson (2003) showed that when transcription factors act cooperatively to activate downstream genes, the behaviour of each interaction becomes switch like. Above some threshold, further increases in the expression of an upstream regulator have little effect on downstream expression. In this model, steady-state transcript levels do not dictate network dynamics. Instead, it is primarily the rates of initial activation of each gene that determine the progression of network states during development. Sea urchin development does accelerate at higher temperatures. However, previous studies have shown that this acceleration is constant across stages such that the timing of emergence of different structures is reliable (Fujisawa 1993). Thus, switch-like regulation of genes in this network may buffer temperature effects on the steady-state transcript levels as long as the relative rates of induction among genes do not change.
It is perhaps not surprising that these sea urchins are highly capable of buffering the effects of thermal stress on development: given this species’ enormous effective population size, abundant genetic variation and the extensive gene flow among populations that experience widely varying temperatures (Palumbi & Wilson 1990; Pespeni et al. 2011), selection for alleles that promote robust development across temperatures may be very efficient. However, while temperature effects and female parent effects (which include nongenetic maternal influences) did not appear to perturb network function, male parent (purely genetic) effects did. This was not simply owing to the fact that male parent effects were larger than temperature effects; female parent effects were larger still and yet were buffered in the network. Population genetic models predict that a large population size and high genetic variation should also promote the evolution of genetic robustness. In these models, only pleiotropy among the genes involved should limit the evolution of environmental robustness, but genetic robustness may not be favoured with strong stabilizing selection (Wagner et al. 1997; Rice 2000). Under these conditions, deleterious alleles are rapidly purged before buffering (canalization) mechanisms can evolve. Given the critical role that the endomesoderm and ectoderm genetic network plays in sea urchin development, and the conservation of modules of the network over long evolutionary periods (Hinman & Davidson 2007), it is reasonable to assume that the network’s function is under strong stabilizing selection. Thus, it may be that the observed genetic variation in the network is at this canalization limit.
This contrast between genetic effects and temperature effects provides insight into a long-standing unresolved question regarding the evolution of robustness: whether environmental and genetic perturbations are buffered by the same mechanisms (Meiklejohn & Hartl 2002; Visser et al. 2003). In this comparison at least, it appears that the effects of temperature on expression were buffered out by the system almost immediately, while genetic effects persisted and influenced downstream developmental events. Nevertheless, the overall developmental system was robust to both forms of perturbation: embryos at all temperatures and from all parents developed successfully through embryogenesis to the larval stage. At this very local level of regulation, the mechanisms of robustness appear distinct.
The robustness of our network to female parent effects seems to contradict this hypothesis as the same diversity of genetic effects passed on by male parents will also contribute to female parent effects. However, a dominant source of maternal effect variation in sea urchin development is thought to be variability in the quality and quantity of nutritional stores provided in the egg. Variation in egg provisioning is known to be common in echinoderms—between females from deep and shallow water (Bertram & Strathmann 1998), or from sheltered and exposed habitats (George 1999). Thus, buffers may have evolved to compensate for variability in egg quality, swamping the signal of the remaining female-associated genetic perturbations. In support of this hypothesis, we observed variation in the heat shock response (Hsp70 and Hsp90 expression) among embryos with different female parents, but not male parents. Confirming these trends will require further study across networks, species and environments.
Finally, despite abundant functional genetic variation for gene expression among genes in the network, we observed very little evidence that genetic effects were altered by environmental stress. Such GEIs are important for evolving phenotypic plasticity and tolerance of novel environments (Via & Lande 1985; Schlichting & Pigliucci 1993). We found only five examples of parent-by-temperature variation in gene expression, and all involved differences in female effect responses to temperature. Thus, we could not rule out the possibility that all such apparent GEIs were entirely owing to non-genetic maternal effects. We can only conclude that, at least in this network, the higher temperatures that southern populations of S. purpuratus will soon experience (according to climate models of near-future climate change) are unlikely to lead to a sudden release of previously cryptic genetic variation and a boost in evolutionary innovation (Masel 2006). This scenario may stem directly from effective buffering of temperature effects in the network: if regulatory function (rather than just expression level) is robust to perturbations, there may be little opportunity for GEIs to arise.
The buffering of environmental and genetic perturbations we have documented is a system-level feature of this gene regulatory network; they could not have been detected by studying individual genes, nor studied without detailed prior knowledge of gene interrelations. Analyses of similar networks will be crucial to placing our results in context. Based on our RNA-seq analysis, the genes of the endomesodermal and ectodermal developmental network in the embryo were typical in both their frequency and magnitude of plasticity to temperature stress relative to most other genes transcriptome-wide. As in our focal network, higher temperatures only subtly altered the expressions of the majority of genes in the embryo. Instead, major temperature responses appeared restricted to a limited number of stress response, metabolic and signalling pathways. Thus, it seems likely that the patterns of functional genetic variation and GEIs in gene expression that we observed in the endomesodermal and ectodermal developmental network will be typical of many other regulatory networks in the embryos. However, it will be interesting to assess levels of genetic and GEI variation in networks directly related to thermal stress or immunity and defence in embryos, as evolution in these pathways may be the most critical for near-term adaptation. Importantly, the organization of the gene regulatory network may influence the course of evolution by allowing populations to tolerate rapid temperature changes, or by channelling the pleiotropy of genetic variants in ways that could affect responses of the developmental system to natural selection. Tracking this system in the future as marine climates continue to change will be broadly informative for understanding the evolution of robustness during development and the behaviour of genetic systems under environmental change.
Supplementary Material
Acknowledgements
This work was supported by the NSF (DEB 0614509 to GAW), NIH (5P-50-GM-081883 to the Duke Center for Systems Biology) and the Sigma Xi (Grant-in-Aid to DER). The authors would like to thank the staff of the Duke University Phytotron for technical assistance and L Warner, J Tung, D Des Marais, D McCandlish and two anonymous reviewers for helpful comments on this manuscript.
Footnotes
The authors are broadly interested in linking population genetics, developmental biology, ecology and genetics in the study of evolution and climate change. D.E.R. studies quantitative genetics and ecological developmental biology, primarily focusing on climate change factors in sea urchins. D.A.G. studies the population genetics of developmental evolution. C.C.B. studies human and primate evolution and is interested in the evolution of gene regulation. J.A.W. studies sea urchin development and morphogenesis. SM studies statistical approaches to high dimensional data. G.A.W. focuses on the evolution of evolution of development and genomes.
Data accessibility
qPCR and morphology data are in the supporting online materials. DASL and RNA-seq gene expression data are deposited in NCBI GEO: Series GSE29504. R scripts for all analyses have been deposited in the Dryad repository: http://dx.doi:10.5061/dryad.36md8.
DR designed research, performed research, analysed data and wrote the paper. DG designed research and performed research. CB performed RNA-seq. JW performed qPCR. SM analysed data. GW designed research.
Supporting information
Additional supporting information may be found in the online version of this article.
Table S1 Primer sequences, amplification efficiencies and original citations for the qPCR assays used to assay Hsp 70 and Hsp 90 expression.
Table S2 Raw qPCR data on Hsp70 and Hsp90.
Table S3 Sample info for RNA-seq.
Table S4 Annotation information for developmental genes assayed by DASL.
Table S5 Gene network relationships among target developmental genes.
Table S6 Female parent effects on heat shock gene expression at 18C but not 12C.
Table S7 Embryo morphology data.
Table S8 Prior hyperparameters used in this study.
Fig. S1 Dendrograms showing relationships among significantly enriched PANTHER categories for up- (A) and down-(B) regulated genes.
Fig. S2 Estimates of temperature responses by RNA-seq and DASL were consistent.
Fig. S3 Percentage of variation in gene expression explained by temperature, stage and parental effects.
Fig. S4 Cultures were slightly more variable at higher temperatures, and differences in sampling time by temperature was small.
Appendix S1 Supplemental methods.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Abramoff MD, Magelhaes PJ, Ram SJ. Image processing with ImageJ. Biophotonics international. 2004;11:36–42. [Google Scholar]
- Andronikov V. Heat resistance of gametes of marine invertebrates in relation to temperature conditions under which the species exist. Marine Biology. 1975;30:1–11. [Google Scholar]
- Angerer LM, Angerer RC. Patterning the Sea urchin embryo: gene regulatory networks, signaling pathways, and cellular interactions. Current Topics in Developmental Biology. 2003;53:159–198. doi: 10.1016/s0070-2153(03)53005-8. [DOI] [PubMed] [Google Scholar]
- Azad AK, Pearce CM, McKinley RS. Influence of stocking density and temperature on early development and survival of the purple sea urchin, Strongylocentrotus purpuratus (Stimpson, 1857) Aquaculture Research. 2011:1–15. [Google Scholar]
- Barry JP, Baxter CH, Sagarin RD, Gilman SE. Climate-related, long-term faunal changes in a California rocky intertidal community. Science. 1995;267:672–675. doi: 10.1126/science.267.5198.672. [DOI] [PubMed] [Google Scholar]
- Basch L, Tegner M. Reproductive responses of purple sea urchin (Strongylocentrotus purpuratus) populations to environmental conditions across a coastal depth gradient. Bulletin of Marine Science. 2007;81:255–282. [Google Scholar]
- Bédard PA, Brandhorst BP. Translational activation of maternal mRNA encoding the heat-shock protein hsp90 during sea urchin embryogenesis. Developmental Biology. 1986;117:286–293. doi: 10.1016/0012-1606(86)90371-4. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate—a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. Series B (Methodological) 1995;57:289–300. [Google Scholar]
- Bertram D, Strathmann R. Effects of maternal and larval nutrition on growth and form of planktotrophic larvae. Ecology. 1998;79:315–327. [Google Scholar]
- Bolouri H, Davidson EH. Transcriptional regulatory cascades in development: initial rates, not steady state, determine network kinetics. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:9371–9376. doi: 10.1073/pnas.1533293100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürger R, Lynch M. Evolution and extinction in a changing environment: a quantitative-genetic analysis. Evolution. 1997;49:151–163. doi: 10.1111/j.1558-5646.1995.tb05967.x. [DOI] [PubMed] [Google Scholar]
- Byrne M, Selvakumaraswamy P, Ho MA, Woolsey E, Nguyen HD. Sea urchin development in a global change hotspot, potential for southerly migration of thermotolerant propagules. Deep Sea Research Part II: Topical Studies in Oceanography. 2011;58:712–719. [Google Scholar]
- Chen C, Chen B. Effects of high temperature on larval development and metamorphosis of Arachnoides placenta (Echinodermata: Echinoidea) Marine Biology. 1992;122:445–449. [Google Scholar]
- Cochran RC, Engelmann F. Environmental regulation of the annual reproductive season of Strongylocentrotus purpuratus (Stimpson) Biological Bulletin. 1975;148:393–401. doi: 10.2307/1540516. [DOI] [PubMed] [Google Scholar]
- Comstock R, Robinson H. The components of genetic variance in populations of biparental progenies and their use in estimating the average degree of dominance. Biometrics. 1948;4:254–266. [PubMed] [Google Scholar]
- Davidson EH, Rast JP, Oliveri P, et al. A genomic regulatory network for development. Science. 2002;295:1669–1678. doi: 10.1126/science.1069883. [DOI] [PubMed] [Google Scholar]
- Delworth TL, Broccoli AJ, Rosati A, et al. GFDL’s CM2 global coupled climate models. Part I: formulation and simulation characteristics. Journal of Climate. 2006;19:643–674. [Google Scholar]
- Edmands S, Moberg P, Burton R. Allozyme and mitochondrial DNA evidence of population subdivision in the purple sea urchin Strongylocentrotus purpuratus. Marine Biology. 1996;126:443–450. [Google Scholar]
- Farmanfarmaian A, Giese A. Thermal tolerance and acclimation in the western Purple Sea urchin, Strongylocentrotus purpuratus. Physiological Zoology. 1963;36:237–243. [Google Scholar]
- Feder ME, Hofmann GE. Heat-shock proteins, molecular chaperones, and the stress response: evolutionary and ecological physiology. Annual Review of Physiology. 1999;61:243–282. doi: 10.1146/annurev.physiol.61.1.243. [DOI] [PubMed] [Google Scholar]
- Fujisawa H. Differences in temperature dependence of early development of sea urchins with different growing seasons. Biological Bulletin. 1989;176:96. [Google Scholar]
- Fujisawa H. Temperature sensitivity of a hybrid between two species of sea urchin differing in thermotolerance. Development Growth and Differentiation. 1993;35:395–401. doi: 10.1111/j.1440-169X.1993.00395.x. [DOI] [PubMed] [Google Scholar]
- Fujisawa H, Shigei M. Correlation of embryonic temperature sensitivity of sea-urchins with spawning season. Journal of Experimental Marine Biology and Ecology. 1990;136:123–139. [Google Scholar]
- Gasch AP, Spellman PT, Kao CM, et al. Genomic expression programs in the response of yeast cells to environmental changes. Molecular Biology of the Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George SB. Egg quality, larval growth and phenotypic plasticity in a forcipulate seastar. Journal of Experimental Marine Biology and Ecology. 1999;237:203–224. [Google Scholar]
- Gibson G, Dworkin I. Uncovering cryptic genetic variation. Nature Reviews Genetics. 2004;5:681–690. doi: 10.1038/nrg1426. [DOI] [PubMed] [Google Scholar]
- Gienapp P, Teplitsky C, Alho JS, Mills JA, Merila J. Climate change and evolution: disentangling environmental and genetic responses. Molecular Ecology. 2008;17:167–178. doi: 10.1111/j.1365-294X.2007.03413.x. [DOI] [PubMed] [Google Scholar]
- Giese A, Farmanfarmaian A. Resistance of the purple sea urchin to osmotic stress. Biological Bulletin. 1963;124:182. [Google Scholar]
- Hammond LM, Hofmann GE. Thermal tolerance of Strongylocentrotus purpuratus early life history stages: mortality, stress-induced gene expression and biogeographic patterns. Marine Biology. 2010;157:2677–2687. doi: 10.1007/s00227-010-1528-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellemans J, Mortier G, Paepe AD, Speleman F, Vandesompele J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biology. 2007;8:R19. doi: 10.1186/gb-2007-8-2-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinman VF, Davidson EH. Evolutionary plasticity of developmental gene regulatory network architecture. Proceedings of the National Academy of Sciences. 2007;104:19404–19409. doi: 10.1073/pnas.0709994104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochachka PW, Somero GN. Biochemical Adaptation: Mechanism and Process in Physiological Evolution. 1st edn. New York, NY, USA: Oxford University Press; 2002. [Google Scholar]
- Hoffmann AA, Hercus M. Environmental stress as an evolutionary force. BioScience. 2000;50:217–226. [Google Scholar]
- Huey RB. Behavioral thermoregulation in lizards— importance of associated costs. Science. 1974;184:1001–1003. doi: 10.1126/science.184.4140.1001. [DOI] [PubMed] [Google Scholar]
- Huey RB, et al. Why tropical forest lizards are vulnerable to climate warming. Proceedings of the Royal Society. B, Biological Sciences. 2009;276:1939–1948. doi: 10.1098/rspb.2008.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IPCC. Climate Change 2007: Synthesis Report. Cambridge, UK: IPCC, Cambridge University Press; 2007. [Google Scholar]
- Kellermann V, Heerwaarden BV, Sgro C, Hoffmann AA. Fundamental evolutionary limits in ecological traits drive Drosophila species distributions. Science. 2009;325:1244–1246. doi: 10.1126/science.1175443. [DOI] [PubMed] [Google Scholar]
- Kenner MC. Population dynamics of the sea urchin Strongylocentrotus purpuratus in a Central California kelp forest: recruitment, mortality, growth, and diet. Marine Biology. 1992;112:107–118. [Google Scholar]
- Koehn RK, Bayne BL. Towards a physiological and genetical understanding of the energetics of the stress response. Biological Journal of the Linean Society. 1989;37:157–171. [Google Scholar]
- Kuhn K, Baker S, Chudin E, et al. A novel, high-performance random array platform for quantitative gene expression profiling. Genome Research. 2004;14:2347–2356. doi: 10.1101/gr.2739104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biology. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist S. The heat-shock response. Annual Reviews Biochemistry. 1986;55:1151–1191. doi: 10.1146/annurev.bi.55.070186.005443. [DOI] [PubMed] [Google Scholar]
- Lindsley DE, Poodry CA. A reversible temperature-induced developmental arrest in Drosophila. Developmental Biology. 1977;56:213–218. doi: 10.1016/0012-1606(77)90165-8. [DOI] [PubMed] [Google Scholar]
- Loeb J. The Organism as a Whole, From a Physicochemical Viewpoint. New York, NY: Putnam’s Sons; 1916. [Google Scholar]
- Longabaugh WJR, Davidson EH, Bolouri H. Computational representation of developmental genetic regulatory networks. Developmental Biology. 2005;283:1–16. doi: 10.1016/j.ydbio.2005.04.023. [DOI] [PubMed] [Google Scholar]
- Lynch M, Walsh B. Genetics and Analysis of Quantitative Traits. 1st edn. Sunderland, MA: Sinauer Associates Inc; 1998. [Google Scholar]
- Masel J. Cryptic genetic variation is enriched for potential adaptations. Genetics. 2006;172:1985–1991. doi: 10.1534/genetics.105.051649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Research. 2012;40:4288–4297. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiklejohn C, Hartl D. A single mode of canalization. Trends in Ecology & Evolution. 2002;17:468–473. [Google Scholar]
- Oleksiak MF, Roach JL, Crawford DL. Natural variation in cardiac metabolism and gene expression in Fundulus heteroclitus. Nature Genetics. 2005;37:67–72. doi: 10.1038/ng1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver TA, Garfield DA, Manier MK, Haygood R, Wray GA, Palumbi SR. Whole-genome positive selection and habitat-driven evolution in a shallow and a deep-sea urchin. Genome Biology and Evolution. 2010;2:800–814. doi: 10.1093/gbe/evq063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveri P, Tu Q, Davidson E. Global regulatory logic for specification of an embryonic cell lineage. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:5955–5962. doi: 10.1073/pnas.0711220105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osovitz C, Hofmann GE. Thermal history-dependent expression of the hsp70 gene in purple sea urchins: biogeographic patterns and the effect of temperature acclimation. Journal of Experimental Marine Biology and Ecology. 2005;327:134–143. [Google Scholar]
- Palumbi SR, Wilson AC. Mitochondrial DNA diversity in the sea urchins Strongylocentrotus purpuratus and S. droebachiensis. Evolution. 1990;44:403–415. doi: 10.1111/j.1558-5646.1990.tb05208.x. [DOI] [PubMed] [Google Scholar]
- Parmesan C, Yohe G. A globally coherent fingerprint of climate change impacts across natural systems. Nature. 2003;421:37–42. doi: 10.1038/nature01286. [DOI] [PubMed] [Google Scholar]
- Pearse JS. Ecological role of purple sea urchins. Science. 2006;314:940–941. doi: 10.1126/science.1131888. [DOI] [PubMed] [Google Scholar]
- Pearse JS, Pearse VB, Davis KK. Photoperiodic regulation of gametogenesis and growth in the sea urchin Strongylocentrotus purpuratus. Journal of Experimental Zoology. 1986;237:107–118. [Google Scholar]
- Pespeni MH, Garfield DA, Manier MK, Palumbi SR. Genome-wide polymorphisms show unexpected targets of natural selection. Proceedings of the Royal Society. B, Biological Sciences. 2011;279:1412–1420. doi: 10.1098/rspb.2011.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter IS, Davidson EH. The endoderm gene regulatory network in sea urchin embryos up to mid-blastula stage. Developmental Biology. 2010;340:188–199. doi: 10.1016/j.ydbio.2009.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl M. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Research. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfennig DW, Wund MA, Snell-Rood EC, Cruickshank T, Schlichting CD, Moczek AP. Phenotypic plasticity’s impacts on diversification and speciation. Trends in Ecology & Evolution. 2010;25:459–467. doi: 10.1016/j.tree.2010.05.006. [DOI] [PubMed] [Google Scholar]
- R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2010. [Google Scholar]
- Rice SH. The evolution of developmental interactions: epistasis, canalization, and integration. In: Wolf JBW, Brodie ED, Wade MJ, editors. Epistasis and the Evolutionary Process. New York, NY USA: Oxford University Press; 2000. pp. 1–17. [Google Scholar]
- Richter K, Haslbeck M, Buchner J. The heat shock response: life on the verge of death. Molecular Cell. 2010;40:253–266. doi: 10.1016/j.molcel.2010.10.006. [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roepke TA, Snyder MJ, Cherr GN. Estradiol and endocrine disrupting compounds adversely affect development of sea urchin embryos at environmentally relevant concentrations. Aquatic Toxicology. 2005;71:155–173. doi: 10.1016/j.aquatox.2004.11.003. [DOI] [PubMed] [Google Scholar]
- Root TL, Price JT, Hall KR, Schneider SH, Rosenzweig C, Pounds JA. Fingerprints of global warming on wild animals and plants. Nature. 2003;421:57–60. doi: 10.1038/nature01333. [DOI] [PubMed] [Google Scholar]
- Sanders B, Martin L. Copper inhibits the induction of stress protein synthesis by elevated temperatures in embryos of the sea urchin Strongylocentrus purpuratus. Comparative Biochemistry and Physiology, Part C, Pharmacology, Toxicology & Endocrinology. 1994;109C(3):295–307. [PubMed] [Google Scholar]
- Schlichting CD. Hidden reaction norms, cryptic genetic variation, and evolvability. Annals of the New York Academy of Sciences. 2008;1133:187–203. doi: 10.1196/annals.1438.010. [DOI] [PubMed] [Google Scholar]
- Schlichting CD, Pigliucci M. Control of phenotypic plasticity via regulatory genes. The American Naturalist. 1993;142(2):366–370. doi: 10.1086/285543. [DOI] [PubMed] [Google Scholar]
- Sewell M, Young C. Temperature limits to fertilization and early development in the tropical sea urchin Echinometra lucunter. Journal of Experimental Marine Biology and Ecology. 1999;236:291–305. [Google Scholar]
- Sodergren E, Weinstock G, Davidson E, et al. The genome of the sea urchin Strongylocentrotus purpuratus. Science. 2006;314:941–952. doi: 10.1126/science.1133609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somero GN. The physiology of climate change: how potentials for acclimatization and genetic adaptation will determine ‘winners’ and ”losers”. Journal of Experimental Biology. 2010;213:912–920. doi: 10.1242/jeb.037473. [DOI] [PubMed] [Google Scholar]
- Strathmann MF. Reproduction and Development of Marine Invertebrates of the Northern Pacific Coast. Seattle, WA, USA: Univ of Washington Pr; 1987. data and methods for the study of eggs, embryos, and larvae. [Google Scholar]
- Todgham AE, Hofmann GE. Transcriptomic response of sea urchin larvae Strongylocentrotus purpuratus to CO2-driven seawater acidification. Journal of Experimental Biology. 2009;212:2579–2594. doi: 10.1242/jeb.032540. [DOI] [PubMed] [Google Scholar]
- Tomanek L. Variation in the heat shock response and its implication for predicting the effect of global climate change on species’ biogeographical distribution ranges and metabolic costs. Journal of Experimental Biology. 2010;213:971–979. doi: 10.1242/jeb.038034. [DOI] [PubMed] [Google Scholar]
- Tu Q, Brown CT, Davidson EH, Oliveri P. Sea urchin Forkhead gene family: phylogeny and embryonic expression. Developmental Biology. 2006;300:49–62. doi: 10.1016/j.ydbio.2006.09.031. [DOI] [PubMed] [Google Scholar]
- Via S, Lande R. Genotype-environment interaction and the evolution of phenotypic plasticity. Evolution. 1985;39:505–522. doi: 10.1111/j.1558-5646.1985.tb00391.x. [DOI] [PubMed] [Google Scholar]
- Visser J, Hermisson J, Wagner G. Perspective: evolution and detection of genetic robustness. Evolution. 2003;57:1959–1972. doi: 10.1111/j.0014-3820.2003.tb00377.x. [DOI] [PubMed] [Google Scholar]
- Wagner G, Booth G, Bagheri H. A population genetic theory of canalization. Evolution. 1997;51:329–347. doi: 10.1111/j.1558-5646.1997.tb02420.x. [DOI] [PubMed] [Google Scholar]
- Wray G, Kitazawa C, Miner B. Culture of echinoderm larvae through metamorphosis. Methods in Cell Biology. 2004;74:75–86. doi: 10.1016/s0091-679x(04)74004-2. [DOI] [PubMed] [Google Scholar]
- Yuh C-H, Dorman ER, Davidson EH. Brn1/2/4, the predicted midgut regulator of the endo16 gene of the sea urchin embryo. Developmental Biology. 2005;281:286–298. doi: 10.1016/j.ydbio.2005.02.034. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.