Abstract
Ribonuclease L (RNase L) is a Type I Interferon regulated factor that can significantly inhibit retroviruses in vitro and may activate cytoplasmic sensing pathways to augment adaptive immunity. However, the antiretroviral activity of RNase L remains to be validated in vivo. We investigated the role of RNaseL in counteracting Friend retrovirus (FV) infection relative to a well-described restriction factor, Apobec3. C57BL/6 wild-type (WT) and RNaseL knock-out (KO) mice exhibited similar acute FV infection levels despite significant transcriptional induction of Oligoadenylate Synthetase 1, which produces activators of RNase L. Apobec3 KO mice showed higher FV infection levels relative to WT mice, but deletion of RNaseL in Apobec3 KO mice did not augment FV infection. Moreover, RNaseL did not influence FV-specific IgG responses and recovery from viremia by 28 days post-infection. The results suggest that RNase L is not an evolutionarily-conserved host defense mechanism to counteract retroviruses in vivo.
Keywords: Ribonuclease L, Apobec3, Friend retrovirus, restriction factor, humoral immunity
INTRODUCTION
The long-standing evolutionary conflict between retroviruses and mammalian hosts led to the emergence of host factors that have intrinsic antiretroviral activity (Duggal and Emerman, 2012). These ‘restriction factors’ could directly inhibit specific steps of the retrovirus life cycle and appear to act as effectors of the Type I interferon (IFN) response. Several restriction factors that include APOBEC3G, Tetherin/BST-2, TRIM5α and SAMHD1 that can inhibit HIV-1 replication were identified in the past decade and have since been the subject of intense study (Malim and Bieniasz, 2012). However, prior to the identification of these antiviral factors, the Type I IFN antiviral response was associated with double-stranded RNA (dsRNA)-dependent ribonuclease activity (Lengyel et al., 1980). This ribonuclease activity was later attributed to the sequential action of two enzymes, Oligoadenylate Synthetase (OAS) and its main downstream effector, Ribonuclease L (RNase L) (Chakrabarti et al., 2011).
RNase L is an 84 kDa latent enzyme composed of ankyrin repeats, a kinase homology domain and a ribonuclease domain (Zhou, Hassel and Silverman, 1993). Following dsRNA activation, OAS synthesizes 2’-5’ oligoadenylates (2-5A) which bind to RNase L, leading to RNase L dimerization (Dong and Silverman, 1995). RNase L dimers cleave both viral and host RNAs preferably at UU or UA dinucleotides (Floyd-Smith, Slattery and Lengyel, 1981; Wreschner et al, 1981) in single-stranded RNA loops of hairpin structures (Han et al., 2004). Cleavage of viral RNA is expected to directly inhibit virus replication, whereas cleavage of host RNAs (Silverman et al., 1983) was linked to the ability of RNase L to mediate apoptosis (Castelli et al., 1998). In addition, the cleavage of viral and host RNAs by RNase L may liberate RNA species that could activate cytoplasmic RNA sensing pathways. In particular, the generation of small self-RNAs (Malathi et al., 2007), as well as the Hepatitis C virus svRNA fragment (Malathi et al., 2010), results in the activation of the RNA sensor RIG-I to trigger IFN-β production. Thus, RNase L may further augment innate immunity, which in turn could prime a more effective adaptive immune response.
Since retroviruses harbor two RNA genomes per virion that form extensive secondary structures (for example, see Watts et al., 2009), RNase L was considered as a potential antiretroviral factor. Several in vitro studies supported this hypothesis. Upregulation of 2-5A in HIV infected T cell lines correlated with the detection of HIV RNA cleavage products (Schroder et al., 1989). Treatment of HIV-infected target cells ex vivo with nuclease-resistant 2-5AN6B resulted in inhibition of HIV-1 replication (Dimitrova et al., 2007). Substituting RNase L for Nef in HIV-1, as well as co-transfecting RNase L expression plasmids with HIV-1 provirus, diminished HIV-1 replication (Maitra and Silverman, 1998).
For several RNA viruses such as West Nile virus and encephalomyocarditis virus, the antiviral role of RNase L was strengthened following experimental infections revealing higher virus replication in RNaseL knock-out (KO) versus wild-type (WT) mice (Zhou et al., 1997; Samuel et al., 2006). Thus, investigating the role of RNase L in mouse retrovirus infection models may provide insights on the antiretroviral properties of RNase L from a more physiological context. Friend retrovirus (FV) infection of mice represents one of the most well-described in vivo models of retroviral pathogenesis, paving the way for the initial identification of host antiretroviral genes and understanding salient features of retrovirus adaptive immune responses (Nair et al., 2011; Halemano et al., 2013). FV is an ecotropic gammaretrovirus complex that consists of Friend Murine Leukemia Virus (F-MuLV) and Spleen Focus Forming Virus (SFFV) that establishes chronic infection in mice. F-MuLV encodes a fully functional provirus, whereas the pathogenic component, SFFV, is replication-defective and is present in the complex at >100-fold lower levels than F-MuLV (Steeves et al., 1971; He et al., 2008). Given that F-MuLV is essential for replication of the FV complex, detection assays for FV infection are directed primarily against F-MuLV proteins or nucleic acids (Robertson et al., 1991; Dittmer et al., 2002; Santiago et al., 2011; Smith et al., 2011). Thus, the FV infection levels determined from most in vivo infections, as well as this study, refer specifically to the F-MuLV component.
Recently, the FV infection model was used to validate the physiological importance and evolutionary conservation of intrinsic restriction factors and innate sensing pathways. FV infects multiple cells in immune compartments, including erythroblasts, myeloid cells, B cells and to a lesser extent, T cells (Dittmer et al., 2002; Santiago et al., 2010). Thus, intrinsic resistance factors expressed in these susceptible cells could potentially inhibit FV infection. Using knockout mice, our group and others demonstrated a significant role for an enzyme known as Apobec3 in inhibiting acute FV replication (Santiago et al., 2008; Takeda et al., 2008; Tsuji-Kawahara et al., 2010; Santiago et al., 2011; Smith et al., 2011). Apobec3 is a deoxycytidine deaminase that could get packaged into retrovirus particles and inhibits retrovirus replication in the next target cell (Malim and Bieniasz, 2012). Apobec3 likely inhibits FV virion infectivity by blocking early reverse transcription rather than inducing lethal G-to-A hypermutation (Smith et al., 2011). Interestingly, Apobec3 maps within the region in chromosome 15 encoding a classical resistance gene, Recovery from Friend virus gene 3 or Rfv3, which dictates recovery from FV infection by promoting a stronger neutralizing antibody response (Chesebro and Wehrly, 1979; Hasenkrug et al., 1995). Subsequently, our group and others reported that mice deficient in the B6 Apobec3 gene developed weaker FV-specific antibody responses (Santiago et al., 2008; Tsuji-Kawahara et al., 2010; Smith et al., 2011), supporting the notion that Rfv3 is encoded by Apobec3. Thus, the FV infection model was also instrumental in demonstrating that an innate restriction factor could modulate an adaptive immune response.
The expression of restriction factors in virus-susceptible target cells provides a rationale for testing a potential inhibitory role. Similar to Apobec3 (Takeda et al., 2008; Santiago et al., 2010; Tsuji-Kawahara et al., 2011), RNase L is expressed highly in immune compartments that include the spleen (Zhou et al., 1997), a primary site of FV replication in vivo (Dittmer et al., 2002; Santiago et al., 2010). In microarray studies, RNase L mRNA was detected in multiple immune cell subpopulations in the bone marrow (BM) including erythroblasts, myeloid cells, B cells and T cells (Konuma et al., 2011; Gene Expression Omnibus Profile 75447109 and 75447110). Similarly, Apobec3 expression was documented in these immune cell types (Santiago et al., 2010; Tsuji-Kawahara et al., 2011). Thus, the FV infection model represents a suitable experimental platform to compare the role of RNase L versus Apobec3 in counteracting retrovirus infection and influencing adaptive immunity in vivo.
MATERIALS AND METHODS
Mice
C57BL/6 (B6) WT mice were purchased from The Jackson Laboratory. RNaseL KO mice were generated by targeted gene disruption (Zhou et al., 1997) and backcrossed for 10 generations into B6. Apobec3 KO mice were generated from the XN450 gene-trap embryonic stem cell line (BayGenomics) (Santiago et al., 2008) and backcrossed for 9 generations into B6. B6 Apobec3 and B6 RNaseL KO mice were intercrossed to generate double KO mice. This study was approved by the Institutional Animal Care and Use Committee at the University of Colorado [Permit Number B-89709(10)1E].
Cell culture
Mus dunni and 293T cells were maintained in DMEM (Mediatech) with 2% penicillin/streptomycin/glutamine (Mediatech) and 10% fetal bovine serum (FBS; Gemini).
Co-transfection of FV, RNase L and Apobec3
500 ng of the F-MuLV molecular clone pLRB302 was co-transfected with 500 ng of p3 × FLAG-CMV-14 (Sigma) vector or expression constructs encoding B6 Apobec3 (Santiago et al., 2008) or B6 RNaseL (pFS164, from Thomas Michiels, Universite Catholique de Louvain) into 293T cells using Fugene 6 (Promega). The final concentration of DNA was held constant at 1.5 µg, using the vector control as ‘filler’ DNA. After 2 days, supernatants were collected from the transfected 293T cells. To quantify FV infectious titers, serial dilutions of the supernatants were used to infect Mus dunni cells plated in 48-well plates in the presence of 4 µg/ml polybrene (Sigma). At 2 days post-infection (dpi), the cells were fixed with 95% ethanol and stained with F-MuLV envelope-specific MAb 720 (Robertson et al., 1991). Foci were developed using a horseradish-peroxidase conjugated anti-mouse IgG and 3-amino-9-ethylcarbazole (Sigma) substrate. Foci were counted to determine the infectious titer and normalized to input ml of supernatant.
Virus stocks and FV infection
Two types of B-tropic FV were used in this study: (1) an FV stock containing only F-MuLV and SFFV (also referred to as ‘LDV-free FV’ in prior publications); and (2) the classical ‘FV/LDV’ stock containing F-MuLV, SFFV, and a co-infecting endemic RNA virus, Lactate Dehydrogenase-elevating Virus (LDV) (Robertson et al., 2008; Halemano et al., 2013). FV stocks were prepared and titered in BALB/c mice as described (Santiago et al., 2008). Mice (2 to 4 months old) were infected intravenously via the retro-orbital route with 104 Spleen Focus Forming Units (SFFU) of FV complex and mice were sacrificed at indicated timepoints for analysis.
Viral load quantifications
The quantitative PCR (qPCR) methods for measuring proviral DNA copy numbers and plasma infectious viremia were specific for the F-MuLV helper virus and described previously (Smith et al., 2011; Harper et al., 2013). Proviral DNA levels provide an estimate of the number of integrated proviruses per cell (Smith et al., 2011). Plasma infectious viremia corresponds to the levels of reverse transcripts produced in target Mus dunni cells following inoculation with 5 µl of plasma (Harper et al., 2013). Cellular viral RNA levels indicate the abundance of retroviral RNA transcripts in productively infected cells, which may be susceptible to RNase L cleavage. Cellular viral RNA load was measured from RNA extracted from BM cells using the RNeasy Kit (Qiagen) and eluted in RNase-free water. qPCR was performed as described for the plasma viral load assay (Santiago et al., 2011; Smith et al., 2011). Data were normalized to beta-actin RNA, detected using Mouse.B-actin.sense: 5’-GGCACCACACCTTCTACAATG; Mouse.B-actin.antisense: 5’-GGGGTGTTGAAGGTCTCAAAC; and Mouse.B-actin.probe: FAMTGTGGCCCCTGAGGAGCACCC-TAMRA.
Flow cytometry
BM and spleen cells were stained with FV Glyco-Gag specific MAb 34 (Chesebro et al., 1993) for flow cytometry as described (Smith et al., 2011) and adapted from a previous study (Dittmer et al., 2002). The levels of FV Glyco-Gag+ cells measured by flow cytometry provides a measure of cellular infection and spread based on viral protein production. PBS (Mediatech) with 1% FBS was used to wash the cells. Cells were co-stained with Ter119-FITC (clone TER-119, BD Biosciences), CD3-Alexa700 (17A2, BD Biosciences), CD19-PerCPCy5.5 (6D5) (Biolegend), CD11b–PE (CF594, BD Biosciences), and anti-mouse IgG2b-APC (Columbia Biosciences). The stained cells were analyzed in an LSR-II flow cytometer (BD Biosciences), collecting 200,000–250,000 events per sample. Data was analyzed using the Flowjo software (Treestar), gating on live cells followed by singlet discrimination. Cells from uninfected mice were used to gate FV+ cells.
OAS1a induction
RNA from 1 dpi BM and spleen samples using the RNAEasy kit (Qiagen) and cDNA was synthesized using the Quantiscript kit (Qiagen). cDNA (20 ng/25 µl reaction) was added to 1× Quantitect SYBR qPCR Mastermix (Qiagen) with a final concentration of 5 pmol each of the following primers: Oas1a.Forward-5’-CTGCATCAGGAGGTGGAGTT-3’ and Oas1a.Reverse-5’-GGATGGCATAGATTCTGGGA-3’ or Actin.Forward 5’-CTCCATCGTGGGCCGC-3’ and Actin.Reverse 5’-CCAGTTGGTAACAATGCCATGTTC-3’. qPCR was performed in a Bio-rad CFX96 cycler using the following conditions: 95°C for 15 min, 40 cycles of 95°C for 15 s and 60°C for 1 min. Melt curve analysis was performed to verify specific amplification. Fold-inductions were calculated using the ΔΔCt method.
ELISA
Enzyme-linked immunosorbent assays were performed as described (Smith et al., 2011). Endpoint IgG titers were calculated as the plasma dilution corresponding to 2× the PBS background from each plate using best-fit nonlinear regression (GraphPad Prism 5.0).
Statistical analysis
Data were first tested for normality using the Kolmogorov-Smirnov test. Data with a normal distribution were evaluated using a 2-tailed unpaired Student’s t test. Otherwise, the nonparametric 2-tailed Mann-Whitney U test was used. Differences with p<0.05 were considered statistically significant. Multi-cohort datasets were first subjected to 1-way Analysis of Variance (ANOVA). Only datasets with p<0.05 by ANOVA were analyzed for differences between pairs of groups using a 2-tailed Student’s t test. All calculations were performed using Prism 5.0 (GraphPad).
RESULTS
RNase L potently restricts F-MuLV in vitro
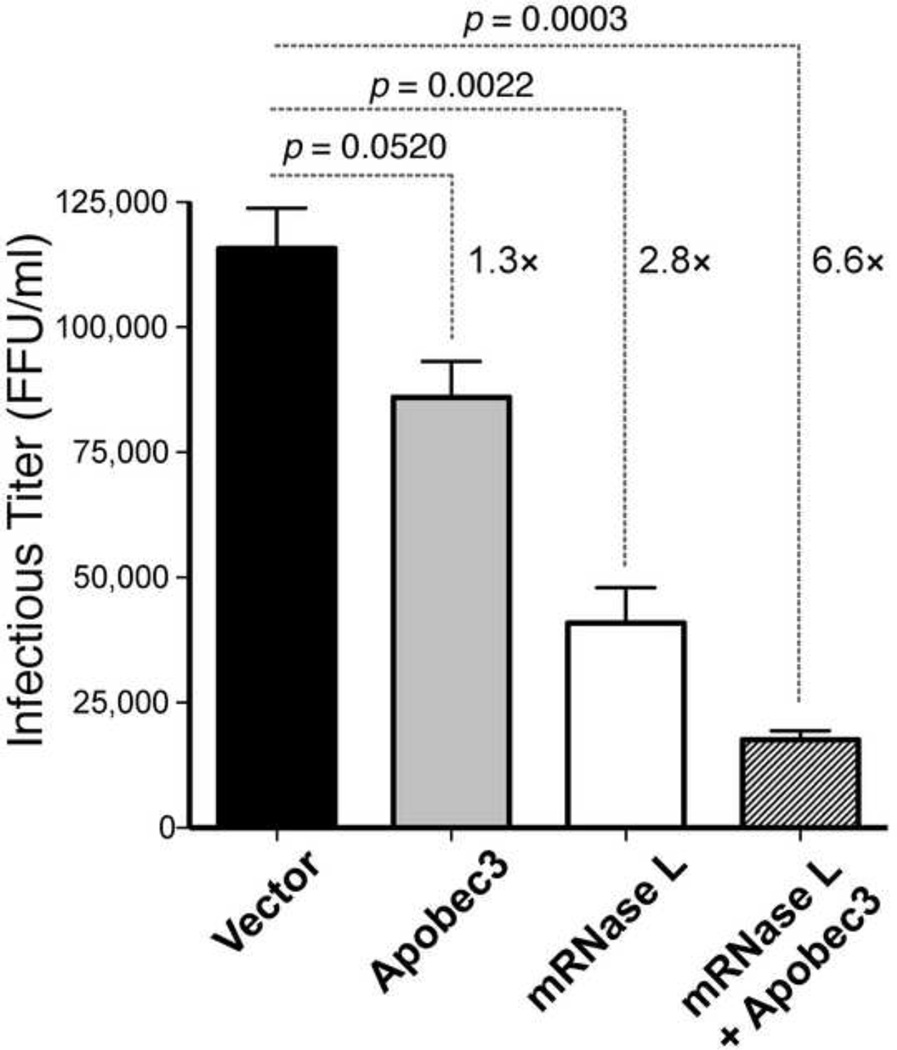
We initially tested if murine RNase L has in vitro activity against F-MuLV, the component of FV complex that is essential for replication. Expression plasmids containing RNaseL and/or Apobec3 from B6 mice were co-transfected with an F-MuLV molecular clone, pLRB302 (Portis et al., 1992), at an equimolar ratio in 293T cells. After 48 h, supernatants were harvested and titered in Mus dunni cells using a focal infectivity assay. The focal infectivity assay would detect not only a decrease in virion infectivity due to Apobec3, but also a decrease in viral production, as would be expected with RNase L activity in the producer cell. As shown in Fig. 1, co-transfection with RNaseL expression plasmid led to a significant 2.8-fold decrease in viral titer relative to the vector control, whereas Apobec3 inhibited by only 1.3-fold. Thus, RNase L significantly inhibited F-MuLV to a greater degree than Apobec3. Interestingly, co-transfection of F-MuLV with both Apobec3 and RNaseL expression constructs led to a 6.6-fold inhibition, higher than the product of the individual fold-inhibition values (3.6-fold). This finding raised the possibility of synergism between Apobec3 and RNase L antiretroviral activity in vitro.
Fig. 1. Mouse RNase L potently inhibits FV in vitro.
Expression plasmids encoding murine Apobec3, RNaseL or empty vector were co-transfected with an F-MuLV molecular clone, pLRB302, into 293T cells. Serial 3-fold dilutions of the supernatants were used to infect Mus dunni cells, and after 48 h, cells were fixed and stained with a monoclonal antibody (MAb 720) against the FV envelope protein. Foci were counted and infectious titers were calculated per ml of supernatant. Mean fold-differences relative to vector control are shown. P values were calculated using 2-tailed Student’s t test, with p<0.05 considered statistically significant.
RNase L is not a potent antiretroviral factor compared to Apobec3 in vivo
To determine whether the in vitro findings in Fig. 1 apply in vivo, B6 WT and RNaseL KO mice were infected with FV and analyzed 7 days later. Since B6 Apobec3 KO mice were more susceptible to FV infection than B6 WT mice (Santiago et al., 2008; Takeda et al., 2008; Tsuji-Kawahara et al., 2010; Smith et al., 2011), B6 Apobec3 KO mice were also infected in parallel as an internal positive control. Recently, we reported that B6 Apobec3 might mask the activity of a potent Tetherin variant (Barrett et al., 2012). To exclude the possibility that B6 Apobec3 may mask the antiretroviral activity of RNaseL, and to test whether Apobec3 and RNase L could synergistically inhibit FV as suggested in Fig. 1, mice doubly deficient in Apobec3 and RNaseL (double KO) were also generated and infected with FV.
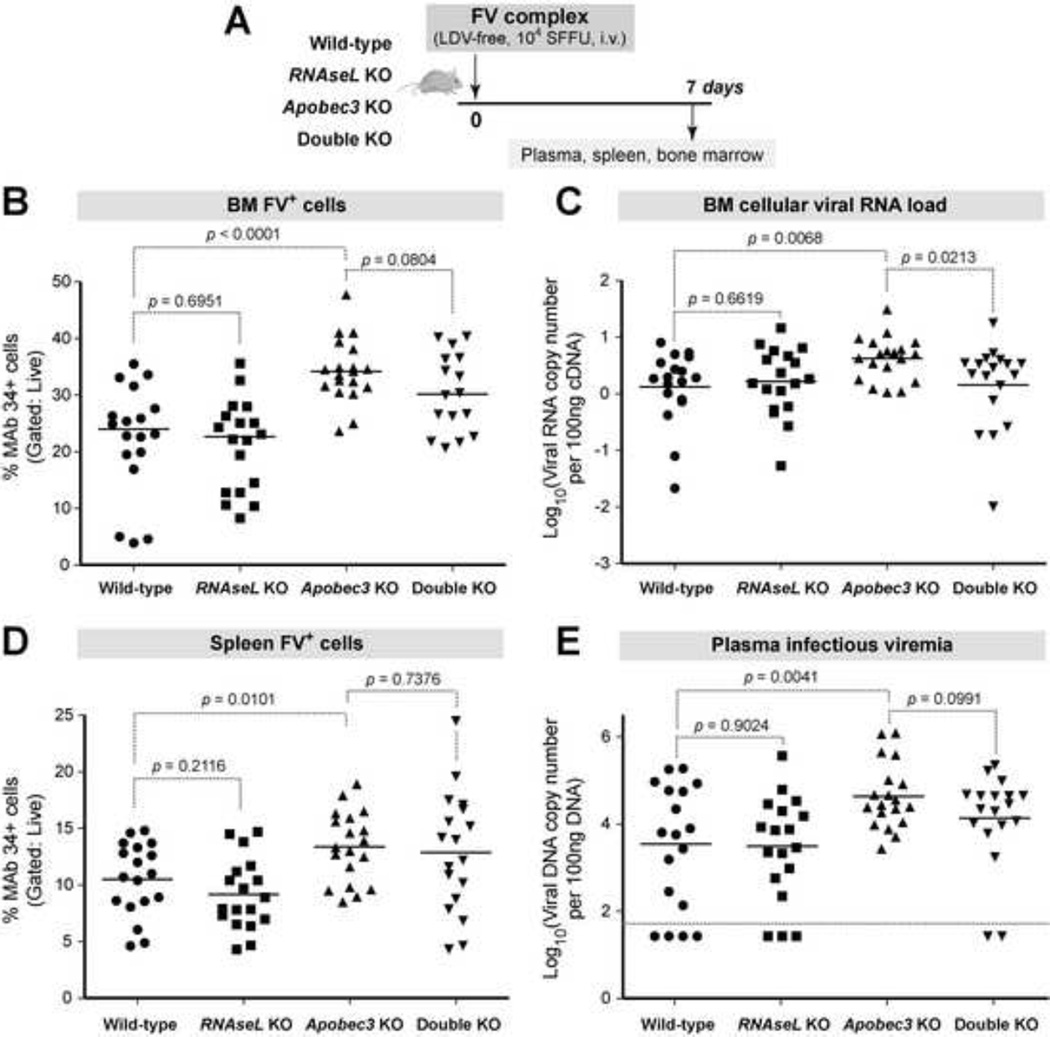
WT, RNaseL KO, Apobec3 KO and double KO mice were inoculated with FV and samples were analyzed at 7 dpi (Fig. 2A). Several virological assays were utilized in an effort to capture an effect of RNase L in different steps of the retroviral life cycle as outlined in the Materials and Methods section. FV infection levels based on flow cytometry, cellular viral RNA and infectious viremia were significantly different between the 4 groups of mice based on ANOVA (Fig. 2B–E). In contrast, spleen mass and BM proviral loads were not significantly different (Supplementary Figure 1). As expected, B6 Apobec3 KO mice had significantly higher FV infection levels in the BM (Fig. 2B–C) and spleen (Fig. 2D) compared to WT mice. B6 Apobec3 KO mice also had higher infectious viremia in the plasma (Fig. 2E). In contrast, RNaseL KO mice had similar BM FV Glyco-Gag+ cells (Fig. 3B), BM cellular viral RNA load (Fig. 2C), spleen FV Glyco-Gag+ cells (Fig. 2D) and plasma infectious viremia (Fig. 2E) compared to WT mice. Removal of RNaseL in Apobec3 KO mice likewise did not result in higher FV replication relative to B6 Apobec3 KO mice (Fig. 2B–E). Intriguingly, BM cellular viral RNA levels were significantly lower in double-KO versus B6 Apobec3 KO mice (Fig. 2C). However, this was not observed with other measures of FV infection (Fig. 2B, 2D–E). Altogether, these data reveal that in contrast to Apobec3, RNaseL does not significantly inhibit acute FV infection in vivo.
Fig. 2. RNaseL does not significantly impact acute FV infection even in the absence of Apobec3.
(A) Infection timeline. WT, RNaseL KO, Apobec3 KO, and RNaseL/Apobec3 double KO mice were sacrificed at 7 dpi for analyses. Data were pooled from 3 independent infection cohorts. (B) BM FV+ cells. Glyco-Gag+ cells were stained with MAb 34 and the percent of FV infected cells was determined by flow cytometry. (C) BM cellular viral RNA load. RNA was extracted from BM and qPCR was used to determine FV RNA copies, normalized to beta-actin mRNA copies. Log10 values are shown. (D) FV Glyco-Gag+ splenocytes, stained for flow cytometry as in (B). (E) Infectious plasma viremia. Plasma (5 µl) were inoculated into Mus dunni cells, and after 48 h, DNA was extracted and qPCR was used to determine the amount of FV DNA. Data was normalized to 100 ng of DNA input. Log10 values are shown. Dashed lines correspond to the detection limit of the assay. For all panels, each dot corresponds to an infected mouse, bars correspond to mean values. Differences between pairs of experimental groups were evaluated using 2-tailed Student’s t test, with p<0.05 considered statistically significant.
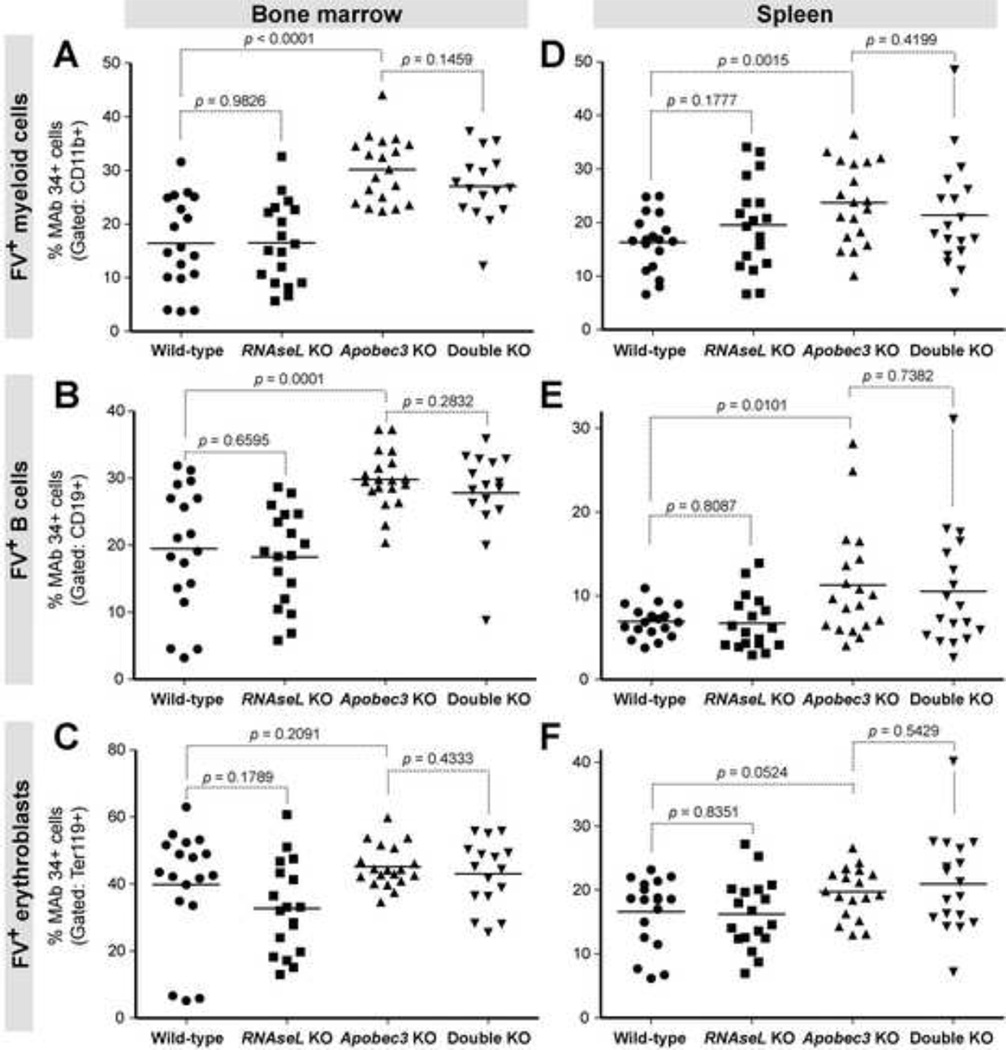
Fig. 3. RNaseL does not significantly inhibit acute FV infection in target cell subpopulations.
WT, RNaseL KO, Apobec3 KO, and double KO mice were infected with 104 SFFU of FV, and at 7 dpi, BM and splenocytes were subjected to flow cytometric analyses. Data were pooled from 3 independent infection cohorts. BM (panels A to C) and spleen (panels D to F) were stained with antibodies against myeloid cells (CD11b; panels A and D), B cells (CD19; panels B and E), and erythroid cells (Ter119; panels C and F). FV expression was detected using MAb 34. For all panels, each dot corresponds to an infected mouse, and bars correspond to mean values. Differences between pairs of experimental groups were evaluated using 2-tailed Student’s t test, with p<0.05 considered statistically significant.
RNase L does not significantly inhibit FV in specific immune cell subpopulations
RNase L did not have a significant effect on FV infection of total BM and spleen cells. However, an effect of RNase L on specific cell subpopulations cannot be excluded. We therefore determined the levels of FV infection in myeloid (CD11b+), B cell (CD19+), erythroblast (Ter119+) and T cell (CD3+) subsets by flow cytometry. In contrast to myeloid and B cell subpopulations (Fig. 3), no significant differences between each genotype in FV infection levels were observed for T cells by ANOVA (Supplementary Figure 2). As expected, B6 Apobec3 KO mice had significantly higher FV infection levels in myeloid and B cells in both the BM and spleen (Fig. 3A–B, D–E). Of note, B6 Apobec3 had no significant impact on FV replication in BM erythroblasts (Fig. 3C), and only approached statistical significance for splenic erythroblasts (p=0.052; Fig. 3F). Thus, the antiretroviral impact of B6 Apobec3 in bulk BM and spleen was likely due to an effect on myeloid and B cell subpopulations. The data on B cells is consistent with another study (Tsuji-Kawahara et al., 2011). The erythroblast data contrasted from results obtained with FV/LDV infections of (B6 × BALB/c)F1 and (B6 × A.BY)F1 mice. In these splenomegaly-susceptible strains, B6 Apobec3 had a significant effect on FV Glyco-Gag+ erythroblasts (Santiago et al., 2010; Smith et al., 2011). Importantly, RNaseL deficiency did not significantly influence FV replication in myeloid (Fig. 3A,D), B cell (Fig. 3B,E), erythroblast (Fig. 3C,F) and T cell (Supplementary Figure 2) subsets, either in the pure B6 or B6 Apobec3 KO backgrounds in both the BM and the spleen. Thus, RNase L does not significantly inhibit acute FV infection in specific cell subsets.
RNase L does not significantly inhibit FV in the context of LDV co-infection
The FV stock utilized in Figs. 2 and 3 lacked LDV, an endemic mouse RNA virus found in the classical FV stocks (Robertson et al., 2008). LDV is a potent inducer of Type I IFNs (Ammann et al., 2009; Gerlach et al., 2006; Duley et al., 2012), and could modulate the immune response against FV as revealed in comparative studies of FV versus FV/LDV infections (Robertson et al., 2008; Marques et al., 2008; Duley et al., 2012). Since RNase L and its activator, OAS1, could be induced by Type I IFNs (Zhou et al., 1993; Harper et al., 2013), FV/LDV infection of WT versus RNaseL KO mice may yield a different acute FV infection outcome. We therefore infected B6 WT and RNaseL KO mice with FV/LDV and evaluated FV infection levels at 7 dpi. The results revealed that B6 WT and RNaseL KO mice had similar FV infection levels (Supplementary Figure 3). In addition, no significant differences in FV infection were found in erythroblasts, myeloid cells, B cells and T cells, although a trend for higher FV infection in RNaseL KO mice was observed in myeloid cells (p=0.08; Supplementary Figure 4). Thus, in the context of augmented Type I IFN induction due to LDV co-infection, RNase L still did not have a significant impact on acute FV replication in vivo.
FV infection induces OAS1a transcription independent of RNase L and LDV co-infection
RNase L is highly expressed in multiple immune compartments (Zhou et al., 1993), but RNase L activity requires dimerization through cross-linking by 2-5A, which is synthesized by OAS1. Insufficient RNase L activation, rather than expression, may therefore account for the lack of inhibitory activity of RNase L against FV in vivo. However, 2-5A is highly unstable in cells due to endogenous phosphodiesterase and phosphatase activities (Player and Torrence, 1998; Kubota et al., 2004). Thus, as an indirect indicator for RNase L activation, we measured the induction of OAS1 following FV infection.
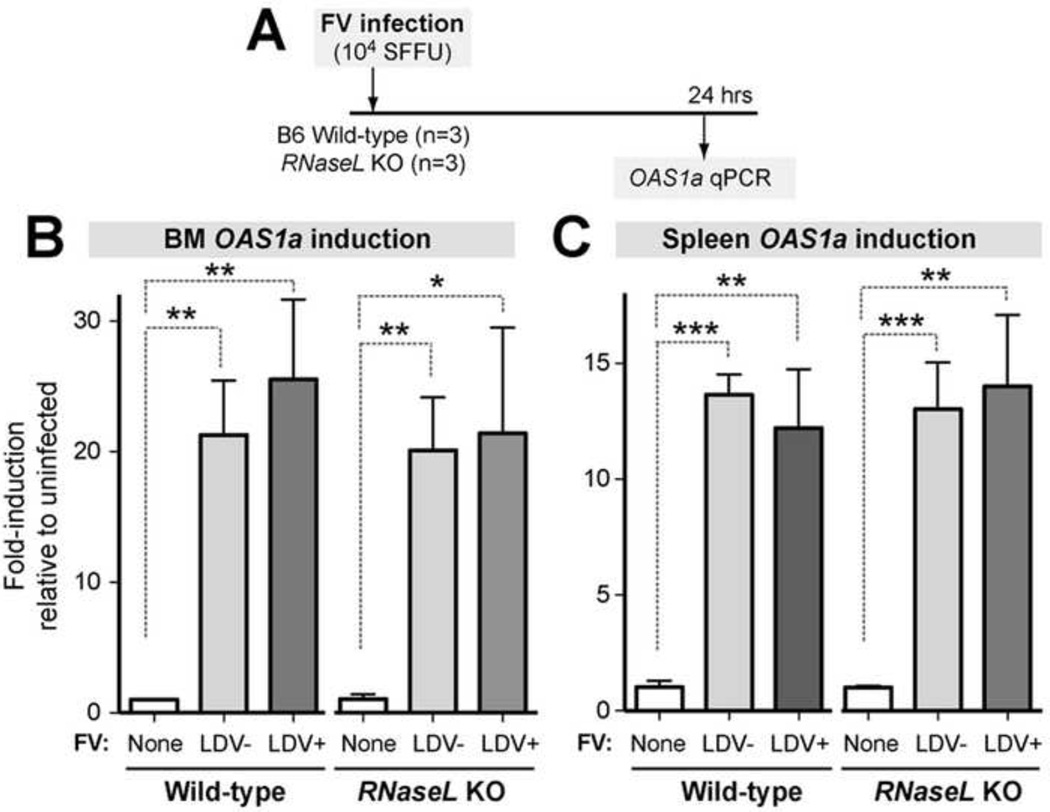
Murine OAS1 is comprised of several related genes, of which only OAS1a and OAS1g were reported to synthesize 2-5A (Kakuta, Shibata and Iwakura, 2002). B6 WT and RNaseL KO mice were infected with either FV/LDV or FV (n=3 mice each), and after 24 h, OAS1a levels were compared to mock-infected controls (Fig. 4A). In the BM, FV infection induced OAS1a expression ~20-fold independent of LDV-coinfection and the presence/absence of RNaseL (Fig. 4B). In the spleen, FV induced OAS1a transcription to a lesser extent (~12-fold), but the OAS1a induction levels relative to mock were statistically significant. No difference was observed between the OAS1a induction levels in WT versus RNaseL KO mice. These data suggest that the lack of antiretroviral effect by RNaseL was not due to the lack of OAS1a expression.
Fig. 4. Transcriptional induction of OAS1a following FV infection in WT and RNaseL-deficient mice.
(A) Timeline. Relative OAS1a transcript levels were measured by qPCR, normalizing for actin levels. Fold-induction levels were normalized against the mean OAS1a expression levels in uninfected mice. (B) BM OAS1a mRNA levels. (C) Spleen OAS1a mRNA levels. Statistical tests were performed using a 2-tailed Student’s t test, with *, p<0.05, **, p<0.01, and ***, p<0.001. Bars correspond to standard deviations from mean OAS1a levels from 3 mice each. For each mouse, mean OAS1a levels were obtained from 3 technical replicates.
RNase L does not influence FV-specific IgG responses
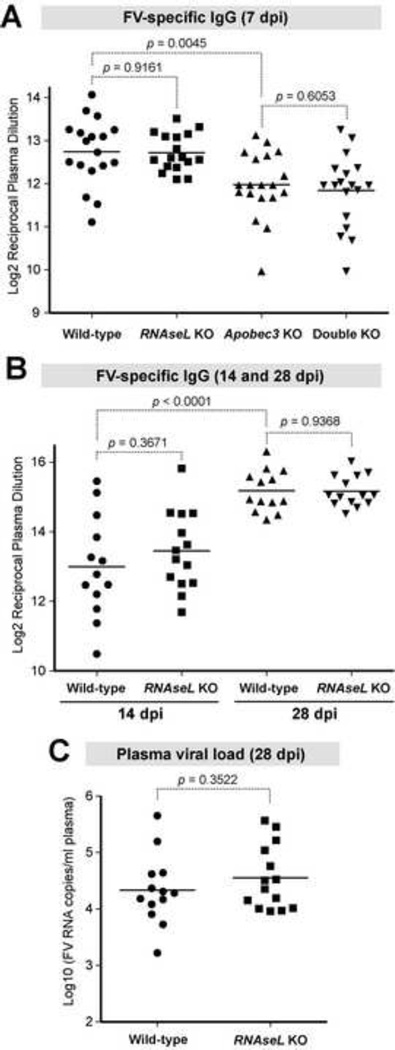
The results above revealed that RNase L is not a potent innate antiretroviral restriction factor. However, even low-level RNase L activity could liberate RNA fragments that could activate innate cytoplasmic sensing pathways (Malathi et al., 2007; Malathi et al., 2010) and enhance adaptive immunity. Thus, we investigated if RNase L could promote adaptive immunity. We focused on FV-specific IgG antibodies against native virions since this is a phenotype that was influenced by B6 Apobec3 (Santiago et al., 2008; Tsuji-Kawahara et al., 2010; Smith et al., 2011). Thus, B6 Apobec3 KO mice could again serve as an internal positive control. At 7 dpi, we observed significantly lower FV-specific IgG endpoint titers in B6 Apobec3 KO compared to WT mice (Fig. 5A). However, in both the WT and Apobec3 KO genetic backgrounds, RNaseL deficiency had no effect on early FV-specific IgG titers (Fig. 5A).
Fig. 5. RNaseL did not significantly influence the FV-specific IgG response or recovery from FV viremia.
Mice were infected with 104 SFFU of FV and plasma FV-specific IgG titers at indicated timepoints were analyzed by endpoint ELISA against native FV virions. (A) 7 dpi IgG titers from groups of mice described in Figures 3–4. (B) Endpoint IgG titers at 14 dpi and 28 dpi. The data pooled from two independent experiments. (C) Plasma viral RNA load at 28 dpi from the same mice in (B) were analyzed by qPCR. For all panels, each dot corresponds to an infected mouse, and bars correspond to mean values. Differences between pairs of experimental groups were evaluated using 2-tailed Student’s t test, with p<0.05 considered statistically significant.
We next compared the FV-specific IgG response at 14 and 28 dpi in 2 new cohorts of B6 WT and RNaseL KO mice. As shown in Fig. 5B, there was a significant increase in FV-specific IgG titer from 14 to 28 dpi, consistent with the development of FV-specific antibody responses. However, in both timepoints, there was no significant difference in FV-specific IgG titers between WT and RNaseL KO mice (Fig. 5B).
RNase L does not promote recovery from FV viremia
It is possible that a significant impact of RNaseL on adaptive immunity, particularly T cell responses, was missed. However, if these adaptive immune responses were significantly augmented by RNase L, a concomitant increase in viremia at later timepoints post-infection should be expected in RNaseL KO mice. We therefore evaluated the plasma viral RNA load at 28 dpi, and found no significant difference between WT and RNaseL KO mice (Fig. 5C). Similar results were obtained at 14 dpi (data not shown). Thus, RNase L did not promote recovery from FV viremia.
DISCUSSION
RNase L was among the first effectors of the Type I IFN antiviral response that was molecularly cloned (Zhou, Hassel and Silverman, 1993) but its role in restricting retroviruses remains surprisingly unclear. The recent identification of potent retrovirus restriction factors that are antagonized by retrovirus-encoded proteins (Malim and Bieniasz, 2012) may have shifted focus away from RNase L, which currently does not have a bona fide retroviral antagonist. However, in theory, the highly folded retroviral RNA genome should be susceptible to RNase L, and results from in vitro studies (Schroder et al, 1989; Maitra et al., 1998; Dimitrova et al., 2007) supported this notion. However, none of these studies directly tested RNaseL antiretroviral activity in vivo.
In this study, we utilized the FV infection model and RNaseL KO mice to investigate the in vivo antiretroviral potency of RNase L. As expected, RNase L significantly inhibited F-MuLV in vitro. In contrast, RNase L did not significantly inhibit acute FV replication in vivo, using a variety of assays that were specific to the F-MuLV component of the FV complex. The disconnect between the in vitro versus in vivo antiretroviral potency of RNase L against F-MuLV mirrored our recent findings with Tetherin. In backcrossed mice, a Tetherin variant that potently restricted F-MuLV in vitro significantly inhibited FV complex in vivo only in the context of weak Apobec3 activity (Barrett et al., 2012). Moreover, IFN-α treatment inhibited acute FV replication primarily through Apobec3, despite the fact that Tetherin and OAS1 were highly induced (Harper et al., 2013). These findings led us to propose a ‘restriction factor hierarchy’, whereby potent restriction factors such as B6 Apobec3 may mask the activity of other innate restriction factors (Barrett et al., 2012). The notion of a restriction factor hierarchy provided a rationale for testing the impact of RNase L in a B6 Apobec3 KO background: by removing B6 Apobec3, the antiretroviral activity of RNaseL may be unleashed. However, even in B6 Apobec3 KO mice, RNaseL did not significantly inhibit FV replication. Co-infection with LDV, a potent Type I IFN inducer, likewise did not result in a significant effect of RNase L against acute FV infection in vivo. Altogether, these data suggest that RNase L is not a significant antiretroviral factor in the FV infection model. Alternatively, RNase L may occupy a lower position in the restriction factor hierarchy than Apobec3 or Tetherin. The results highlight the need for caution in extrapolating data on the in vitro antiviral activity of restriction factors to more physiologically relevant biological systems.
The data revealed that RNaseL did not have a significant impact on acute FV infection. However, minor restriction effects could have a cumulative impact if sustained during the course of infection. RNase L may also liberate self and FV RNA fragments that could potentiate adaptive immunity (Malathi et al., 2007; Malathi et al., 2010). However, at later timepoints, FV infection and FV-specific IgG responses were similar between WT and RNaseL KO mice. Thus, RNase L did not significantly influence FV recovery and humoral immunity in vivo, raising doubts on the biological significance of subtle inhibitory effects that may have been missed by the battery of virological tests utilized in this study.
It should be noted that the lack of RNase L activity against FV infection of mice might not necessarily extend to other murine retrovirus infections such as Mouse Mammary Tumor Virus (MMTV), Moloney MLV, CasFrKP MLV and LP-BM5 (Murine AIDS). In addition to FV infection, Apobec3 exhibited a significant impact on acute infection levels of MMTV, Moloney MLV, CasFrKP MLV and LP-BM5 (Okeoma et al., 2007; Low et al., 2009; Kolokithas et al., 2010; Jones et al., 2012). Together, these studies strengthen the importance of Apobec3 as a potent antiretroviral restriction factor in mice. Thus, it would be of interest to test the impact of RNase L against other murine retroviruses. However, even if RNase L had an impact against these other retroviral infections, the lack of RNase L activity against FV would imply that RNase L would have a more retrovirus-specific effect.
Along the same lines, we note the limitations of extrapolating the FV infection results to pathogenic human retrovirus infections such as HIV-1 and HTLV-I. Sequence differences that affect RNA folding between these retroviruses may confer different sensitivities for RNase L cleavage. Amino acid differences between human and mouse RNase L may likewise limit extrapolation of these results between species. However, human and mouse RNase L could significantly inhibit HIV-1 and F-MuLV in vitro, respectively, and the RNaseL KO mice used in this study exhibited higher susceptibility to RNA viruses (Zhou et al., 1997; Samuel et al., 2006). These findings argue against the notion that mouse RNase L may have an inherent defect that would invalidate its relevance to human RNase L function. Retroviruses have been co-evolving with mammalian hosts for millennia, and many innate antiviral mechanisms, including Apobec3 as confirmed in this study, are conserved between mice and humans. Thus, the data suggests that RNase L is not an evolutionarily conserved host defense mechanism against retroviruses in vivo.
Insights on the evolutionary conservation of antiviral mechanisms may have practical implications for the rapidly expanding field of innate retrovirus restriction factor biology. The list of Type I IFN effectors have significantly grown through large-scale in vitro screens (Schoggins et al., 2011; Liu et al., 2012), and making sense of these diverse restriction mechanisms — all of which appear to have potent antiretroviral activities in vitro — has become an important challenge. In this regard, the FV infection model may provide critical insights on how putative antiretroviral factors operate in an in vivo context. The identification of potent and evolutionarily conserved antiretroviral mechanisms may inform which innate factors to exploit for novel therapies and basic vaccine discovery against pathogenic human retroviruses.
Supplementary Material
ACKNOWLEDGMENTS
We thank Kim Hasenkrug (Rocky Mountain Laboratories) for the FV stocks, monoclonal antibodies and protocols for FV quantification, Jessica Diaz-Rivera (University of Puerto Rico-Cayey) for assistance with ELISAs, Brent Palmer (University of Colorado Denver; UCD) for flow cytometry advice, David Barton (UCD) for helpful discussions, Peter DeWitt (UCD Biostatistics, Epidemiology and Research Design Core) for statistical advice and the UCD Animal Core Facility for technical assistance. This work was supported by the National Institutes of Health R01 AI090795 (M.L.S), NIH R01 CA044059 (R.H.S.), The Colorado Clinical and Translational Sciences Institute TL1 TR000155 (S.X.L.), the Tim Gill Foundation (M.S.H.), The Robert D. Watkins Fellowship of the American Society for Microbiology (K.H.), and the UCD Early Career Scholar Program (M.L.S.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ammann CG, Messer RJ, Peterson KE, Hasenkrug KJ. Lactate dehydrogenase-elevating virus induces systemic lymphocyte activation via TLR7-dependent IFNalpha responses by plasmacytoid dendritic cells. PLOS ONE. 2009;4:e6105. doi: 10.1371/journal.pone.0006105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett BS, Smith DS, Li SX, Guo K, Hasenkrug KJ, Santiago ML. A single nucleotide polymorphism in Tetherin promotes retrovirus restriction in vivo. PLOS Pathog. 2012;8:e1002596. doi: 10.1371/journal.ppat.1002596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelli JC, Hassel BA, Maran A, Paranjape J, Hewitt JA, Li XL, Hsu YT, Silverman RH, Youle RJ. The role of 2'-5' oligoadenylate-activated ribonuclease L in apoptosis. Cell Death Differ. 1998;5:313–320. doi: 10.1038/sj.cdd.4400352. [DOI] [PubMed] [Google Scholar]
- Chakrabarti A, Jha BK, Silverman RH. New insights into the role of RNase L in innate immunity. J Interferon Cytokine Res. 2011;31:49–57. doi: 10.1089/jir.2010.0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesebro B, Wehrly K. Identification of a non-H-2 gene (Rfv-3) influencing recovery from viremia and leukemia induced by Friend virus complex. Proc. Natl. Acad. Sci. U. S. A. 1979;76:425–429. doi: 10.1073/pnas.76.1.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesebro B, Britt W, Evans L, Wehrly K, Nishio J, Cloyd M. Characterization of monoclonal antibodies reactive with murine leukemia viruses: use in analysis of strains of Friend MCF and Friend ecotropic murine leukemia virus. Virology. 1983;127:134–148. doi: 10.1016/0042-6822(83)90378-1. [DOI] [PubMed] [Google Scholar]
- Dimitrova DI, Reichenbach NL, Yang X, Pfleiderer W, Charubala R, Gaughan JP, Suh B, Henderson EE, Suhadolnik RJ, Rogers TJ. Inhibition of HIV type 1 replication in CD4+ and CD14+ cells purified from HIV type 1-infected individuals by the 2-5A agonist immunomodulator 2-5A(N6B) AIDS Res. Hum. Retroviruses. 2007;23:123–134. doi: 10.1089/aid.2005.0091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmer U, Race B, Peterson KE, Stromnes IM, Messer RJ, Hasenkrug KJ. Essential roles for CD8+ T cells and gamma interferon in protection of mice against retrovirus-induced immunosuppression. J. Virol. 2002;76:450–454. doi: 10.1128/JVI.76.1.450-454.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong B, Silverman RH. 2-5A-dependent RNase molecules dimerize during activation by 2-5A. J. Biol. Chem. 1995;270:4133–4137. doi: 10.1074/jbc.270.8.4133. [DOI] [PubMed] [Google Scholar]
- Duggal NK, Emerman M. Evolutionary conflicts between viruses and restriction factors shape immunity. Nat. Rev. Immunol. 2012;12:687–695. doi: 10.1038/nri3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duley AK, Ploquin MJ, Eksmond U, Ammann CG, Messer RJ, Myers L, Hasenkrug KJ, Kassiotis G. Negative impact of IFN-γ on early host immune responses to retroviral infection. J. Immunol. 2012;189:2521–2529. doi: 10.4049/jimmunol.1201125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floyd-Smith G, Slattery E, Lengyel P. Interferon action: RNA cleavage pattern of a (2'-5')oligoadenylate--dependent endonuclease. Science. 1981;212:1030–1032. doi: 10.1126/science.6165080. [DOI] [PubMed] [Google Scholar]
- Gerlach N, Schimmer S, Weiss S, Kalinke U, Dittmer U. Effects of type Iinterferons on Friend retrovirus infection. J. Virol. 2006;80:3438–3444. doi: 10.1128/JVI.80.7.3438-3444.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halemano K, Harper MS, Guo K, Li SX, Heilman KJ, Barrett BS, Santiago ML. Humoral immunity in the Friend retrovirus infection model. Immunol Res. 2013;55:249–260. doi: 10.1007/s12026-012-8370-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasenkrug KJ, Valenzuela A, Letts VA, Nishio J, Chesebro B, Frankel WN. Chromosome mapping of Rfv3, a host resistance gene to Friend murine retrovirus. J. Virol. 1995;69:2617–2620. doi: 10.1128/jvi.69.4.2617-2620.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han JQ, Wroblewski G, Xu Z, Silverman RH, Barton DJ. Sensitivity of hepatitis C virus RNA to the antiviral enzyme Ribonuclease L is determined by a subset of efficient cleavage sites. J Interferon Cytokine Res. 2004;24:664–676. doi: 10.1089/jir.2004.24.664. [DOI] [PubMed] [Google Scholar]
- Harper MS, Barrett BS, Smith DS, Li SX, Gibbert K, Dittmer U, Hasenkrug KJ, Santiago ML. IFN-alpha treatment inhibits acute Friend retrovirus replication primarily through the antiviral effector molecule Apobec3. J. Immunol. 2013;190:1583–1590. doi: 10.4049/jimmunol.1202920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He JY, Cheng HJ, Wang YF, Zhu YT, Li GQ. Development of a real-time quantitative reverse transcriptase PCR assay for detection of the Friend leukemia virus load in murine plasma. J. Virol. Methods. 2008;147:245–350. doi: 10.1016/j.jviromet.2007.10.009. [DOI] [PubMed] [Google Scholar]
- Jones PH, Mehta HV, Okeoma CM. A novel role for APOBEC3: susceptibility to sexual transmission of murine acquied immunodeficiency virus (mAIDS) is aggravated in APOBEC3 deficient mice. Retrovirology. 2012;9:50. doi: 10.1186/1742-4690-9-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakuta S, Shibata S, Iwakura Y. Genomic structure of the mouse 2',5'-oligoadenylate synthetase gene family. J. Interferon Cytokine Res. 2002;22:981–993. doi: 10.1089/10799900260286696. [DOI] [PubMed] [Google Scholar]
- Konuma T, Nakamura S, Miyagi S, Negishi M, Chiba T, Oguro H, Yuan J, Mochizuki-Kashio M, Ichikawa H, Miyoshi H, Vidal M, Iwama A. Forced expression of the histone demethylase Fbxl10 maintains self-renewing hematopoietic stem cells. Exp. Hematol. 2011;39:697–709. doi: 10.1016/j.exphem.2011.03.008. [DOI] [PubMed] [Google Scholar]
- Kolokithas A, Rosenke K, Malik F, Hendrick D, Swanson L, Santiago ML, Portis JL, Hasenkrug KJ, Evans LH. The glycosylated Gag protein of a murine leukemia virus inhibits the antiretroviral function of APOBEC3. J. Virol. 2010;84:10933–10936. doi: 10.1128/JVI.01023-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota K, Nakahara K, Ohtsuka T, Yoshida S, Kawaguchi J, Fujita Y, Ozeki Y, Hara A, Yoshimura C, Furukawa H, Haruyama H, Ichikawa K, Yamashita M, Matsuoka T, Iijima Y. Identification of 2’-phophodiesterase, which plays a role in the 2-5A system regulated by interferon. J. Biol. Chem. 2004;279:37832–37841. doi: 10.1074/jbc.M400089200. [DOI] [PubMed] [Google Scholar]
- Lengyel P, Samanta H, Pichon J, Dougherty J, Slattery E, Farrell P. Double-stranded RNA and the enzymology of interferon action. Ann. N. Y. Acad. Sci. 1980;350:441–447. doi: 10.1111/j.1749-6632.1980.tb20647.x. [DOI] [PubMed] [Google Scholar]
- Liu SY, Sanchez DJ, Aliyari R, Lu S, Cheng G. Systematic identification of type I and type II interferon-induced antiviral factors. Proc. Natl. Acad. Sci. U. S. A. 2012;109:4239–4244. doi: 10.1073/pnas.1114981109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low A, Okeoma CM, Lovsin N, de las Heras M, Taylor TH, Peterlin BM, Ross SR, Fan H. Enhanced replication and pathogenesis of Moloney murine leukemia virus in mice defective in the murine APOBEC3 gene. Virology. 2009;385:455–463. doi: 10.1016/j.virol.2008.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maitra RK, Silverman RH. Regulation of human immunodeficiency virus replication by 2',5'-oligoadenylate-dependent RNase L. J. Virol. 1998;72:1146–1152. doi: 10.1128/jvi.72.2.1146-1152.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malathi K, Dong B, Gale MJr, Silverman RH. Small self-RNA generated by RNase L amplifies antiviral innate immunity. Nature. 2007;448:816–819. doi: 10.1038/nature06042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malathi K, Saito T, Crochet N, Barton DJ, Gale MJr, Silverman RH. RNase L releases a small RNA from HCV RNA that refolds into a potent PAMP. RNA. 2010;16:2108–2119. doi: 10.1261/rna.2244210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malim MH, Bieniasz PD. HIV restriction factors and mechanisms of evasion. Cold Spring Harb. Perspect. Med. 2012;2:a006940. doi: 10.1101/cshperspect.a006940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques R, Antunes I, Eksmond U, Stoye J, Hasenkrug KJ, Kassiotis G. Lymphocyte activation by coinfection prevents immune control of Friend virus infection. J. Immunol. 2008;181:3432–3440. doi: 10.4049/jimmunol.181.5.3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair S, Bayer W, Ploquin MJ, Kassiotis G, Hasenkrug KJ, Dittmer U. Distinct roles of CD4+ T cell subpopulations in retroviral immunity: lessons from the Friend virus mouse model. Retrovirology. 2011;8:76. doi: 10.1186/1742-4690-8-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okeoma CM, Lovsin N, Peterlin BM, Ross SR. APOBEC3 inhibits mouse mammary tumuor virus replication in vivo. Nature. 2008;445:927–930. doi: 10.1038/nature05540. [DOI] [PubMed] [Google Scholar]
- Player MR, Torrence PF. The 2-5A system: modulation of viral and cellular processes through acceleration of RNA degradation. Pharmacol. Ther. 1998;78:55–113. doi: 10.1016/S0163-7258(97)00167-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portis JL, McAtee FJ, Kayman SC. Infectivity of retroviral DNA in vivo. J. AIDS. 1992;5:1272–1273. doi: 10.1097/00126334-199212000-00011. [DOI] [PubMed] [Google Scholar]
- Robertson MN, Miyazawa M, Mori S, Caughey B, Evans LH, Hayes SF, Chesebro B. Production of monoclonal antibodies reactive with a denatured form of the Friend murine leukemia virus gp70 envelope protein: use in a focal infectivity assay, immunohistochemical studies, electron microscopy and western blotting. J. Virol. Methods. 1991;34:255–271. doi: 10.1016/0166-0934(91)90105-9. [DOI] [PubMed] [Google Scholar]
- Robertson SJ, Ammann CG, Messer RJ, Carmody AB, Myers L, Dittmer U, Nair S, Gerlach N, Evans LH, Cafruny WA, Hasenkrug KJ. Suppression of acute anti-Friend virus CD8+ T cell responses by coinfection with lactate-dehydrogenase elevating virus. J. Virol. 2008;82:408–418. doi: 10.1128/JVI.01413-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel MA, Whitby K, Keller BC, Marri A, Barchet W, Williams BR, Silverman RH, Gale MJr, Diamond MS. PKR and RNase L contribute to protection against lethal West Nile Virus infection by controlling early viral spread in the periphery and replication in neurons. J. Virol. 2006;80:7009–7019. doi: 10.1128/JVI.00489-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago ML, Montano M, Benitez R, Messer RJ, Yonemoto W, Chesebro B, Hasenkrug KJ, Greene WC. Apobec3 encodes Rfv3, a gene influencing neutralizing antibody control of retrovirus infection. Science. 2008;321:1343–1346. doi: 10.1126/science.1161121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago ML, Benitez RL, Montano M, Hasenkrug KJ, Greene WC. Innate retroviral restriction by Apobec3 promotes antibody affinity maturation in vivo. J. Immunol. 2010;185:1114–1123. doi: 10.4049/jimmunol.1001143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago ML, Smith DS, Barrett BS, Montano M, Benitez RL, Pelanda R, Hasenkrug KJ, Greene WC. Persistent Friend virus replication and disease in Apobec3-deficient mice expressing functional B-cell-activating factor receptor. J Virol. 2011;85:189–199. doi: 10.1128/JVI.01838-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, Rice CM. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder HC, Wenger R, Kuchino Y, Muller WE. Modulation of nuclear matrix-associated 2',5'-oligoadenylate metabolism and Ribonuclease L activity in H9 cells by human immunodeficiency virus. J. Biol. Chem. 1989;264:5669–5673. [PubMed] [Google Scholar]
- Silverman RH, Skehel JJ, James TC, Wreschner DH, Kerr IM. rRNA cleavage as an index of ppp(A2'p)nA activity in interferon-treated encephalomyocarditis virus-infected cells. J. Virol. 1983;46:1051–1055. doi: 10.1128/jvi.46.3.1051-1055.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DS, Guo K, Barrett BS, Heilman KJ, Evans LH, Hasenkrug KJ, Greene WC, Santiago ML. Noninfectious retrovirus particles drive the Apobec3/Rfv3 dependent neutralizing antibody response. PLOS Pathog. 2011;7:e1002284. doi: 10.1371/journal.ppat.1002284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steeves RA, Eckner RJ, Mirand EA, Priore RL. Rapid assay of murine leukemia virus helper activity for Friend spleen focus-forming virus. J. Natl. Cancer Inst. 1971;46:1219–1228. [PubMed] [Google Scholar]
- Takeda E, Tsuji-Kawahara S, Sakamoto M, Langlois MA, Neuberger MS, Rada C, Miyazawa M. Mouse APOBEC3 restricts Friend leukemia virus infection and pathogenesis in vivo. J Virol. 2008;82:10998–11008. doi: 10.1128/JVI.01311-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji-Kawahara S, Chikaishi T, Takeda E, Kato M, Kinoshita S, Kajiwara E, Takamura S, Miyazawa M. Persistence of viremia and production of neutralizing antibodies differentially regulated by polymorphic APOBEC3 and BAFF-R loci in Friend virus-infected mice. J. Virol. 2010;84:6082–6095. doi: 10.1128/JVI.02516-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts JM, Dang KK, Gorelick RJ, Leonard CW, Bess JW, Jr, Swanstrom R, Burch CL, Weeks KM. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature. 2009;460:711–716. doi: 10.1038/nature08237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wreschner DH, McCauley JW, Skehel JJ, Kerr IM. Interferon action--sequence specificity of the ppp(A2'p)nA-dependent ribonuclease. Nature. 1981;289:414–417. doi: 10.1038/289414a0. [DOI] [PubMed] [Google Scholar]
- Zhou A, Hassel BA, Silverman RH. Expression cloning of 2-5A-dependent RNAase: a uniquely regulated mediator of interferon action. Cell. 1993;72:753–765. doi: 10.1016/0092-8674(93)90403-d. [DOI] [PubMed] [Google Scholar]
- Zhou A, Paranjape J, Brown TL, Nie H, Naik S, Dong B, Chang A, Trapp B, Fairchild R, Colmenares C, Silverman RH. Interferon action and apoptosis are defective in mice devoid of 2',5'-oligoadenylate-dependent RNase L. EMBO J. 1997;16:6355–6363. doi: 10.1093/emboj/16.21.6355. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.