Abstract
The identification and quantitative evaluation of lung tumors in mouse models is challenging and an unmet need in preclinical arena. In this study, we developed a noninvasive contrast-enhanced microCT (μCT) method to longitudinally evaluate and quantitate lung tumors in mice. Commercially available μCT contrast agents were compared to determine the optimal agent for visualization of thoracic blood vessels and lung tumors in naïve mice and in non-small-cell lung cancer models. Compared with the saline control, iopamidol and iodinated lipid agents provided only marginal increases in contrast resolution. The inorganic nanoparticulate agent provided the best contrast and visualization of thoracic vascular structures; the density contrast was highest at 15 min after injection and was stable for more than 4 h. Differential contrast of the tumors, vascular structures, and thoracic air space by the nanoparticulate agent enabled identification of tumor margins and accurate quantification. μCT data correlated closely with traditional histologic measurements (Pearson correlation coefficient, 0.995). Treatment of ELM4–ALK mice with crizotinib yielded 65% reduction in tumor size and thus demonstrated the utility of quantitative μCT in longitudinal preclinical trials. Overall and among the 3 agents we tested, the inorganic nanoparticulate product was the best commercially available contrast agent for visualization of thoracic blood vessels and lung tumors. Contrast-enhanced μCT imaging is an excellent noninvasive method for longitudinal evaluation during preclinical lung tumor studies.
Abbreviations: μCT, microCT; HU, Hounsfield units; RECIST, response evaluation criteria in solid tumors
Lung cancer is the leading cause of cancer death worldwide, and non-small–cell lung cancer is the most common form of lung cancer that is diagnosed.19 Various animal models have been used to mimic these cancers, understand their biology, and evaluate potential therapeutics.14,20,27 Traditionally, the method to study the disease in vivo has been to inject human tumor derived cell lines subcutaneously in immunodeficient mice (xenografts) or to orthotopically implant tumors in tissues of interest. Xenografts, despite being of human origin, are not ideal models because tumor cell–stroma interactions cannot be reconstituted in the system. Moreover, the tumor microenvironment has been shown to be essential in predicting cancer cell survival, progression, metastasis, and response to therapy.14,20,27 To overcome these issues, several investigators have propagated tumors orthotopically by either direct injection of tumor cells into the organ of choice, intravenous injection of tumor cell lines, or implantation of patient-derived tumor biopsy samples into immunodeficient mice. These models simulate the tumor microenvironment and bear a closer resemblance to clinical cancer than do xenografts.11,27 In the past decade, tremendous progress has been made in development of genetically engineered mouse models that reconstitute facets of human disease in the organ of choice. In these models, the tumors are developed in immunocompetent hosts with intact tumor–stroma interactions, and the tumors can be controlled temporally and spatially.7,14,25,26
Despite the many advantages of genetically engineered and orthotopic models, their use has been limited, primarily due to the heterogeneity and technical difficulties associated with monitoring disease progression.17 Conventional optical imaging is not widely used with genetically engineered and orthotopic models, because these modalities provide low spatial resolution, limited tissue penetration,1,17 and rarely accomdate reporter genes like luciferase or fluorescent proteins.30 Nuclear imaging (for example, positron-emission tomography and MRI) has been used in evaluating lung tumor models,7,13,33 but these modalities are not readily available in all facilities, require specialized laboratories (for example, radionucleotide synthesis), and do not achieve accurate quantification of nodular tumors.33 X-ray CT has been used in human clinical practice to assess lung tumor nodules to predict likelihood of malignancy and to monitor the response of tumors to treatment.18,23 Response Evaluation Criteria in Solid Tumors (RECIST) scores based on CT imaging data are used routinely in clinical trials and practice.32 Similarly, high-resolution microCT (μCT) scanners have been used successfully to image lung tumors6,15,17,24,25,34 in small animals such as rodents. The investigators in the aforementioned studies took advantage of the natural air–tissue contrast within the thorax to identify tumors. However, this method was unable to distinguish tumor and soft tissue from nearby vascular structures.25,29 This limitation decreased the accuracy of tumor margin demarcation and tumor volumetric measurements.
The goal of our study was to compare 3 commercially available μCT contrast agents (products containing iopamidol, iodinated lipid, and inorganic nanoparticulate) to evaluate and accurately quantify lung tumors in preclinical models. Our results showed that, among those we tested, the nanoparticulate product was the best contrast agent for tumor visualization and quantitation. In addition, we demonstrated the utility of contrast-enhanced μCT in a preclinical evaluation of the efficacy of crizotinib (PF-2341066), a small-molecule multikinase inhibitor,9,10 in a genetically modified mouse model of lung cancer.
Materials and Methods
Contrast agents.
We used the commercially available CT contrast agents Fenestra VC (iodinated lipid; Advanced Research Technologies, Montreal, Quebec, Canada), Isovue-370 (iopamidol; Bracco Diagnostics, Princeton, NJ), and Viscover ExiTron Nano 12,000 (inorganic nanoparticulate; Miltenyi Biotec, Bergisch-Gladbach, Germany) in the current study. Fenestra VC consists of contrast-enhancing iodinated lipids (oil-in-water emulsion of glyceryl-2-oleoyl-1,3-di-7-[3-amino-2,4,6-triiodophenyl]-heptanoate in a vehicle containing methoxy polyethylene glycol 1-2-distearyl phosphatidylethanolamine) that are designed to enable visualization of blood vasculature over extended periods. The Fenestra VC formulation has an iodine concentration of 50 mg/mL.8 Each mL of Isovue-370 provides 755 mg iopamidol with 1 mg tromethamine and 0.48 mg edentate calcium disodium. This agent provides a dose of 370 mg iodine per milliliter. ExiTron Nano 12,000 is an inorganic nanoparticle-based blood-pooling contrast agent. The contrast density of 100 µL of ExiTron Nano 12,000 per mouse (25 g) is equivalent to 12,000 Hounsfield units (HU), which corresponds to a dose equivalent of 1200 mg iodine per kilogram of body weight. In our studies, we used 200 µL each of the iodinated lipid and iopamidol products, whereas only 100 µL of the nanoparticulate agent was injected for each imaging session. Volumes were limited to maximum of 200 µL to meet the general animal welfare standards and institutional guidelines.
Viral vector production.
Adenovirus.
Adenovirus expressing Cre recombinase was purchased from the University of Iowa Gene Transfer Vector Core. Viral titers were determined by using Adenoviral Titration Kits (Clontech, CA) according to the manufacturer's protocol. The modified adenovirus was prepared in media (Modified Eagle Medium, Gibco BRL; Life Sciences, CA) containing 10 mM CaCl2 followed by incubation at room temperature for 20 min.
EML4–ALK lentivirus.
A replication-incompetent HIV-based lentivirus was engineered to express the human EML4–ALK fusion oncogene31 driven by the cytomegalovirus promoter. The LTR-PACK system (Biosettia, San Diego, CA) and Lipofectamine 2000 transfection system (Invitrogen, Grand Island, NY) were used to generate virus. At 48 h after transfection, viral supernatant was filtered (Steriflip tubes, Millipore, Billerica, MA). Viral titers were approximated by generating a Cre-expressing lentivirus in the same vector at the same time and functionally testing Cre activity by transducing a reporter cell line engineered with a lox-stop-lox green-fluorescent protein.
Ethics statement and animal husbandry.
All animal experimentation followed protocols approved by the Pfizer IACUC. Mice were housed under a standard 12:12-h light:dark cycle in ventilated racks at a room temperature of 72 °F (22.1 °C) and relative humidity of 30% to 70%.
Animal models.
C57BL/6 mice were obtained from Charles River Labs (San Diego, CA). KrasG12D-LSL/+ (Kras) and KrasG12D-LSL/+ p53fl/fl (Kras–p53) mice were obtained from Jackson Laboratories (Jax West, Sacramento, CA) at 3 to 4 wk of age. Lung tumors in Kras and Kras–p53 mice were generated by intranasal inoculation of the Cre-containing adenovirus (2.5 × 107 infectious units) under ketamine–xylazine anesthesia as described previously.12 EML4–ALK mice were generated by intranasal inoculation of C57BL/6 mice with lentivirus (2.5 × 105 infectious units) under ketamine–xylazine anesthesia. The number of mice used for each experiment is given in the figure legends.
Animal experimentation
μCT imaging.
Contrast agents were injected slowly via the tail vein by using a disposable syringe fitted with a 27-gauge luer slip-type needle. Mice were anesthetized with isoflurane (3% to 4% for induction; 1% to 2% for maintenance) by using a COMPAC5 anesthesia machine (VetEquip, Pleasanton, CA) and then transferred to the μCT imaging chamber. The thorax of each mouse (which included lungs and heart) was imaged (VivaCT-75, Scanco Medical, Brüttisellen, Switzerland). For determining the time-course of contrast enhancement, baseline (0 min) images were acquired from naïve C57BL/6 mice. These mice were injected with different contrast agents and imaged at 15 min, 1 h, 2 h, 4 h, and 24 h after administration. For all other studies, images were acquired at 15 min after injection of saline (control) or contrast agent. Contrast agents were compared in the lung tumors of the Kras–p53 model at 16 wk after the inhalation of adenovirus expressing Cre recombinase. This genetically engineered mouse model has been well characterized and develops lung adenocarcinomas in the lungs by 16 wk after inhalation of virus.21,22 In our experience, mice bearing tumors at this stage are very sensitive to handling and do not survive multiple manipulations; we therefore evaluated 2 cohorts of mice (saline compared with iopamidol compared with the nanoparticulate agent and saline compared with the iodinated lipid and nanoparticulate agents) and compared the imaging agents in 2 phases. After baseline scans, mice were injected with saline followed by contrast agent (iopamidol or iodinated lipid) and scanned. After a washout period of 7 d, mice were injected with the nanoparticulate agent and images acquired.
Images were acquired at standard resolution by using the following parameters: 70 kVp; 57 to 114 µA; integration time, 200 to 300 ms; and voxel size, 41 µm. To curtail the effects of animal movement due to respiration, imaging was performed with respiratory gating according to the manufacturer's (Scanco Medical) protocol. A small plastic pressure pad was positioned under the animal's sternum and connected to a pressure transducer. The rotation of the scanner ceased temporarily whenever a respiratory event crossed the user-defined threshold for gating. The scanning protocol was programmed to acquire images via a rotating gantry, resulting in a total of 500 projections per scan. The projections were reconstructed with a matrix of 1024 × 1024 × 1024 by using the software provided by the manufacturer (Scanco Medical). During each imaging session, the mice in our study each received a radiation dose of approximately 0.07 mGy, which is lower than the doses reported to be safe in tumor models.4,28 The dose received during our longitudinal µCT imaging did not have any effect on the health of the mice or tumor size.
Preclinical trial.
The therapeutic efficacy of the multikinase inhibitor crizotinib was evaluated in ELM4–ALK mice. This model has been well characterized in our labs (data not shown), and mice develop tumor nodules by 26 wk after inoculation. Mice were enrolled in the study when tumor nodules reached 1.5 to 2.5 mm in diameter. Mice (n = 3) carrying multiple tumor nodules were treated with crizotinib (100 mg/kg daily) for 7 d; animals were injected with the nanoparticulate agent on days 0 and 7 after treatment and were imaged at 15 min after injection of the contrast agent.
Radiodensity measurements and image analysis.
The 2D binary digital imaging communication files obtained by using a µCT evaluation program (version 6.5-1, Scanco Medical) were imported into a 64-bit (OsiriX version 3.9.2, Pixmeo, Geneva, Switzerland) for further analysis. Contrast enhancement (density) was measured by designating regions of interest in axial slices of the left heart ventricle, as suggested previously.3 Contrast enhancement at different time points was expressed in Hounsfield units and compared with baseline values; data were reported as mean ± SEM. Volumetric tumor burden of nodular tumors was quantified by using a 3D region-growing algorithm after 2D threshold segmentation of tumors by using seed points or morphing of manually contoured tumors. The region-growing technique in OsiriX allowed us to extract a connected region of an image according to user-defined criteria, such as contrast intensity. Seed points were planted within the identified tumor. Repositioning of seed points was often necessary because establishing the threshold was sensitive to noise and intensity differences. Morphing of segmented tumors was accomplished by drawing multiple contours of the tumors on different slices and using a morphing algorithm to generate the missing contours across the series to ascertain tumor volume. Once the morphing was done, we individually checked each slice to ensure that no adjoining anatomic regions were included in the tumor during the automated process. This quality-control step allowed us to obtain accurate tumor volume measurements for nodular tumors.
Histology.
EML4–ALK mice were euthanized, and the lungs and the trachea were excised en bloc. Photographs were taken to identify the surface lung tumors (data not shown). Briefly, lungs were gently inflated through the tracheal lumen by using 10% neutral buffered formalin, after which the entire lung block was immersed in 10% neutral buffered formalin. After fixation for 16 h, the lungs were placed into 70% ethanol prior to paraffin embedding. Longitudinal sections (4 μm) of the entire lung were obtained every 100 μm. Sections were deparaffinized and stained with hematoxylin and eosin. Each section was evaluated by a pathologist to identify lesions and to differentiate lung tumors from other space-occupying alterations (for example, hemorrhage, consolidation, inflammation). Each slide was digitally imaged (model C9600-02, Nanozoomer Digital Pathology Slide Scanner, Hamamatsu Photonics, Hamamatsu City, Japan) and a cross-section of each tumor was measured for area. Coronal μCT slices (representing the same plane of histology section) at 82-μm increments were compared the serial longitudinal histology sections.
Results
Characterization of contrast enhancement in the thorax.
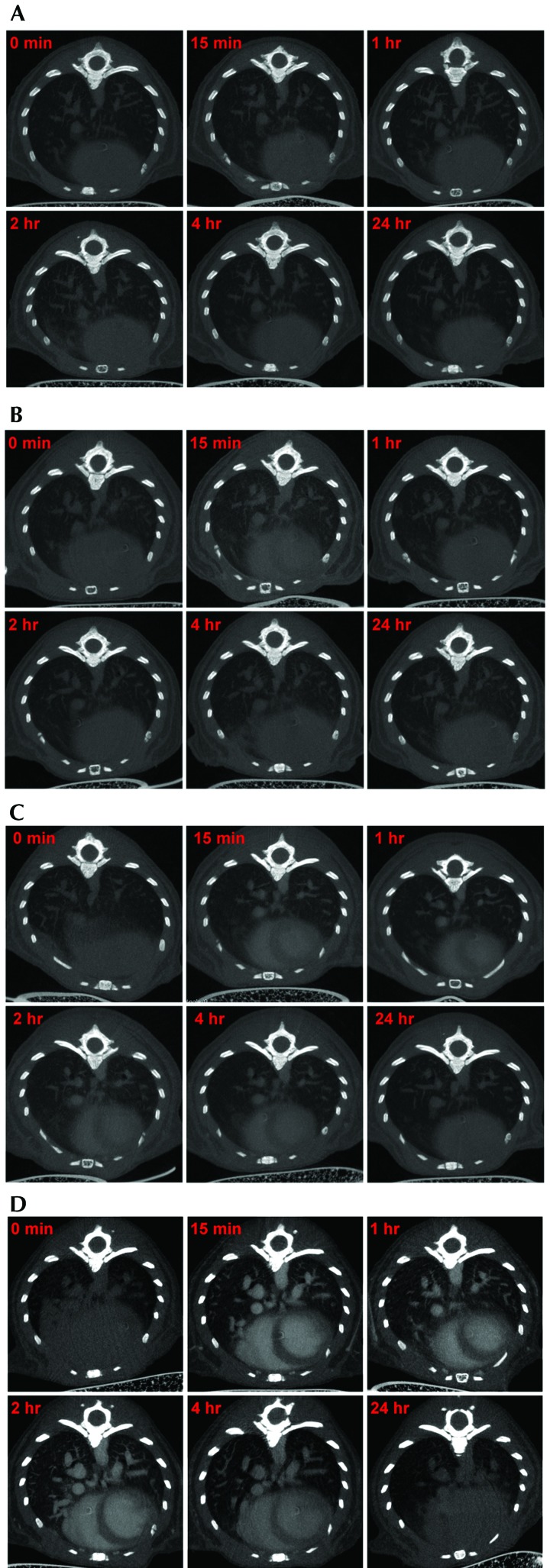
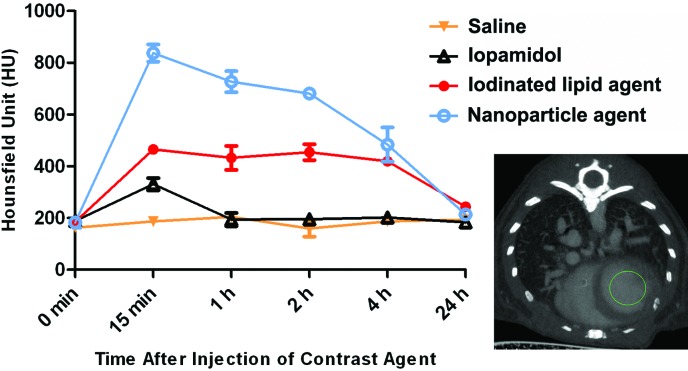
All mice tolerated injection of the various contrast agents and showed no adverse effects. The 2D images (axial view) of the thorax from representative mice in each contrast agent group at the various time points are shown in Figure 1. In saline-injected mice, the air spaces in the lungs, major blood vessels, and cardiac outline were all clearly visible, and no changes in the contrast density was noted at any of the time points evaluated (Figure 1 A). The iopamidol agent provided only a marginal increase in the quality of images compared with that of the saline control (Figure 1 B). The chambers of the heart were barely distinguishable at 15 min after iopamidol injection. The iodinated lipid product exhibited a greater ability to distinguish the chambers of the heart and blood vessels than did iopamidol (Figure 1 C). The nanoparticulate agent provided the best contrast for distinguishing the chambers of heart and anatomic structures of the blood vasculature in thorax (Figure 1 D). The contrast density in the left ventricle at 15 min after the injection of saline was approximately 186.7 HU and was similar to that of baseline (0 min) scans (Figure 2). Injection of the iodinated contrast agent iopamidol improved the density marginally (approximately 330 HU at 15 min after injection), but the density decreased to baseline by 1 h after injection (Figure 2). The iodinated lipid agent increased the density contrast by 2.5-fold (approximately 465 HU, 15 min after injection) relative to baseline, and the signal was stable for as long as 4 h (Figure 2). The best contrast enhancement was provided by the nanoparticulate agent, which yielded a peak of approximately 837 HU at 15 min after injection (Figure 2); the contrast intensity signal in the left ventricle reduced gradually and was similar to that of iodinated lipid by 4 h. All contrast agents were cleared from the heart and lungs by 24 h after injection. These data established 15 min after injection as the optimal time point for imaging thoracic structures for all contrast agents used in the study. All subsequent scans for the comparison of the contrast agents in lung tumor models were performed at 15 min after injection.
Figure 1.
Contrast enhancement of thorax by various agents; 2D axial images of the thorax after administration of different agents are shown. Preinjection control is shown at 0 min and compared with the postinjection time points of 15 min, 1 h, 2 h, 4 h, and 24 h. (A) Kinetics of saline injection: no significant enhancement of contrast was observed between control (0 min) and the saline-injected images at different time points. (B) Kinetics of iopamidol injection: the iopamidol-based agent gave a marginal increase in the overall contrast and the chambers of heart were hardly distinguishable at 15 min postinjection. Images obtained beyond 1 h postinjection were similar to the control (0 min). (C) Kinetics of iodinated lipid injection: the iodinated lipid product yielded marked contrast enhancement relative to saline and iopamidol, and the chambers of heart were distinguishable for as long as 4 h after injection. (D) Kinetics of nanoparticulate injection: the nanoparticle-based agent gave the best contrast enhancement among the contrast agents tested. The chambers of heart and the blood vessels were easily distinguishable until 4 h after injection. The images are representative of data from 3 or 4 mice per group for all figures.
Figure 2.
Kinetics of the contrast enhancement from different agents. Density contrast (in Hounsfield units) of the left ventricle by the agents was measured at different time points postinjection. Saline afforded no contrast enhancement, whereas contrast intensity due to the 3 commercially available agents peaked at 15 min postinjection. All contrast agents were cleared from the thorax by 24 h after injection. The inset μCT image shows the area (left ventricle in green circle) used to measure the contrast density. Error bars represent the SEM of means from 3 or 4 mice.
Effect of contrast enhancement on tumor visualization.
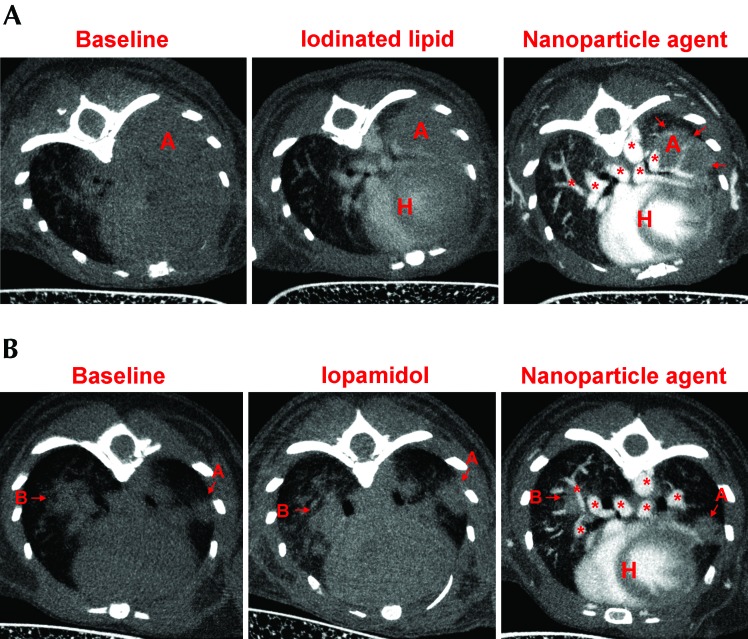
We next compared the ability of each of the agents to provide contrast enhancement in the lung tumors of Kras–p53 mice (Figure 3 A and B). A single tumor (labeled A) in the left thorax at approximately the same anatomic location was monitored for each of the contrast agents (Figure 3 A). In the saline-injected mouse, a large mass contiguous with the heart occupied the left thorax; the tumor margins and blood vessels of this mass could not be identified. A region of interest was drawn manually that excluded the heart and yielded a tumor volume of approximately 165 mm3. In the same mouse, injection of iodinated lipid slightly improved visualization of major blood vessels, the cardiac outline, and tumor margins, resulting in a smaller region of interest and a measured tumor volume of approximately 125 mm3. When the same mouse was imaged by using the nanoparticulate product, the tumor margins were distinct and resulted in a still smaller tumor volume of approximately 27.5 mm3. The improvement in defining the tumor margins was possible due to the reduced contrast intensity of tumor relative to the surrounding blood vessels and heart but increased contrast intensity of tumor relative to air spaces in the lungs. Using the nanoparticulate agent revealed the tumor to have 2 well-defined nodular areas separated by a blood vessel that appeared to run longitudinally within the tumor (Figure 3 A). In the second example (Figure 3 B), we have depicted the comparison between saline, iopamidol, and the nanoparticulate agent by monitoring the progress of 2 different tumors (A, left lung; B, right lung; Figure 3 B). Again with saline infusion, tumor margins could not be clearly identified. Iopamidol increased the radiodensity of the vascular structures but without clear demarcation of the tumor margins, thus preventing accurate measurement of tumor volume. In comparison, the nanoparticulate product clearly demarcated the tumor margins, making them easily distinguishable from the surrounding vasculature and heart. As determined by using the nanoparticulate agent, tumor volumes were 4.24 mm3 for tumor A and 4.22 mm3 for tumor B (Figure 3 B).
Figure 3.
Comparison of contrast agents in the Kras–p53 mouse model. Saline, iopamidol, and iodinated lipid agents were compared with an inorganic nanoparticulate product in regard to contrast enhancement of lung tumors in Kras–p53 mice (n = 3). (A) The iodinated lipid contrast agent faintly distinguished between the heart (H), major blood vessels (asterisks), and tumor (arrows) but was unable to accurately outline the tumor nodule. The nanoparticle-based agent highlighted the chambers of heart and major and minor blood vessels and clearly demarcated the tumor nodule from heart and blood vessels (tumor nodule A). (B) Saline and iopamidol failed to differentiate between blood vessels and tumor nodules, whereas the nanoparticule-based product readily differentiated the heart, blood vessels, and tumor nodules (nodules A and B).
Comparison of contrast-enhanced tumor visualization in different models of lung cancer.
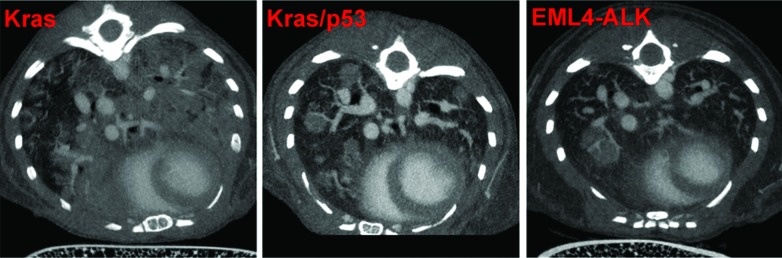
Once we determined that the nanoparticulate product was the best contrast agent for visualizing lung tumors in mice, we performed imaging studies in other lung cancer models to evaluate whether this methodology was applicable across different tumor phenotypes (that is, nodular compared with diffuse). For this purpose, we used the lentivirus-induced EML4–ALK model and the adenovirus-induced Kras (mixed tumor) and Kras–p53 (nodular) models. Data from representative images are shown in Figure 4. Tumors induced by Kras–p53 and EML4–ALK mutations were of the nodular type, with clearly demarcated borders. Quantitation of tumor volumes in these models was straightforward, in that we could contour the individual tumors and use a region-growing algorithm to propagate the region of interest (that is, tumor margins) throughout multiple slices to obtain a 3D volume. We also obtained the largest diameter of each tumor as a measure of tumor size.
Figure 4.
Identification of tumor phenotypes in various lung cancer models by using the nanoparticulate contrast agent. Kras–p53 (n = 5) and EML4–ALK (n = 3) mice had only nodular tumors, whereas Kras mice (n = 5) had both diffuse and nodular tumors (mixed phenotype).
Tumors induced by adenovirus-induced Kras expression were diffuse in nature. The nanoparticulate contrast agent helped in the visualization of blood vessels as expected but failed to clearly define tumors margins. As a result, the seeding and region-growing techniques could not be used to measure tumor volume. To overcome this challenge, we created a mask outlining the bilateral lungs and propagated it across the entire series of the scan. We then planted seed points in the aerated part of the lungs and used a 3D region-growing algorithm to segment the healthy lung tissue. Finally, multifocal tumors were ascertained by subtracting the volume of aerated lung from the overall lung volume as determined from the mask to determine overall tumor burden.
Comparison of μCT with histopathology.
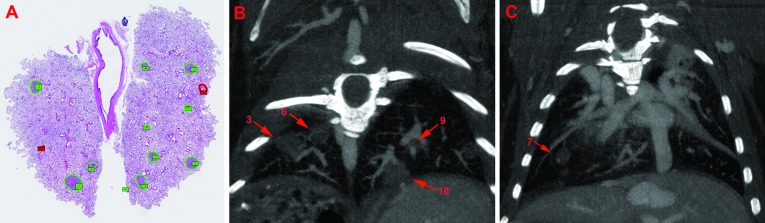
Tumor measurement and volume estimation in genetically modified mouse models of non-small–cell lung cancer typically are done through histologic analysis of serial sections of the lung. We wanted to compare our contrast-enhanced μCT method with histologic analysis to assess the accuracy of μCT-based tumor-burden estimation. We ascertained tumor diameter and 2D tumor size in each slice or section by using both methods (Figure 5 A through C). Our μCT scans successfully identified the same total number of tumors that was identified through histology (9 in right lung and 4 in left lung). The μCT quantitation of each tumor was generally higher than that obtained through histology. Table 1 compares the 2D tumor measurements of 3 tumor nodules, which yielded a Pearson correlation coefficient (r) of 0.9952 between histology and μCT. The smallest diameter of a single tumor that was determined through histology was 232 μm whereas that obtained through μCT of the same tumor was 325 μm. These data indicate that contrast-based μCT imaging is a fairly accurate method for identifying and quantifying lung tumors in mice.
Figure 5.
Comparison of μCT and histology methodology in EML4–ALK mice. Coronal views of histologic sections and μCT images are represented here. (A) Standard histology identified 13 possible tumor nodules (green) in the represented lung image. (B and C) Representative coronal μCT images in different planes from the same lung. μCT detected all of the tumor nodules (arrows) that were determined by histology
Table 1.
Comparison of tumor measurements (maximal area, mm2) obtained by μCT and histology
| Tumor nodule | Histology | μCT |
| 9 | 0.999 | 1.240 |
| 10 | 1.205 | 1.591 |
| 11 | 0.999 | 1.198 |
Pearson coefficient (r) between histology and μCT was –0.9952.
Monitoring therapeutic efficacy by using μCT imaging.
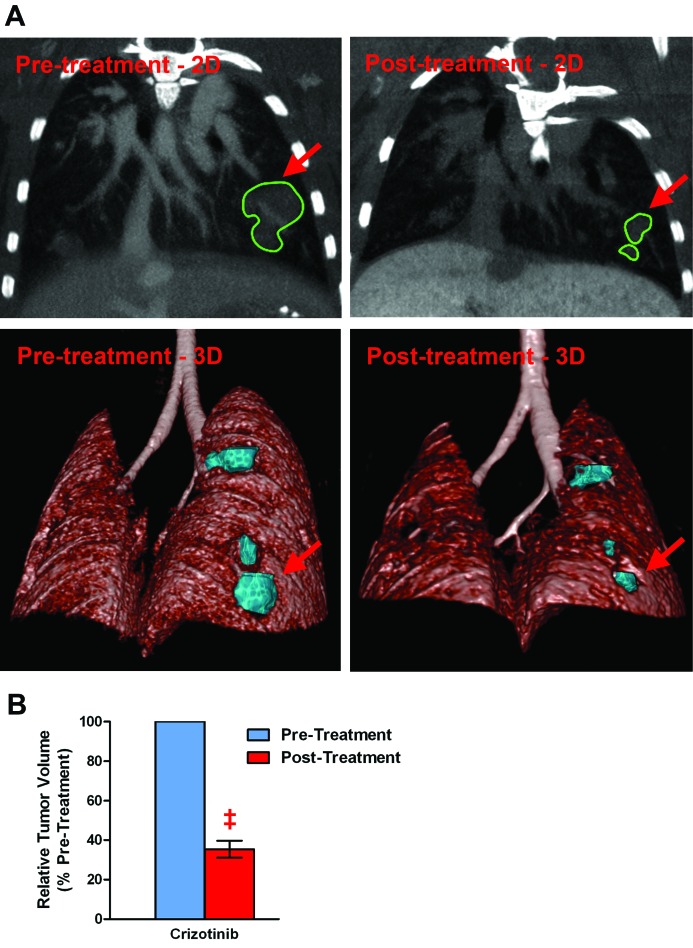
To demonstrate the utility of the above discussed μCT methodology, we decided to evaluate therapeutic efficacy of crizotinib in a mouse model of non-small–cell lung cancer that is induced by the EML4–ALK oncogene. Crizotinib therapy reduced the levels of the ALK and phosphoALK proteins (data not shown) and markedly decreased the size of the tumors (Figure 6 A). Volumetric quantitation of the tumor nodules revealed that crizotinib therapy reduced the tumor volume to 35.4% ± 4.3% of the pretreatment size (Figure 6 B).
Figure 6.
Monitoring therapeutic efficacy in a mouse lung-cancer model by using μCT imaging. EML4–ALK mice treated with crizotinib (100 mg/kg daily) for 7 d demonstrated marked reduction in tumor size according to μCT imaging. (A) Pre- and posttreatment μCT images (2D and 3D views) are shown; arrows indicate the tumor nodules depicted in both the 2D and 3D views. (B) Relative comparison (before and after treatment) of tumor regression due to crizotinib therapy (n = 7 tumor nodules from 3 mice); ‡, P < 0.001.
Discussion
X-ray CT provides good spatial resolution of anatomic structures in 3D format. The image data obtained through this modality is based on the differential absorption of the X-rays by the tissues and thereby the density of the tissues examined. In vivo tissues that are most amenable to imaging by this method are bone and lung, because of their innate differences in density relative to other tissue types. The air spaces and soft-tissue structures of thoracic blood vessels and lung provide natural differences in density that are amenable to CT imaging, which has been used extensively in human patients for the diagnosis of thoracic diseases, including lung tumors.5,18,29
Lung tumors in patients are clinically assessed through CT measurement to assess changes in size over time to predict malignancy and response to therapy.18 According to the maximal diameter of tumor nodules on serial scans, tumor response in patients is classified into complete response, partial response, stable disease, and disease progression, according to RECIST scores.32 To evaluate tumors according to RECIST scores, size measurements of the tumor nodules need to be accurate and consistent to enable the assessment of change over time.16 Although lung tumors in rodents have been evaluated by using CT imaging, methods for classifying or estimating tumor responses to therapy have not been developed. μCT detection and monitoring of lung tumors in mice have been described for orthotopic tumors and the Kras, Kras–p53, and Kras–Lkb1 mouse mutant models.7,15,25 These studies described methods that used the natural contrast of air, lung, and heart tissue to visualize the tumors. A distinct disadvantage of these studies was the inability to distinguish between vascular and tumor tissue.7,15 To reduce radiation exposure, we performed our studies at a resolution of 41 μm, which corresponded to a well-tolerated radiation dose of 0.07mGy for each imaging session. At this resolution, unless the tumor nodule was distinct and well separated from blood vessels or the adjoining heart, the margins of the tumors were very difficult to discriminate clearly (Figure 3, baseline images). Measuring tumor volumes therefore required the best judgment of the investigator to outline tumor boundaries. Another consequence of the 41-μm resolution was that smaller tumors (less than 500 μm) were often difficult to discern. This kind of approximate analysis does not provide the rigor needed to develop a scoring system for tumor evaluation, such as the RECIST guidelines used in human patients.
Contrast-enhanced μCT could help overcome these technical challenges. A contrast-enhanced μCT method that used a proprietary liposomal–iodinated contrast agent to evaluate lung tumors was reported recently.1 In comparison, we here show the utility of commercially available contrast agents including an iodinated contrast agent (iopamidol) and blood-pool contrast agents (iodinated lipid and nanoparticle-based products). All 3 contrast agents evaluated differed markedly in their iodine concentrations, which is the component that is responsible for the tissue contrast. At the concentrations used, the nanoparticulate contrast agent was the product that provided the greatest contrast enhancement for blood vessels (Figures 1 and 2) and clearest demarcation of tumor margins (Figure 3). Another advantage of this nanoparticulate material is that only 100 μL was needed to obtain sufficiently high contrast enhancement to distinguish the fine structures like blood vessels and tumors. The lower iodine concentration of the iodinated lipid agent could explain its associated lack of contrast enhancement in our study. However, we could not evaluate the use of a higher concentration of iodine (600 μL to achieve an iodine dose of 30 mg) because the necessary volume would have exceeded the intravenous dosing limit guidelines of our institutional IACUC. Therefore, use of the nanoparticulate agent addresses the ‘refinement’ aspect of the 3Rs principle of the use of laboratory animals.
Our data demonstrate the huge inaccuracies in the volume measurements of nodular tumors that result when tumor margins are not clearly demarcated from adjoining soft-tissue structures (Figure 3). μCT using the nanoparticulate contrast agent detected tumors as small as 300 μm in diameter. Measurements by μCT method were slightly (approximately 20%) greater than the histology measurements. This difference in the tumor size according to histology may be due to the shrinkage of tissue during fixation and processing.2 In addition, histologic analysis (tumor area in a 4-μm section) of just 11 sections of lung took about 4 h. But tumor detection and tumor volume measurement by μCT was relatively easy and took only about 60 min per mouse. This result shows the accuracy and adaptability of contrast-based μCT imaging in preclinical studies. One disadvantage of the μCT method is the inability to distinguish between adenomas compared with adenocarcinomas30 or the type of inflammatory responses that are generated in the tumor microenvironment. For those kinds of measures and as a terminal confirmation of lesions, histology can still be used.
Furthermore our study shows the utility of μCT contrast agents in tumors of different phenotypes (nodular compared with diffuse). The accuracy of tumor measurement was dependent on the phenotype of the tumors. In addition, using a seed-growing algorithm in the OsiriX software, we show that accurate volumetric quantification of the tumors can be done. Although nodular tumors could be measured accurately by using this method, diffuse tumors that were distributed throughout the lungs, like those seen in the advanced stages of the Kras model, could not be discerned as accurately as were the nodular tumors. In such diffuse tumor models, incorporating pulmonary blood vessels into the tumor volume measurement may be inevitable. However, using contrast agent still provided benefit in these diffuse tumor models because the margins of the heart were more clearly visualized and could be excluded from the region-of-interest.
Finally, to validate the contrast enhanced μCT methodology, we conducted a preclinical trial with a small sample size. Crizotinib (PF-2341066) is an oral competitive inhibitor of the ALK receptor tyrosine kinase that recently was approved for the treatment of non-small–cell lung cancer caused by a translocation that generates an EML4–ALK fusion gene.9,10 Using the ELM4–ALK mouse model, we were able to monitor the treatment efficacy in individual tumors in each mouse longitudinally (before and after treatment), thereby reducing the number of animals required to obtain statistically meaningful data. Contrast-enhanced μCT imaging clearly demonstrated the efficacy of crizotinib in this preclinical trial and therefore is an excellent tool for noninvasively and accurately monitoring tumor progression and response to therapy in rodents. This methodology likely can be adapted to evaluate and quantify other orthotopic tumor models.
Acknowledgments
We thank Dr Theodore Simon (Pfizer) for critically reviewing the manuscript and David Paterson (Pfizer) for his technical assistance during these studies. This study was fully funded by Pfizer.
All authors on this manuscript are current or past employees of Pfizer. The financial support for the study was provided by Pfizer.
References
- 1.Badea CT, Athreya KK, Espinosa G, Clark D, Ghafoori AP, Li Y, Kirsch DG, Johnson GA, Annapragada A, Ghaghada KB. 2012. Computed tomography imaging of primary lung cancer in mice using a liposomal-iodinated contrast agent. PLoS ONE 7:e34496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blasdale C, Charlton FG, Weatherhead SC, Ormond P, Lawrence CM. 2010. Effect of tissue shrinkage on histological tumour-free margin after excision of basal cell carcinoma. Br J Dermatol 162:607–610 [DOI] [PubMed] [Google Scholar]
- 3.Boll H, Nittka S, Doyon F, Neumaier M, Marx A, Kramer M, Groden C, Brockmann MA. 2011. MicroCT based experimental liver imaging using a nanoparticulate contrast agent: a longitudinal study in mice. PLoS ONE 6:e25692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carlson SK, Classic KL, Bender CE, Russell SJ. 2007. Small animal absorbed radiation dose from serial microcomputed tomography imaging. Mol Imaging Biol 9:78–82 [DOI] [PubMed] [Google Scholar]
- 5.Caulo A, Mirsadraee S, Maggi F, Leccisotti L, van Beek EJ, Bonomo L. 2012. Integrated imaging of non-small cell lung cancer recurrence: CT and PET-CT findings, possible pitfalls and risk of recurrence criteria. Eur Radiol 22:588–606 [DOI] [PubMed] [Google Scholar]
- 6.Cavanaugh D, Johnson E, Price RE, Kurie J, Travis EL, Cody DD. 2004. In vivo respiratory-gated microCT imaging in small-animal oncology models. Mol Imaging 3:55–62 [DOI] [PubMed] [Google Scholar]
- 7.Chen Z, Cheng K, Walton Z, Wang Y, Ebi H, Shimamura T, Liu Y, Tupper T, Ouyang J, Li J, Gao P, Woo MS, Xu C, Yanagita M, Altabef A, Wang S, Lee C, Nakada Y, Pena CG, Sun Y, Franchetti Y, Yao C, Saur A, Cameron MD, Nishino M, Hayes DN, Wilkerson MD, Roberts PJ, Lee CB, Bardeesy N, Butaney M, Chirieac LR, Costa DB, Jackman D, Sharpless NE, Castrillon DH, Demetri GD, Janne PA, Pandolfi PP, Cantley LC, Kung AL, Engelman JA, Wong KK. 2012. A murine lung cancer coclinical trial identifies genetic modifiers of therapeutic response. Nature 483:613–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chouker A, Lizak M, Schimel D, Helmberger T, Ward JM, Despres D, Kaufmann I, Bruns C, Lohe F, Ohta A, Sitkovsky MV, Klaunberg B, Thiel M. 2008. Comparison of Fenestra VC contrast-enhanced computed tomography imaging with gadopentetate dimeglumine and ferucarbotran magnetic resonance imaging for the in vivo evaluation of murine liver damage after ischemia and reperfusion. Invest Radiol 43:77–91 [DOI] [PubMed] [Google Scholar]
- 9.Christensen JG. 2011. Proof of principle for crizotinib in anaplastic lymphoma kinase-positive malignancies was achieved in ALK-positive nonclinical models. Mol Cancer Ther 10:2024. [DOI] [PubMed] [Google Scholar]
- 10.Cui JJ, Tran-Dube M, Shen H, Nambu M, Kung PP, Pairish M, Jia L, Meng J, Funk L, Botrous I, McTigue M, Grodsky N, Ryan K, Padrique E, Alton G, Timofeevski S, Yamazaki S, Li Q, Zou H, Christensen J, Mroczkowski B, Bender S, Kania RS, Edwards MP. 2011. Structure based drug design of crizotinib (PF-02341066), a potent and selective dual inhibitor of mesenchymal–epithelial transition factor (c-MET) kinase and anaplastic lymphoma kinase (ALK). J Med Chem 54:6342–6363 [DOI] [PubMed] [Google Scholar]
- 11.Doki Y, Murakami K, Yamaura T, Sugiyama S, Misaki T, Saiki I. 1999. Mediastinal lymph node metastasis model by orthotopic intrapulmonary implantation of Lewis lung carcinoma cells in mice. Br J Cancer 79:1121–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DuPage M, Dooley AL, Jacks T. 2009. Conditional mouse lung cancer models using adenoviral or lentiviral delivery of Cre recombinase. Nat Protoc 4:1064–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Engelman JA, Chen L, Tan X, Crosby K, Guimaraes AR, Upadhyay R, Maira M, McNamara K, Perera SA, Song Y, Chirieac LR, Kaur R, Lightbown A, Simendinger J, Li T, Padera RF, Garcia-Echeverria C, Weissleder R, Mahmood U, Cantley LC, Wong KK. 2008. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat Med 14:1351–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frese KK, Tuveson DA. 2007. Maximizing mouse cancer models. Nat Rev Cancer 7:654–658 [DOI] [PubMed] [Google Scholar]
- 15.Fushiki H, Kanoh-Azuma T, Katoh M, Kawabata K, Jiang J, Tsuchiya N, Satow A, Tamai Y, Hayakawa Y. 2009. Quantification of mouse pulmonary cancer models by microcomputed tomography imaging. Cancer Sci 100:1544–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gavrielides MA, Kinnard LM, Myers KJ, Petrick N. 2009. Noncalcified lung nodules: volumetric assessment with thoracic CT. Radiology 251:26–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haines BB, Bettano KA, Chenard M, Sevilla RS, Ware C, Angagaw MH, Winkelmann CT, Tong C, Reilly JF, Sur C, Zhang W. 2009. A quantitative volumetric microcomputed tomography method to analyze lung tumors in genetically engineered mouse models. Neoplasia 11:39–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hasegawa M, Sone S, Takashima S, Li F, Yang ZG, Maruyama Y, Watanabe T. 2000. Growth rate of small lung cancers detected on mass CT screening. Br J Radiol 73:1252–1259 [DOI] [PubMed] [Google Scholar]
- 19.Herbst RS, Heymach JV, Lippman SM. 2008. Lung cancer. N Engl J Med 359:1367–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hirst GL, Balmain A. 2004. Forty years of cancer modelling in the mouse. Eur J Cancer 40:1974–1980 [DOI] [PubMed] [Google Scholar]
- 21.Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R, Jacks T, Tuveson DA. 2001. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev 15:3243–3248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jackson MA, Lea I, Rashid A, Peddada SD, Dunnick JK. 2006. Genetic alterations in cancer knowledge system: analysis of gene mutations in mouse and human liver and lung tumors. Toxicol Sci 90:400–418 [DOI] [PubMed] [Google Scholar]
- 23.Jaffe CC. 2006. Measures of response: RECIST, WHO, and new alternatives. J Clin Oncol 24:3245–3251 [DOI] [PubMed] [Google Scholar]
- 24.Kennel SJ, Davis IA, Branning J, Pan H, Kabalka GW, Paulus MJ. 2000. High-resolution computed tomography and MRI for monitoring lung tumor growth in mice undergoing radioimmunotherapy: correlation with histology. Med Phys 27:1101–1107 [DOI] [PubMed] [Google Scholar]
- 25.Kirsch DG, Grimm J, Guimaraes AR, Wojtkiewicz GR, Perez BA, Santiago PM, Anthony NK, Forbes T, Doppke K, Weissleder R, Jacks T. 2010. Imaging primary lung cancers in mice to study radiation biology. Int J Radiat Oncol Biol Phys 76:973–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kucherlapati R. 2012. Genetically modified mouse models for biomarker discovery and preclinical drug testing. Clin Cancer Res 18:625–630 [DOI] [PubMed] [Google Scholar]
- 27.Moro M, Bertolini G, Tortoreto M, Pastorino U, Sozzi G, Roz L. 2012. Patient-derived xenografts of non-small cell lung cancer: resurgence of an old model for investigation of modern concepts of tailored therapy and cancer stem cells. J Biomed Biotechnol 2012:568567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Osborne DR, Yan S, Stuckey A, Pryer L, Richey T, Wall JS. 2012. Characterization of X-ray dose in murine animals using microCT, a new low-dose detector and nanodot dosimeters. PLoS ONE 7:e49936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schambach SJ, Bag S, Schilling L, Groden C, Brockmann MA. 2010. Application of microCT in small animal imaging. Methods 50:2–13 [DOI] [PubMed] [Google Scholar]
- 30.Simmons BH, Lee JH, Lalwani K, Giddabasappa A, Snider BA, Wong A, Lappin PB, Eswaraka J, Kan JL, Christensen JG, Shojaei F. 2012. Combination of a MEK inhibitor at sub-MTD with a PI3K/mTOR inhibitor significantly suppresses growth of lung adenocarcinoma tumors in Kras(G12D-LSL) mice. Cancer Chemother Pharmacol 70:213–220 [DOI] [PubMed] [Google Scholar]
- 31.Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, Fujiwara S, Watanabe H, Kurashina K, Hatanaka H, Bando M, Ohno S, Ishikawa Y, Aburatani H, Niki T, Sohara Y, Sugiyama Y, Mano H. 2007. Identification of the transforming EML4–ALK fusion gene in non-small-cell lung cancer. Nature 448:561–566 [DOI] [PubMed] [Google Scholar]
- 32.Therasse P, Arbuck SG, Eisenhauer EA, Wanders J, Kaplan RS, Rubinstein L, Verweij J, Van Glabbeke M, van Oosterom AT, Christian MC, Gwyther SG. 2000. New guidelines to evaluate the response to treatment in solid tumors. European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J Natl Cancer Inst 92:205–216 [DOI] [PubMed] [Google Scholar]
- 33.Tidwell VK, Garbow JR, Krupnick AS, Engelbach JA, Nehorai A. 2012. Quantitative analysis of tumor burden in mouse lung via MRI. Magn Reson Med 67: 572–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Winkelmann CT, Figueroa SD, Rold TL, Volkert WA, Hoffman TJ. 2006. Microimaging characterization of a B16-F10 melanoma metastasis mouse model. Mol Imaging 5:105–114 [PubMed] [Google Scholar]