Abstract
Objective
Circulatory metabolites are important biomarkers for many diseases, especially metabolic disorders. The biological mechanism regulating circulatory levels of metabolites remains incompletely understood. Focusing on the liver as the central organ controlling metabolic homeostasis, we investigated the potential function of nine polymorphisms associated with serum metabolomic traits in a recent GWAS.
Materials/Methods
The mRNA levels of the associated genes were measured by real-time PCR and correlated with genotypes in normal liver tissue (n=42). Genotype and mRNA data were also correlated with total hepatic lipid content (HLC). Our findings were also compared with the previously published gene expression quantitative traits loci (eQTL) data in the liver.
Results
We found that seven out of nine genes were highly expressed in hepatic tissue, while expression of four genes were significantly or marginally associated with genotypes (SPTLC3 vs. rs168622, p=0.002; ACADS vs. rs2014355, p=0.016; PLEKHH1 vs. rs7156144, p=0.076; ACADL vs. rs2286963, p=0.068). The SNP rs168622 at the SPTLC3 locus was also significantly correlated with HLC (p=0.02). HLC was significantly correlated with FADS1 (r=−0.45; p=0.003) and ETFDH (r=0.33; p=0.037) expression. When compared with published eQTL data, SNPs in SPTLC3, ACADS, ELOVL2 and FADS1 were also in strong linkage disequilibrium (R2≥0.41, D’≥0.96) with eQTLs significantly affecting expression of these genes (p≤1.74×10−5).
Conclusions
Our study suggests that genetic variants affecting serum metabolites levels may play a functional role in the liver. This may help elucidate the mechanism by which genetic variants are involved in metabolic diseases.
Keywords: serum metabolites, hepatic fat, mRNA, polymorphism
Introduction
The dysregulation of the homeostasis of metabolism is involved in the pathogenesis of many diseases. It has long been recognized that the deviation of the circulatory levels of lipids, carbohydrates or amino acids from a normal range is highly correlated with various metabolic perturbations. Understanding the mechanism underlying the quantitative regulation of the circulatory metabolites is of critical importance to delineate the disease pathogenesis.
A recent GWAS was performed for 163 metabolite traits including amino acids, sugars, acylcarnitines and phospholipids in serum measured by ionization tandem mass spectrometry. This study revealed nine single nucleotide polymorphisms (SNPs) located within or around FADS1, ELOVL2, ACADS, ACADM, ACADL, SPTLC3, ETFDH, SLC16A9 and PLEKHH1 genes which were associated with population variability of serum levels of different metabolites [1]. This integrated metabolomic and genomic approach has significantly furthered our understanding of the key pathways involved in the regulation of human metabolism. However, questions still remain in which organ or tissue these genes and pathways mainly function, and how the function of genes and pathways in one tissue affects the metabolic signatures in the circulatory system. More importantly, whether these GWAS-identified variants and the nearby genes are causative to the alteration of the metabolite traits is largely unknown. Addressing these questions will not only ravel the detailed mechanism involved in the regulation of metabolism in humans, but also help define diagnostic markers and therapeutic targets for various diseases.
Given the central role of the liver in controlling the metabolic homeostasis of carbohydrates, lipids and amino acids in the body, we hypothesize that the regulation of metabolite levels in the blood may be largely attributable to the genes and metabolic pathways in the liver. To test this hypothesis, we investigated the potential role of the aforementioned 9 genetic variants [1] in modulating the gene expression in the liver using tissue samples from healthy donors. Furthermore, since most of these genes are involved in lipid metabolism, and hepatic lipids are one of the major forms of energy storage in the human body, we also tested the relationship between these genes/variants and the lipid accumulation in the tissue samples.
Methods
Detailed methods and materials were included as a supplementary appendix.
Liver tissue samples
Liver tissue samples from 42 healthy Caucasian donors were used in this study. Purdue University and the University of Chicago IRB have approved their use for the purpose of this study.
RNA preparation and quantitative real-time PCR
Total RNA was extracted from the liver tissues and real-time PCR was used to quantify the mRNA expression of each gene.
Genotyping
SNPs were genotyped using 5′ nuclease allelic discrimination TaqMan® SNP genotyping assays (Applied Biosystems, Foster City, CA) (Table S1).
Hepatic lipid extraction and quantification
Total lipids were extracted from 50 mg of liver tissue using a hexane:isopropanol (3:2) solvent mixture as described in our previous study [2].
Data analysis and Statistics
SNP genotypes were associated with mRNA or hepatic lipid levels using t-test. Correlations between mRNA and lipid levels were performed using Pearson correlation.
Comparison between our findings and previously published eQTL data
A previously published liver eQTL database [3] was searched for the genes investigated. Linkage disequilibrium levels between investigated SNPs and significant cis-eQTLs were assessed.
Results
Total lipid extraction and quantification
Total lipid content was quantified in 42 normal livers. The median total lipid content in these livers was 2.9% (range 1.1%–7.9%). Population variability (Coefficient of Variation, CV) in total lipid content was 48.5% (data not shown).
Association between SNPs and gene expression and hepatic lipid content (HLC)
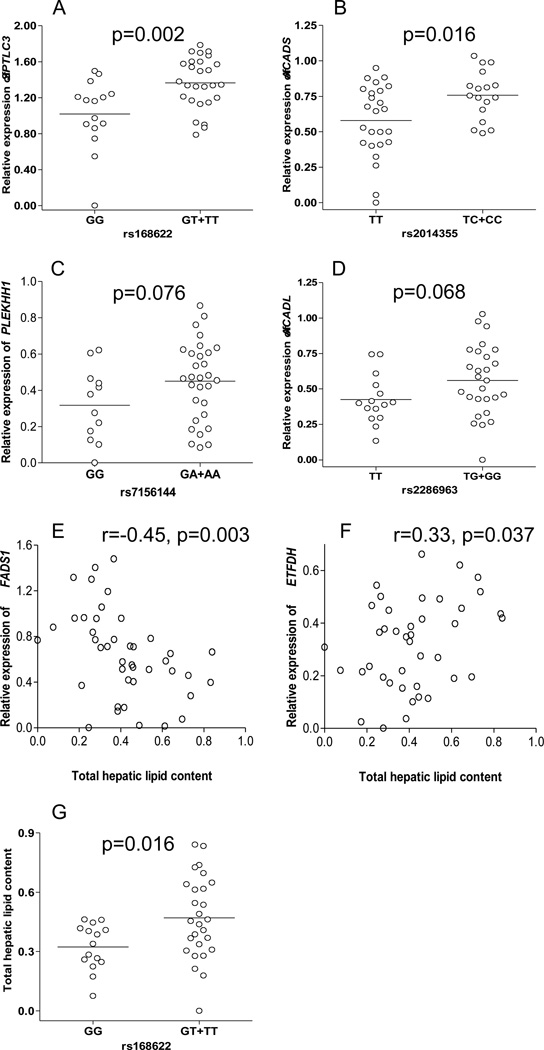
Seven out of the nine genes investigated were highly expressed in the liver tissue. The mRNA levels of two genes (ACADM and SLC16A9) were not detectable or quantifiable. We found a significant association between rs168622 and increased SPTLC3 gene expression (Figure 1A). The same SNP was also nominally associated with increased total hepatic lipids, but was not significant after Bonferroni correction (Figure 1G). SNP rs2014355 was also associated with increased ACADS mRNA levels (Figure 1B). Another two variants rs7156144 and rs2286963 were marginally associated (p<0.1) with increased mRNA levels of PLEKHH1 and ACADL, respectively (Figures 1C and 1D, respectively). No significant correlation was found between rs174547 and rs8396 and either the corresponding mRNA levels or total lipids (data not shown).
Figure 1. Correlations between SNPs, mRNA and hepatic lipid content (HLC) in liver tissue.
A) rs168622 and SPTLC3; B) rs2014355 and ACADS; C) rs7156144 and PLEKHH1; D) rs2286963 and ACADL; E) FADS1 and HLC; F) ETFDH and HLC; and G) rs168622 and HLC. HLC and all gene expression data (ratio between each gene and the internal control TBP gene) were log transformed (+log10).
Association between hepatic lipid content and gene expression levels
Hepatic lipid content was significantly correlated with decreased FADS1 but increased ETFDH gene expression (Figures 1E and 1F, respectively). The association between FADS1 gene expression and HLC remained significant after Bonferroni correction.
Relationship between the SNPs investigated and eQTLs regulating these genes
Statistically significant cis-eQTLs were found in ELOVL2, ACADS, SPTLC3 and FADS1 genes. Interestingly, these eQTLs were physically close to and in strong LD with the SNPs investigated in our study (Table 1).
Table 1.
Relationship between metabolite-associated SNPs and eQTLs in the liver.
| Gene | SNP | eQTL | Distance (bp) |
R2 | D' | p-value |
|---|---|---|---|---|---|---|
| ELOVL2 | rs9393903 | rs1323739 | 38348 | 0.41 | 1.00 | 1.12X10−10 |
| ACADS | rs2014355 | rs10431384 | 48177 | 0.78 | 0.98 | 1.01X10−7 |
| SPTLC3 | rs168622 | rs680379 | 3311 | 0.97 | 1.00 | 1.95X10−6 |
| FADS1 | rs174547 | rs174548 | 565 | 0.80 | 0.96 | 1.74X10−5 |
Discussion
Genome-wide association studies in recent years have successfully identified a large number of genetic loci conferring risk for many diseases and complex traits. However, understanding the biological pathways and mechanism mediating the genotype-phenotype correlation remains a major challenge. Focusing on nine genes and polymorphisms identified in a recent GWAS of serum metabolomic traits [1], our study suggests that four SNPs (rs168622, rs2014355, rs7156144 and rs2286963) identified in this study may affect the transcription of nearby genes in liver tissue, and two genes (FADS1 and ETFDH) may influence the deposition of lipids in the liver. Combined with the previously published liver eQTL data [3], a total of 6 SNPs or their proxies are associated with the expression of the nearby gene in the liver. These findings highlight the importance of liver tissue in the genetic control of human metabolism and energy balance.
Our findings may also help understand the detailed mechanism by which genetic variants convey risk for abnormalities in metabolism and the associated clinical perturbations. Hepatic fat accumulation in humans is a common example of imbalance in lipid and energy homeostasis. Excessive fat accumulation in the liver (steatosis) represents an important clinical marker for type 2 diabetes mellitus and cardiovascular disease [4–6]. Our study suggests that three genes (SPTLC3, FADS1 and ETFDH) may play a functional role in the hepatic lipid accumulation. These three genes encode important enzymes involved in lipid metabolism [7–9]. Our results demonstrate that the functional allele increasing SPTLC3 expression may also increase the lipid deposition in the liver. Meanwhile, the total lipids accumulation in these livers is associated with increased ETFDH but decreased FADS1 expression. The correlation between the FADS1 expression level and HLC remains significant even after adjusting the multiple testing. This is in agreement with a recent study that showed an association between increased expression of FADS1 and resistance to liver steatosis in a mouse model [10]. Moreover, although the correlation between ETFDH and HLC is only suggestive in our study, it has been demonstrated that this gene is directly regulated by PPAR-alpha, an important transcription factor with well-known function in hepatic lipid metabolism [11].
We recognize that the small sample size is a limitation in our study, which may reduce the power for genetic correlations. When compared to the liver eQTL data where a much larger sample set were used, proxies of SNPs in an additional two genes, FADS1 and ELOVL2, were demonstrated to regulate gene expression. No correlation was observed between rs8396 (or its proxies) and ETFDH in either our study or the previous eQTL study. This may indicate that either this SNP is not causative to the altered gene function, or the ETFDH gene may not be the causative gene for the specific trait, since it has been suggested that over 30% eQTLs fall greater than 100kb away from the gene locus [3]. Furthermore, we noticed that there are several splicing variants (http://genome.ucsc.edu) for this gene, which may also confound the measurement for its expression level and consequently affect its correlation with the SNPs. In addition, it is also possible that this SNP changes the gene function in other organs/tissues. This is particularly consistent with the low or non-detectable level of the two low-expression genes (SLC16A9 and ACADM) investigated in this study. Previous studies indicated that SLC16A9 and ACADM are highly expressed in adrenocortical and intestinal tissue, respectively [12, 13]. Taken together, these possibilities further reflect the complexity of the human metabolism and the underlying regulatory mechanism, for which systems approaches involving larger sample sets and different tissues/organs will be particularly necessary to achieve a better understanding.
Supplementary Material
Acknowledgments
Funding
This study was supported by the NIH/NIDDK grant (R21 DK090437) (W.L.) and a Pilot and Feasibility award (W.L.) from the Diabetes Research and Training Center (P60 DK-20595) at the University of Chicago.
Abbreviations
- SNP
single nucleotide polymorphism
- GWAS
genome-wide association studies
- HLC
hepatic lipid content
- eQTL
expression quantitative trait loci
- LD
linkage disequilibrium
- FADS1
fatty acid desaturase 1
- ELOVL2
ELOVL fatty acid elongase 2
- ACADS
acyl-CoA dehydrogenase C-2 to C-3 short chain
- ACADM
acyl-CoA dehydrogenase C-4 to C-12 straight chain
- ACADL
acyl-CoA dehydrogenase, long chain
- SPTLC3
serine palmitoyltransferase, long chain base subunit 3
- ETFDH
electron-transferring-flavoprotein dehydrogenase
- SLC16A9
solute carrier family 16, member 9
- PLEKHH1
pleckstrin homology domain containing, family H
- TBP
TATA box binding protein
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure statement
The authors declare that they have no competing interests.
Author’s contributions
SM carried out the gene expression experiments, quantified hepatic lipids, performed statistical analysis and drafted the manuscript. JLM carried out the genotyping, helped with statistical analysis and revised the final manuscript. JR helped to draft and revise the final manuscript. WL conceived of the study and its design, performed statistical analysis and helped to draft the manuscript. All authors approved the final manuscript.
References
- Illig T, Gieger C, Zhai G, et al. A genome-wide perspective of genetic variation in human metabolism. Nat Genet. 2010;42:137–141. doi: 10.1038/ng.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Song J, Mirkov S, et al. Comparing morphometric, biochemical, and visual measurements of macrovesicular steatosis of liver. Hum Pathol. 2011;42:356–360. doi: 10.1016/j.humpath.2010.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadt EE, Molony C, Chudin E, et al. Mapping the genetic architecture of gene expression in human liver. PLoS Biol. 2008;6(5):e107. doi: 10.1371/journal.pbio.0060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith BW, Adams LA. Nonalcoholic fatty liver disease and diabetes mellitus: pathogenesis and treatment. Nat Rev Endocrinol. 2011;7:456–465. doi: 10.1038/nrendo.2011.72. [DOI] [PubMed] [Google Scholar]
- Cox AJ, Wing MR, Carr JJ, et al. Association of PNPLA3 SNP rs738409 with liver density in African Americans with type 2 diabetes mellitus. Diabetes Metab. 2011;37(5):452–455. doi: 10.1016/j.diabet.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nseir W, Shalata A, Marmor A, et al. Mechanisms Linking Nonalcoholic Fatty Liver Disease with Coronary Artery Disease. Dig Dis Sci. 2011;56(12):3439–3449. doi: 10.1007/s10620-011-1767-y. [DOI] [PubMed] [Google Scholar]
- Hicks AA, Pramstaller PP, Johansson A, et al. Genetic determinants of circulating sphingolipid concentrations in European populations. PLoS Genet. 2009;5:e1000672. doi: 10.1371/journal.pgen.1000672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornemann T, Penno A, Rütti MF, et al. The SPTLC3 subunit of serine palmitoyltransferase generates short chain sphingoid bases. J Biol Chem. 2009;284:26322–26330. doi: 10.1074/jbc.M109.023192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen RK, Olpin SE, Andresen BS, et al. ETFDH mutations as a major cause of riboflavin-responsive multiple acyl-CoA dehydrogenation deficiency. Brain. 2007;130:2045–2054. doi: 10.1093/brain/awm135. [DOI] [PubMed] [Google Scholar]
- Hall D, Poussin C, Velagapudi VR, et al. Peroxisomal and microsomal lipid pathways associated with resistance to hepatic steatosis and reduced pro-inflammatory state. J Biol Chem. 2010;285:31011–31023. doi: 10.1074/jbc.M110.127159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakhshandehroo M, Sanderson LM, Matilainen M, et al. Comprehensive analysis of PPAR-alpha-dependent regulation of hepatic lipid metabolism by expression profiling. PPAR Res. 2007;2007:26839. doi: 10.1155/2007/26839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Ranvier GG, Weng J, Yeh RF, et al. Identification of biomarkers of adrenocortical carcinoma using genomewide gene expression profiling. Arch Surg. 2008;143:841–846. doi: 10.1001/archsurg.143.9.841. [DOI] [PubMed] [Google Scholar]
- Simula MP, Cannizzaro R, Canzonieri V, et al. PPAR signaling pathway and cancer-related proteins are involved in celiac disease-associated tissue damage. Mol Med. 2010;16:199–209. doi: 10.2119/molmed.2009.00173. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.