Abstract
RAF and MEK (mitogen-activated or extracellular signal–regulated protein kinase kinase) inhibitors are effective in treating patients with BRAF-mutant melanoma. However, most responses are partial and short-lived, and many patients fail to respond at all. We found that suppression of TORC1 activity in response to RAF or MEK inhibitors, as measured by decreased phosphorylation of ribosomal protein S6 (P-S6), effectively predicted induction of cell death by the inhibitor in BRAF-mutant melanoma cell lines. In resistant melanomas, TORC1 activity was maintained after treatment with RAF or MEK inhibitors, in some cases despite robust suppression of mitogen-activated protein kinase (MAPK) signaling. In in vivo mouse models, suppression of TORC1 after MAPK inhibition was necessary for induction of apoptosis and tumor response. Finally, in paired biopsies obtained from patients with BRAF-mutant melanoma before treatment and after initiation of RAF inhibitor therapy, P-S6 suppression predicted significantly improved progression-free survival. Such a change in P-S6 could be readily monitored in real time by serial fine-needle aspiration biopsies, making quantitation of P-S6 a valuable biomarker to guide treatment in BRAF-mutant melanoma.
INTRODUCTION
BRAF mutations occur in ∼7% of all cancers and in more than half of melanomas and lead to constitutive BRAF kinase activity and downstream activation of the mitogen-activated protein kinase (MAPK) signaling pathway (1). Selective RAF and MEK (mitogen-activated or extracellular signal–regulated protein kinase kinase) inhibitors have profoundly changed the treatment of BRAF-mutant melanoma (2, 3). Selective RAF inhibitors—such as vemurafenib (PLX4032), which was recently approved by the Food and Drug Administration, and dabrafenib (GSK2118436)—can induce responses in 50 to 80% of BRAF-mutant melanoma patients, leading to improved survival compared to standard chemotherapy (4–7). Similarly, the MEK inhibitor trametinib (GSK1120212), which acts downstream of mutant BRAF, induces responses in ∼20% of BRAF-mutant melanoma patients and leads to improved survival (8, 9). Despite these successes, a substantial percentage of patients show minimal or no tumor regression on these therapies, and the time to disease progression for many patients is only a few months (5–7).
Recent studies by several groups have identified multiple potential mechanisms of acquired and intrinsic resistance to MAPK inhibitors in BRAF-mutant melanoma (10–14). The large number of resistance mechanisms identified to date suggests that clinical resistance to RAF or MEK inhibition can arise through a complex array of diverse events that affect several signaling pathways. This complexity presents a challenge in predicting which tumors are most likely to demonstrate resistance and in identifying alternate therapeutic strategies for such patients. Thus, robust biomarkers that predict resistance in BRAF-mutant melanomas and guide the choice of therapies to overcome resistance will be of critical importance.
RESULTS
Phosphorylated extracellular signal – regulated kinase suppression is not sufficient to predict sensitivity to RAF or MEK inhibitors
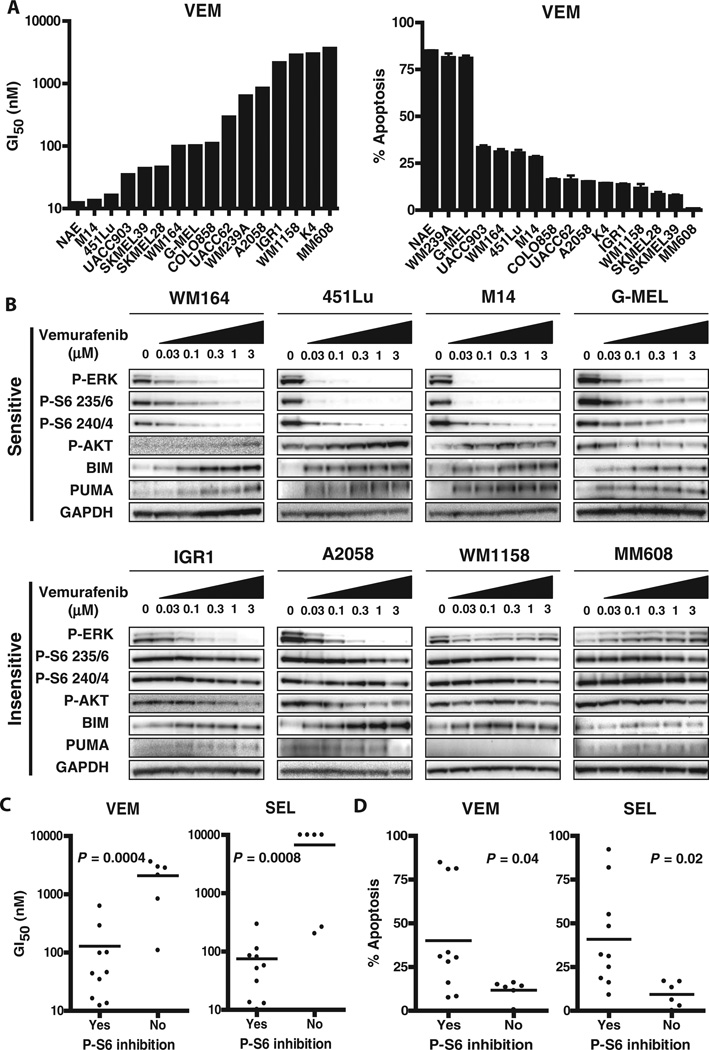
To better understand the determinants of sensitivity to MAPK inhibition among BRAF-mutant melanomas, we evaluated a panel of 16 BRAF V600–mutant melanoma cell lines (table S1) for sensitivity to the selective RAF inhibitor vemurafenib (PLX4032) and the selective MEK inhibitor selumetinib (AZD6244) using measures of growth inhibition (GI50) and apoptosis induction (Fig. 1A and fig. S1). Clinical pharmacodynamic studies of paired patient biopsies taken before and 2 weeks after vemurafenib treatment have demonstrated that substantial (typically >80%) inhibition of extracellular signal–regulated kinase (ERK) phosphorylation was required to induce a tumor response (15). Consistent with these findings, we found that cell lines in which vemurafenib or selumetinib failed to substantially decrease the amount of phosphorylated ERK1 and ERK2 (P-ERK) (for example, WM1158 and MM608) were less sensitive to vemurafenib (Figs. 1B and fig. S2 and S3).
Fig. 1. Reduction of TORC1 activity by RAF or MEK inhibition in sensitive BRAF-mutant melanoma cell lines.
(A) The indicated BRAF-mutant melanoma cell lines were treated for 72 hours with vemurafenib (VEM). Left: Vemurafenib concentrations ranging from 1 nM to 10 µM were tested to determine Gl50. Right: Induction of apoptosis was determined after treatment with 3 µM vemurafenib for 72 hours (average of at least three independent experiments). Error bars represent SD. (B) Cell lines were treated for 24 hours with the indicated concentration of vemurafenib, and lysates were probed with the indicated antibodies. (C) Comparison of the Gl50 values for cell lines treated with vemurafenib (VEM) or selumetinib (SEL) in which P-S6 was inhibited and cell lines in which P-S6 was not inhibited. P-S6 inhibition was determined using quantitative analyses of Western blots as described in Materials and Methods. (D) Apoptosis induction in cell lines in which P-S6 was inhibited and cell lines in which P-S6 was not inhibited. P values in (C) and (D) were calculated with two-tailed Student's t test.
Nevertheless, we also observed a lack of sensitivity to vemurafenib or selumetinib in several cell lines (for example, IGR1 and A2058) despite robust P-ERK inhibition that was comparable to that achieved in sensitive cell lines (for example, WM164 and 451Lu) (Fig. 1B and figs. S2 and S3). These findings suggest that, although inhibition of P-ERK is clearly necessary, it alone is not sufficient to predict sensitivity to MAPK inhibition, and some BRAF melanoma cell lines may therefore have ERK-independent survival signals.
RAF or MEK inhibition reduces TORC1 activity in drug-sensitive cell lines
Analysis of other signaling changes after RAF or MEK inhibition revealed that a decrease in phosphorylated ribosomal protein S6 (P-S6) levels after vemurafenib or selumetinib treatment correlated well with sensitivity to these agents (Fig. 1, B to D). In this cell line panel, P-S6 suppression was a more effective predictor of sensitivity than several other candidate biomarkers previously reported to predict sensitivity in BRAF-mutant melanoma cells, including loss of PTEN, increased basal levels of P-AKT, or induction of P-AKT levels after MAPK inhibition (fig. S4, A to E) (16, 17). S6 phosphorylation is a marker of mammalian target of rapamycin (mTOR) complex 1 (TORC1) activity (18, 19). TORC1 is a critical mediator of cellular growth and metabolism and integrates signals from multiple upstream pathways including the phosphatidylinositol 3-kinase (PDK)–AKT, MAPK, and liver kinase Bl (LKBl)–adenosine monophosphate– activated protein kinase (AMPK) pathways (19–22). Despite these multiple upstream regulators, our data indicate that, in sensitive BRAF-mutant melanomas, TORC1 activity is primarily regulated by the MAPK pathway. This finding is noteworthy because the PI3K–AKT pathway, rather than the MAPK pathway, is often believed to be the predominant upstream regulator of TORC1 activity (23,24). We observed a reduction in P-S6 after MAPK inhibition in a sensitive melanoma cell line (WM239A) that harbors loss of the phosphatase and tensin homolog (PTEN) tumor suppressor gene, which has constitutive activation of the PI3K–AKT pathway (figs. S2 to S4A). Moreover, in some sensitive melanoma cells, we observed a reduction in P-S6 upon RAF or MEK inhibition, even though feedback induction of P-AKT was also observed (for example, WM164, 451Lu, and M14) (Fig. 1B and figs. S2 and S3), again suggesting that MAPK is the dominant regulator of TORC1 signaling in BRAF-mutant melanomas sensitive to vemurafenib.
The phosphorylation sites on S6 are usually regulated by p70 S6 kinase 1, a direct target of TORC1, and are thus often used as pharmacodynamic markers of TORC1 signaling. However, occasionally, the MAPK pathway can also affect S6 phosphorylation directly (via a TORC1 -independent pathway) by regulating the activity of p90 ribosomal S6 kinase (RSK), which can phosphorylate S6 selectively on the Ser235/236 phosphorylation sites (25). However, we observed concordant down-regulation of P-S6 on both the Ser235/236 and Ser240/244 sites. Because the latter sites are regulated exclusively by TORC1, the decrease in P-S6 is consistent with suppression of TORC1 rather than suppression of only RSK activity (Fig. 1B and figs. S2 and S3). Additionally, we found that maintenance of P-S6 in some insensitive cells occurred despite robust suppression of RSK phosphorylation by vemurafenib, further supporting that persistence of P-S6 is not due to RSK activity (fig. S5A). Consistent with this hypothesis, a reduction in 4EBP1 phosphorylation, another marker of TORC1 activity (18, 19), was also observed in sensitive BRAF-mutant melanomas after RAF or MEK inhibition (fig. S5B).
Maintenance of S6 phosphorylation after RAF or MEK inhibitor treatment was observed in insensitive BRAF-mutant cell lines. Not only were P-S6 levels maintained in insensitive cell lines that failed to suppress P-ERK after vemurafenib or selumetinib treatment, but they were also maintained in insensitive cell lines in which robust P-ERK suppression was achieved (for example, IGR1 and A2058) (Fig. 1B and figs. S2 and S3). Thus, in these cells, an ERK-independent signal maintains TORC1 activity despite MAPK pathway inhibition and may drive resistance. These data suggest that failure to down-regulate P-S6 levels may be universally associated with resistance, whether it is a result of failure to inhibit P-ERK or of ERK-independent mechanisms.
Down-regulation of TORC1 is required for maximal apoptotic response to MAPK inhibitors
We next determined whether TORC1 down-regulation is necessary for BRAF-mutant melanoma cells to respond to MAPK inhibition.
We constitutively activated TORC1 in the sensitive BRAF-mutant melanoma cell line WM164 by knocking down TSC2, an integral component of the tuberous sclerosis complex (TSC) that negatively regulates TORC1 activity (26). Cells with TSC2 knockdown maintained TORC1 activity and P-S6 levels after treatment with vemurafenib or selumetinib and showed a significant reduction in the apoptotic response to each inhibitor (Fig. 2A and table S3).
Fig. 2. Contribution of TORC1 suppression to the apoptotic response in BRAF-mutant melanomas.
(A) WM164 cells were infected with short hairpin RNA (shRNA) targeting TSC2 (shTSC2) or control shRNA (3shGFP). After puromycin selection, cells were treated in triplicate with 3 µM vemurafenib (VEM) or 1 µM selumetinib (SEL) for 24 hours before analysis by Western blot or for 72 hours before assessment of apoptosis (minimum of three independent experiments). **P = 0.03 (for VEM) and P = 0.001 (for SEL) by two-tailed Student's t test. (B) WM164 or IGR1 cells were treated with or without 3 µM vemurafenib (+VEM) in the presence (8055) or absence (con) of 300 nM AZD8055. Cells were lysed for Western blots after 24 hours and were analyzed for apoptosis after 72 hours of treatment. **P = 0.001 (versus VEM) and P < 0.0001 (versus 8055); N.S., not significant, by one-way analysis of variance (ANOVA) with Bonferroni posttest. (C) Induction of apoptosis was measured by annexin V staining in WM164 and IGR1 cells treated in triplicate for 72 hours in the presence or absence of 3 µM vemurafenib without (CON) or with 300 nM AZD8055, 1 µM GDC0941, or 500 nM BEZ235. **P < 0.0001 for combination relative to each single agent alone by one-way ANOVA with Bonferroni posttest. (D) Cells were treated in triplicate for 72 hours with 3 µM vemurafenib, 1 µM ABT-263, or both inhibitors in combination and were assessed for apoptosis, as in (C). **P < 0.0001 by one-way ANOVA with Bonferroni posttest for combination treatment relative to each single agent alone. Error bars represent SD for all experiments.
We also observed that inhibition of the persistent TORC1 signaling in resistant cells restored an apoptotic response to vemurafenib. In a resistant BRAF-mutant melanoma cell line (IGR1), inhibition of TORC1 activity with the catalytic TORC inhibitor AZD8055 (27) in combination with vemurafenib markedly increased the apoptotic response (Fig. 2B table S4, and fig. S6). In contrast, in the sensitive WM164 cell line, the addition of a TORC inhibitor did not further increase the activity of vemurafenib, presumably because TORC1 activity is already substantially inhibited by vemurafenib alone in this cell line (Fig. 2B). Because the PI3K–AKT pathway is a well-known regulator of TORC1 activity (18, 19, 28, 29), we tested whether inhibition of PI3K in combination with vemurafenib could also lead to TORC1 down-regulation and to increased apoptosis. When combined with vemurafenib, the TORC inhibitor AZD8055, the pan-PI3K inhibitor GDC0941 (30), and the dual PI3K–TORC inhibitor BEZ235 (31) were all able to induce a similar degree of apoptosis in some melanoma cell lines exhibiting ERK-independent resistance to MAPK pathway inhibitors (Fig. 2C table S5, and fig. S6A). Nevertheless, even in these cells, PI3K inhibition alone failed to suppress TORC1 signaling (fig. S7), supporting the notion that the MAPK pathway still provides important signaling input for TORC1 regulation even in drug-resistant BRAF-mutant melanoma cells. Collectively, these results suggest that modulation of TORC1 activity is critical for the apoptotic response to MAPK inhibitors in BRAF-mutant melanomas.
Next, we investigated the mechanism by which TORC1 signaling mediates the apoptotic response to MAPK inhibition in BRAF-mutant melanomas. Consistent with previous studies, we observed that the proapoptotic BH3-only protein BIM (a well-known target of MAPK signaling) (32–35) was more substantially induced by vemurafenib or selumetinib in cell lines in which these agents led to inhibition of P-ERK (for example, WM164) than in cell lines in which P-ERK levels were maintained despite the presence of these inhibitors (for example, MM608) (Fig. 1B and figs. S2, S3, and S8A). However, BIM levels were induced in all cell lines in which P-ERK inhibition was achieved, including the resistant cell lines in which P-S6 levels were maintained (for example, IGR1 and A2058). This suggests that BIM regulation is MAPK-dependent, but independent of TORC1, and that BIM up-regulation alone is not sufficient to promote maximal amounts of apoptosis. Although expression of most apoptotic proteins tested did not change upon treatment with vemurafenib (fig. S8B), expression of the BH3-only protein PUMA was specifically induced by vemurafenib or selumetinib in cell lines with P-S6 inhibition (Fig. 1B and figs. S2 and S3). Accordingly, constitutive activation of TORC1 by TSC2 knockdown blocked PUMA induction but did not impair BIM induction (Fig. 2A). Conversely, in the resistant IGR1 cells, addition of TORC1 or PI3K inhibitors to vemurafenib led to the induction of PUMA (Fig. 2B and fig. S7). PUMA knockdown significantly reduced the apoptotic response to vemurafenib, but not to the cytotoxic chemotherapy agent paclitaxel, suggesting a specific role for PUMA in the apoptotic response to RAF inhibition (fig. S9 and table S6). Collectively, these observations suggest that TORC activity contributes to the regulation of PUMA in BRAF-mutant melanoma cells. Although TORC1 may also contribute to apoptosis through additional PUMA-independent mechanisms, the induction of PUMA appears to be a major effector of apoptosis induced by TORC1 suppression. Consistent with this hypothesis, we observed that the combination of vemurafenib and the BH3 mimetic ABT263—a compound that, like PUMA and other BH3 proteins, can bind and inhibit antiapoptotic proteins (36)—was able to promote apoptosis in vemurafenib-insensitive melanoma cells (Fig. 2D, table S7, and fig. S6B).
In summary, we found that combining vemurafenib with a TORC inhibitor, a PI3K inhibitor, or a BH3 mimetic can improve cell killing in BRAF-mutant melanomas exhibiting ERK-independent resistance to MAPK inhibition. Thus, the identification of patients whose tumors are resistant to MAPK inhibition through this mechanism could lead to specific therapeutic interventions.
P-S6 suppression predicts sensitivity to MAPK inhibition in vivo
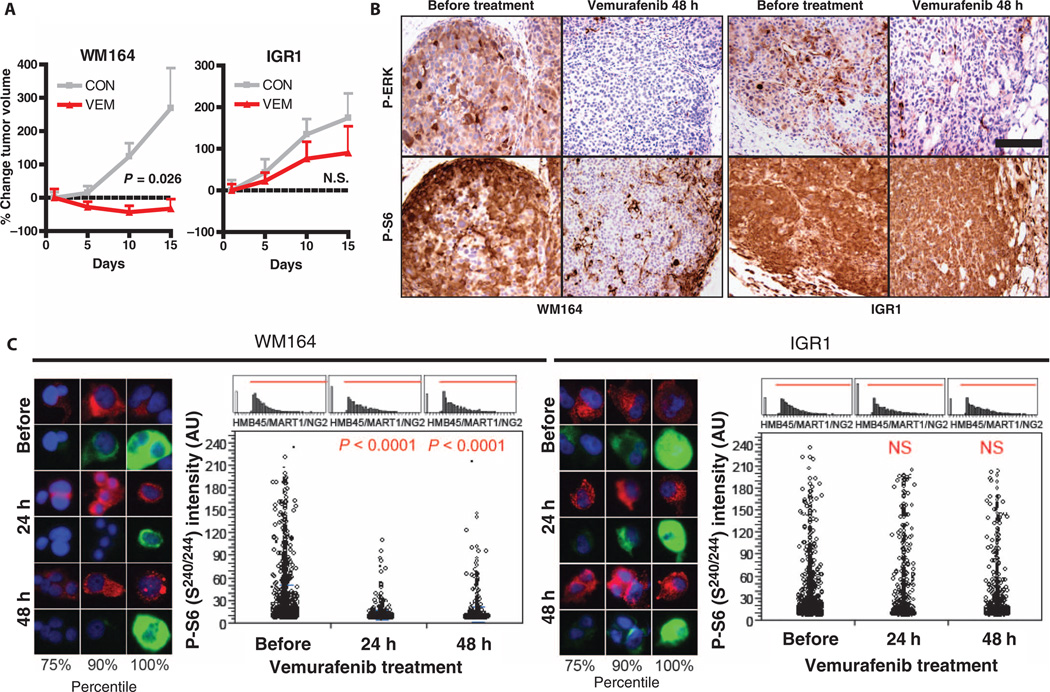
Consistent with our in vitro studies, treatment of WM164 xenografts with vemurafenib or selumetinib led to tumor regression in vivo (Fig. 3A and fig. S10). Suppression of P-S6 levels was observed 24 hours, 48 hours, and 2 weeks after vemurafenib or selumetinib treatment (Fig. 3B and figs. S11 and S12). Conversely, IGR1 tumor cell xenografts progressed despite treatment with either agent, with no significant tumor inhibition. In these xenografts, the levels of P-S6 did not change with treatment at any time point assessed. Although P-S6 levels effectively distinguished these two classes of cells, we saw robust suppression of P-ERK in both xenografts throughout the treatment period. Thus, a reduction in P-S6 after RAF or MEK inhibitor treatment predicted drug sensitivity in vivo and distinguished between sensitive and resistant melanoma xenografts, both of which showed suppression of ERK signaling in response to RAF or MEK inhibition.
Fig. 3. Correlation of P-S6 suppression with sensitivity to vemurafenib in BRAF mutant melanoma xenografts.
(A) Tumor xenografts generated from WM164 and IGR1 cells were treated with vehicle (CON) or vemurafenib (VEM) (75 mg/kg) twice daily (individual tumor measurements shown in fig. S10). Error bars represent SEM. P value calculated by two-tailed t test for vehicle versus vemurafenib treatment. (B) P-ERK and P-S6 (s240/244) staining by immunohistochemistry in xenografts harvested before or after 48 hours of treatment with vemurafenib, as in (A). Scale bar, 100 µM. (C) Serial FNAs were performed on xenograft tumors before treatment and after 24 and 48 hours of vemurafenib treatment and were processed, stained, and analyzed as described in Materials and Methods. Images of representative cells in the indicated percentiles of P-S6 (s240/244) staining intensity are shown. Green, P-S6; red, melanoma markers (HMB45/MART1/NG2); blue, 4’,6-diamidino-2-phenylindole (DAPI) nuclear stain. For quantification of P-S6 staining by automated fluorescence microscopy, each open circle represents an individual tumor cell. Histograms showing the HMB45/MART1/NG2 staining intensities of tumor cells used in the analysis are shown above each quantitation. A minimum of 960 cells was analyzed per condition (range, 960 to 2500 cells per analysis). P values calculated by Student's t test (unequal variances) are shown relative to the before treatment FNA. NS, no significant decrease; AU, arbitrary units.
We next evaluated whether treatment-induced changes in P-S6 levels could be assessed in real time in intact tumors using a quantitative imaging assay. Because decreased P-S6 levels are detectable within 24 to 48 hours of treatment initiation, this approach could facilitate rapid evaluation of the likely response to drugs, allowing for rapid selection of appropriate therapies (Fig. 3B and figs. S11 and S12). Fine-needle aspiration (FNA) biopsies are a minimally invasive means of obtaining tumor cells for ex vivo analysis and are routinely performed as part of clinical care (37). Serial FNAs from the same tumor were performed on WM164 and IGR1 xenografts before treatment and at various times after initiation of therapy. Tumor cells were fixed, dissociated, and costained with melanoma markers (HMB45, MART1, and NG2) and with antibodies against P-ERK or P-S6. P-ERK or P-S6 staining in melanoma marker–positive cells was assessed by quantitative immunofluorescence microscopy. Consistent with our previous results, a significant decrease in the number of cells with high P-ERK levels was observed in FNAs from both xenografts after treatment with either vemurafenib or selumetinib (figs. S13 and S14). Although P-S6 levels in individual tumor cells decreased significantly within 3 days of MAPK inhibitor treatment in WM164 xenografts, no significant decrease in P-S6 was detected in IGR1 xenografts (Fig. 3C and fig. S14). Thus, the predictive changes in P-S6 can be rapidly and quantitatively assessed by microscopic imaging in tumors sampled by minimally invasive FNA biopsies.
P-S6 can predict responsiveness to RAF inhibition in melanoma patients
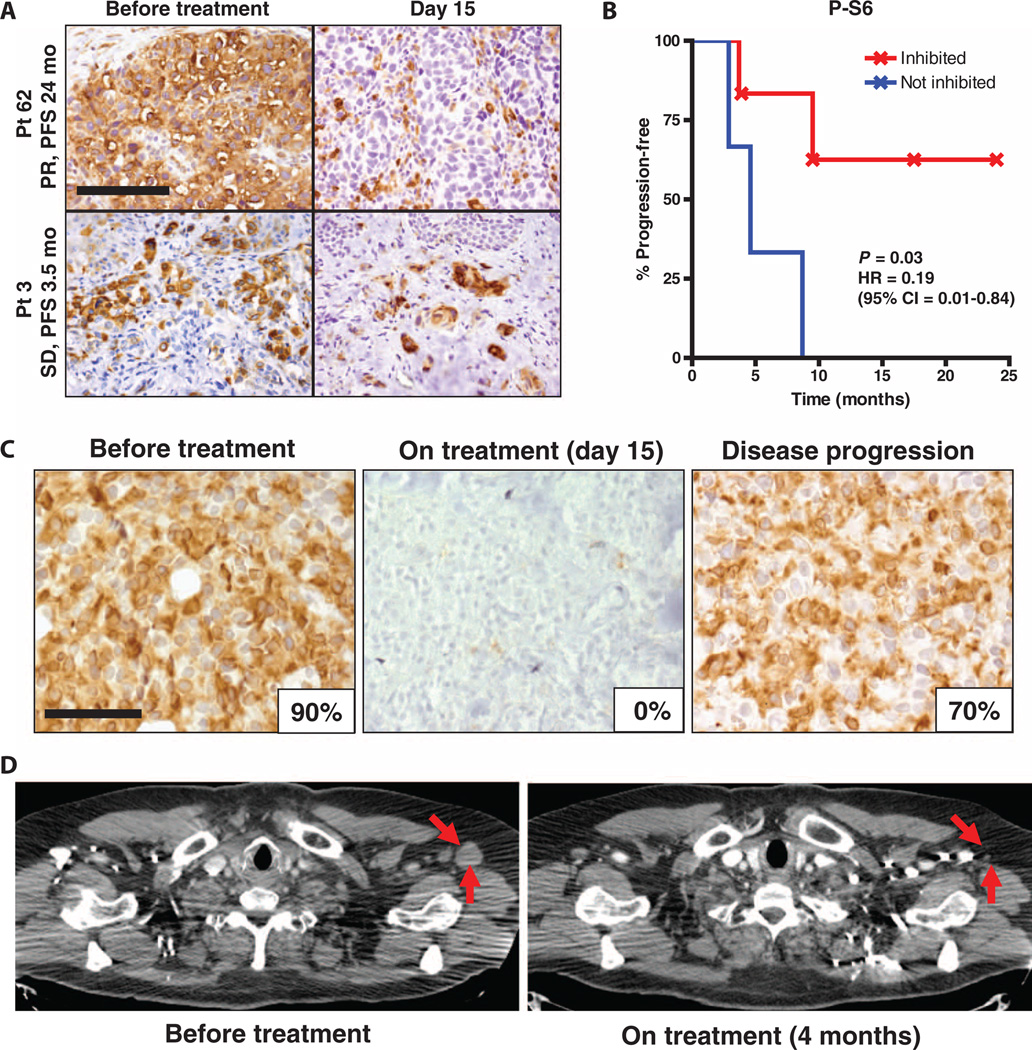
To establish the feasibility of real-time P-S6 and P-ERK assessment in BRAF-mutant melanoma patients, we prospectively evaluated patients with BRAF-mutant melanoma treated with RAF inhibitor therapy. Two patients, one treated with vemurafenib only and the other treated with an experimental combination of RAF and MEK inhibitors (dabrafenib and trametinib) (38), were evaluated by serial FNA of the same tumor, before and 15 days after initiation of therapy. In each patient, we observed a significant decrease in P-S6 and P-ERK after initiation of therapy (Fig. 4, A to C). Both patients went on to have tumor responses as measured radiographically at the time of their first restaging assessment after 2 months of therapy (Fig. 4, B to D).
Fig. 4. Real-time detection of P-S6 suppression in response to RAF inhibitor therapy.
(A to D) Serial FNA biopsies were performed on two BRAF-mutant melanoma patients receiving RAF inhibitor–based regimens. One patient (A and B) was treated with vemurafenib and was biopsied before treatment and on day 8 of vemurafenib treatment, and the second patient (C and D) was treated with dabrafenib and trametinib and was biopsied before treatment and on day 15 of therapy. In each patient, the same lesion was biopsied at each time point. (A and C) Quantification of P-ERK and P-S6 (s240/244) staining in the before treatment and on-treatment biopsies for each patient by automated fluorescence microscopy (as in Fig. 3C). A minimum of 360 cells was analyzed per condition (range, 360 to 2300 cells per analysis). P values calculated by Student's t test (unequal variances) are shown relative to before treatment FNA. (B) Positron emission tomography (PET) scans of the same patient from (A) before treatment and after 2 months of vemurafenib therapy. Note marked improvement in tumor nodules in the thorax, liver, and spleen, indicated by red arrows. Normal physiologic PET tracer uptake persists in the heart (H) and kidneys (K) in the 2-month restaging scan. (D) Computed tomography (CT) scans of the same patient from (C) before treatment and after 2 months of therapy with dabrafenib and trametinib. Tumor is indicated by red arrows.
Having established the feasibility of real-time minimally invasive monitoring of P-S6 and P-ERK in patients with BRAF-mutant melanoma, we next tested whether a reduction in P-S6 levels after treatment with a RAF inhibitor–based regimen correlated with patient outcome. We evaluated the change in P-S6 levels in paired biopsies taken from the same patients before and after 15 days of treatment with a RAF inhibitor. Although the availability of paired pretreatment and on-treatment biopsies from BRAF-mutant melanoma patients is limited because these paired biopsies are not routinely obtained as part of clinical care, we were able to obtain 21 such paired biopsies for evaluation, 9 of which had sufficient tissue for analysis in both the pretreatment and on-treatment biopsy. The change in P-S6 staining by immunohistochemistry between pretreatment and on-treatment biopsies was evaluated by two independent dermatopathologists, who were blinded to the patient outcome data. A reduction in P-S6 levels after treatment was correlated with a significant improvement in patient outcome, with a nearly fivefold improvement in progression-free survival seen in patients demonstrating a decrease in P-S6 after treatment (Fig. 5, A and B, and table S2). Although further clinical studies will be required to validate these observations, assessment of P-S6 levels before and after treatment with RAF inhibitors may thus help predict which patients with BRAF-mutant melanoma derive significant benefit from current regimens and which may have a more limited response, necessitating a different line of therapy.
Fig. 5. Association of P-S6 suppression with response to RAF inhibition in BRAF-mutant melanoma patients.
(A)Representative images of paired before treatment and day 15 biopsies from two patients treated with vemurafenib and stained for P-S6 by immunohistochemistry. Note that P-S6 staining is maintained in the day 15 biopsy for patient 3. However, P-S6 staining is markedly diminished in tumor cells in the day 15 biopsy of patient 62, although some residual P-S6 staining is still present in macrophages and stromal cells within the tumor. Best objective tumor response by RECIST (SD, stable disease; PR, partial response) and progression-free survival (PFS) for each patient are also shown. Scale bar, 100 µm. (B) Progression-free survival curves for patients demonstrating inhibition (n = 6) and no inhibition (n = 3) of P-S6 (s240/244) in day 15 treatment biopsies, analyzed by log-rank test. HR, hazard ratio; 95% CI, 95% confidence interval. Patients remaining on study without evidence of progression at the time of the data cutoff are indicated by crosshatches. (C) P-S6 immunohistochemistry was performed on paired tumor biopsies from the same patient taken before treatment, on day 15 of treatment, and after disease progression but before discontinuation of therapy. The percent of tumor cells positive for P-S6 is shown for each biopsy. Scale bar, 50 µm. (D) CT scans of the same patient from (C) before treatment and after 4 months of therapy with dabrafenib and trametinib. Tumor is indicated by red arrows.
We also interrogated biopsies from a patient who initially responded to the combination of dabrafenib and trametinib but later developed resistance to treatment We saw complete suppression of P-S6 in a tumor biopsy obtained 15 days after initiation of treatment (Fig. 5C). This patient went on to achieve a complete response (total regression of target tumor lesions) by Response Evaluation Criteria in Solid Tumors (RECIST) (Fig. 5D). Ultimately, the patient developed a progressing subcutaneous nodule, and in a biopsy taken from this lesion at disease progression, while the patient was still on therapy, we observed recovery of P-S6 levels (Fig. 5C). The tight correlation between P-S6 and tumor response evident in these clinical specimens suggests that P-S6 may prove to be a robust biomarker of sensitivity in BRAF-mutant melanoma.
DISCUSSION
Here, we identified a correlation between sensitivity to MAPK inhibition and suppression of TORC1 in BRAF-mutant melanoma. Several additional factors have previously been suggested to predict resistance to MAPK pathway inhibition in BRAF-mutant melanoma cells, including loss of PTEN, increased basal levels of P-AKT, or induction of P-AKT levels after MAPK inhibition (16,17). Although a tendency toward resistance was observed for some melanoma cell lines bearing these markers, we failed to detect a significant correlation for most markers across our BRAF-mutant melanoma cell line panel after treatment with either vemurafenib or selumetinib. Although each of these mechanisms may play a role in resistance to MAPK inhibition, the interaction of these mechanisms and others that influence sensitivity may be sufficiently complex that evaluation of any of these alone may be inadequate to predict responsiveness. Indeed, a multitude of resistance mechanisms to RAF and MEK inhibitors has been identified in recent studies, including activation of receptor tyrosine kinases, amplification or alternative splicing of BRAF, mutation or overexpression of MAPK pathway signaling components (for example, NRAS and COT), or activation of the PI3K pathways (10–14, 39–42). This suggests that sensitivity to MAPK inhibition can be influenced by a complex network of signaling events, either through reactivation of the MAPK pathway or through ERK-independent resistance signals in the presence of sustained MAPK inhibition. Because TORC1 activity, which modulates S6 phosphorylation, is a point of convergence that integrates multiple important upstream signaling pathways in BRAF-mutant melanoma, it may be a critical downstream signaling node that can indicate sensitivity or resistance by capturing all of these diverse resistance mechanisms (18,19). Indeed, TORC1 remains active and P-S6 levels are maintained when resistance is mediated by any mechanism that leads to sustained MAPK signaling in the presence of MAPK inhibitors, as well as by mechanisms that lead to ERK-independent survival, such as activation of the PI3K–AKT pathway. Thus, by integrating these multiple upstream inputs, TORC1 activity and P-S6 levels may serve as a global functional indicator of sensitivity in BRAF-mutant melanoma. Additionally, because TORC1 serves as a key signaling node that integrates signals from several important upstream pathways, it is possible that suppression of TORC1 activity may be an effective indicator of sensitivity in other oncogene-addicted cancers with mutations in pathways upstream of TORC1. Work by Elkabets and colleagues, published in this issue of Science Translational Medicine (43), suggests that suppression of TORC1 activity can predict sensitivity of PIK3CA-mutant breast cancers to PI3K p110α inhibitors.
Despite the association of P-S6 levels with drug sensitivity, not every melanoma cell line with a treatment-induced decrease in P-S6 was sensitive to RAF or MEK inhibitors. Three of 10 cell lines with a treatment-induced decrease in P-S6 (UACC62, SKMEL28, and SKMEL39) showed a limited apoptotic response to vemurafenib or selumetinib. A likely explanation is that, although TORC1 effectively integrates multiple upstream signals that can lead to resistance, a change in TORC1 activity and P-S6 levels cannot account for resistance because of downstream events. Indeed, in all three of these cell lines that showed a decrease in P-S6 after treatment with RAF inhibition despite exhibiting drug resistance, we observed decreased expression of the downstream proapoptotic protein BIM, a known mechanism of resistance to MAPK inhibition, despite adequate suppression of P-ERK (35). Thus, P-S6 may need to be evaluated in the context of additional biomarkers (such as BIM) to optimize its predictive value. Still, all melanoma cells that failed to down-regulate P-S6 universally showed decreased sensitivity.
A potential limitation of this approach is that measuring a change in P-S6 after initiation of treatment requires a second tumor biopsy while the patient is on treatment. Still, if the information obtained through this approach can more effectively predict who is most or least likely to benefit from therapy, the ability to spare patients the added cost and potential toxicity of an ineffective therapy and the opportunity to switch to a potentially more active agent or combination of agents may justify the additional procedure. Moreover, the technique described herein can rapidly and effectively assess multiple markers, including P-S6 and P-ERK, in individual melanoma cells from FNAs, which are less invasive and safer than surgically obtained excisional biopsies and are routinely used in clinical practice. Additionally, in melanoma patients, roughly one-third of patients present with easily accessible subcutaneous nodules or lymph nodes that can be readily accessed for FNA biopsy with minimal risk (44).
Combinations of targeted therapies are under development to overcome specific mechanisms of resistance in BRAF-mutant melanoma. As noted above, a RAF and MEK inhibitor combination that overcomes multiple mechanisms of resistance as a result of reactivation of the MAPK pathway (10, 11, 39, 42) was recently reported to demonstrate promising activity in BRAF-mutant melanoma patients (38). We speculate that this RAF/MEK inhibitor combination may be most effective in patients whose melanomas exhibit resistance stemming from a failure to suppress P-ERK by single-agent RAF inhibitors. Dual RAF/MEK inhibition would be less effective in patients with ERK-independent resistance, in which adequate P-ERK suppression is achieved with a RAF inhibitor alone and alternative pathways are activated. In such cases, P-S6 measurement can effectively identify tumors with ERK-independent resistance, and our data suggest that alternative combinations of targeted therapies, such as a RAF inhibitor with a TORC inhibitor, a PI3K inhibitor, or a BH3 mimetic, may be effective. P-S6 may therefore constitute an important clinical biomarker whose integration into clinical trials could help optimize the targeted therapy of BRAF-mutant melanoma.
MATERIALS AND METHODS
Study design
The objective of this study was to identify potential biomarkers that may predict which BRAF-mutant melanomas are most likely to be sensitive or resistant to RAF or MEK inhibitors. Changes in pharmacodynamic markers after treatment with vemurafenib or selumetinib were evaluated in a panel of BRAF-mutant melanoma cell lines with varying sensitivities to these inhibitors. A decrease in S6 phosphorylation after treatment with RAF or MEK inhibitors was assessed by quantitative immunofluorescence microscopy in individual tumor cells obtained by FNA biopsies before and after treatment initiation from BRAF-mutant melanoma xenografts or from patients with BRAF-mutant melanoma. Previously obtained paired biopsies taken before and after initiation of RAF inhibitor therapy in BRAF-mutant melanoma patients were analyzed for a decrease in S6 phosphorylation after treatment initiation by immunohistochemistry, and these measurements were correlated with clinical outcome. Quantitation of P-S6 signal by immunohistochemistry before and after treatment was performed by two independent dermatopathologists who were blinded to the clinical data.
Cell lines and reagents
All cell lines were grown in Dulbecco’s modified Eagle’s medium (DMEM)/F12 (Gibco) with 10% fetal bovine serum (FBS), assayed in DMEM/F12 with 5% FBS, and obtained from the Massachusetts General Hospital Center for Molecular Therapeutics, which performs routine cell line authentication testing by single-nucleotide polymorphism and short tandem repeat analysis. Chemical inhibitors from the following sources were dissolved in dimethyl sulfoxide (DMSO) for in vitro studies: vemurafenib, ABT-263, and AZD8055 (Active Biochem); selumetinib (Otava Chemicals); GDC0941 (Selleck Chemicals); and BEZ235 (Novartis). Paclitaxel was obtained from the Massachusetts General Hospital Pharmacy and was diluted in saline.
Western blot analysis, immunoprecipitation, and antibodies
Western blotting was performed with standard methods. After treatment with indicated drugs, cells were washed with cold phosphate-buffered saline (PBS) and lysed in the following lysis buffer: 20 mM tris (pH 7.4), 150 mM NaCl, 1% NP-40,10% glycerol, 1 mM EDTA, 1 mM EGTA, 5 mM sodium pyrophosphate, 50 mM NaF, 10 nM β-glycerophosphate, 1 mM sodium vanadate, 0.5 mM dithiothreitol, leupeptin (4 µg/ml), pepstatin (4 µg/ml), aprotinin (4 µg/ml), 1 mM phenylmethylsulfonyl fluoride. Lysates were centrifuged at 16,000g for 5 min at 4°C Protein concentrations were determined by BCA assay (Thermo Scientific). Proteins were resolved by SDS–polyacrylamide gel electrophoresis and transferred onto a polyvinylidene difluoride membrane (Hybond-P, Amersham). Immunoblotting was performed per antibody manufacturer’s specifications. The following antibodies were purchased from Cell Signaling (catalog number, dilution): P-ERK (9101, 1:2000), P-S6 S235/236 (2211, 1:4000), P-S6 S240/244 (2215, 1:1000), P-AKT S473 (9271, 1:1000), BIM (2819, 1:1000), PUMA (4976, 1:1000), TSC2 (3612, 1:1000), PTEN (9552, 1:1000), P-p90RSK (9341, 1:1000), P-p70S6K (9205, 1:1000), 4EBP1 (9452, 1:1000), BCL2 (2872, 1:1000), and BCL-XL (2764, 1:1000). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibody was purchased from Millipore (MAB374, 1:1000), and MCL-1 antibody was purchased from Santa Cruz Biotechnology (sc-819, 1:500). P-S6 suppression by Western blot (minimum of three independent experiments) was assessed by quantitative chemiluminescence with the Syngene G:BOX camera (Synoptics) and Syngene GeneTools software (Synoptics). Suppression was defined as a 75% or greater reduction in P-S6 band intensity after vemurafenib or selumetinib treatment.
Determination of cell titer and Gl50
Cells were seeded at 2000 cells per well in parallel 96-well plates. The following day, cells were treated at least in triplicate with 3 µM vemurafenib or 1 µM selumetinib for 72 hours, and cell titer was determined with CellTiter-Glo assay (Promega) according to the manufacturer’s protocol. GI50 values (the concentration of inhibitor required to inhibit viable cell titer relative to control by 50%) were calculated with GraphPad Prism software.
Annexin V apoptosis assays
Cells were seeded at ∼30 to 40% confluence in 6-cm plates. After overnight incubation, medium was aspirated and replaced with medium with or without various concentrations of indicated drugs. After 72 hours, medium was collected. Cells were washed with PBS and trypsinized. PBS wash and trypsinized cells were added to the collected medium in a single tube. Cells were pelleted, washed once with PBS, and resuspended in annexin binding buffer (BD Biosciences) at ∼1 × 106 cells/ml. Cells were stained with propidium iodide (BD Biosciences) and annexin V–Cy5 (BioVision) according to the manufacturer’s protocol and assayed on an LSRII flow cytometer (BD Biosciences). The percentage of apoptotic cells was measured as the percentage of annexin V–positive cells (minimum of 10,000 cells analyzed per condition).
Lentiviral shRNA experiments
shRNA constructs in the pLKO.1 lentiviral vector containing the following targeting sequences were used: shGFP, 5’-GCAAGCTGACCCT-GAAGTTCAT-3’; shTSC2, 5’-CGACGAGTCAAACAAGCCAAT-3’; shPUMA, 5’-GAGGGTCCTGTACAATCTCAT-3’.
Lentiviral particles were generated, and target cells were infected as described previously (45). The day before infection, cells were seeded in six-well plates at 1 × 105 to 2 × 105 cells per well. The morning after infection, cells were treated with puromycin (2 µg/ml) for 48 hours to eliminate uninfected cells. Medium without puromycin containing the indicated concentrations of drug was then added for 24 hours for Western blot analysis or for 72 hours for apoptosis assays.
Immunohistochemistry
Immunohistochemistry on formalin-fixed, paraffin-embedded tissue was performed for P-ERK as previously described (46). Immunohistochemistry for P-S6 S240/244 was performed with Cell Signaling antibody #5364 [1:400 dilution in 1% bovine serum albumin (BSA)/PBS overnight at 4°C] according to the manufacturer’s protocol.
P-S6 staining of human tumor biopsies was scored by two independent dermatopathologists (A.P. and R.M.N.), who were blinded to the clinical outcomes. The percentage of tumor cells that were positive for P-S6 was scored by each dermatopathologist independently, and the average percentage score between the two independent measurements was used. The relative change in P-S6 score for each patient was calculated by subtracting the on-treatment P-S6 score from the before treatment P-S6 score and dividing the difference by the before treatment P-S6 score. P-S6 was classified as “inhibited” after treatment if a relative decrease in P-S6 score of at least 50% was observed from pre-treatment to on-treatment biopsy for a given patient.
Mouse treatment studies
WM164 or IGR1 cells were injected (5 × 106 cells per injection) into the flanks of athymic nude mice (Charles River Laboratories). Once tumors reached an average volume of ∼100 to 200 mm3, mice were randomized into treatment arms, and tumor volume was assessed by caliper measurements. Vemurafenib for mouse studies was obtained from the Massachusetts General Hospital Pharmacy, formulated in 5% DMSO and 1% methylcellulose, and dosed at 75 mg/kg twice daily by oral gavage. Selumetinib was formulated in 0.5% methylcellulose and 0.4% polysorbate and dosed at 25 mg/kg twice daily by oral gavage. For pharmacodynamic studies, tumor tissue was harvested and formalin-fixed 3 hours after the morning doses of drug on the specified day of treatment. All mouse studies were conducted through Institutional Animal Care and Use Committee–approved animal protocols in accordance with institutional guidelines.
FNA of mouse subcutaneous tumor xenografts was performed in isoflurane-anesthetized mice with a 21-gauge needle two to three times. The cells were fixed with 4% formaldehyde, treated with collagenase IV, and deposited onto glass slides by centrifugation at 800 rpm for 10 min in a CytoSpin cytocentrifuge (Thermo Scientific).
Excisional and FNA biopsy
All human tumor biopsy specimens were obtained through Institutional Review Board–approved protocols. Tumor biopsies were obtained from patients with BRAF V600 mutation–positive melanoma 0 to 21 days before initiation of therapy (with vemurafenib or with the combination of dabrafenib and trametinib) and at day 15 of therapy (±8 days) or additionally at the time of recurrence. Excisional biopsy specimens from patients #59 to #97 were procured and provided by Roche. All remaining biopsy specimens were obtained at the Massachusetts General Hospital and were performed by an experienced surgeon (J.A.W.). FNAs were performed with a 21-gauge needle two to three times, fixed with 4% formaldehyde, treated with collagenase IV, and deposited onto glass slides by centrifugation at 800 rpm for 10 min in a CytoSpin cytocentrifuge (Thermo Scientific). Progression-free survival data and tumor response measurements by RECIST were provided by Roche for patients #59 to #97 or were obtained from the clinical record.
Immunofluorescence microscopy, quantitation, and analysis
FNA samples processed as above were blocked/permeabilized with 5% goat serum/0.3% Triton X-100 for 1 hour at room temperature. Primary antibody staining was performed overnight at 4°C in 1% BSA/0.3% Triton X-100 with antibodies against melanoma antigens NG2 (R&D Systems, MAB2585, 1:250), MART1 (Dako, m7196, 1:50), and HMB45 (Santa Cruz Biotechnology, sc-59305, 1:100); CD45 (for mouse, BD Biosciences, BD553078, 1:250; for human, Santa Cruz Biotechnology, sc-59305, 1:100) to discriminate white blood cells; and P-S6 S240/244 (5364, 1:200) and P-ERK T202/Y204 (4370, 1:50), both from Cell Signaling Technology. Secondary antibodies conjugated to appropriate fluorophores were from Invitrogen. Slides were visualized and scanned at ×20 magnification with an automated scanning microscope and customized cell segmentation algorithms (BioView).
All images of tumor cells were reviewed manually to verify cellular morphology. Data for cellular events were displayed as histograms of mean fluorescence intensity for each measured fluorophore per cellular event, and tumor cells were identified as the CD45-negative, melanoma antigen–positive, DAPI-positive fraction in each sample. The distribution of P-ERK or P-S6 mean fluorescence in each population of tumor cells was compared across different specimens obtained from the same mouse or patient with a Student’s t test (unequal variances).
Statistical analyses
Two-tailed Student’s t test was used for Figs. 1, C and D, 2A, and 3A and figs. S4, C to E, and S10. Statistical significance was established for P < 0.05. Student’s t test (unequal variances) was used as described above for Figs. 3C and Fig. 4, A and C, and figs. S13 and S14. One-way ANOVA with Bonferroni posttest was used for Fig. 2, B to D, and fig. S9. Progression-free survival data were analyzed by log-rank test (Fig. 5B) with GraphPad Prism software. Linear regression analysis (fig. S4B) was also performed with GraphPad Prism software.
Supplementary Material
Acknowledgments
We thank Roche for providing paired melanoma biopsy samples from patients treated with vemurafenib.
Funding:
This work was funded by a Damon Runyon Clinical Investigator Award (to R.B.C.), NIH/National Institute of Dental and Craniofacial Research grant 1K08DE020139 (to S.M.R.), NIH grant R01CA137008 (to J.A.E.), National Cancer Institute grant CA129933 (to D.A.H.).
Footnotes
SUPPLEMENTARY MATERIALS
www.sciencetranslationalmedicine.org/cgi/content/full/5/196/196ra98/DC1
Fig. S1. Sensitivity of BRAF-mutant melanoma cell lines to selumetinib.
Fig. S2. Signaling changes after vemurafenib treatment.
Fig. S3. Signaling changes after selumetinib treatment.
Fig. S4. Correlation of sensitivity to MAPK inhibitors with PTEN status and AKT phosphorylation.
Fig. S5. TORC1 inhibition by vemurafenib in vemurafenib-sensitive BRAF-mutant melanoma cells.
Fig. S6. Induction of apoptosis by inhibitor combinations in COLO858 cells.
Fig. S7. Signaling effects of vemurafenib in combination with TORC or PI3K inhibitors.
Fig. S8. Effect of vemurafenib on apoptotic proteins.
Fig. S9. Decreased apoptotic response to vemurafenib resulting from PUMA knockdown.
Fig. S10. Change in volume of individual tumors in BRAF-mutant melanoma xenografts treated with vemurafenib or selumetinib.
Fig. S11. Effect of vemurafenib on P-ERK and P-S6 in melanoma xenografts.
Fig. S12. Effect of selumetinib on P-ERK and P-S6 in melanoma xenografts.
Fig. S13. Serial FNAs from vemurafenib-treated melanoma xenografts.
Fig. S14. Serial FNAs from selumetinib-treated melanoma xenografts.
Table S1. Melanoma cell lines used in the study.
Table S2. P-S6 scores in paired pretreatment and day 15 biopsies.
Table S3. Data for Fig. 2A.
Table S4. Data for Fig. 2B.
Table S5. Data for Fig. 2C.
Table S6. Data for fig. S9.
Table S7. Data for Fig. 2D.
Author contributions:
R.B.C., S.M.R., A.N.H., A.C.F., A.P., R.M.N., R.D.B., J.T.G., D.W., J.W., M.M.-K., and J.A.W. performed the experiments. R.B.C, S.M.R., A.N.H., A.C.F., A.P., R.M.N., M.M.-K, JA.W., S.M, J.S., KT.F., D.A.H, and J.A.E. designed the experiments and analyzed the data. R.B.C., S.M.R, K.T.F., D.A.H., and J.A.E. wrote the manuscript.
Competing interests:
J.S. is an employee of Genentech Inc. and equity holder of Roche Pharmaceuticals. J.A.E. has paid consulting relationships with Novartis, Sanofi-Aventis, AstraZeneca, GlaxoSmithKline, Genentech, Abbott, Intellikine, Cell Signaling, Pathway Therapeutics, and Chugai.
REFERENCES
- 1.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, Davis N, Dicks E, Ewing R, Floyd Y, Gray K, Hall S, Hawes R, Hughes J, Kosmidou V, Menzies A, Mould C, Parker A, Stevens C, Watt S, Hooper S, Wilson R, Jayatilake H, Gusterson BA, Cooper C, Shipley J, Hargrave D, Pritchard-Jones K, Maitland N, Chenevix-Trench G, Riggins GJ, Bigner DD, Palmieri G, Cossu A, Flanagan A, Nicholson A, Ho JW, Leung SY, Yuen ST, Weber BL, Seigler HF, Darrow TL, Paterson H, Marais R, Marshall CJ, Wooster R, Stratton MR, Futreal PA. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 2.Flaherty KT. BRAF inhibitors and melanoma. Cancer J. 2011;17:505–511. doi: 10.1097/PPO.0b013e31823e5357. [DOI] [PubMed] [Google Scholar]
- 3.Flaherty KT, Hodi FS, Fisher DE. From genes to drugs: Targeted strategies for melanoma. Nat. Rev. Cancer. 2012;12:349–361. doi: 10.1038/nrc3218. [DOI] [PubMed] [Google Scholar]
- 4.Hauschild A, Grob JJ, Demidov LV, Jouary T, Gutzmer R, Millward M, Rutkowski P, Blank CU, Miller WH, Jr., Kaempgen E, Martfn-Algarra S, Karaszewska B, Mauch C, Chiarion-Sileni V, Martin AM, Swann S, Haney P, Mirakhur B, Guckert ME, Goodman V, Chapman PB. Dabrafenib in BRAF-mutated metastatic melanoma: A multicentre open-label phase 3 randomised controlled trial. Lancet. 2012;380:358–365. doi: 10.1016/S0140-6736(12)60868-X. [DOI] [PubMed] [Google Scholar]
- 5.Flaherty KT, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA, O'Dwyer PJ, Lee RJ, Grippo JF, Nolop K, Chapman PB. Inhibition of mutated activated BRAF in metastatic melanoma. N. Engl. J. Med. 2010;363:809–819. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, Dummer R, Garbe C, Testori A, Maio M, Hogg D, Lorigan P, Lebbe C, Jouary T, Schadendorf D, Ribas A, O'Day SJ, Sosman JA, Kirkwood JM, Eggermont AM, Dreno B, Nolop K, Li J, Nelson B, Hou J, Lee RJ, Flaherty KT, McArthur GA. BRIM-3 Study Group Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl. J. Med. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sosman JA, Kim KB, Schuchter L, Gonzalez R, Pavlick AC, Weber JS, McArthur GA, Hutson TE, Moschos SJ, Flaherty KT, Hersey P, Kefford R, Lawrence D, Puzanov I, Lew’s KD, Amaravadi RK, Chmielowski B, Lawrence HJ, Shyr Y, Ye F, Li J, Nolop KB, Lee RJ, Joe AK, Ribas A. Survival in BRAF V600-mutant advanced melanoma treated with vemurafenib. N Engl. J. Med. 2012;366:707–714. doi: 10.1056/NEJMoa1112302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Falchook GS, Lewis DK, Infante JR, Gordon MS, Vogelzang NJ, DeMarini DJ, Sun P, Moy C, Szabo SA, Roadcap LT, Peddareddigari VG, Lebowitz PF, Le NT, Burris HA, III, Messersmith WA, O'Dwyer PJ, Kim KB, Flaherty K, Bendell JC, Gonzalez R, Kurzrock R, Fecher LA. Activity of the oral MEK inhibitor trametinib in patients with advanced melanoma: A phase 1 dose-escalation trial. Lancet Oncol. 2012;13:782–789. doi: 10.1016/S1470-2045(12)70269-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flaherty KT, Robert C, Hersey P, Nathan P, Garbe C, Milhem M, Demidov LV, Hassel JC, Rutkowski P, Mohr P, Dummer R, Trefzer U, Larkin JM, Utikal J, Dreno B, Nyakas M, Middleton MR, Becker JC, Casey M, Sherman LJ, Wu FS, Ouellet D, Martin AM, Patel K, Schadendorf D. METRIC; Study Group Improved survival with MEK inhibition in BRAF-mutated melanoma. N Engl. J. Med. 2012;367:107–114. doi: 10.1056/NEJMoa1203421. [DOI] [PubMed] [Google Scholar]
- 10.Corcoran RB, Settleman J, Engelman JA. Potential therapeutic strategies to overcome acquired resistance to BRAF or MEK inhibitors in BRAF mutant cancers. Oncotarget. 2011;2:336–346. doi: 10.18632/oncotarget.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Poulikakos PI, Persaud Y, Janakiraman M, Kong X, Ng C, Moriceau G, Shi H, Atefi M, Titz B, Gabay MT, Salton M, Dahlman KB, Tadi M, Wargo JA, Flaherty KT, Kelley MC, Misteli T, Chapman PB, Sosman JA, Graeber TG, Ribas A, Lo RS, Rosen N, Solit DB. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E) Nature. 2011;480:387–390. doi: 10.1038/nature10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi H, Moriceau G, Kong X, Lee MK, Lee H, Koya RC, Ng C, Chodon T, Scolyer RA, Dahlman KB, Sosman JA, Kefford RF, Long GV, Nelson SF, Ribas A, Lo RS. Melanoma whole-exome sequencing identifies (V600E)B-RAF amplification-mediated acquired B-RAF inhibitor resistance. Nat. Commun. 2012;3:724. doi: 10.1038/ncomms1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilson TR, Fridlyand J, Yan Y, Penuel E, Burton L, Chan E, Peng J, Lin E, Wang Y, Sosman J, Ribas A, Li J, Moffat J, Sutherlin DP, Koeppen H, Merchant M, Neve R, Settleman J. Widespread potential for growth-factor-driven resistance to anticancer kinase inhibitors. Nature. 2012;487:505–509. doi: 10.1038/nature11249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Straussman R, Morikawa T, Shee K, Barzily-Rokni M, Qian ZR, Du J, Davis A, Mongare MM, Gould J, Frederick DT, Cooper ZA, Chapman PB, Solit DB, Ribas A, Lo RS, Flaherty KT, Ogino S, Wargo JA, Golub TR. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature. 2012;380:358–365. doi: 10.1038/nature11183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bollag G, Hirth P, Tsai J, Zhang J, Ibrahim PN, Cho H, Spevak W, Zhang C, Zhang Y, Habets G, Burton EA, Wong B, Tsang G, West BL, Powell B, Shellooe R, Marimuthu A, Nguyen H, Zhang KY, Artis DR, Schlessinger J, Su F, Higgins B, Iyer R, D'Andrea K, Koehler A, Stumm M, Lin PS, Lee RJ, Grippo J, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA, Chapman PB, Flaherty TK, Xu X, Nathanson KL, Nolop K. Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature. 2010;467:596–599. doi: 10.1038/nature09454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gopal YN, Deng W, Woodman SE, Komurov K, Ram P, Smith PD, Davies MA. Basal and treatment-induced activation of AKT mediates resistance to cell death by AZD6244 (ARRY-142886) in BRAF-mutant human cutaneous melanoma cells. Cancer Res. 2010;70:8736–8747. doi: 10.1158/0008-5472.CAN-10-0902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xing F, Persaud Y, Pratilas CA, Taylor BS, Janakiraman M, She QB, Gallardo H, Liu C, Merghoub T, Hefter B, Dolgalev I, Viale A, Heguy A, De Stanchina E, Cobrinik D, Bollag G, Wolchok J, Houghton A, Solit DB. Concurrent loss of the PTEN and RB1 tumor suppressors attenuates RAF dependence in melanomas harboring V600EBRAF. Oncogene. 2012;31:446–457. doi: 10.1038/onc.2011.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Magnuson B, Ekim B, Fingar DC. Regulation and function of ribosomal protein S6 kinase (S6K) within mTOR signalling networks. Biochem. J. 2012;441:1–21. doi: 10.1042/BJ20110892. [DOI] [PubMed] [Google Scholar]
- 19.Laplante M, Sabatini DM. mTOR signaling at a glance. J. Cell Sci. 2009;122:3589–3594. doi: 10.1242/jcs.051011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roux PP, Ballif BA, Anjum R, Gygi SP, Blenis J. Tumor-promoting phorbol esters and activated Ras inactivate the tuberous sclerosis tumor suppressor complex via p90 ribosomal S6 kinase. Proc. Natl. Acad. Sci. U.S.A. 2004;101:13489–13494. doi: 10.1073/pnas.0405659101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma L, Chen Z, Erdjument-Bromage H, Tempst P, Pandolfi PP. Phosphorylation and functional inactivation of TSC2 by Erk implications for tuberous sclerosis and cancer pathogenesis. Cell. 2005;121:179–193. doi: 10.1016/j.cell.2005.02.031. [DOI] [PubMed] [Google Scholar]
- 22.Shaw RJ, Bardeesy N, Manning BD, Lopez L, Kosmatka M, DePinho RA, Cantley LC. The LKB1 tumor suppressor negatively regulates mTOR signaling. Cancer Cell. 2004;6:91–99. doi: 10.1016/j.ccr.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 23.Dancey J. mTOR signaling and drug development in cancer. Nat. Rev. Clin. Oncol. 2010;7:209–219. doi: 10.1038/nrclinonc.2010.21. [DOI] [PubMed] [Google Scholar]
- 24.Liu P, Cheng H, Roberts TM, Zhao JJ. Targeting the phosphoinositide 3-kinase pathway in cancer. Nat. Rev. Drug Discov. 2009;8:627–644. doi: 10.1038/nrd2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roux PP, Shahbazian D, Vu H, Holz MK, Cohen MS, Taunton J, Sonenberg N, Blenis J. RAS/ERK signaling promotes site-specific ribosomal protein S6 phosphorylation via RSK and stimulates cap-dependent translation. J. Biol. Chem. 2007;282:14056–14064. doi: 10.1074/jbc.M700906200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Inoki K, Li Y, Xu T, Guan KL. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 2003;17:1829–1834. doi: 10.1101/gad.1110003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chresta CM, Davies BR, Hickson I, Harding T, Cosulich S, Critchlow SE, Vincent JP, Ellston R, Jones D, Sini P, James D, Howard Z, Dudley P, Hughes G, Smith L, Maguire S, Hummersone M, Malagu K, Menear K, Jenkins R, Jacobsen M, Smith GC, Guichard S, Pass M. AZD8055 is a potent selective and orally bioavailable ATP-competitive mammalian target of rapamycin kinase inhibitor with in vitro and in vivo antitumor activity. Cancer Res. 2010;70:288–298. doi: 10.1158/0008-5472.CAN-09-1751. [DOI] [PubMed] [Google Scholar]
- 28.Engelman JA. Targeting PI3K signalling in cancer: Opportunities challenges and limitations. Nat. Rev. Cancer. 2009;9:550–562. doi: 10.1038/nrc2664. [DOI] [PubMed] [Google Scholar]
- 29.Courtney KD, Corcoran RB, Engelman JA. The PI3K pathway as drug target in human cancer. J. Clin. Oncol. 2010;28:1075–1083. doi: 10.1200/JCO.2009.25.3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Folkes AJ, Ahmadi K, K Alderton W, Alix S, Baker SJ, Box G, Chuckowree IS, Clarke PA, Depledge P, Eccles SA, Friedman LS, Hayes A, Hancox TC, Kugendradas A, Lensun L, Moore P, Olivero AG, Pang J, Patel S, Pergl-Wilson GH, Raynaud FI, Robson A, Saghir N, Salphati L, Sohal S, Ultsch MH, Valenti M, Wallweber HJ, Wan NC, Wiesmann C, Workman P, Zhyvoloup A, Zvelebil MJ, Shuttleworth SJ. The identification of 2-(1H–indazol-4-yl)-6-(4-methanesulfonyl-piperazin-1-ylmethyl)-4-morpholin-4-yl-thieno[3,2-d]pyrimidine (GDC-0941) as a potent selective orally bioavailable inhibitor of class I PI3 kinase for the treatment of cancer. J. Med. Chem. 2008;51:5522–5532. doi: 10.1021/jm800295d. [DOI] [PubMed] [Google Scholar]
- 31.Maira SM, Stauffer F, Brueggen J, Furet P, Schnell C, Fritsch C, Brachmann S, Chene P, De Pover A, Schoemaker K, Fabbro D, Gabriel D, Simonen M, Murphy L, Finan P, Sellers W, Garcfa-Echeverrfa C. Identification characterization of NVP-BEZ235 a new orally available dual phosphatidylinositol 3-kinase/mammalian target of rapamycin inhibitor with potent in vivo antitumor activity. Mol. Cancer Ther. 2008;7:1851–1863. doi: 10.1158/1535-7163.MCT-08-0017. [DOI] [PubMed] [Google Scholar]
- 32.Ley R, Balmanno K, Hadfield K, Weston C, Cook SJ. Activation of the ERK1/2 signaling pathway promotes phosphorylation proteasome-dependent degradation of the BH3-only protein, Bim. J. Biol. Chem. 2003;278:18811–18816. doi: 10.1074/jbc.M301010200. [DOI] [PubMed] [Google Scholar]
- 33.Wang YF, Jiang CC, Kiejda KA, Gillespie S, Zhang XD, Hersey P. Apoptosis induction in human melanoma cells by inhibition of MEK is caspase-independent mediated by the Bcl-2 family members PUMA, Bim and Mcl-1. Clin. Cancer Res. 2007;13:4934–4942. doi: 10.1158/1078-0432.CCR-07-0665. [DOI] [PubMed] [Google Scholar]
- 34.Faber AC, Li D, Song Y, C Liang M, Yeap BY, Branson RT, Lifshits E, Chen Z, Maira SM, Garcfa-Echeverrfa C, Wong KK, Engelman JA. Differential induction of apoptosis in HER2 and EGFR addicted cancers following PI3K inhibition. Proc. Natl. Acad. Sci. USA. 2009;106:19503–19508. doi: 10.1073/pnas.0905056106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Faber AC, Corcoran RB, Ebi H, Sequist LV, Waltman BA, Chung E, Incio J, Digumarthy SR, Pollack SF, Song Y, Muzikansky A, Lifshits E, Roberge S, Coffman EJ, Benes CH, Gomez HL, Baselga J, Arteaga CL, Rivera MN, Dias-Santagata D, Jain RK, Engelman JA. BIM expression in treatment-naTve cancers predicts responsiveness to kinase inhibitors. Cancer Discov. 2011;1:352–365. doi: 10.1158/2159-8290.CD-11-0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tse C, Shoemaker AR, Adickes J, Anderson MG, Chen J, Jin S, Johnson EF, Marsh KC, Mitten MJ, Nimmer P, Roberts L, Tahir SK, Xiao Y, Yang X, Zhang H, Fesik S, Rosenberg SH, Elmore SW. ABT-263: A potent and orally bioavailable Bcl-2 family inhibitor. Cancer Res. 2008;68:3421–3428. doi: 10.1158/0008-5472.CAN-07-5836. [DOI] [PubMed] [Google Scholar]
- 37.Murali R, Thompson JF, Uren RF, Scolyer RA. Fine-needle biopsy of metastatic melanoma: Clinical use and new applications. Lancet Oncol. 2010;11:391–400. doi: 10.1016/S1470-2045(09)70332-8. [DOI] [PubMed] [Google Scholar]
- 38.Flaherty KT, Infante JR, Daud A, Gonzalez R, Kefford RF, Sosman J, Hamid O, Schuchter L, Cebon J, Ibrahim N, Kudchadkar R, Burris HA, III, Falchook G, Algazi A, Lewis K, Long GV, Puzanov I, Lebowitz P, Singh A, Little S, Sun P, Allred A, Ouellet D, Kim KB, Patel K, Weber J. Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. N. Engl. J. Med. 2012;367:1694–1703. doi: 10.1056/NEJMoa1210093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Corcoran RB, Dias-Santagata D, Bergethon K, lafrate AJ, Settleman J, Engelman JA. BRAF gene amplification can promote acquired resistance to MEK inhibitors in cancer cells harboring the BRAF V600E mutation. Sci. Signal. 2010;3 doi: 10.1126/scisignal.2001148. ra84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nazarian R, Shi H, Wang Q, Kong X, Koya RC, Lee H, Chen Z, Lee KM, Attar N, Sazegar H, Chodon T, Nelson SF, McArthur G, Sosman JA, Ribas A, Lo RS. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–977. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Villanueva J, Vultur A, Lee JT, Somasundaram R, Fukunaga-Kalabis M, Cipolla AK, Wubbenhorst B, Xu X, Gimotty PA, Kee D, Santiago-Walker AE, Letrero R, D'Andrea K, Pushparajan A, Hayden JE, Brown KD, Laquerre S, McArthur GA, Sosman JA, Nathanson KL, Herlyn M. Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF-1R/PI3K. Cancer Cell. 2010;18:683–695. doi: 10.1016/j.ccr.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Johannessen CM, Boehm JS, Kim SY, Thomas SR, Wardwell L, Johnson LA, Emery CM, Stransky N, Cogdill AP, Barretina J, Caponigro G, Hieronymus H, Murray RR, Salehi-Ashtiani K, Hill DE, Vidal M, Zhao JJ, Yang X, Alkan O, Kim S, Harris JL, Wilson CJ, Myer VE, Finan PM, Root DE, Roberts TM, Golub T, Flaherty KT, Dummer R, Weber BL, Sellers WR, Schlegel R, Wargo JA, Hahn WC, Garraway LA. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature. 2010;468:968–972. doi: 10.1038/nature09627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Elkabets M, Vora S, Juric D, Morse N, Mino-Kenudson M, Muranen T, Tao J, Campos AB, Rodon J, Ibrahim YH, Serra V, Rodrik-Outmezguine V, Hazra S, Singh S, Kim P, Quadt C, Liu M, Huang A, Rosen N, Engelman JA, Scaltriti M, Baselga J. mTORG inhibition is required for sensitivity to PI3K p110a inhibitors in Plk3CA-mutant breast cancer. Sci. Transl. Med. 2013;5:196ra99. doi: 10.1126/scitranslmed.3005747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Testori A, Rutkowski P, Marsden J, Bastholt L, Chiarion-Sileni V, Hauschild A, Eggermont AM. Surgery and radiotherapy in the treatment of cutaneous melanoma. Ann. Oncol. 2009;20(Suppl. 6):Vi22–vi29. doi: 10.1093/annonc/mdp257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, Piqani B, Eisenhaure TM, Luo B, Grenier JK, Carpenter AE, Foo SY, Stewart SA, Stockwell BR, Hacohen N, Hahn CW, Lander ES, Sabatini DM, Root DE. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 46.Engelman JA, Chen L, Tan X, Crosby K, Guimaraes AR, Upadhyay R, Maira M, McNamara K, Perera SA, Song Y, Chirieac LR, Kaur R, Lightbown A, Simendinger J, Li T, Padera RF, Garcfa-Echeverrfa C, Weissleder R, Mahmood U, Cantley LC, Wong KK. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat. Med. 2008;14:1351–1356. doi: 10.1038/nm.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.