Abstract
Regulatory T cells (Tregs) play a crucial role in the immune system by preventing autoimmunity, limiting immunopathology, and maintaining immune homeostasis1. However, they also represent a major barrier to effective anti-tumor immunity and sterilizing immunity to chronic viral infections1. The transcription factor Foxp3 plays a major role in the development and programming of Treg cells2,3. The relative stability of Tregs at inflammatory disease sites has been highly contentious4-6. There is considerable interest in identifying pathways that control Treg stability as many immune-mediated diseases are characterized by either exacerbated or limited Treg function. Here we show that the immune cell-expressed ligand semaphorin-4a (Sema4a) and the Treg-expressed receptor neuropilin-1 (Nrp1) interact to potentiate Treg function and survival in vitro and in inflammatory sites in vivo. Nrp1 is dispensable for suppression of autoimmunity and maintenance of immune homeostasis, but is required by Tregs to limit anti-tumor immune responses and to cure established inflammatory colitis. Sema4a ligation of Nrp1 restrained Akt phosphorylation cellularly and at the immunologic synapse (IS) via phosphatase and tensin homolog (PTEN), which increased nuclear localization of the transcription factor Foxo3a. The Nrp1-induced transcriptome promoted Treg stability by enhancing quiescence/survival factors while inhibiting programs that promote differentiation. Importantly, this Nrp1-dependent molecular program is evident in intratumoral Tregs. Our data support a model in which Treg stability can be subverted in certain inflammatory sites, but is maintained by a Sema4a:Nrp1 axis, highlighting this pathway as a potential therapeutic target that could limit Treg-mediated tumor-induced tolerance without inducing autoimmunity.
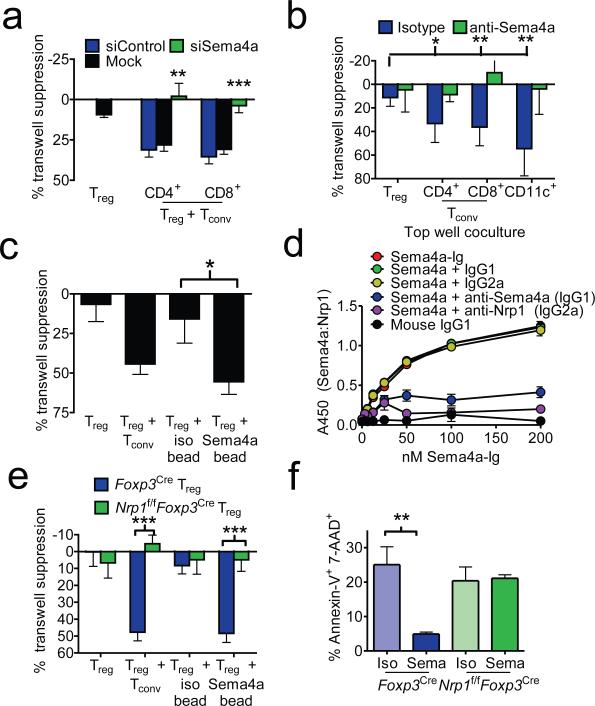
It was initially suggested that Treg-mediated suppression was contact-dependent, and soluble factors were insufficient, as purified Treg cells failed to suppress across a permeable transwell membrane3,7. However, we have previously shown that additional signals, derived from co-cultured T cells, are required to potentiate maximal Treg suppression via soluble factors across a transwell membrane8. Indeed, Tregs stimulated in the presence of fixed or live CD4+ conventional T cells (Tconv) in the top chamber of a transwell plate can suppress Tconv in the bottom chamber (Supplementary Fig. 1a). To determine the Tconv-derived signals responsible for potentiating/boosting Treg-mediated suppression, we compared the Tconv and Treg transcriptome and identified Sema4a as a potential ligand that could boost Treg function (Supplementary Fig. 1b-d). Sema4a is involved in neural patterning and has been implicated in modulating immune function, with hematopoietic cell expression restricted to CD4+ and CD8+ T cells, natural killer cells and dendritic cells (DCs)9. Using RNA interference, loss-variant selection and overexpression in hybridomas, and antibody blockade, we confirmed that Sema4a was necessary for Tconv and DCs to potentiate Treg-mediated transwell suppression (Fig. 1a, b and Supplementary Fig. 2a-e). Sema4a is also sufficient to potentiate suppression as Tregs cultured in the presence of Sema4a-Ig-coated beads, instead of fixed Tconv potentiated transwell suppression (Fig. 1c). In contrast, soluble Sema4a-Ig inhibited Tconv-potentiated, Treg-mediated transwell suppression (Supplementary Fig. 2f). These data demonstrate that Sema4a is required and sufficient to potentiate Treg function via soluble factors in vitro.
Figure 1. Sema4a binds Nrp1 to potentiate Treg function and survival in vitro.
a, Transwell suppression assay in which Treg cells were cocultured in the top chamber of a transwell plate with anti-CD3/anti-CD28 coated beads in the presence or absence of CD4+ or CD8+ Tconv cells that had been previously transfected with scrambled or siRNA to Sema4a. Proliferation of Tconv cells stimulated with anti-CD3/anti-CD28 coated beads in the bottom chambers was measured by [3H]-thymidine uptake. b, Transwell suppression assay with Treg cells cocultured in top chamber with CD4+, CD8+, or CD11c+ cells including anti-Sema4a or its isotype control. c, Transwell suppression assay in which Treg were cocultured in the absence of Tconv but in the presence of beads coated with Sema4a-Ig or its isotype control. d, ELISA based binding assay in which plates coated with Nrp1 were incubated with Sema4a-Ig or IgG1 in the presence of various blocking antibodies. Sema4a-Ig was detected using an isotype specific antibody. e, Transwell suppression assay where Treg were purified by flow cytometry from Foxp3Cre or Nrp1f/fFoxp3Cre mice. f, Annexin V-7-AAD staining of Treg stimulated for 48 h in vitro in the presence of Sema4a-Ig or its isotype control. Results represent the mean of five independent experiments. *, p < 0.05, **, p < 0.01, ***, p < 0.001 by unpaired t-test. Error bars indicate s.e.m.
Nrp1 is a homogeneously expressed marker of thymically-derived Tregs, and has also been shown to bind Sema3a, which shares homology with Sema4a, and vascular endothelial growth factor (VEGF) in mediating neural axon growth and angiogenesis (Supplementary Fig. 3a)10-13. Thus, we hypothesized that Nrp1 functions as a receptor for Sema4a on Tregs. Sema4a could directly and specifically bind to Nrp1, albeit with slightly lower avidity to Sema3a, as demonstrated in an ELISA-based binding assay with recombinant Nrp1 and Sema family proteins and blocking antibodies to Nrp1 or Sema4a (Fig. 1d and Supplementary Fig. 3b-d). Sema4a-Ig also bound to littermate control (Foxp3YFP-iCre, labeled henceforth Foxp3Cre) but not Nrp1-deficient (Foxp3YFP-iCre × Nrp1f/f, labeled henceforth Nrp1f/fFoxp3Cre) Tregs in a flow cytometric assay (Supplementary Fig. 3e). These data support a direct interaction between Nrp1 and Sema4a.
While Nrp1-deficient Tregs develop normally and can suppress in a “classical” contact-dependent suppression assay (Supplementary Fig. 4a), they were unable to suppress across a transwell membrane in the presence of either Tconv or Sema4a-Ig-coated beads (Fig. 1e). Transwell suppression can be blocked by anti-Nrp1 and, as previously reported, is dependent on IL-10 and IL-35 (Supplementary Fig. 4b-d).8 Sema4a:Nrp1 ligation substantially reduced cell death and modestly increased proliferation, and while single-cell levels of IL-10 and IL-35 did not change, increased survival resulted in greater amounts of IL-10 and IL-35 in Sema4a-treated Treg cultures (Fig. 1f and Supplementary Fig. 4e-i). Expression NRP1 on human Tregs has been contentious14,15. We observed sustained NRP1 expression, albeit modest, was observed on optimally suppressive Tregs that could mediate transwell suppression in response to human SEMA4A in a NRP1-dependent manner (Supplementary Fig. 5). Taken together, these data suggest Sema4a:Nrp1 interaction promotes Treg survival and function.
Nrp1f/fFoxp3Cre mice did not exhibit any autoimmune phenotype up to 16 months old (data not shown). In addition, the capacity of Foxp3Cre and Nrp1f/fFoxp3Cre Tregs to limit the development of the autoimmune sequelae caused by Foxp3 deletion was comparable (Supplementary Fig. 6), suggesting that maintenance of immune homeostasis and prevention of autoimmunity may not require Nrp1 signaling.
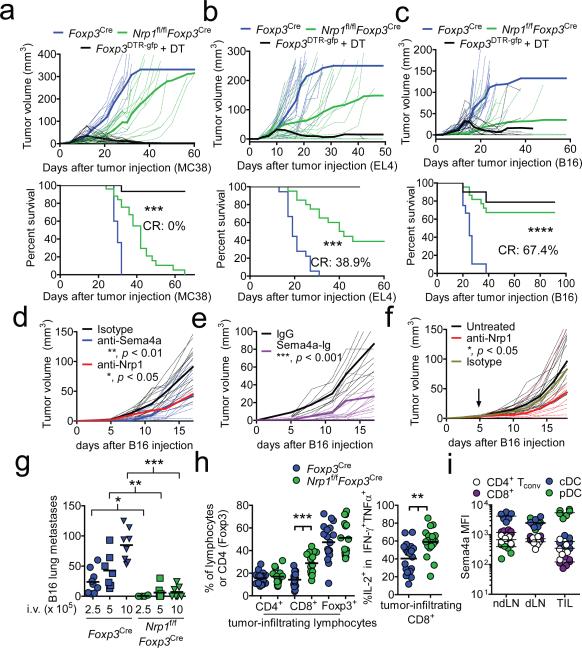
We hypothesized that Nrp1 signaling may regulate Treg function under inflammatory conditions. Tregs are recruited to and are induced by tumor cells, which consequently hamper protective anti-tumor immunity16. Foxp3DTR.gfp mice, which allow for conditional Treg deletion following diphtheria toxin (DT) treatment17, can clear MC38 adenocarcinoma, EL4 thymoma, and B16 melanoma tumors when treated with DT at the time of inoculation, although they invariably succumb to autoimmune disease (Fig. 2a-c). Strikingly, Nrp1f/fFoxp3Cre mice exhibited reduced, delayed tumor growth and increased survival, especially with B16 melanoma, without any detectable autoimmune consequences (Fig. 2a-c and data not shown). In wild-type C57BL/6 mice, blockade of this pathway using Sema4a mAb, Nrp1 mAb (which does not block Nrp1:VEGF interaction), and Sema4a-Ig (which acts as a soluble antagonist), significantly decreased tumor growth (Fig. 2d-f and Supplementary Fig. 2f, 3b, c). Remarkably, Nrp1-deficient Tregs also failed to suppress clearance of B16 lung metastases, even with very high tumor cell inoculates (Fig. 2g and Supplementary Fig. 7a). Nrp1f/fFoxp3Cre mice displayed increased intratumoral CD8+ T cells, especially in the IFN-γ+IL-2+TNF-α+ tumoricidal subset (Fig. 2h)18. While we originally presumed the source of Sema4a would be tumor-infiltrating T cells and conventional DCs (cDCs), the majority of Sema4a+ cells infiltrating tumors were plasmacytoid DCs (pDCs; 57.4% of intratumoral Sema4a+ cells), consistent with previous suggestions highlighting mechanistic links between Tregs and pDCs in mediating tumor-induced tolerance (Fig. 2i and Supplementary Fig. 7b, c)19. Indeed, pDCs can potentiate Treg function in vitro in a Sema4a:Nrp1-dependent manner (Supplementary Fig. 7d). Treatment with Sema4a-Ig also resulted in an increased number of intratumoral CD8+ T cells, consistent with observations with Nrp1f/fFoxp3Cre mice (Supplementary Fig. 7e). Nrp1-deficient Treg also fail to cure established inflammatory colitis, suggesting that the utilization of this pathway is not restricted to the tumor microenvironment (Supplementary Fig. 8). Thus, while Nrp1 appears to be dispensable for regulating immune homeostasis, it is required for maximal Treg-mediated control of inflammatory environments.
Figure 2. Nrp1-deficient Treg fail to suppress antitumor immune responses.
a, Tumor growth curve (top) and survival plot (bottom) of Foxp3Cre, Nrp1f/fFoxp3Cre, or Foxp3DTR.gfp mice receiving 1.25 × 105 MC38 melanoma cells s.c. and (for Foxp3DTR.gfp) 100 μg diphtheria toxin (DT) i.p. twice weekly. b, As in a, but mice received 1.25 × 105 EL4 thymoma i.d. c, As in a, but mice received 1.25 × 105 B16 melanoma i.d. d, Tumor growth curve of C57/BL6 mice receiving 1.25 × 105 B16 melanoma i.d. concomitant with injections of isotype control, anti-Sema4a, or anti-Nrp1 (100 μg) twice weekly. e, Tumor growth curve as in d except mice received Sema4a-Ig twice weekly. f, Tumor growth curve of C57/BL6 mice receiving 1.25 × 105 B16 melanoma i.d. When tumors were palpable (day 5, indicated by arrow), mice began receiving injections of anti-Nrp1 or its isotype control (400 μg initially, 200 μg every 3 d). g, Lung metastasis counts from Foxp3Cre or Nrp1f/fFoxp3Cre mice injected with 2.5-10 × 105 B16 cells i.v. 17-20 d earlier. h, Tabulation of flow cytometric analysis of tumor-infiltrating lymphocytes from Foxp3Cre or Nrp1f/fFoxp3Cre mice injected i.d. with B16 18 d earlier. i, Sema4a expression of various immune cells in ndLN, dLN, or TIL. Results represent the mean of five (a-c, n=10-25 mice), three (d-h n=8-20 mice), or three (i) experiments. *, p < 0.05, **, p < 0.01, ***, p < 0.001, by (a-f) one-way ANOVA or (g-i) unpaired t-test. Error bars indicate s.e.m.
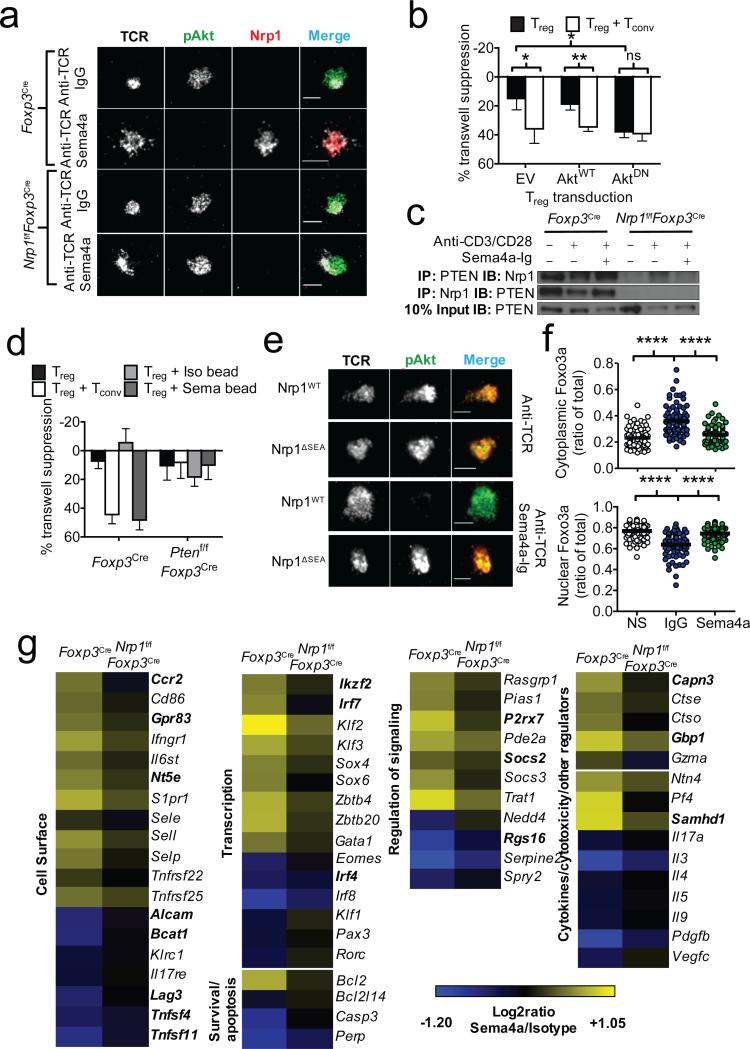
We next sought to determine the signaling pathway downstream of Nrp1. Given the importance of limiting Akt-mechanistic target of rapamycin (mTOR) signaling in Treg function and previous suggestions that Nrp1 modulates Akt signaling, we hypothesized that Nrp1 inhibits Akt function in Tregs20-22. Indeed, TCR/CD28-induced whole-cell Akt-mTOR signaling (as determined by phosphorylation of Akt and S6K1) was reduced to baseline levels in freshly isolated Treg cells (as well as 47-54% in IL-2-expanded Treg cells) by Sema4a-mediated Nrp1 ligation (Supplementary Fig. 9a, b). Furthermore, Treg activation on stimulatory lipid bilayers containing Sema4a-Ig, but not an isotype control, recruited Nrp1 to the immunologic synapse and inhibited IS Akt phosphorylation, while sparing global IS tyrosine phosphorylation (Fig. 3a and Supplementary Fig. 9c, d). Retroviral overexpression of a dominant-negative Akt mutant23 in Tregs expanded with IL-2 limits the requirement for Nrp1 ligation, at least in transwell suppression assays, suggesting that Akt may be a dominant pathway that limits Treg transwell suppression (Fig. 3b and Supplementary Fig. 9e). As IS pAkt diminution in response to Sema4a was rapid, we hypothesized that Nrp1 recruits the phosphatase PTEN to the IS, restraining Akt-mTOR signaling24. Nrp1 constitutively bound PTEN in resting Tregs, which was reduced upon activation but maintained in the presence of Sema4a:Nrp1 interaction (Fig. 3c). PTEN-deficient Treg cells failed to inhibit Akt phosphorylation at the IS in the presence of Sema4a, and failed to suppress across a transwell membrane in response to Sema4a- or Tconv-mediated potentiation (Fig. 3d and Supplementary Fig. 10a, b). Nrp1 has a small cytoplasmic domain consisting of an evolutionarily conserved PDZ domain-binding motif (C-terminal amino acid sequence: SEA)25. Nrp1 mutants lacking this motif could not inhibit Akt phosphorylation at the IS or recruit PTEN (Fig. 3e and Supplementary Fig. 10c, d). Activated Akt can phosphorylate Foxo transcription factors, promoting their exclusion from the nucleus26. Foxo transcription factors are critical for Treg development, through interaction with Foxp3 as well as inducing several Treg signature genes26-28. Indeed, Sema4a-Ig limited the TCR/Akt-induced nuclear export of Foxo3a (Fig. 3f). We propose that Nrp1 ligation restrains Akt phosphorylation via PTEN, facilitates Foxo nuclear localization, and thereby intra-tumoral potentiates Treg function.
Figure 3. Ligation of Nrp1 by Sema4a promotes Treg stability through modulation of Akt-mTOR signaling.
a, TIRF microscopic analysis of IS Akt phosphorylation in Tregs stimulated 20 min on a lipid bilayer coated with anti-TCR antibodies in the presence or absence of Sema4a-Ig. b, Transwell suppression assay using Tregs retrovirally expressing WT or DN-Akt. Transductants were selected and expanded using puromycin and IL-2. c, Immunoprecipitation (IP) analysis of Nrp1 using in vitro expanded Tregs serum starved for 3h, then stimulated as indicated for 6 h prior to IP. d, Transwell suppression assay using Foxp3Cre or Ptenf/fFoxp3Cre Tregs. e, TIRF microscopic analysis of IS Akt phosphorylation as in a of Nrp1f/fFoxp3Cre Treg cells retrovirally reconstituted with WT or ΔSEA Nrp1. f, Foxo3a cytoplasmic (top) and nuclear (bottom) localization signals, as defined by masking using actin and DNA staining. Arbitrary Units (AU) represent fluorescence intensity calculated volumetrically through 20-30 slices of Treg cells. n=70-93 g, Heat map (right) of genes regulated by Nrp1. Foxp3Cre and Nrp1f/fFoxp3Cre CD45RBlo Foxp3 (YFP)+ CD4+ T cells were stimulated for 48h with anti-CD3, anti-CD28, 100 U/mL rhIL-2, and immobilized IgG1 or Sema4a-Ig. RNA was subjected to Affymetrix gene profiling. Treg signature genes are in bold. All genes shown met FDR < 0.10 and compared using two-way ANOVA. Results represent at least three independent experiments (a, c, e, f) or represent means of three (b, d) or seven (g) experiments. *, p < 0.05, ** p < 0.01 by unpaired t-test. Error bars indicate s.e.m.
Gene expression analysis revealed an Nrp1-induced transcriptional profile consistent with promoting Treg survival, stability and quiescence, and was similar to a Foxo-dependent transcriptional signature (Fig. 3g)26-28. Gene Ontology (GO) and Gene Set Enrichment Analysis (GSEA) revealed that Nrp1 ligation modulated multiple pathways and programs, including the IL-2- and IL-7-related transcriptional programs, repression of cytokine transcripts and modulated Foxp3 targets (Fig. 3g, Supplementary Fig. 11-13 and Supplementary Tables 1, 2). Of particular interest was the stabilization of the transcription factor Kruppel-like Factor 2 (KLF2) (and its targets Sell, Ccr7, Il7r), the Treg regulator Helios (Ikzf2), and the anti-apoptotic protein Bcl2, accompanied by a concomitant repression of the lineage defining transcription factors Eomes, IRF4 and RORγt, suggesting a role for Nrp1 in stabilizing the Treg program and repressing terminal differentiation (Fig. 3g and Supplementary Fig. 13b-d).
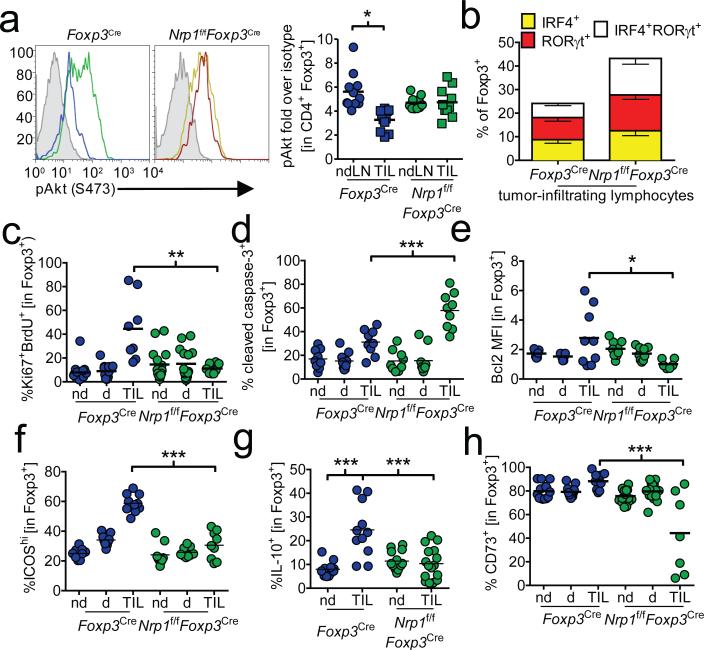
Finally, we sought to determine if the molecular fingerprints of Nrp1 signaling could be observed in vivo. Tumor-infiltrating Tregs restrained Akt phosphorylation in an Nrp1-dependent manner (Fig. 4a). Likewise, tumor-infiltrating Tregs exhibited Nrp1-dependent upregulation of Helios and downregulation of IRF4 and RORγt (Fig. 4b and Supplementary Fig. 14a, b). This was associated with an increase in intratumoral Treg proliferation, as revealed by Ki67-BrdU staining, reduced caspase-3-dependent programmed cell death, and increased expression of the anti-apoptotic protein Bcl2 (Fig. 4c-e and Supplementary Fig. 14c-e). We also observed an Nrp1-dependent increase in the percentage of ICOS+, IL-10+ and CD73+ intratumoral Tregs (Fig. 4f-h and Supplementary Fig. 14f-h). Taken together, our data support a role for Nrp1 in modulating Treg cell stability, survival and function in certain tumor microenvironments.
Figure 4. Tumor-infiltrating Treg bear a signature similar to Sema4a:Nrp1 ligation.
a, Akt phosphorylation in Treg. Tumor-bearing Foxp3Cre or Nrp1f/fFoxp3Cre mice were sacrificed and ndLN and TIL were harvested. Cells were immediately fixed and stained for phospho-Akt. Shaded histogram indicates isotype control. Results are tabulated normalized to isotype control staining. IRF4/RORγt (b), Ki67/BrdU (c), cleaved caspase-3 (d), Bcl2 (e), ICOS (f), IL-10 (g) and CD73 (h) staining from ndLN, dLN, or TIL from tumor-bearing Foxp3Cre or Nrp1f/fFoxp3Cre mice. Ki67/BrdU analysis included injection with BrdU 14 h prior to harvest. IL-10 staining included restimulation with PMA and ionomycin for 16h in the presence of brefeldin A. Results represent the mean of 3-5 independent experiments. * p < 0.05, ** p < 0.01, *** p < 0.001 by paired t-test (a, n=7) or unpaired t-test (b-h n=9-25). Error bars indicate s.e.m.
Our data demonstrate that cell contact-dependent potentiation of Treg cell stability and function is mediated by Sema4a-Nrp1 ligation via a PTEN:Akt:Foxo axis (Supplementary Fig. 15). This pathway enhances Treg cell function indirectly by enforcing stability and promoting survival, most evident in inflammatory sites such as certain tumors and colitic intestinal mucosa. However, Nrp1 signaling may also boost Treg cell function directly by enhancing some suppressive mechanisms (eg. CD73). Apart from hematopoietic lineages, Sema4a appears to have a pattern of expression consistent with sites in which Treg cell tolerogenic activity would be desired, such as the nervous system, eye, and intestine29. The issue of Treg stability has been highly contentious, and the mechanisms that maintain Treg stability remain elusive4-6. As Foxo family members enhance Foxp3 function and promote Treg homeostasis and function,26-28 it is intriguing that Nrp1 signaling may counteract the negative impact of Akt on Foxo nuclear localization, resulting in substantial overlap between the transcriptional profiles induced by Foxo and Nrp1 signaling28. It is possible that the Nrp1:Sema4a pathway may be perturbed genetically or under certain pathologic circumstances; this could also provide a basis for the seemingly contradictory perceptions of Treg stability in a variety of normal and diseased states.
Previous studies have shown that pDCs promote tolerance, Treg differentiation and function, and their ablation has been shown to correlate with enhanced antitumor immunity19,30. Given that the dominant intratumoral source of Sema4a was pDCs, this raises the possibility that Nrp1-induced Treg stability and survival provides a mechanistic explanation for these observations. As Tregs represent a major barrier to anti-tumor immunity in many cancers, a clinically relevant and critical question is whether it is possible to limit Treg function in tumors while preventing inflammatory or autoimmune adverse events. Recently, a role for Nrp1 in Tregs was proposed to limit tumor growth in the MT/ret melanoma model29, although contrary to their findings we did not observe any differences in Treg prevalence in B16 melanoma tumors (Fig. 2h). However, further studies will be required to delineate in which tumors and under what conditions these two disparate functions, namely regulation of Treg migration and the maintenance of Treg survival and stability, of Nrp1 in Tregs are utilized. Our identification of Nrp1:Sema4a as a pivotal pathway required for intratumoral Treg stability, but dispensable for the maintenance of immune homeostasis, suggests that Sema4a:Nrp1 blockade via antibodies or soluble antagonists may be a viable therapeutic strategy to limit tumor-induced tolerance without evoking autoimmunity.
METHODS
Antibodies
Sema4a staining antibody was purchased from MBL (clone 5E3), and conjugated to biotin or Alexa Fluor 647 in-house. Polyclonal anti-Nrp1 was purchased from R&D Systems (AF566). Monoclonal antibodies were obtained from R&D Systems (Sema4a, 757129; Nrp1, 761704, MAB59941). Most flow cytometric antibodies were purchased from BioLegend. Anti-Foxp3 and anti-Eomes were purchased from eBioscience. KLF2 antibody was purchased from Millipore. Phospho-Akt (S473), phospho-S6K1 (T421/S424), Foxo3a, and pan Akt antibodies were purchased from Cell Signaling Technologies. PTEN-HRP antibody was purchased from Santa Cruz Biotechnology.
RNA interference
Control siRNA (Catalog # 4390843) and pools of Sema4a (Catalog # 4390771, siRNA # s73547) siRNA were purchased from Life Technologies and resuspended per the manufacturer's instructions. CD4+ and CD8+ conventional T cells were sorted magnetically by negative selection and transfected by Amaxa (Lonza) with 300 pMol siRNA and 2 μg of pMaxGFP control plasmid, rested overnight in Amaxa nucleofector media. Cells were then sorted based on GFP, CD25, and CD45RB expression and cocultured with Treg cells in the top well of a transwell suppression assay.
Plasmids
Nrp1.mCherry was obtained from Addgene and used as a template to generate retroviral overexpresion constructs. Nrp1WT was generated by adding the native signal sequence and cloned into pMICherry. Nrp1ΔSEA was generated from the WT construct, deleting the terminal SEA motif by mutation of the serine codon to a stop codon. AktWT, AktDN (dominant-negative kinase dead K179M; originally described by Franke et al23), and pBabe empty vector were obtained from D.R. Green.
Human T cell populations
Human umbilical cord samples were provided by B. Triplett, M. Howard and M. McKenna at the St. Louis Cord Blood Bank, and were obtained from the umbilical vein immediately after vaginal delivery with the informed consent of the mother and approved by St. Louis Cord Blood Bank Institutional Review Board (IRB). Research use approved by the St. Jude IRB.
Transwell suppression
1.25 × 104 Treg purified flow cytometrically purified (CD4+ CD45RBlo Foxp3YFP-iCre+) were stimulated in the top chamber of a Millipore Millicell 96 (0.4 μm pore size) in the presence of flow cytometrically purified Tconv (CD45RBhi CD25– CD4+ or CD8+), B cells (B220+), or Treg at a 1:4 ratio, Sema4a-Ig- or IgG-conjugated latex beads (1:1 ratio), anti-CD3ε (145.2C11) and anti-CD28 (37.51) conjugated latex beads (1:1 ratio), and/or neutralizing antibodies. In some experiments, the top well cocultured cells were fixed with 2% PFA for 15 min and washed extensively before co-culture with Treg. 5 × 104 purified Tconv were stimulated in the bottom well with anti-CD3/anti-CD28 beads at a 1:1 ratio. Cells were cultured for 72 h and pulsed with 3[H]-thymidine for the final 8 h. The bottom chambers were harvested and read with a beta counter.
For human studies, flow cytometrically purified umbilical cord blood Tconv (CD4+CD25–) and Treg (CD4+CD25+) were activated with 3 μg/mL plate-bound anti-CD3 (clone OKT3), 2 μg/mL soluble anti-CD28 (clone CD28.1), and 100 U/mL rhIL-2 for 7-9 d. After harvesting and washing, Treg were stimulated at a 1:10 ratio with fixed autologous Tconv or IgG/Sema4a-Ig coated latex beads in the top well of a transwell plate. 2.5 × 104 Tconv were stimulated in the bottom well at a 1:1 ratio with OKT3/CD28.1 coated latex beads. Cells were cultured for 5 d and pulsed with 3[H]-thymidine for the final 8 h. The bottom chambers were harvested and read with a beta counter.
“Percent transwell suppression” is defined as 100 – 100 × [(CPM of a particular well) / (Average CPM of unsuppressed cells)] to normalize across experiments.
Fusion Proteins
The sequence encoding the extracellular domains of Sema4a and Nrp1 were cloned in-frame to pX-Ig to create a Sema4a- or Nrp1-mouse IgG1 fusion protein construct (Sema4a-Ig and Nrp1-Ig). J558L B cells were electroporated with this construct, and high producing clones were selected by single-cell sorting. High producing clones were seeded into Sartorious Bioreactors and harvested for protein G purification and concentration. Sulfate latex 4 μm beads (Life Technologies) were conjugated with isotype control (mouse IgG1, MOPC21, R&D Systems) or Sema4a-Ig overnight with 3 pg protein per bead, blocked with 10% FBS, and stored in media. Mouse Sema-3a-Fc, Sema4a-Fc, mouse Nrp1, and human Sema4a-Fc was purchased from R&D Systems.
Binding assays
High protein binding plates were coated with 500 ng/mL recombinant murine Nrp1 (R&D Systems) overnight in PBS. After a 1-2h block in 1% BSA in PBS at room temperature, coated plates were incubated with various concentrations of Sema4a-Ig or mouse IgG1 for 2-4 h in the presence of anti-Sema4a, anti-Nrp1, or isotype control antibodies. Plates were then washed with PBS + 0.05% TWEEN-20 10 times and incubated with 500 ng/mL biotinylated anti-mouse IgG1 antibody (BD Biosciences) to bind the fusion protein (or mouse IgG1 control). After 7 washes, Strepdavidin-HRP (GE Healthcare) was added at 500 ng/mL to detect the biotinylated antibody. After another 7 washes, TMB substrate (Thermo Scientific) was added and stopped with 1N H2SO4.
For VEGF binding, the same protocol was followed, except rather than Sema4a-Ig being used, VEGF165 (R&D Systems) was used at 50 ng/mL in PBS and detected with 500 ng/mL anti-VEGF-biotin (R&D Systems) followed by SA-HRP for detection.
For comparisons across Sema family members, plates were coated with varying concentrations of Sema3a-Fc, Sema4d-Fc, Sema4a-Ig, or isotype control overnight. Biotinylated Nrp1-Ig was added and incubated for 3 h, and SA-HRP was used for detection.
mRNA analysis
RNA was extracted from cells lysed in TRIzol reagent (Life Technologies) and reverse transcribed with the High Capacity Reverse Transcription kit (Applied Biosystems). Real-time PCR was performed using primers and probes and TaqMan master mix or SYBR green chemistry (Applied Biosystems).
Rescue of Foxp3-deficient autoimmunity
CD45.1 × Foxp3+/– female mice were bred to CD45.1 male mice in timed breedings. Male progeny were genotyped at birth for Foxp3– status. 1 × 106 purified Foxp3Cre or Nrp1f/fFoxp3Cre CD45.2+ Tregs, purified by flow cytometry, were injected intraperitoneally into Foxp3– male pups within 3 days of birth. Mice were monitored for the scurfy phenotype (scaly skin, eye inflammation, runted phenotype, and lack of mobility). For some experiments, all mice were sacrificed at 5 weeks for histological analysis of the ear pinna, liver, and lung.
Tumor Models
Foxp3Cre, Nrp1f/fFoxp3Cre, or Foxp3DTR.gfp mice were injected with B16.F10 melanoma (1.25 × 105 cells i.d.), EL4 thymoma (1.25 × 105 cells i.d.), or MC38 colon carcinoma (2.5 × 105 cells s.c.). Tumors were measured regularly with digital calipers and tumor volume calculated blinded but unrandomized. Tumors and lymph nodes were harvested for analysis. TILs were prepared using a Percoll gradient from tumor samples after mechanical disruption. For metastasis studies, B16.F10 was injected intravenously at various doses. After 17-20 d, lungs were harvested, inflated with H2O2, and metastases were counted. Therapeutic B16 experiments were conducted by injecting 1.25 × 105 B16 melanoma cells i.d. and waiting until tumors were palpable (5 days). On day 5, mice began receiving intraperitoneal injections of either rat IgG2a, or anti-Nrp1 (R&D Systems clone 761704, MAB59941) (400 μg initial dose and 200 μg every three days). Prophylactic experiments included anti-Sema4a (R&D Systems clone 757129) and Sema4a-Ig consisting of twice weekly injections of 100 μg of protein starting on the day of tumor inoculation. To achieve reasonable power, at least 15 mice were used in each group, at least five mice per experiment. Additional mice were added to experiments as appropriate.
Experimental colitis
6-to-8 week old RAG2−/− mice were injected intraperitoneally with 4 × 105 congenically marked CD45RBhi CD25– Tconv cells. 21 to 28 days later (when the majority of the mice had lost 10% body weight and had colitis symptoms), 1 × 106 Foxp3Cre or Nrp1f/fFoxp3Cre Treg were injected intraperitoneally. Mice that did not lose 10% body weight at this time received no injection and were excluded from analysis, and mice received randomized injections (different genotypes of transferred cells per cage). Body weight was measured daily in a blinded fashion, and 28 days after Treg rescue, sections were stained for histology. To achieve reasonable power, at least 15 mice were used in each group, at least five mice per experiment. Additional mice were added to experiments as appropriate.
Signaling analysis
For flow cytometry, Treg were stimulated with anti-CD3e/anti-CD28 coated beads and either purified Teff or Sema4a-Ig beads for various times, then fixed with 1% PFA for 15 min at 37 deg. Cells were then permeabilized in ice-cold 90% MeOH for 20 min at −20 deg. After extensive washing in PBS, cells were blocked with 10% normal mouse serum in PBS for 10 min at RT. Cells were then stained with antibodies in 1% BSA in PBS (pS6K1 (T421/S424), pAkt (S473)) for 1 h at RT in the dark. Finally, cells were stained with appropriate secondary antibodies for 30 min at RT in the dark, then washed and analyzed. For immunoblot analysis, Treg were expanded with 1 ng/mL phorbol-13-myristol acetate and 10 ng/mL ionomycin with 500U rhIL-2 for 3 d, then washed extensively with media, and expanded to 10X volume in 500U rhIL-2. After an overnight rest with no IL-2, Treg were stimulated with plate-bound anti-CD3, soluble anti-CD28, and bead-bound Sema4a-Ig for 6 h, and lysed in whole cell lysis buffer (1% NP40, 5 mM EDTA, 5mM EGTA, TWEEN-20) for 15 min on ice, then subjected to immunoblotting. In some experiments, 1-3 × 106 Treg were lysed in a larger volume, and cleared. Nrp1 was immunoprecipitated using a polyclonal anti-Nrp1 antibody (R&D Systems, AF566) 6-16h followed by a 3 h incubation with Protein G beads. Beads were washed with lysis buffer before elution and reduction prior to immunoblotting. Briefly, precipitates or input lysates were incubated at 96° with 0.1M DTT and 4X LDS sample buffer (Life Technologies), then loaded into 4-12% Bis-Tris NuPAGE gels (Life Technologies), and run for 1 h at 200V. Separated gels were electrotransferred to PVDF membranes using the Criterion Gel Blotting System (Biorad), and blocked for 1h at RT with 3% BSA in TBS supplemented with 0.1% TWEEN20. Blocked membranes were incubated overnight with anti-PTEN directly conjugated to HRP (Santa Cruz Biotechnologies), washed three times with TBS-TWEEN, and imaged using Western Lightning ECL. For other immunoblotting, blocked membranes were incubated with varying primary antibodies (anti-Nrp1, anti-phospho/total Akt, anti-phospho-S6K1) overnight, washed 3 times with TBS-TWEEN, and incubated with appropriate HRP-conjugated secondary antibody controls. After 3 additional washes membranes were imaged using Western Lightning ECL.
Retroviral transduction
293T cells were transfected with pPAM-EQ and pVSV-G packaging plasmids with various retroviral constructs to transduce GPE86 retroviral producer cells. Treg cells were purified flow cytometrically. Treg were activated and cycled with PMA and ionomycin in the presence of 500U/mL rhIL-2 for 24h in 96 well flat bottom plates at 5 × 104 per well in 100 μL. Viral supernatants were concentrated using 100 kDa MWCO concentrators (Millipore) 10 fold and added in equal volume to cycling Treg cells in the presence of 500U/mL rhIL-2 and 6 μg/mL polybrene and centrifuged at 2500 rpm for 60 min at 37 deg, then incubated for 24h. The spinduction process was repeated twice every 24h, removing 100 μL of supernatant from the cultured Treg each day to keep the culture volume at 200 μL per well. Treg cells were then washed in media and sorted based on fluorescent protein expression or selected with 1 μg/mL puromycin and expanded further in IL-2. Fluorescent protein or intracellular epitope staining (anti-HA, Sigma) was confirmed prior to use. Functional assays were performed after a 24 h rest without IL-2.
Microscopy
TIRF illumination of IS activation was performed as previously described. Briefly, lipid bilayers containing anti-TCR and an anti-mouse IgG1 capture antibody loaded with Sema4a-Ig or isotype control were prepared. Treg cells were stimulated on the bilayer for 20 minutes, then fixed, permeabilized, and stained for phospho-Akt (S473), global phosphotyrosine (4G10), or Nrp1. “Percentage of pAkt+ TCR clusters ” represents the ratio of phosphorylated Akt (S473) positive synapses to the total number of synapses formed as read-out by TCR clustering.
Foxo3a was performed on freshly isolated Treg left unstimulated in media overnight or stimulated with immobilized anti-CD3/anti-CD28 in the presence or absence of immobilized Sema4a-Ig or its isotype control. Cells were harvested, fixed in 1% PFA, and permeabilized with 0.1% Triton X-100 in TBS. After blocking with normal mouse serum, cells were stained with anti-Foxo3a (Cell Signaling Technologies) overnight in Tris-buffered 1% BSA. After several washes, cells were stained with Alexa Fluor 647 conjugated anti-rabbit IgG (Life Technologies), and then washed several times. Cells were then loaded with DAPI and phalloidin-Alexa Fluor 546 or 488 prior to microscopy. Random fields of 10-30 cells were visualized using spinning-disc laser-scanning confocal microscopy. Blinded masks were generated using phalloidin and DAPI staining to determine cytoplasmic and nuclear volume, respectively, and only then was the Foxo3a staining visualized. The nuclear and cytoplasmic volumes of Foxo3a fluorescence of 20-30 stacks were calculated using Slidebook (3i, Inc.) software in arbitrary fluorescence units and analyzed in Graphpad Prism.
Affymetrix array and analysis
Foxp3Cre or Nrp1f/fFoxp3Cre Treg were flow cytometrically sorted to 99.0% purity from 6-8 week old mice, and stimulated 48 h with plate-bound anti-CD3, anti-CD28, 100 U/mL rhIL-2, and either isotype or Sema4a-Ig coated latex beads. Cells were harvested, washed three times with PBS, and lysed in TRIzol reagent (Life Technologies). Quality was confirmed by UV spectrophotometry and by analysis on an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). Total RNA (100ng) was processed and labelled in the Hartwell Center for Biotechnology & Bioinformatics according to the Affymetrix 3’ IVT Express protocol and arrayed on a mouse high throughput 430 PM GeneChip array. Signal data was RMA summarized, visualized, quality checked by principal component analysis (PCA) (Partek Genomics Suite 6.6 St Louis MO, USA ). Batch correction was applied as needed to correct differences in completely replicated experiments scanned on distinct dates. To compare Tconv cells to resting Tregs and unequal variance t test was applied to each probeset and the log2ratio calculated. This same analysis was used to compare T conv cells to activated Treg cells. To compare the effect of Sema4a treatment in wild-type Treg cells to the effect of sema treatment in Nrp1-deficient cells a two factor ANOVA interaction of treatment and genotype was applied to each probeset and the Storey q value was found to correct for multiple comparisons. The categorical mean of each probeset was found , transformed to a Z-score, hierarchically clustered and visualized by heat-map in Spotfire DecisionSite 9.1 (Tibco, Somerville MA, USA). The heat map in Figure 5f was composed of the top named genes that had the passed p value interaction FDR at 10%, had a minimum mean expression of 6 in one class and a minimum absolute value logratio difference of at least .5. The volcano plots were generated using STATA/SE 11.1 (College Station TX, USA ). For all volcano plots genes without official symbols or names were removed. In these plot score refrs to the −log base 10 transfomed p value. For the interaction volcano plot genes a metric for distance from the origin was applied to color code the graph |(score/10 + | logratio difference|)/2| > .5. Statistical tests and multiple comparison corrections were performed using Partek Genomics Suite 6.6 (St Louis MO, USA). Sequences were retrieved for probesets that had at least a 3 fold difference between Tconv and activated Treg cells and a p value of .01 and these sequences were then tested with SignalP 3.0 software to identify transmembrane domains.
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to thank E.J. Wherry and H.Chi for advice, A. Rudensky, D. Cheresh, K. Yang, T. Geiger and H. Chi for mice, and D.R. Green for plasmids, B. Triplett, M. Howard and M. McKenna at St. Louis Cord Blood Bank for cord blood samples. We also thank K. Forbes, and A. McKenna for maintenance, breeding and genotyping of mouse colonies, A. Castellaw for preparation of human cord blood samples, K. M. Vignali for assistance with cloning, A. Herrada for generating Nrp1-Ig, A. L. Szymczak-Workman for assistance with histological analysis, S. Morgan, G. Lennon, and R. Cross of the St. Jude Immunology Flow Lab for cell sorting, the staff of the Shared Animal Resource Center at St Jude for the animal husbandry, the Hartwell Center for Biotechnology and Bioinformatics at St Jude for Affymetrix microarray analysis, the Veterinary Pathology Core for histological preparation, and the Immunology Department at St. Jude for helpful discussions. This work was supported by the National Institutes of Health (R01 AI091977 and AI039480 to D.A.A.V.; F32 AI098383 to G.M.D.), NCI Comprehensive Cancer Center Support CORE grant (CA21765, to D.A.A.V.), and ALSAC (to D.A.A.V.).
Footnotes
AUTHOR CONTRIBUTIONS
G.M.D. designed and performed most of the experiments and wrote the manuscript. S-R.W. performed critical initial experiments and identified Sema4a and Nrp1 as the ligand-receptor pair. M.E.T. conducted many of the tumor experiments. D.M.G. performed a substantial portion of the colitis experiments. C.G. performed TIRF microscopy. M.L.B. assisted with the Foxp3-deficiency rescue experiments. A.E.O. assisted with several experiments. P.V. performed histological analysis. D.F. performed computational analysis of the microarray data. J.B. provided the blocking monoclonal antibodies to Sema4a and Nrp1. C.J.W. conducted and curated the initial microarray analysis. D.A.A.V. conceived the project, directed the research and wrote the manuscript. All authors edited and approved the manuscript.
AUTHOR INFORMATION
The data discussed in this publication have been deposited in the NCBI Gene Expression Omnibus and are accessible through GEO Series accession number GSE41185.
The authors declare competing financial interests. J. Bonnevier is an employee of R&D Systems.
REFERENCES
- 1.Vignali DA, Collison LW, Workman CJ. How regulatory T cells work. Nat Rev Immunol. 2008;8:523–532. doi: 10.1038/nri2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fontenot JD, Gavin MA, Rudensky AY. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat Immunol. 2003;4:330–336. doi: 10.1038/ni904. [DOI] [PubMed] [Google Scholar]
- 3.Hori S, Nomura T, Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science. 2003;299:1057–1061. [PubMed] [Google Scholar]
- 4.Miyao T, et al. Plasticity of Foxp3(+) T cells reflects promiscuous Foxp3 expression in conventional T cells but not reprogramming of regulatory T cells. Immunity. 2012;36:262–275. doi: 10.1016/j.immuni.2011.12.012. [DOI] [PubMed] [Google Scholar]
- 5.Rubtsov YP, et al. Stability of the regulatory T cell lineage in vivo. Science. 2010;329:1667–1671. doi: 10.1126/science.1191996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou X, et al. Instability of the transcription factor Foxp3 leads to the generation of pathogenic memory T cells in vivo. Nat Immunol. 2009;10:1000–1007. doi: 10.1038/ni.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qiao M, Thornton AM, Shevach EM. CD4+ CD25+ [corrected] regulatory T cells render naive CD4+ CD25- T cells anergic and suppressive. Immunology. 2007;120:447–455. doi: 10.1111/j.1365-2567.2007.02544.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collison LW, Pillai MR, Chaturvedi V, Vignali DA. Regulatory T cell suppression is potentiated by target T cells in a cell contact, IL-35- and IL-10-dependent manner. J Immunol. 2009;182:6121–6128. doi: 10.4049/jimmunol.0803646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kumanogoh A, et al. Class IV semaphorin Sema4A enhances T-cell activation and interacts with Tim-2. Nature. 2002;419:629–633. doi: 10.1038/nature01037. [DOI] [PubMed] [Google Scholar]
- 10.Bruder D, et al. Neuropilin-1: a surface marker of regulatory T cells. Eur J Immunol. 2004;34:623–630. doi: 10.1002/eji.200324799. [DOI] [PubMed] [Google Scholar]
- 11.Kolodkin AL, et al. Neuropilin is a semaphorin III receptor. Cell. 1997;90:753–762. doi: 10.1016/s0092-8674(00)80535-8. [DOI] [PubMed] [Google Scholar]
- 12.Weiss JM, et al. Neuropilin 1 is expressed on thymus-derived natural regulatory T cells, but not mucosa-generated induced Foxp3+ T reg cells. J Exp Med. 2012 doi: 10.1084/jem.20120914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yadav M, et al. Neuropilin-1 distinguishes natural and inducible regulatory T cells among regulatory T cell subsets in vivo. J Exp Med. 2012 doi: 10.1084/jem.20120822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Milpied P, et al. Neuropilin-1 is not a marker of human Foxp3+ Treg. Eur J Immunol. 2009;39:1466–1471. doi: 10.1002/eji.200839040. [DOI] [PubMed] [Google Scholar]
- 15.Piechnik A, et al. The VEGF receptor, neuropilin-1 (NRP1) represents a promising novel target for chronic lymphocytic leukemia patients. Int J Cancer. 2013 doi: 10.1002/ijc.28135. [DOI] [PubMed] [Google Scholar]
- 16.Nishikawa H, Sakaguchi S. Regulatory T cells in tumor immunity. Int J Cancer. 2010;127:759–767. doi: 10.1002/ijc.25429. [DOI] [PubMed] [Google Scholar]
- 17.Kim JM, Rasmussen JP, Rudensky AY. Regulatory T cells prevent catastrophic autoimmunity throughout the lifespan of mice. Nat Immunol. 2007;8:191–197. doi: 10.1038/ni1428. [DOI] [PubMed] [Google Scholar]
- 18.Wilde S, et al. Human antitumor CD8+ T cells producing Th1 polycytokines show superior antigen sensitivity and tumor recognition. J Immunol. 2012;189:598–605. doi: 10.4049/jimmunol.1102165. [DOI] [PubMed] [Google Scholar]
- 19.Demoulin S, Herfs M, Delvenne P, Hubert P. Tumor microenvironment converts plasmacytoid dendritic cells into immunosuppressive/tolerogenic cells: insight into the molecular mechanisms. J Leukoc Biol. 2013;93:343–352. doi: 10.1189/jlb.0812397. [DOI] [PubMed] [Google Scholar]
- 20.Castro-Rivera E, Ran S, Brekken RA, Minna JD. Semaphorin 3B inhibits the phosphatidylinositol 3-kinase/Akt pathway through neuropilin-1 in lung and breast cancer cells. Cancer Res. 2008;68:8295–8303. doi: 10.1158/0008-5472.CAN-07-6601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crellin NK, Garcia RV, Levings MK. Altered activation of AKT is required for the suppressive function of human CD4+CD25+ T regulatory cells. Blood. 2007;109:2014–2022. doi: 10.1182/blood-2006-07-035279. [DOI] [PubMed] [Google Scholar]
- 22.Haxhinasto S, Mathis D, Benoist C. The AKT-mTOR axis regulates de novo differentiation of CD4+Foxp3+ cells. J Exp Med. 2008;205:565–574. doi: 10.1084/jem.20071477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Franke TF, et al. The protein kinase encoded by the Akt proto-oncogene is a target of the PDGF-activated phosphatidylinositol 3-kinase. Cell. 1995;81:727–736. doi: 10.1016/0092-8674(95)90534-0. [DOI] [PubMed] [Google Scholar]
- 24.Stambolic V, et al. Negative regulation of PKB/Akt-dependent cell survival by the tumor suppressor PTEN. Cell. 1998;95:29–39. doi: 10.1016/s0092-8674(00)81780-8. [DOI] [PubMed] [Google Scholar]
- 25.Pellet-Many C, Frankel P, Jia H, Zachary I. Neuropilins: structure, function and role in disease. Biochem J. 2008;411:211–226. doi: 10.1042/BJ20071639. [DOI] [PubMed] [Google Scholar]
- 26.Merkenschlager M, von Boehmer H. PI3 kinase signalling blocks Foxp3 expression by sequestering Foxo factors. J Exp Med. 2010;207:1347–1350. doi: 10.1084/jem.20101156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kerdiles YM, et al. Foxo transcription factors control regulatory T cell development and function. Immunity. 2010;33:890–904. doi: 10.1016/j.immuni.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ouyang W, et al. Foxo proteins cooperatively control the differentiation of Foxp3+ regulatory T cells. Nat Immunol. 2010;11:618–627. doi: 10.1038/ni.1884. [DOI] [PubMed] [Google Scholar]
- 29.Heng TS, Painter MW. The Immunological Genome Project: networks of gene expression in immune cells. Nat Immunol. 2008;9:1091–1094. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
- 30.Sawant A, et al. Depletion of plasmacytoid dendritic cells inhibits tumor growth and prevents bone metastasis of breast cancer cells. J Immunol. 2012;189:4258–4265. doi: 10.4049/jimmunol.1101855. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.