Abstract
Purpose.
Transforming growth factor β1 (TGFβ1), TGFβ receptor (TGFβR2), and connective tissue growth factor (CTGF) are key regulators of fibrosis in the cornea and in other tissues, including liver, skin, and kidney. We developed an antifibrotic treatment targeting these three critical scarring genes by using a combination of small interfering RNAs (siRNAs) and assessed its effect on downstream scarring genes, collagen I, and α smooth muscle actin (SMA).
Methods.
Up to six individual siRNAs for each of the three target gene mRNAs were transfected into cultures of rabbit corneal fibroblasts at concentrations from 15 to 90 nM. The knockdown of target gene proteins was measured by ELISA, and the two most effective siRNAs were tested in dual combinations. Knockdown percentages of both individual and dual siRNA combinations were analyzed for synergy by using combination index to predict “effective” and “ineffective” triple siRNA combinations. Effects of both triple siRNA combinations on target and downstream mRNAs were measured by using quantitative RT-PCR, and levels of SMA protein were assessed by immunohistochemistry.
Results.
Single and dual siRNA combinations produced a wide range of protein knockdown of target genes (5%–80%). The effective triple siRNA combination significantly reduced mRNA levels of target genes (>80%) and downstream scarring genes (>85%), and of SMA protein (>95%), and significantly reduced cell migration without reducing cell viability.
Conclusions.
Simultaneous targeting of TGFβ1, TGFβR2, and CTGF genes by effective triple siRNA combination produced high knockdown of target and downstream scarring genes without cell toxicity, which may have clinical applications in reducing corneal fibrosis and scarring in other tissues.
Keywords: siRNA, RNA interference, combination therapy, CTGF, TGFβ1
An optimized triple combination of siRNAs targeting TGFβ1, TGFβR2, and CTGF genes produced very high knockdown of target and downstream scarring genes (collagen and alpha smooth muscle actin) without cell toxicity, suggesting a new approach for reducing fibrosis in the cornea and other tissues.
Introduction
Corneal scarring, which is described clinically as corneal haze, is a major cause of impaired vision. In an injured cornea, a complex signaling cascade of molecular events is initiated by transforming growth factor β (TGFβ1), which binds to the transforming growth factor receptor II (TGFβR2) and induces the synthesis of connective tissue growth factor (CTGF).1–3 Connective tissue growth factor then directly activates quiescent keratocytes to transform into activated fibroblasts that synthesize collagen and further transform into myofibroblasts that are characterized by their high concentration of α smooth muscle actin (SMA) and elevated expression of cadherins and TGFβ1 receptors.4,5 In vivo confocal microscopy has revealed that the major reflective (light-scattering) structures in corneas after excimer laser ablation were activated fibroblasts and myofibroblast in the wound area.6–8
Addition of neutralizing antibodies to CTGF to cultures of TGFβ-stimulated corneal fibroblasts blocks the synthesis of collagen and SMA, indicating that CTGF is the major mediator of TGFβ effects on collagen synthesis by corneal fibroblasts.9,10 Thus, these in vitro data support our hypothesis that the TGFβ1/TGFβR2/CTGF system is the major regulator of corneal wound healing and that agents that selectively inhibit these key genes would reduce formation of corneal scar and fibrosis.
There currently are no Food and Drug Administration–approved drugs that selectively reduce the expression of genes causing corneal scarring. Mitomycin C is used during some ocular surgeries, but it may have very damaging side effects, including death of endothelial cells,11 stromal melting, and conjunctival thinning.12–14 Alternative strategies such as antibodies15 and antisense oligonucleotides (ASOs)16 have also been used to block the TGFβ1 pathway. However, these techniques have some challenges for the treatment of scar formation following injury, surgery, or infection. For example, use of ASOs can induce an interferon response in some patients. Monoclonal antibody–based therapies can lead to the production of auto-antibodies against the antigen-binding domain of a monoclonal antibody or the constant region of a nonhumanized mouse monoclonal antibody. Other challenges for topical monoclonal antibody therapies to reduce corneal scarring is the very short lifetime of monoclonal antibodies in tears of patients, thereby requiring multiple doses a day to reduce TGFβ1 protein in tears. Thus, there is a need for an effective antiscarring drug that can be given topically and specifically targets fibrotic genes while also avoiding nonspecific serious side effects that threaten vision.
In this study, we developed an RNA interference–based treatment to reduce corneal fibrosis by targeting three critical scarring genes: TGFβ1, TGFβR2, and CTGF. The RNA interference (RNAi) pathway begins with exogenous double-stranded RNA (dsRNA) being processed by the RNAse III family member Dicer into small interfering RNA (siRNA) molecules that are 20 to 25 nucleotides long.17 The resulting siRNA is incorporated into the multiprotein complex called RNA-induced silencing complex (RISC), with the degradation of the sense strand of the double-stranded RNA.18 The resulting siRNA recognizes the target mRNAs with sequence complementarity, which is then cleaved by Argonaute 2, within the RISC complex.19
It is often difficult to achieve a significant therapeutic effect by using a one-target, one-drug paradigm on such a complex, multifactorial, amplifying, signaling pathway. Hence, a multitarget approach that can interrupt a complex, amplifying, signaling network at multiple points should be able to affect the cell in ways that an individual component cannot. In 1983, Chou and Talaly20 developed the combination index (CI) calculation that can quantify potential synergism or antagonism when two or more drug treatments are combined.21
In the field of antifibrotic research, there have been recent articles published on using multiple siRNAs that target a single mRNA gene target22 or combining siRNA treatment with other drugs.23 However, there have been no reports of combining multiple siRNAs that target different genes within the same pathway to reduce corneal scarring. We hypothesized that simultaneously targeting three key scarring genes within the same major wound-healing pathway, namely, TGFβ1, TGFβR2, and CTGF, would have a cascade effect on reducing the expressions of downstream mediators such as collagen I and SMA in corneal scarring.
Methods
Cell Culture
Cultures of rabbit corneal fibroblasts (RCFs) were established by outgrowth from rabbit whole corneas, as described previously.24,25 Briefly, epithelial and endothelial cells were removed from corneas, the stroma was cut into cubes of approximately 1 mm3, placed in culture medium consisting of equal parts Dulbecco's modified Eagle's medium (DMEM), with 4.5 g/L glucose and 1 g/L l-glutamine. Medium was supplemented with 10% heat-inactivated normal calf serum and antibiotic-antimycotic cocktail (Life Technologies, Carlsbad, CA). Cells from cultures between passages 2 and 5 were used for all experiments. To increase the expression of TGFβ1, TGFβR2, and CTGF, RCFs were placed in serum-free media for 48 hours, and then the medium was replaced by 8 μg/mL estradiol (Sigma-Aldrich, St. Louis, MO) and 4 ng/mL TGFβ1 (R&D Systems, Minneapolis, MN) in DMEM. The cells were dosed twice with the estradiol before transfection of the siRNA and then once after.26,27
Protein Concentration
Protein levels for all three proteins (TGFβ1, CTGF, and TGFβR2) were measured by using capture sandwich enzyme-linked immunosorbent assay (ELISA). The levels of TGFβ1 were determined by using the TGFβ1 ELISA kit (R&D Systems). The assay was performed by following the manufacturer's recommended procedure. The level of CTGF protein was analyzed by using the CTGF polyclonal antibodies from US Biological (Swapscott, MA), following our previously published procedure.24,28 TGFβR2 levels were determined by using the same procedure as that for the CTGF protein except that the capture and detection antibodies were TGFβR2 antibodies (R&D Systems). Knockdown percentages were calculated by comparing the protein levels of the treated samples to the samples treated with the scrambled siRNA.
Reverse Transcription–Polymerase Chain Reaction
Total RNA was extracted by using the Qiagen RNeasy Mini Isolation Kit (Qiagen, Inc., Valencia, CA) and used according to the manufacturer's directions. Complementary DNA was synthesized by using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Carlsbad, CA) according to manufacturer's procedure. The level of mRNA for TGFβ1, CTGF, TGFβR2, SMA, collagen II, and 18S ribosomal RNA was determined by using the Real-Time PCR TaqMan Assay (Applied Biosystems, Carlsbad, CA). The primers and probes for each gene are described in Table A1. The endogenous control, 18S ribosomal RNA, was used to normalize target genes. Primers, probes, and cDNA were combined with TaqMan Universal PCR Master Mix (Applied Biosystems) and amplification was performed by the Applied Biosystems 7300HT Fast Real Time PCR System. The thermal cycling conditions were as follows: 2 minutes at 50°C, 10 minutes at 95°C, 40 cycles of 15 seconds at 95°C, and 1 minute at 60°C. The relative gene expression of the growth factors was calculated by using the 2-ΔΔCt method.
In Vitro Knockdown Experiments
Small interfering RNAs targeting the rabbit mRNA sequences of TGFβ1, CTGF, and TGFβR2 were purchased from Dharmacon (Dharmacon Products, Thermo Fisher Scientific, Lafayette, CO) (Table 1). Rabbit corneal fibroblasts were transfected with individual siRNA sequences (15 nM, 30 nM, 60 nM, and 90 nM) by using TransIT-TKO Transfection Reagent (Mirus, Madison, WI) following the manufacturer's instructions. A transfection optimization experiment was performed to ensure maximal transfection of the siRNAs. Six hours after transfection, the medium was changed to full serum growth medium and then it was changed back to serum-free medium 24 hours post transfection.
For the dual siRNA combination transfection, the siRNAs targeting TGFβ1 (designated T1 and T2), TGFβR2 (designated R1 and R2), and CTGF (C1 and C2) were added at 30 nM for a final combined concentration of 60 nM. For the triple combination, the siRNAs were added at 30 nM for a final combined concentration of 90 nM. A commercially available scrambled siRNA sequence purchased from Dharmacon was used as the negative control. Forty-eight hours after transfection, protein or total RNA samples were collected. Cell lysates were collected with the addition of phosphate-buffered saline solution (PBS) supplemented with 0.1% Triton X-100 with protease inhibitors cocktail III (Calbiochem, Darmstadt, Germany) with the addition of 0.5 mM EDTA. The RNA samples were stabilized by using an RNAlater RNA Stabilization Reagent (Qiagen, Inc.). The samples were frozen at −20°C until further use.
Synergism and Generation of Triple siRNA Combination
Combination index is a numerical parameter to quantify the degree of synergism between siRNA sequences. At the extremes, synergism is CI = 0 to 1 and antagonism is CI = 1 to infinity. It is given by the following equation21,29–31:
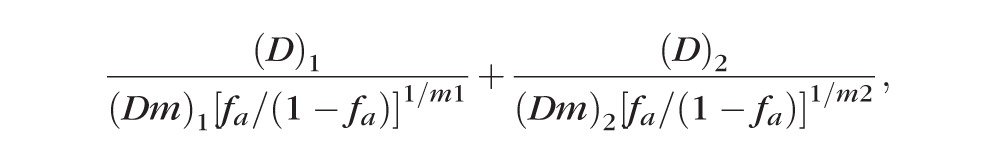 |
where fa is the fraction affected by siRNA sequences (1/100 * knockdown %), D is the individual concentration of the drug in the combination, Dm is the individual drug dose that inhibits system by m%, and m is the coefficient signifying the shape of the dose-effect relationship.
All the data analysis for synergism was performed by using the software Compusyn (Biosoft, Cambridge, UK).28,32
For each target, two individual siRNA sequences were tested, which led to a total of eight different dual combinations. The Compusyn software was then used to compute CI values for each combination, which would give the synergy of individual siRNA sequences within dual combinations at 60 nM. The CI values of 24 different combinations were processed to find two dual combinations with maximal synergy for each target that was made up of the same individual siRNA sequences (Table 2). These individual siRNA sequences were then combined to generate a triple siRNA combination and were deemed to be “effective” as they have maximal synergy in knocking down their respective targets in the presence of the other two siRNA sequences. The same methodology was repeated again to generate an “ineffective” triple siRNA combination whose sequences were identified to have poor synergy among the individual siRNA sequences. These two triple siRNA combinations, effective and ineffective, were further tested in in vitro knockdown experiments to confirm their potency.
Table 1.
Two siRNA Sequences Identified for Each Target
|
Target |
No. |
Target Sequences |
| TGFβ1 | T1 | GCUGACACCCAGUGACACA |
| T2 | GCUGAGAGGUGGAGAGGAA | |
| TGFβR2 | R1 | GCAGAGAACUUGAAAGCAU |
| R2 | CCAUAUGCGGUGUGAAAUA | |
| CTGF | C1 | GUGAUGAGCCCAAGGACCA |
| C2 | GCGAGGAGUGGGUGUGUGA |
Cell Viability and Migration Assays
Forty-eight hours after treatment with the siRNAs, cell viability was assessed by MTS assay, using Cell Titer 96 Aqueous One Solution Reagent (Promega Corp., Madison, WI) and following manufacturer's protocol. Relative absorbance was determined by comparing untreated cells to the siRNA-treated cells. For the migration assay, RadiusTM Cell Migration Plate (Cell Biolabs, San Diego, CA) was used according to manufacturer's protocol. Images were taken at ×10 magnification and analyzed with Adobe Photoshop (Adobe Systems, San Jose, California).
Table 2.
Generation of an Effective and Ineffective Triple Combination by Using CI Values, Calculated as Described in Methods
|
Effective Triple Combination (T1R2C1) |
Ineffective Triple Combination (T2R1C2) |
|||
| Target | Groups | CI Values | Groups | CI Values |
| TGFβ1 | T1C1 | 0.2060 | T2C2 | 3.1394 |
| T1R2 | 0.3684 | T2R1 | 1.3926 | |
| TGFβR2 | T1R2 | 1.0191 | T2R1 | 2.8760 |
| R2C1 | 0.7261 | R1C2 | 3.5837 | |
| CTGF | T1C1 | 1.3384 | T2C2 | 0.9603 |
| R2C1 | 0.1410 | R1C2 | 0.8715 | |
A value from 0 to 1 indicates synergism, while a value of 1 and beyond indicates antagonism.
Immunohistochemistry
To stain for SMA, the siRNA transfection experiment was conducted on a glass slide with four chambers (HLab-Tek, Hatfield, PA). Forty-eight hours after transfection, the chambers were removed, and the cells were fixed in 4% paraformaldehyde for 10 minutes. The slides were then treated with methanol for 15 minutes at −20°C and blocked in goat serum for 1 hour. Finally, they were incubated with SMA antibody-Cy3 (Sigma-Aldrich) for 1 hour. The cells were mounted with 4′,6-diamidino-2-phenylindole, dihydrochloride (DAPI) and imaged by using in vitro confocal microscope.
Image Analysis by ImageJ and Adobe Photoshop
ImageJ software (National Institutes of Health, Bethesda, MD) was used for analyzing the images. Briefly, the image was split into its respective red and blue channels. A constant color threshold of 25-255 was set for all the red channel images, which was used for measuring the area of staining. The number of cells stained with DAPI was counted in the blue channel image by using a constant color threshold of 18-87 and a size value from 8 to infinity. The percentage area of cells stained with SMA was then normalized to the number of cells in each corresponding image. This value was then used to calculate the reduction in SMA in the treated images with respect to the untreated controls.
For the image analysis in migration assay, Photoshop was used to analyze the images. The magic wand feature was used to carefully select and measure the percentage area of cells. This value was used as a measure of the migration of cells.
Statistical Analysis
All experiments were performed in triplicate and all statistical analyses were conducted with GraphPad prism (San Diego, CA). Student's t-test or analyses of variances with Tukey's post hoc assessments were accordingly used to test for significance between the groups. Results were considered statistically significant when P < 0.05.
Results
Single and Dual siRNA Combination Knockdown Experiments
Since the TGFβ pathway is a key mediator of the fibrotic response to corneal wounding, we designed and tested a series of individual siRNA sequences targeting three key scarring genes involved in this pathway (Supplementry Fig. S1). The goal of these experiments was to identify two effective siRNAs for each of the three targets that were capable of knocking down their expression. Following that, we evaluated the effect of different dual combinations of these siRNA sequences on RCFs after stimulation with TGFβ1 and estradiol. Figure 1 represents the knockdown efficiency of all possible dual siRNA combinations that can be obtained by combining three individual siRNA sequences: T1, R2, and C1. The knockdown percentages of both single and dual siRNA combinations were then used in CI calculations to predict a triple siRNA combination with optimal synergism among its individual sequences.
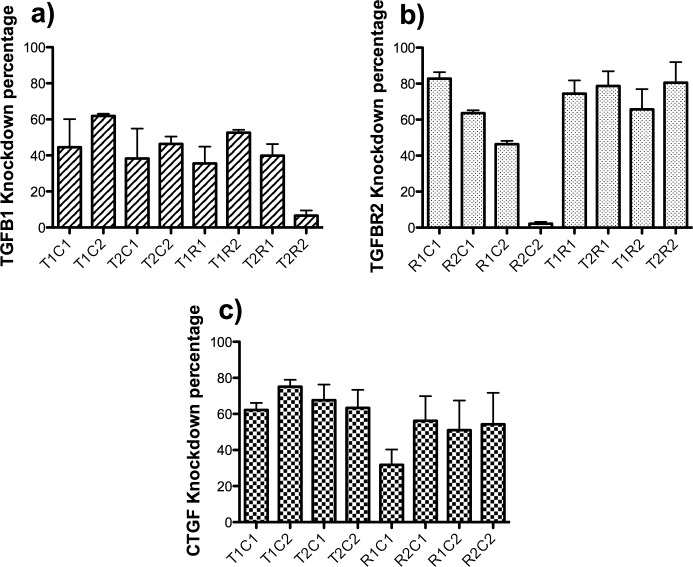
Figure 1.
Knockdown of TGFβ1, TGFβR2, and CTGF by different dual siRNA combinations. Cultures of rabbit corneal fibroblasts stimulated with TGFβ1 and/or estradiol were transfected with different dual combinations of siRNAs targeting TGFβ1 (designated T1 and T2), TGFβR2 (designated R1 and R2), and CTGF (C1 and C2). Each siRNA was added at 30 nM concentration for a final combined concentration of 60 nM. Forty-eight hours after transfection, the cell extract was collected by using a cell lysis buffer and analyzed by using ELISA. The protein level knockdown percentages of (a) TGFβ1, (b) TGFβR2, and (c) CTGF for all dual siRNA combinations were calculated with respect to a scrambled siRNA control.
The combination of TGFβ1 siRNA-1 and CTGF siRNA-2 (T1C2) produced an approximately 60% knockdown of TGFβ1 protein and 79% of CTGF protein (Figs. 1a, 1c). Greater than 75% of TGFβR2 protein was reduced when the TGFβ1 siRNA-1 and TGFβR2–siRNA-2 (T1R2) were combined. Interestingly, there was a reduction of only approximately 37% of TGFβ1 protein in this combination (Figs. 1a, 1b). The combination TGFβR2–siRNA-2 and CTGF siRNA-1 (R2C1) produced a reduction of approximately 58% of CTGF protein and a reduction of approximately 65% of TGFβR2 (Figs. 1b, 1c). A knockdown of CTGF protein of approximately 60% was observed by combining TGFβ1 siRNA-1 with CTGF siRNA-1 (T1C1), while the TGFβ1 protein knockdown was approximately 44% (Figs. 1a, 1c). In some cases, we observed negative interaction between specific pairs of siRNAs. For example, TGFβR2 siRNA R2 led to 60% reduction in TGFβR2 receptor in combination with CTGF siRNA-1 (C1), but less than 10% reduction in the same protein when combined with CTGF siRNA-2 (C2). Similarly, CTGF knockdown by siRNA-1 (C1) was reduced in combination with TGFβ1 receptor siRNA-1 (R1). We believe the reason for this negative interaction may be because of a competition for critical RNAi components, such as the RISC complex, or the preference of Ago2 for a certain configured dsRNA over another as was reported in other experiments.33,34
Synergism and Generation of a Triple siRNA Combination
For each target, two individual siRNA sequences were tested, which led to a total of eight different dual combinations for each distinct target. The Compusyn software was then used to compute CI values for each combination, which gave the synergy of the individual siRNA sequences within dual combinations at 60 nM. At the extremes, synergism is defined as CI = 0 to 1 and antagonism is CI = 1 to infinity. Analyzing the computed CI values of 24 different dual combinations, the sequences T1, C1, and R2 were finalized for the effective siRNA triple combination. This was because the dual combinations involving these siRNA sequences had the maximum synergy based on the computed CI values. Similarly, an ineffective siRNA triple combination was finalized with the sequences T2, C2, and R1 (Table 2). It is noteworthy that each of the individual siRNAs in the low-synergy combination was effective against its intended target when used alone (Supplementary Fig. S1).
Knockdown of TGFβ1, TGFβR2, and CTGF by the Effective Triple Combination (T1R2C1)
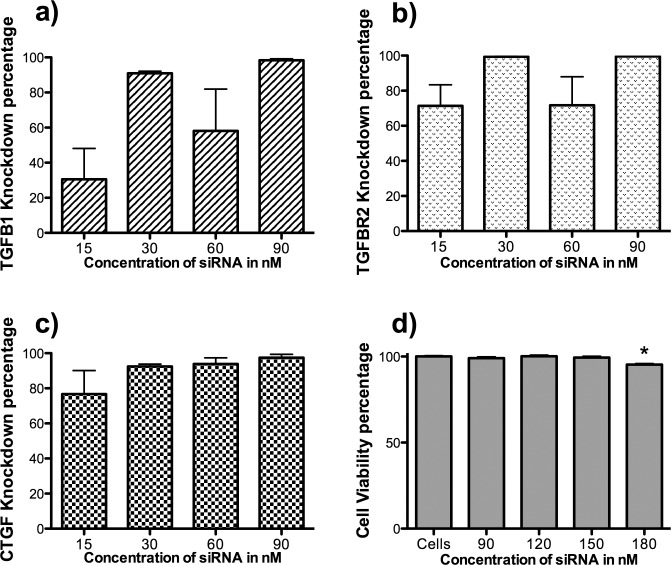
At a total concentration of 90 nM, the effective combination of TGFβ1 siRNA-1, TGFβR2–siRNA-2, and CTGF siRNA-1 (T1R2C1) resulted in approximately 80% knockdown in the expression of the three targets: TGFβ1, TGFβR2, and CTGF (Figs. 2a–c). The knockdown percentages were calculated relative to transduction with a scrambled siRNA control, and all expression levels were normalized to 18S rRNA. The toxicity of the treatment was tested by adding increasing concentrations (90 nM–180 nM) of the effective siRNA triple combination (T1R2C1) to low-passage cultures of RCFs. There was no significant reduction in the cell viability at a final concentration of 90 nM, indicating minimal toxicity (Fig. 2d). However, when compared to cells that were not treated with siRNA, there was a 10% decrease in cell viability when the total siRNA concentration was 180 nM. It should be noted that delivery of 180 nM siRNA required twice the level of moderately toxic transfection reagent. Therefore, we concluded that the reduction in levels of target proteins and mRNAs was not caused by the general toxicity of the siRNAs. Therefore, optimal knockdown of target mRNAs without cytotoxicity was achieved at 90 nM siRNA concentration.
Figure 2.
Knockdown of TGFβ1, TGFβR2, and CTGF by effective triple combination (T1R2C1). Cultures of rabbit corneal fibroblasts stimulated with TGFβ1 and estradiol were transfected with the triple siRNA combination (15 nM, 30 nM, 60 nM, and 90 nM) by using Mirus transfection reagent. The RNA samples were collected 48 hours after transfection and were analyzed by using quantitative RT-PCR (qRT-PCR). On a similar experiment, cells were dosed with increasing concentrations of siRNA triple combination (90 nM–180 nM), and cell viability was assessed by using the MTS assay. The knockdown percentages of (a) TGFβ1, (b) TGFβR2, and (c) CTGF are plotted for the identified effective triple combination (T1R2C1). All expressions were normalized to 18S rRNA. The knockdown percentages were calculated with respect to a scrambled siRNA control. (d) The percentage of viable rabbit corneal epithelial cells after treating with increasing concentrations of the effective triple combination (T1R2C1) (P < 0.05).
The Effective Triple Combination (T1R2C1) Is More Potent Than Any of the Single siRNA Sequence or the Ineffective Triple Combination (T2R1C2) in Blocking Downstream Mediators
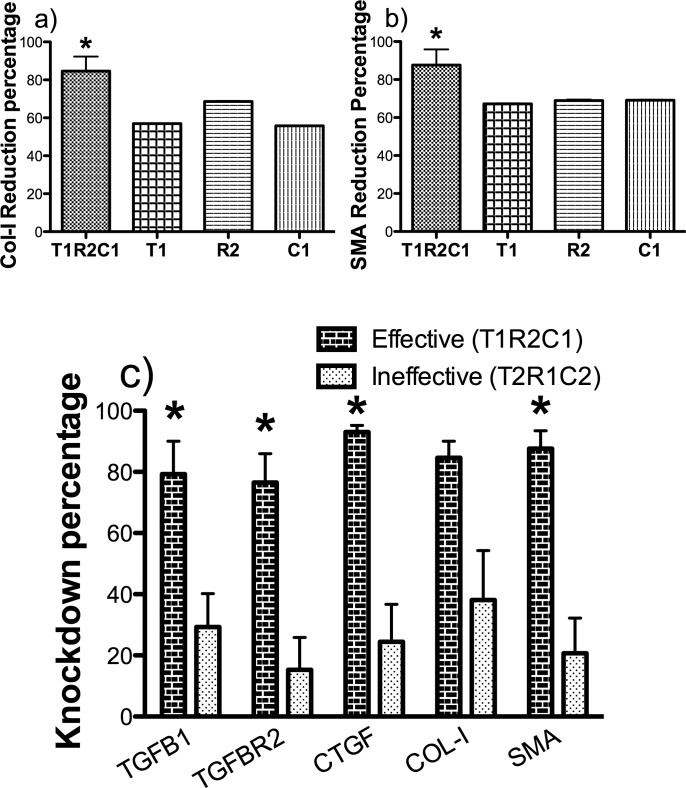
Since the effective triple combination was efficient at reducing the target mRNAs, we next wanted to test the efficacy of this combination on the downstream targets of the TGFβ pathway, which are collagen and SMA. At a total siRNA concentration of 90 nM, the effective triple combination (T1R2C1) was efficient in reducing the mRNA expression of collagen I and SMA by approximately 80%, while individual siRNA sequences at 90 nM were able to achieve a maximum knockdown of only approximately 60% (Figs. 3a, 3b).
Figure 3.
The effective triple combination (T1R2C1) is more potent than single siRNA sequences or ineffective triple combination (T2R1C2) in blocking downstream mediators. Cultures of rabbit corneal fibroblasts stimulated with TGFβ1 and estradiol were transfected with effective (T1R2C1) triple siRNA combination, ineffective (T2R1C2) triple siRNA combination, and individual siRNA sequences at total siRNA concentration of 90 nM by using Mirus TransIT-TKO transfection reagent. The RNA samples were collected 48 hours after transfection and were analyzed by using qRT-PCR. (a, b) Collagen I and SMA reduction percentages are compared between the effective (T1R2C1) combination and its individual siRNA sequences at 90 nM. (c) The RNA level reduction percentages of five genes (target and downstream) are compared between the effective (T1R2C1) and the ineffective (T2R1C2) triple combination. All expression levels were normalized to 18S rRNA. The knockdown percentages were calculated with respect to a scrambled siRNA control (P < 0.05).
At a concentration of 90 nM, the effective triple combination (T1R2C1) also significantly gave a higher knockdown of both the target and downstream mediators when compared to the ineffective triple combination (T2R1C2). The ineffective combination was especially poor (∼20%) in reducing the expression of SMA when compared to the high reduction percentage of approximately 80% by the effective combination (Fig. 3c). The knockdown percentages were calculated with respect to a scrambled siRNA control and normalized to 18S rRNA.
Immunohistostaining for SMA Shows the Effective Triple Combination (T1R2C1) Was More Effective in Knocking Down Protein Expression of SMA Than the Ineffective Triple Combination (T2R1C2)
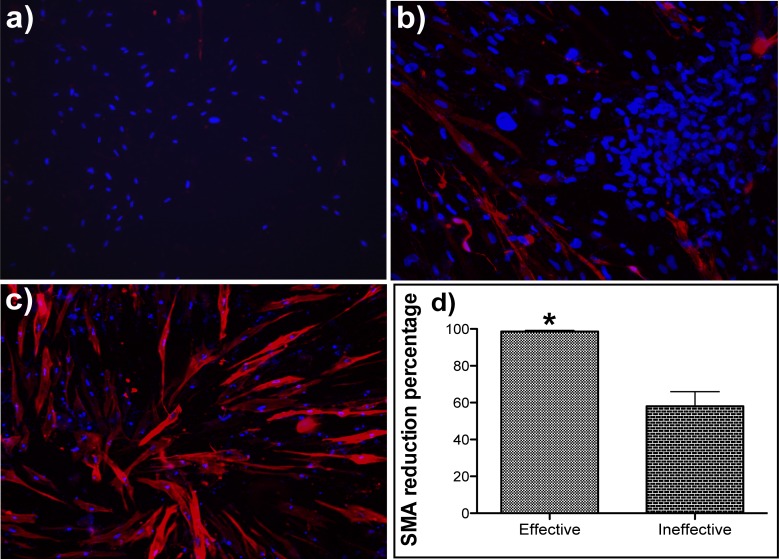
To confirm the protein level knockdown of downstream mediators, immunohistostaining for SMA was performed. The RCFs were treated with TGFβ1 and estradiol to stimulate the expression of SMA. The effect of both effective (Fig. 4a) and ineffective triple combinations (Fig. 4b) was compared to the control, which was transfected with a scrambled siRNA (Fig. 4c). There was dramatic reduction of SMA protein staining when treated with the effective triple combination, while the effect of ineffective triple combination was less evident. Image analysis on 12 random images taken from four different wells showed that the effective triple combination gave a significantly higher reduction of SMA protein staining (∼98%) when compared to the ineffective triple combination (∼55%) (Fig. 4d). The percentage area of cells stained with SMA was normalized to the number of cells in each corresponding image. The SMA reduction percentage in the treated images was calculated with respect to the untreated controls.
Figure 4.
Immunohistostaining for SMA shows that effective triple combination (T1R2C1) is more effective in reducing protein expression of SMA than the ineffective triple combination (T2R1C2). Cultures of rabbit corneal fibroblasts stimulated with TGFβ1 and estradiol were transfected with effective (T1R2C1) and ineffective (T2R1C2) triple siRNA combination (90 nM) by using TransIT-TKO transfection reagent. To stain for SMA, 48 hours after transfection, the cells are fixed in 4% paraformaldehyde, blocked in goat serum, and then incubated with cy3-labeled SMA antibody. Images of SMA protein immunohistostaining (originally ×20) are shown for (a) effective triple combination (T1R2C1), (b) ineffective triple combination (T2R1C2), and (c) scrambled control. (d) The percentage reduction of SMA staining calculated through image analysis from four different areas in three replicate wells. The percentage area of cells stained with SMA was normalized to the number of cells in each corresponding image. The SMA reduction percentage in the treated images was calculated with respect to the untreated controls (P < 0.05).
The Effective Triple Combination (T1R2C1) Inhibited Migration of Rabbit Corneal Fibroblasts
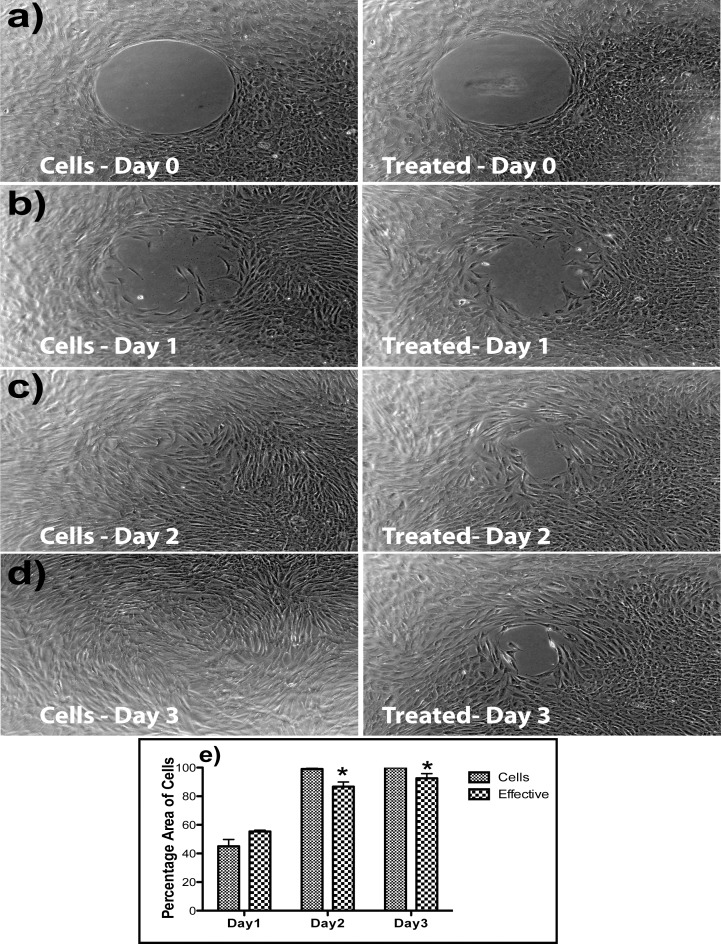
A migration assay was performed to assess the functional effect of the effective triple siRNA combination. Rabbit corneal fibroblasts were cultured in six well plates provided from the Radius cell migration kit. After growing it to confluence, a circular region in the middle of the well was removed by using a gel removal solution. The migration of cells was imaged (Figs. 5a–d) and measured by image analysis with Photoshop. Measured at 2 days after injury, the effective triple combination (T1R2C1) significantly inhibited the migration of cells into the cell-free area. The percentage of cells that migrates was calculated digitally from the three replicates and was used as a measure of migration (Fig. 5e). Linear regression analysis on data from the first 3 days (R2 = −0.8828) was used to predict the time required for complete closure in siRNA-treated wells, approximately 4 days, which is twice that of untreated cells.
Figure 5.
The effective triple combination (T1R2C1) inhibits migration of rabbit corneal fibroblasts. Cultures of rabbit corneal fibroblasts were cultured in six well plates provided with the Radius cell migration kit. After confluence, a circular region in the middle of the well was removed by using a gel removal solution. (a–d) Migration assay results comparing cells alone and effective triple combination (T1R2C1) treatment wells. (e) The percentage area of cells migrating into the circle, based on the migration assays calculated through image analysis from three replicate wells.
Discussion
Corneal wound healing is a complex process in which a variety of cytokines, growth factors, and proteases interact to regulate key phases of corneal healing. However, the TGFβ system has emerged as a key component of the fibrogenic response to wounding by regulating the transformation of quiescent corneal keratocytes into activated fibroblasts that synthesize extracellular matrix and into myofibroblasts that contract corneal matrix.29,30 The TGFβ system has also been implicated in regular scarring of other fibrotic tissues such as lung,32,35 liver,36,37 kidney,35,38 skin,30,39 and pancreas38,40 that are correlated with increased expression of type I and type III collagen, integrins, SMA, and fibronectin.30,39 Apart from scarring, both TGFβ1 and CTGF have roles in angiogenesis,40 glaucoma,41 and proliferative diabetic retinopathy.42
Since TGFβ1, TGFβR2, and CTGF are involved in the scarring mechanism of most fibrotic tissues, the system in this study with a multitarget approach can potentially be modified to regulate scarring in different tissues of the body. The significance of such a system is intensified by the fact that there are currently no FDA-approved drugs to regulate scar formation in any part of the body.
Most of the published literature on siRNAs have used a single target system to reduce fibrosis in eye,43 skin, kidney, and liver.44 We previously developed gene-specific approaches for single targets, using ribozymes and ASOs targeting TGFβ1 or CTGF.24,45 While they are effective in vitro, ribozymes tend to be larger and less stable that siRNAs and ASOs tend to be less potent than siRNAs. Though multiple targets in TGFβ system have been targeted in combination,46 there has not been a multigene siRNA approach in the field of corneal scarring. Multitarget approaches can interrupt the signaling network at several points and affect the corneal fibroblast in ways that reducing the expression of an individual component cannot. Delivery of multiple siRNAs is also no different than that for single siRNAs, and many of the complex in vivo drug interactions that occur between traditional pharmaceuticals do not apply to these agents, thereby facilitating their future use in humans.47
There are several benefits in having three targets in such a complex and interconnected signaling pathway. For example, not only does TGFβ1 auto-induce itself, it also combines with TGFβR2 to stimulate the expression of CTGF, which has been shown to upregulate the expression of SMA and collagen I. Transforming growth factor β1 has been show to stimulate the transition of keratocytes to myofibroblasts that are characterized by the production of SMA. Additionally, the ability of TGFβ1 to elicit multiple, and sometimes opposing, cellular responses in different cell types indicates the complexity of signaling in the wound-healing pathway. For example, TGFβ1 promotes fibroblast cell proliferation and migration but retards epithelial cell migration. It is difficult to obtain an effect by using a single target in a complex signaling pathway where the action of one growth factor can be compensated by another and thus justifies the need for a triple combination.
Combined siRNA sequences targeting multiple mRNAs can have unexpected antagonistic effects.48 There are a number of key determinants of RNAi efficacy that can adversely affect the efficiency of mRNA knockdown by an siRNA combination.33,34 In this study, the triple siRNA combination with optimal synergy between the individual siRNA sequences had significantly better results than the ineffective triple combination, even though each of the siRNAs in the low-synergy group was potent against its intended target mRNA. The effective triple siRNA combination, apart from knocking down the expressions of TGFβ1, TGFβR2, and CTGF by approximately 80% (Fig. 2) through a cascade effect, also knocked down the mRNA level expression of downstream mediators such as collagen I and SMA, which led to a significant reduction in the SMA protein level as detected by the immunohistochemistry (Fig. 4).
Another important functional effect achieved by the effective triple siRNA combination was the reduction in cell migration of corneal fibroblasts. Following an injury in the cornea, it has been documented that the expressions of TGFβ1 and CTGF increases in the surrounding tissues and tears.49 In this study, we have shown that the effective triple combination significantly reduced migration of corneal fibroblasts and may prevent myofibroblast transformation by beneficially prohibiting the stromal fibroblasts from contacting the TGFβ1 present in the surrounding tissues and tears. However, if this retardation of corneal fibroblast were permanent, it might lead to reduced stromal cell density resulting in long-term safety concerns. But, the data from our study showed that the cells treated with the triple siRNA combination could still migrate. In fact, using regression analysis, we estimated that the time required for the triple siRNA-treated cells (4 days) to repopulate the wounding region is twice that of untreated cells (2 days). Since superficial stromal hypercellularity is associated with corneal haze formation,50 it is possible that this temporal retardation of corneal fibroblast migration can partially result in the reduction of haze formation.
The limitation to such a multitargeted approach is the likelihood of off-target effects that may arise from having three targets within a single signaling pathway, which have many important roles in the normal functioning of cells. Apart from wound healing, TFGB1 is involved in growth and differentiation of many cell types and accumulation of matrix proteins such as collagen and fibronectin.1 Connective tissue growth factor also has important roles in many biological processes, including cell adhesion, migration, proliferation, angiogenesis, skeletal development, and tissue wound repair. Hence, a permanent knockdown of these targets may result in permanent safety concerns. The use of short-term delivery (such as nanoparticles) as opposed to long-term delivery (such as viruses) systems to deliver the triple combination may help in reducing the impact of such a knockdown, as their supply is limited to a short amount of time and a specific location.
In addition, siRNAs are also well known to suppress the production of unintended target proteins by binding to the 3′ UTR of their mRNAs, causing off-target effects.51 In this study, the target specificity of siRNAs was improved by ensuring that a region targeted by the siRNA sequences did not share any significant homology with other genes or sequences in the genome, using the BLAST software.
The functional stability of conventional siRNAs in whole animals has been reported to span at least 14 to 21 days52 and the introduction of chemical modifications has been shown to extend the duration of their effectiveness.47,53 Our initial in vivo results obtained from testing the effective triple combination in a rabbit model of corneal scarring are promising.54 The treatment resulted in a reduction in the expression of SMA when compared to the contralateral control cornea. Some of the parameters, such as imaging, dose delivery, and intensity of scar formation, in animals need to be optimized. Once all of these issues are perfected, our siRNA triple combination could prove to be a powerful treatment option for knocking down multiple targets within the same corneal wound healing pathway. This may be the first step toward developing a multitarget approach for the reduction of not only corneal fibrosis but also scarring in the entire body.
Supplementary Material
Acknowledgments
Supported by grants from the United States Army Medical Research Acquisition Activity W81XWH-10-2-0917, National Eye Institute Grant EY000587, National Eye Institute T32-EY07132 Training Grant, and the National Eye Institute P30-EY021721 Vision Core Grant.
Disclosure: S. Sriram, None; P. Robinson, None; L. Pi, None; A.S. Lewin, None; G. Schultz, None
Appendix
Table A1.
TaqMan RT-PCR Primers and Probe Sequences
|
Target |
Type |
Sequences |
| CTGF | Forward | AGGAGTGGGTGTGTGATGAG |
| Reverse | CCAAATGTGTCTTCCAGTCG | |
| Probe | ACCACACCGTGGTTGGCCCT | |
| TGFβR2 | Forward | CGTCGAGACTCCATCTCAAA |
| Reverse | AAACAGCCCACAAATGTCAA | |
| Probe | TCAGCTTTGCACAAGGGCCCT | |
| TGFβ1 | Forward | CCTGTACAACCAGCACAACC |
| Reverse | CGTAGTACACGATGGGCAGT | |
| Probe | CTCCAGCGCCTGTGGCACAC | |
| GAPDH | Forward | GAGACACGATGGTGAAGGTC |
| Reverse | ACAACATCCACTTTGCCAGA | |
| Probe | CCAATGCGGCCAAATCCGTT | |
| Collagen I | Forward | TTCTGCAGGGCTCCAATGAT |
| Reverse | TCGACAAGAACAGTGTAAGTGAACCT | |
| Probe | TTGAACTTGTTGCCGAGGGCAACAG | |
| SMA | Forward | AGAGCGCAAATACTCCGTCT |
| Reverse | CCTGTTTGCTGATCCACATC | |
| Probe | CGGCTCCATCCTGGCCTCTC | |
| 18S rRNA | Forward | GCCGCTAGAGGTGAAATTCTTG |
| Reverse | CATTCTTGGCAAATGCTTTCG | |
| Probe | ACCGGCGCAAGACGGACCAG |
GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
References
- 1. Massague J. The transforming growth factor-beta family. Annu Rev Cell Biol. 1990; 6: 597–641 [DOI] [PubMed] [Google Scholar]
- 2. Blalock TD, Duncan MR, Varela JC, et al. Connective tissue growth factor expression and action in human corneal fibroblast cultures and rat corneas after photorefractive keratectomy. Invest Ophthalmol Vis Sci. 2003; 44: 1879–1887 [DOI] [PubMed] [Google Scholar]
- 3. Grotendorst GR. Connective tissue growth factor: a mediator of TGF-beta action on fibroblasts. Cytokine Growth Factor Rev. 1997; 8: 171–179 [DOI] [PubMed] [Google Scholar]
- 4. Sasaki H, Yamamura K, Nishida K, Nakamura J, Ichikawa M. Delivery of drugs to the eye by topical application. Prog Retin Eye Res. 1996; 15: 583–620 [Google Scholar]
- 5. Jester JV, Barry-Lane PA, Cavanagh HD, Petroll WM. Induction of alpha-smooth muscle actin expression and myofibroblast transformation in cultured corneal keratocytes. Cornea. 1996; 15: 505–516 [PubMed] [Google Scholar]
- 6. Tiemann K, Rossi JJ. RNAi-based therapeutics-current status, challenges and prospects. EMBO Mol Med. 2009; 1: 142–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Møller-Pedersen T. Keratocyte reflectivity and corneal haze. Exp Eye Res. 2004; 78: 553–560 [DOI] [PubMed] [Google Scholar]
- 8. Ivarsen A, Laurberg T, Møller-Pedersen T. Characterisation of corneal fibrotic wound repair at the LASIK flap margin. Br J Ophthalmol. 2003; 87: 1272–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Desmoulière A, Geinoz A, Gabbiani F, Gabbiani G. Transforming growth factor-beta 1 induces alpha-smooth muscle actin expression in granulation tissue myofibroblasts and in quiescent and growing cultured fibroblasts. J Cell Biol. 1993; 122: 103–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kong XB, Zhu QY, Ruprecht RM, et al. Synergistic inhibition of human immunodeficiency virus type 1 replication in vitro by two-drug and three-drug combinations of 3′-azido-3′-deoxythymidine, phosphonoformate, and 2′,3′-dideoxythymidine. Antimicrob Agents Chemother. 1991; 35: 2003–2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sihota R, Sharma T, Agarwal HC. Intraoperative mitomycin C and the corneal endothelium. Acta Ophthalmol Scand. 1998; 76: 80–82 [DOI] [PubMed] [Google Scholar]
- 12. Tong Y-C, Chang S-F, Liu C-Y, Kao WWY, Huang CH, Liaw J. Eye drop delivery of nano-polymeric micelle formulated genes with cornea-specific promoters. J Gene Med. 2007; 9: 956–966 [DOI] [PubMed] [Google Scholar]
- 13. la Fuente de M, Seijo B, Alonso MJ Bioadhesive hyaluronan–chitosan nanoparticles can transport genes across the ocular mucosa and transfect ocular tissue. Gene Ther. 2008; 15: 668–676 [DOI] [PubMed] [Google Scholar]
- 14. Raviv T, Majmudar PA, Dennis RF, Epstein RJ. Mytomycin-C for post-PRK corneal haze. J Cataract Refract Surg. 2000; 26: 1105–1106 [DOI] [PubMed] [Google Scholar]
- 15. Mead AL. Evaluation of anti-TGF-2 antibody as a new postoperative anti-scarring agent in glaucoma surgery. Invest Ophthalmol Vis Sci. 2003; 44: 3394–3401 [DOI] [PubMed] [Google Scholar]
- 16. Cordeiro MF, Mead A, Ali RR, et al. Novel antisense oligonucleotides targeting TGF-beta inhibit in vivo scarring and improve surgical outcome. Gene Ther. 2003; 10: 59–71 [DOI] [PubMed] [Google Scholar]
- 17. Bagasra O, Prilliman KR. RNA interference: the molecular immune system. J Mol Histol. 2004; 35: 545–553 [DOI] [PubMed] [Google Scholar]
- 18. Leuschner PJF, Ameres SL, Kueng S, Martinez J. Cleavage of the siRNA passenger strand during RISC assembly in human cells. EMBO Rep. 2006; 7: 314–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Scherer LJ, Rossi JJ. Approaches for the sequence-specific knockdown of mRNA. Nat Biotechnol. 2003; 21: 1457–1465 [DOI] [PubMed] [Google Scholar]
- 20. Chou TC, Talalay P. Analysis of combined drug effects: a new look at a very old problem. Trends Pharmacol Sci. 1983; 4: 450–454 [Google Scholar]
- 21. Chou T-C. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol Rev. 2006; 58: 621–681 [DOI] [PubMed] [Google Scholar]
- 22. Matsumoto Y, Niimi N, Kohyama K. Characterization of fibrosis-promoting factors and siRNA-mediated therapies in C-protein-induced experimental autoimmune myocarditis. Cell Immunol. 2012; 279: 70–77 [DOI] [PubMed] [Google Scholar]
- 23. Zhang J, Gu C, Noble NA, Border WA, Huang Y. Combining angiotensin II blockade and renin receptor inhibition results in enhanced antifibrotic effect in experimental nephritis. Am J Physiol Renal Physiol. 2011; 301: F723–F732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Robinson PM, Blalock TD, Yuan R, Lewin AS, Schultz GS. Hammerhead ribozyme-mediated knockdown of mRNA for fibrotic growth factors: transforming growth factor-beta 1 and connective tissue growth factor. Methods Mol Biol. 2012; 820: 117–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Woost PG, Jumblatt MM, Eiferman RA, Schultz GS. Growth factors and corneal endothelial cells, I: stimulation of bovine corneal endothelial cell DNA synthesis by defined growth factors. Cornea. 1992; 11: 1–10 [DOI] [PubMed] [Google Scholar]
- 26. Wira CR. Antigen presentation by vaginal cells: role of TGF as a mediator of estradiol inhibition of antigen presentation. Endocrinology. 2002; 143: 2872–2879 [DOI] [PubMed] [Google Scholar]
- 27. Takahashi T, Eitzman B, Bossert NL, et al. Transforming growth factors beta 1, beta 2, and beta 3 messenger RNA and protein expression in mouse uterus and vagina during estrogen-induced growth: a comparison to other estrogen-regulated genes. Cell Growth Differ. 1994; 5: 919–935 [PubMed] [Google Scholar]
- 28. Chou TC, Martin N. CompuSyn for Drug Combinations: PC Software and User's Guide: A Computer Program for Quantitation of Synergism and Antagonism in Drug Combinations, and the Determination of IC50 and ED50 and LD50 Values. Paramus, NJ: ComboSyn, Inc. 2005. [Google Scholar]
- 29. Jester JV, Petroll WM, Cavanagh HD. Corneal stromal wound healing in refractive surgery: the role of myofibroblasts. Prog Retin Eye Res. 1999; 18: 311–356 [DOI] [PubMed] [Google Scholar]
- 30. Chen C, Michelini-Norris B, Stevens S, et al. Measurement of mRNAs for TGFss and extracellular matrix proteins in corneas of rats after PRK. Invest Ophthalmol Vis Sci. 2000; 41: 4108–4116 [PubMed] [Google Scholar]
- 31. Chou T-C, Hayball M. CalcuSyn for Windows: Multiple-Drug Dose-Effect Analyzer and Manual. Cambridge, UK: Biosoft; 1996. [Google Scholar]
- 32. Westergren-Thorsson G, Hernnäs J, Särnstrand B, Oldberg A, Heinegård D, Malmström A. Altered expression of small proteoglycans, collagen, and transforming growth factor-beta 1 in developing bleomycin-induced pulmonary fibrosis in rats. J Clin Invest. 1993; 92: 632–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Grimm D, Wang L, Lee JS, et al. Argonaute proteins are key determinants of RNAi efficacy, toxicity, and persistence in the adult mouse liver. J Clin Invest. 2010; 120: 3106–3119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Castanotto D, Sakurai K, Lingeman R, Li H. Combinatorial delivery of small interfering RNAs reduces RNAi efficacy by selective incorporation into RISC. Nucleic Acids Res. 2007; 35: 5154–5164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Border WA, Noble NA, Yamamoto T, et al. Natural inhibitor of transforming growth factor-beta protects against scarring in experimental kidney disease. Nature. 1992; 360: 361–364 [DOI] [PubMed] [Google Scholar]
- 36. Bullard KM, Cass DL, Banda MJ, Adzick NS. Transforming growth factor beta-1 decreases interstitial collagenase in healing human fetal skin. J Pediatr Surg. 1997; 32: 1023–1027 [DOI] [PubMed] [Google Scholar]
- 37. Perkett EA. Role of growth factors in lung repair and diseases. Curr Opin Pediatr. 1995; 7: 242–249 [DOI] [PubMed] [Google Scholar]
- 38. Vogelmann R, Ruf D, Wagner M, Adler G, Menke A. Effects of fibrogenic mediators on the development of pancreatic fibrosis in a TGF-beta1 transgenic mouse model. Am J Physiol Gastrointest Liver Physiol. 2001; 280: G164–G172 [DOI] [PubMed] [Google Scholar]
- 39. Penn JW, Grobbelaar AO, Rolfe KJ. The role of the TGF-β family in wound healing, burns and scarring: a review. Int J Burns Trauma. 2012; 2: 18–28 [PMC free article] [PubMed] [Google Scholar]
- 40. Abreu JG, Ketpura NI, Reversade B, De Robertis EM. Connective-tissue growth factor (CTGF) modulates cell signalling by BMP and TGF-beta. Nat Cell Biol. 2002; 4: 599–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zee Von CL, Langert KA, Stubbs EB Transforming growth factor-β2 induces synthesis and secretion of endothelin-1 in human trabecular meshwork cells. Invest Ophthalmol Vis Sci. 2012; 53: 5279–5286 [DOI] [PubMed] [Google Scholar]
- 42. Hatanaka H, Koizumi N, Okumura N, et al. Epithelial-mesenchymal transition-like phenotypic changes of retinal pigment epithelium induced by TGF-β are prevented by PPAR-γ agonists. Invest Ophthalmol Vis Sci. 2012; 53: 6955–6963 [DOI] [PubMed] [Google Scholar]
- 43. Sugioka K, Yoshida K, Kodama A, Mishima H, Abe K, Shimomura Y. Connective tissue growth factor cooperates with fibronectin in enhancing attachment and migration of corneal epithelial cells. Tohoku J Exp Med. 2010; 222: 45–50 [DOI] [PubMed] [Google Scholar]
- 44. Li G, Xie Q, Shi Y, et al. Inhibition of connective tissue growth factor by siRNA prevents liver fibrosis in rats. Wound Repair Regen. 2006; 8: 889–900 [DOI] [PubMed] [Google Scholar]
- 45. Blalock TD, Yuan R, Lewin AS, Schultz GS. Hammerhead ribozyme targeting connective tissue growth factor mRNAblocks transforming growth factor-beta mediated cell proliferation. Exp Eye Res. 2004; 78: 1127–1136 [DOI] [PubMed] [Google Scholar]
- 46. Zhao R, Yan Q, Huang H, Lv J, Ma W. Transdermal siRNA-TGFβ1-337 patch for hypertrophic scar treatment. Matrix Biol. 2013; 32: 265–276 [DOI] [PubMed] [Google Scholar]
- 47. Bjorge JD, Pang AS, Funnell M, et al. Simultaneous siRNA targeting of Src and downstream signaling molecules inhibit tumor formation and metastasis of a human model breast cancer cell line. PLoS One. 2011; 6: e19309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Straetemans R, O'Brien T, Wouters L, et al. Design and analysis of drug combination experiments. Biom J. 2005; 47: 299–308 [DOI] [PubMed] [Google Scholar]
- 49. Chen T-C, Lai C-H, Chang J-L, Chang S-W. Mitomycin C retardation of corneal fibroblast migration via sustained dephosphorylation of paxillin at tyrosine 118. Invest Ophthalmol Vis Sci. 2012; 53: 1539–1547 [DOI] [PubMed] [Google Scholar]
- 50. Gambato C, Ghirlando A, Moretto E, Busato F, Midena E. Mitomycin C modulation of corneal wound healing after photorefractive keratectomy in highly myopic eyes [discussion in Ophthalmology. 2005;112:219]. Ophthalmology. 2005. ;. 112: 208–218 [DOI] [PubMed] [Google Scholar]
- 51. Jackson AL, Burchard J, Schelter J, et al. Widespread siRNA “off-target” transcript silencing mediated by seed region sequence complementarity. RNA. 2006; 12: 1179–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bartlett DW, Davis ME. Insights into the kinetics of siRNA-mediated gene silencing from live-cell and live-animal bioluminescent imaging. Nucleic Acids Res. 2006; 34: 322–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Dykxhoorn DM, Lieberman J. Running interference: prospects and obstacles to using small interfering RNAs as small molecule drugs. Annu Rev Biomed Eng. 2006; 8: 377–402 [DOI] [PubMed] [Google Scholar]
- 54. Sriram S, Gibson DJ, Robinson P, Tuli S, Lewin A, Schultz G. Reduction of corneal scarring in rabbits by targeting the TGFB1 pathway with a triple siRNA combination. Adv Biosci Biotechnol. 2013; 4: 47–55 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.