Abstract
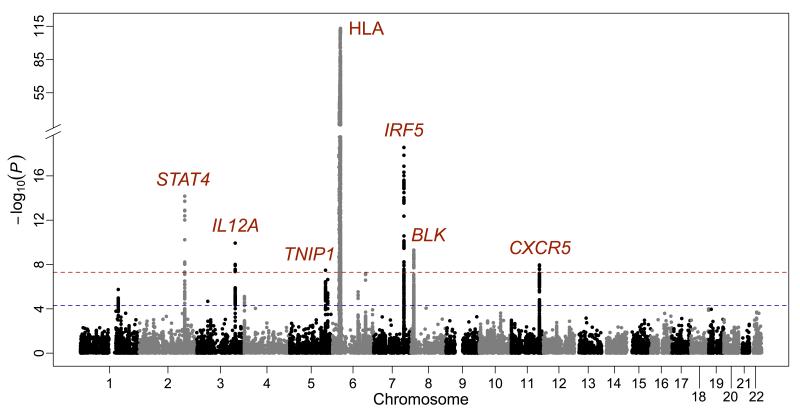
Sjögren’s syndrome is a common autoimmune disease (~0.7% of European Americans) typically presenting as keratoconjunctivitis sicca and xerostomia. In addition to strong association within the HLA region at 6p21 (Pmeta=7.65×10−114), we establish associations with IRF5-TNPO3 (Pmeta=2.73×10−19), STAT4 (Pmeta=6.80×10−15), IL12A (Pmeta =1.17×10−10), FAM167A-BLK (Pmeta=4.97×10−10), DDX6-CXCR5 (Pmeta=1.10×10−8), and TNIP1 (Pmeta=3.30×10−8). Suggestive associations with Pmeta<5×10−5 were observed with 29 regions including TNFAIP3, PTTG1, PRDM1, DGKQ, FCGR2A, IRAK1BP1, ITSN2, and PHIP amongst others. These results highlight the importance of genes involved in both innate and adaptive immunity in Sjögren’s syndrome.
Sjögren’s syndrome is a common (~0.7% of European Americans1), chronic, autoimmune condition characterized by exocrine gland dysfunction resulting in substantial morbidity. Females are affected at a rate of approximately 10 times more than males2,3. Although patients are identified clinically by oral and ocular manifestations, the full disease spectrum may encompass a complex myriad of systemic features, making diagnosis a significant clinical challenge4. Current American-European Consensus Group classification criteria require either the presence of anti-Ro and/or -La autoantibodies or histologic evidence of inflammation within salivary glands plus other subjective and objective measures of keratoconjunctivitis sicca and xerostomia5. Therapeutic options are primarily palliative and do little to prevent irreversible exocrine gland damage6.
Sjögren’s syndrome pathophysiology includes concurrent dysregulation of innate and adaptive immune pathways involving both cell-mediated and humoral disease processes that are incompletely understood7. Labial salivary gland and peripheral blood gene expression microarray studies have demonstrated dysregulation of type I interferon-inducible genes8,9. Previous work in Sjögren’s syndrome genetics is limited to candidate gene studies, yet strong association with several human leukocyte antigen (HLA) molecules, including HLA-DR, HLA-DQB1, and HLA-DQA1, has been established10,11. Several non-HLA regions have been implicated in the disease, such as IRF5 and STAT412-15; however, none of these associations have exceeded the genome-wide significant (GWS) threshold of P = 5×10−8 (Supplementary Table 1). In this study, genome-wide association and large-scale replication approaches were used to identify new risk loci in a case-control cohort of primary Sjögren’s syndrome patients.
Results
To maximize the power of our study, we formed the Sjögren’s Genetics Network (SGENE), a collaborative research effort comprising multiple international sites (Supplementary Figure 1 and Supplementary Table 2). Dataset 1 (DS1) included 395 cases and 1975 population controls of European descent after implementing quality control (QC; see Online Methods and Supplementary Table 2). Genotyping was performed using the Illumina Omni1-Quad array. Dataset 2 (DS2) consisted of 1243 cases and 4779 population controls genotyped on the Illumina ImmunoChip, and Dataset 3 (DS3) included 1158 cases and 3071 population controls genotyped on a supplemental custom array designed to include variants not present on the ImmunoChip (all of European descent and after QC; Supplementary Figure 1b). Dataset 4 (DS4) evaluated 1541 cases and 2634 population controls genotyped on the Illumina ImmunoChip focusing on variants exclusive to DS4 and not common between DS1 and DS2 (Supplementary Figure 1b). Imputation was performed in regions with P<5×10−5 to increase the number of overlapping variants available for analysis (Supplementary Figure 1c and 1d).
Human Leukocyte Antigen (HLA)
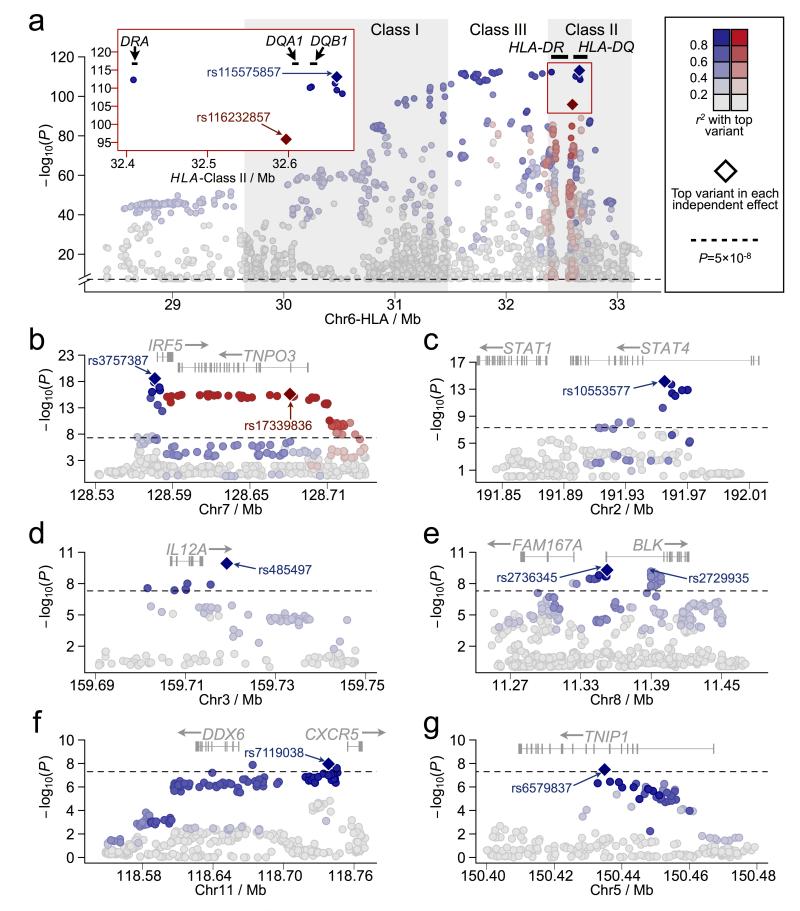
The HLA region at 6p21 was the only association that exceeded the GWS threshold of P<5×10−8 in DS1 (Supplementary Figure 2). After imputation, we found the peak effect at rs112357081 near HLA-DRA with PDS1=1.01×10−32 (Table 1; Supplementary Figure 3). No inflation was observed in the test statistic (λGC=1.0) and no significant deviation from the expected distribution of P-values was detected after removing the HLA region (Supplementary Figure 4). After meta-analysis between the imputed DS1 and DS2 (Figures 1 and 2; Supplementary Figure 1), strong association spanned a region that included the HLA Class I, III, and II loci (Figure 2a; Supplementary Table 3), with peak association observed 5′ of HLA-DQB1 in the HLA Class II region at rs115575857 with Pmeta=7.65×10−114 (Table 1 and Figure 2a). A second set of variants not in linkage disequilibrium (LD) with rs115575857 was also identified peaking at rs116232857 (Pmeta=1.33×10−96; Table 1 and Figure 2a).
Table 1.
Top associations in the HLA region
| SNP | Alleles |
MAFb |
Observed or Imputed in Dataset 1 / 2 |
Dataset 1 |
Dataset 2 |
Meta analysis |
||||
|---|---|---|---|---|---|---|---|---|---|---|
| maj/mina | Case | Control | OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | ||
| HLA-DRA | ||||||||||
| rs112357081 | TCTAA/T | 0.59 | 0.33 | I / - | 2.89 (2.43-3.44) | 1.01×10−32 | - | - | - | - |
| rs3135394 | A/G | 0.27 | 0.11 | I / O | 3.52 (2.83-4.38) | 2.47 ×10−29 | 3.52 (3.10-3.99) | 1.14×10−85 | 3.52 (3.02-4.10) | 5.22×10−113 |
| HLA-DQB1 | ||||||||||
| rs115575857 | A/G | 0.29 | 0.12 | I / I | 3.25 (2.61-4.05) | 4.10 ×10−26 | 3.65 (3.22-4.14) | 3.70×10−90 | 3.53 (3.03-4.11) | 7.65×10−114 |
| rs3129716 | T/C | 0.29 | 0.12 | I / O | 3.29 (2.66-4.09) | 2.11 ×10−27 | 3.51 (3.10-3.97) | 7.74×10−87 | 3.45 (2.97-4.00) | 4.59×10−112 |
| HLA-DQA1 | ||||||||||
| rs116232857 | A/G | 0.64 | 0.42 | I / I | 2.83 (2.37-3.38) | 9.05 ×10−31 | 2.42 (2.19-2.67) | 1.14×10−67 | 2.53 (2.24-2.86) | 1.33×10−96 |
| rs9271588 | T/C | 0.27 | 0.48 | I / O | 0.35 (0.29-0.42) | 1.93 ×10−28 | 0.43 (0.39-0.48) | 4.62×10−59 | 0.41 (0.36-0.46) | 1.37×10−85 |
Major allele / minor allele. Odds ratio for each variant indicates disease risk conferred by the minor allele.
The minor allele frequency (MAF) was calculated using the combined Datasets 1 and 2, except for rs112357081, for which the MAF is derived from Dataset 1 only.
Figure 1.
Summary of genome-wide association results for 27,501 variants overlapping between DS1 and DS2 after imputation and meta-analysis. The −log10(P) for each variant is plotted according to chromosome and base pair position. A total of seven loci (in red font) exceeded the GWS of Pmeta < 5×10−8 (red dashed line). Suggestive threshold (Pmeta < 5×10−5) is indicated by a blue dashed line.
Figure 2.
Zoomed in plots of the meta-analysis results for the seven regions with Pmeta<5×10−8. The HLA region (a) from Mb 28.4 to Mb 33.4 is shown for variants with Pmeta<5×10−8 only. Strong linkage disequilibrium (LD) was observed with variants extending from Class I through Class II (pairwise LD with rs115575857 is given in blue). A second independent effect was identified in Class II peaking at rs116232857 (pairwise LD is depicted in red). The red insert on the left shows a further zoomed in view of the top associated variants in the Class II region. (b) The area of association surrounding IRF5 is shown, including the pairwise LD with the top overall variant, rs3757387, in blue. Pairwise LD with the top SNP, rs17339836, in the second effect is shown in red. STAT4 (c), IL12A (d), BLK (e), CXCR5 (f) and TNIP1 (g) regions are also provided and include pairwise LD with the top listed variant shown in blue. The black dashed lines represent Pmeta = 5×10−8.
Further dissection through logistic regression analysis in DS2 adjusting for rs115575857 identified an ~5 Mb haplotype encompassing the extended HLA region (Supplementary Figure 5). In addition, a narrow region of residual association tagged by rs116232857 (Presidual=7.00×10−26) was observed in the HLA-DRA through HLA-DQA1 regions (Supplementary Figure 5). Logistic regression adjusting for rs115575857 and rs116232857 accounted for most of the association within the HLA region (the residual association in the region at rs115146037 went from 10−80 to 10−9; Supplementary Figure 5), thus identifying the presence of at least two independent effects.
Classical HLA alleles were imputed into our dataset to better understand the relationship between the HLA region variants tested in this study to those reported previously. The top associated classical allele was HLA-DQB1*0201 (Pmeta=1.38×10−95) followed closely by HLA-DQA1*0501 (Pmeta=8.50×10−94; Supplementary Table 4 and Supplementary Figure 6). We used logistic regression adjusting for both the top associated classical HLA alleles and the variants identified through the meta-analysis of DS1 and DS2. As previously described16, our data shows that HLA-DQB1*0201, HLA-DQA1*0501, and HLA-DRB1*0301 are in strong LD (r2>0.97), and adjusted logistic regression using any of the three accounted for the observed association. Moreover, each of these classical alleles accounted for the association at rs115575857. Likewise, rs115575857 explained the association at each of the three classical alleles (Supplementary Figure 6). Adjusting for rs116232857 could account for the association at HLA-DQB1*0501, a previously reported protective allele in Sjögren’s syndrome11. However, HLA-DQB1*0501 could not account for the association observed at rs116232857, suggesting that rs116232857 better tags the functional effect(s) than the classical allele.
Enrichment analysis using all variants with Pmeta<5×10−5 identified a statistically significant association of disease-associated variants within 100 bp of regions found to be cross-linked to the transcription factor (TF) RFX5 (P=1.53×10−14) by ENCODE ChIP-seq studies17. In total, 161 variants contributed to the enrichment in our dataset with all but one located in the HLA region. To further evaluate the influence of the extensive LD in the HLA, we tested for enrichment of Sjögren’s syndrome-associated variants in RFX5 ChIP-seq peaks using only the HLA region variants present in DS1 as the background and continued to observe significant enrichment (P=9.24×10−7; Supplementary Table 5).
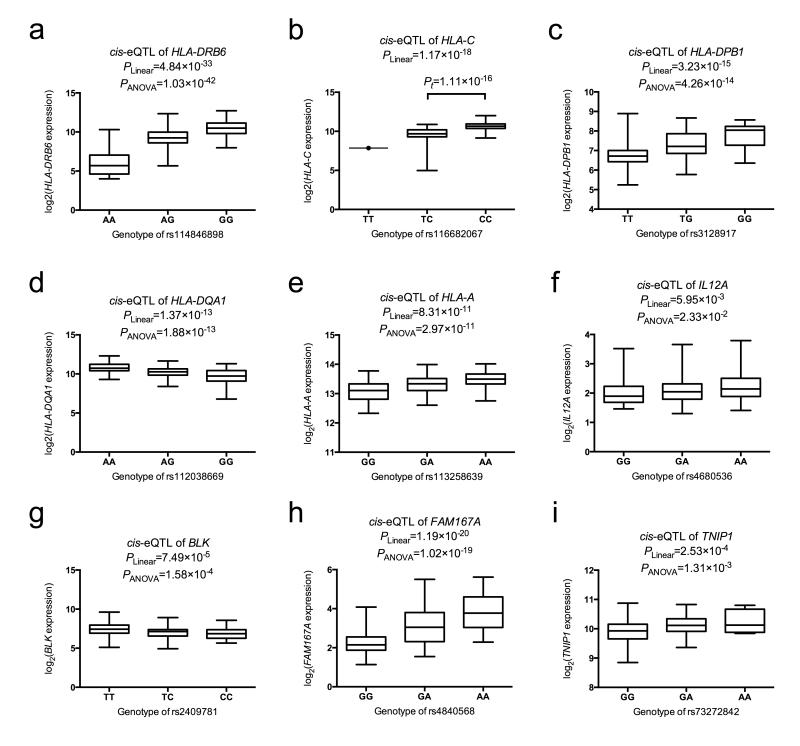
To evaluate the functional mechanism of this RFX5 enrichment within the HLA the variants near RFX5 ChIP-seq peaks were tested as expression quantitative trail loci (eQTL), resulting in several statistically significant results (Figure 3a-e); Supplementary Table 6 and Supplementary Figure 7). The top overall eQTL was observed with rs114846898 and HLA-DRB6 (PANOVA=1.03×10−42; Figure 3a). A modest eQTL was observed for rs116232857 and HLA-DRB6 (PANOVA=1.77×10−12); however, we found that rs116232857 and rs114846898 were nearly in perfect LD with each other (D’=0.98), suggesting that the association at rs116232857 might be due to rs114846898. Looking at the adjusted analysis results, we found that rs115575857 and rs116232857 could explain rs114846898, but adjusting for rs115575857 and rs114846898 left residual association (Presidual=9.44×10−7) at rs116232857, suggesting that rs116232857 likely tags additional functional variant(s).
Figure 3.
Identification of cis-eQTL in Sjögren’s syndrome-associated regions. Sjögren’s syndrome-associated variants in the HLA region within 100bp of RFX5 binding sites were evaluated for cis-eQTL with the expression of HLA genes in 222 European subjects. The top cis-eQTL in HLA-DRB6 (a), HLA-C (b), HLA-DPB1 (c), HLA-DQA1 (d), and HLA-A (e) are shown in the figure. We also identified cis-eQTL for top Sjögren’s syndrome-associated variants in IL12A (f), FAM167A-BLK (g, h), and TNIP1 (i). The risk genotype of each eQTL variant is plotted on the right side of each figure. Data are shown using box-and-whisker plots, in which distribution of the expression values are highlighted (median, upper quantile, lower quantile, maximum, and minimum). Each eQTL was evaluated using both a linear model and Analysis of Variance (ANOVA) model. Panel b only had one subject with the TT genotype; thus, we performed a t-test rather than an ANOVA to compare the TC to the CC genotype.
In addition to the strong association observed in the HLA region, six non-HLA loci surpassed the GWS threshold, including IRF5, STAT4, IL12A, BLK, CXCR5 and TNIP1 (Table 2 and Figure 1).
Table 2.
Non-HLA regions associated with Sjögren’s syndrome exceeding genome-wide significant threshold
| SNP | Alleles |
MAFb |
Observed or Imputed in Dataset 1 / 2 |
Dataset 1 |
Dataset 2 |
Meta analysis |
||||
|---|---|---|---|---|---|---|---|---|---|---|
| maj/mina | Case | Control | OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | ||
| IRF5 (Chr 7) | ||||||||||
| rs3757387 | T/C | 0.54 | 0.45 | I / I | 1.31 (1.11-1.54) | 1.49×10−3 | 1.50 (1.37-1.64) | 7.50 ×10−18 | 1.44 (1.29-1.62) | 2.73 ×10−19 |
| rs4728142 | G/A | 0.53 | 0.44 | O / O | 1.29 (1.09-1.52) | 2.85 ×10−3 | 1.46 (1.33-1.60) | 2.05 ×10−15 | 1.40 (1.25-1.57) | 9.75 ×10−17 |
| rs17339836 | C/T | 0.18 | 0.12 | O / O | 1.65 (1.32-2.06) | 8.53 ×10−6 | 1.55 (1.37-1.75) | 5.73 ×10−12 | 1.58 (1.36-1.84) | 2.43 ×10−16 |
| rs17338998 | C/T | 0.18 | 0.12 | I / I | 1.64 (1.32-2.05) | 9.51 ×10−6 | 1.55 (1.37-1.75) | 5.70 ×10−12 | 1.57 (1.35-1.83) | 2.67 ×10−16 |
| rs10954213 | A/G | 0.34 | 0.38 | O / O | 0.83 (0.70-0.98) | 2.91 ×10−2 | 0.82 (0.74-0.90) | 3.63 ×10−5 | 0.82 (0.73-0.92) | 3.20 ×10−6 |
| STAT4 (Chr 2) | ||||||||||
| rs10553577 | TATA/T | 0.30 | 0.23 | I / I | 1.38 (1.15-1.65) | 5.13 ×10−4 | 1.45 (1.31-1.61) | 2.30 ×10−12 | 1.43 (1.26-1.62) | 6.80 ×10−15 |
| rs13426947 | G/A | 0.24 | 0.19 | O / O | 1.34 (1.11-1.62) | 2.44 ×10−3 | 1.31 (1.18-1.47) | 1.09 ×10−6 | 1.32 (1.16-1.51) | 9.45 ×10−9 |
| IL12A (Chr 3) | ||||||||||
| rs485497 | G/A | 0.54 | 0.48 | O / O | 1.32 (1.12-1.55) | 9.47 ×10−4 | 1.30 (1.18-1.42) | 3.15 ×10−8 | 1.30 (1.16-1.46) | 1.17 ×10−10 |
| rs583911 | A/G | 0.48 | 0.42 | I / I | 1.29 (1.10-1.52) | 2.16 ×10−3 | 1.26 (1.15-1.38) | 1.28 ×10−6 | 1.27 (1.13-1.42) | 9.88 ×10−9 |
| BLK (Chr 8) | ||||||||||
| rs2736345 | A/G | 0.36 | 0.29 | I / I | 1.16 (0.97-1.37) | 1.01 ×10−1 | 1.37 (1.24-1.50) | 2.76 ×10−10 | 1.30 (1.16-1.47) | 4.97 ×10−10 |
| rs2729935 | C/A | 0.41 | 0.35 | I / I | 1.28 (1.08-1.52) | 4.02 ×10−3 | 1.30 (1.19-1.43) | 4.29 ×10−8 | 1.30 (1.16-1.46) | 6.85 ×10−10 |
| rs6998387 | G/A | 0.37 | 0.32 | O / I | 1.34 (1.13-1.59) | 6.75 ×10−4 | 1.23 (1.12-1.36) | 2.65 ×10−5 | 1.26 (1.12-1.42) | 7.96 ×10−8 |
| CXCR5 (Chr 11) | ||||||||||
| rs7119038 | A/G | 0.18 | 0.23 | I / I | 0.79 (0.64-0.98) | 3.32 ×10−2 | 0.72 (0.64-0.81) | 6.33 ×10−8 | 0.74 (0.64-0.86) | 1.10 ×10−8 |
| rs4936443 | T/C | 0.16 | 0.20 | O / O | 0.79 (0.63-0.98) | 3.21 ×10−2 | 0.74 (0.65-0.83) | 5.03 ×10−7 | 0.75 (0.65-0.87) | 6.82 ×10−8 |
| TNIP1 (Chr 5) | ||||||||||
| rs6579837 | G/T | 0.12 | 0.09 | I / I | 1.58 (1.23-2.04) | 3.94 ×10−4 | 1.38 (1.19-1.59) | 1.71 ×10−5 | 1.43 (1.20-1.71) | 3.30 ×10−8 |
| rs7732451 | A/G | 0.15 | 0.12 | O / O | 1.36 (1.09-1.71) | 6.77 ×10−3 | 1.33 (1.17-1.52) | 2.43 ×10−5 | 1.34 (1.14-1.57) | 5.32 ×10−7 |
Major allele / minor allele. Odds ratio for each variant indicates disease risk conferred by the minor allele.
The minor allele frequency (MAF) was calculated using the combined Datasets 1 and 2.
Interferon Regulatory Factor 5 (IRF5)
The most statistically significant association outside the HLA region after meta-analysis was observed in the region of IRF5. In total, 67 SNPs exceeded the Pmeta=5×10−8 threshold, with the peak effect observed in the IRF5 promoter region (rs3757387, Pmeta=2.73×10−19; Figure 2b; Supplementary Table 7 and Supplementary Figure 8). A group of variants, tagged by rs17339836 (Pmeta=2.43×10−16) and spanning IRF5 and TNPO3, were not correlated with the promoter variants (Figure 2b).
Logistic regression adjusting for rs3757387 explained all of the promoter region association, while residual association persisted with variants spanning the IRF5-TNPO3 genes tagged by rs17339836 (Presidual=2.86×10−4; Supplementary Figure 9). Moreover, a group of variants tagged by rs6948875 that were not associated in the unadjusted single marker analysis became statistically significant when adjusting for rs3757387. Adjusting for both rs3757387 and rs17339836 again resulted in residual association of the region tagged by rs6948875 and rs10954213 (Supplementary Figure 9). Since rs10954213 was statistically significant in the single marker analysis (Supplementary Table 7), this SNP was added to the model in lieu of rs6948875. Logistic regression models adjusting for rs3757387, rs17339836, and rs10954213 resulted in no residual association in the regions, indicating the likelihood of three independent effects. Bioinformatics database mining of this region indicated that the associated variants were located within a variety of biologically relevant features encompassing a wide range of potential functional mechanisms (Supplementary Figure 10).
Signal Transducer and Activator of Transcription 4 (STAT4)
Statistically significant association exceeding GWS was observed between Sjögren’s syndrome and 12 variants, 8 of which fall within the third intron of the STAT4 locus, with peak association at an insertion-deletion (indel) polymorphism (rs10553577; Pmeta=6.80×10−15; Figure 2c; Supplementary Table 8 and Supplementary Figures 11 and 12). As expected, the indel rs10553577 accounted for the association within the region of STAT4; however, residual association was observed with rs3024918 (Presidual=2.70×10−4) in the promoter region of STAT1, but did not surpass our required threshold (P<1×10−4) after adjustment for an independent effect (Supplementary Figure 12). Bioinformatics databases revealed a large number of TFs bound to both regions (Supplementary Figure 13); however, neither effect resulted in a statistically significant eQTL (data not shown).
Interleukin 12A (IL12A)
Seven variants spanning the entire region from the promoter demonstrated strong association, with peak association occurring 5.3kb 3′ of IL12A at rs485497 (Pmeta=1.17×10−10; Figure 2d; Supplementary Table 9 and Supplementary Figure 14). Logistic regression analysis adjusting for rs485497 indicated this variant accounted for all the observed association (Supplementary Figure 15). To determine the functional role of the variants associated between IL12A and Sjögren’s syndrome, eQTL analysis was done for all variants showing evidence of association in the region. Of the variants exceeding GWS, only rs485497 was associated with IL12A transcript expression; however, seven variants 3′ of IL12A influenced expression more significantly, with the eQTL peaking at rs4680536 (Figure 3f; Supplementary Table 6). Haplotype analysis of the IL12A associated variants indicates the presence of only one risk haplotype and the possibility that rs485497 may tag functional variants located on either side of this SNP (Supplementary Figure 15), which is supported by the presence of multiple transcription factors and other regulatory elements located in the flanking LD blocks (Supplementary Figure 16).
B Lymphoid Kinase (BLK)
A total of 30 variants on chromosome 8 exceeded GWS in the region of FAM167A and BLK (Figure 2e; Supplementary Table 10 and Supplementary Figure 17). Maximum association after meta-analysis was observed in the shared promoter region of FAM167A-BLK at rs2736345 (Pmeta=4.97×10−10); however, this SNP was not statistically significant in DS1 (Table 2). The second most significant variant after meta-analysis (significant in both DS1 and DS2) was rs2729935 with Pmeta=6.85×10−10, which is located in the first intron of BLK. Logistic regression analysis adjusting for rs2729935 (as this variant was significant in both datasets) indicated residual association (Presidual=1.75×10−3) present at rs2736345. However, adjusting for rs2736345 in the logistic model showed the likely presence of only one effect accounting for all the association in the region (Supplementary Figure 18).
Using expression data for BLK, eQTL analysis of the associated variants revealed several with the potential to influence transcript expression, including the top overall SNP in the region of BLK, rs2736345 (Supplementary Table 6). The most statistically significant eQTL was located in the first intron of BLK at rs2409781 (Figure 3g), a variant loosely correlated with rs2736345 and rs2729935 (r2≈0.5). Evaluation of FAM167A probes with the variants associated with Sjögren’s syndrome yielded greater statistical significance than those with BLK (Figure 3h). Bioinformatics analysis indicated the presence of many transcriptionally relevant markers not only in the shared promoter region, but also in the first intron of BLK, suggesting the possibility that association in this region impacts the transcriptional regulation of BLK and/or FAM167A (Supplementary Figure 19).
Chemokine (C-X-C motif) receptor 5 (CXCR5)
Association with a variant ~16 kb 5′ of the coding region of CXCR5 was observed at rs7119038 (Pmeta=1.10×10−8; Figure 2f; Supplementary Table 11 and Supplementary Figure 20). Three additional variants also exceeded the GWS threshold extending to rs11217033 located as far as ~90 kb 5′ of CXCR5 and as close to ~12 kb 5′ of the neighboring gene, DEAD (Asp-Glu-Ala-Asp) box polypeptide 6 (DDX6) (Figure 2f). Adjusting for rs7119038 explained all the association signals in this region (Supplementary Figure 21). Although no statistically significant eQTLs were present in the region for CXCR5, bioinformatics analysis indicated the presence of many regulatory elements throughout the region (Supplementary Figure 22). Assessment of eQTLs for DDX6 was not possible as the probe failed QC in the gene expression study.
TNFAIP3 interaction protein 1 (TNIP1)
In the region of TNIP1, one SNP, rs6579837, exceeded GWS with Pmeta=3.30×10−8; however, 29 additional variants showed evidence of association with Pmeta<5×10−5 (Figure 2g; Supplementary Table 12 and Supplementary Figure 23). Logistic regression analysis adjusting for rs6579837 accounted for all association observed in the region (Supplementary Figure 24). Several of the variants associated with Sjögren’s syndrome were also eQTLs for the TNIP1 probe, including rs6579837 (Supplementary Table 6). However, the most statistically significant eQTL was observed at rs73272842 located in TNIP1, where multiple regulatory elements have been identified (Figure 3i; Supplementary Figure 25).
Suggestive associations with Sjögren’s syndrome
We identified 8 loci that were significant in meta-analysis (Pmeta<5×10−5) but did not surpass the overall GWS threshold (Supplementary Figure 1b and Table 3). Twenty-one additional regions surpassed suggestive thresholds for significance (Pmeta<5×10−5) using DS4 (evaluated variants were exclusive to DS4; Supplementary Figure 1b; Supplementary Table 13). Further replication of these regions is needed to determine if they are bona fide risk loci.
Table 3.
Suggestive regions associated with Sjögren’s syndrome
| Gene symbol |
Chr | SNP | Alleles |
MAFb |
Observed or Imputed in Dataset 1 / 2 or 3c |
Dataset 1 |
Dataset 2 or 3 |
Meta analysis |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| maj/mina | Case | Control | OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | ||||
| Replicated using Dataset 2 | ||||||||||||
| TNFAIP3 | 6 | rs6933404 | T/C | 0.26 | 0.21 | O / O | 1.18 (0.97-1.43) | 9.27×10−2 | 1.34 (1.20-1.49) | 1.02×10−7 | 1.29 (1.13-1.47) | 6.53×10−8 |
| TNFAIP3 | 6 | rs35926684 | GA/G | 0.27 | 0.22 | I / I | 1.23 (1.01-1.49) | 3.65×10−2 | 1.31 (1.18-1.46) | 4.47×10−7 | 1.29 (1.13-1.47) | 7.21×10−8 |
| PTTG1 | 5 | rs2431098 | G/A | 0.45 | 0.51 | I / I | 0.83 (0.71-0.97) | 1.90×10−2 | 0.80 (0.73-0.88) | 3.54×10−6 | 0.81 (0.73-0.91) | 2.28×10−7 |
| PTTG1 | 5 | rs2431697 | T/C | 0.39 | 0.43 | O / O | 0.85 (0.72-1.00) | 4.75×10−2 | 0.82 (0.75-0.90) | 2.49×10−5 | 0.83 (0.74-0.93) | 3.76×10−6 |
| PRDM1 | 6 | rs526531 | G/A | 0.36 | 0.31 | O / O | 1.25 (1.05-1.49) | 1.10×10−2 | 1.21 (1.10-1.34) | 8.71×10−5 | 1.22 (1.09-1.38) | 2.93×10−6 |
| PRDM1 | 6 | rs498679 | C/T | 0.35 | 0.31 | I / I | 1.26 (1.06-1.50) | 8.29×10−3 | 1.20 (1.09-1.32) | 2.07×10−4 | 1.22 (1.08-1.37) | 5.46×10−6 |
| DGKQ | 4 | rs3733346 | T/C | 0.54 | 0.49 | I / I | 1.18 (1.01-1.39) | 4.00×10−2 | 1.20 (1.32-1.10) | 6.56×10−5 | 1.20 (1.22-1.18) | 7.73×10−6 |
| DGKQ | 4 | rs4690326 | C/A | 0.42 | 0.47 | O / O | 0.85 (0.72-1.00) | 4.47×10−2 | 0.83 (0.76-0.91) | 8.08×10−5 | 0.84 (0.75-0.93) | 1.05×10−5 |
| FCGR2A | 1 | rs6658353 | C/G | 0.45 | 0.49 | I / I | 0.77 (0.66-0.91) | 1.96×10−3 | 0.86 (0.78-0.94) | 1.13×10−3 | 0.83 (0.75-0.93) | 1.06×10−5 |
| FCGR2A | 1 | rs10800309 | G/A | 0.30 | 0.34 | O / O | 0.85 (0.71-1.01) | 6.94×10−2 | 0.84 (0.76-0.92) | 3.62×10−4 | 0.84 (0.75-0.95) | 6.72×10−5 |
| Replicated using Dataset 3 | ||||||||||||
| IRAK1BP1 | 6 | rs1507153 | C/A | 0.41 | 0.36 | O / O | 1.33 (1.13-1.57) | 5.93×10−4 | 1.22 (1.09-1.35) | 2.91×10−4 | 1.26 (1.11-1.43) | 7.09×10−7 |
| ITSN2 | 2 | rs1545257 | C/T | 0.44 | 0.48 | O / O | 0.74 (0.63-0.87) | 2.42×10−4 | 0.85 (0.77-0.94) | 1.71×10−3 | 0.81 (0.71-0.91) | 2.47×10−6 |
| PHIP | 6 | rs10943608 | T/C | 0.41 | 0.37 | O / O | 1.30 (1.11-1.53) | 1.49×10−3 | 1.19 (1.07-1.32) | 1.09×10−3 | 1.23 (1.08-1.40) | 6.22×10−6 |
Major allele / minor allele. Odds ratio for each variant indicates disease risk conferred by the minor allele.
The minor allele frequency (MAF) was calculated using the combined datasets.
Observed (O) or Imputed (I) variant in Dataset 1 / Dataset 2 or 3 dataset. Both the top observed and imputed variants for each locus are given for those replicated using Dataset 2.
Protein-protein interactions and pathway analysis
To assess the potential effects of associated variants on the underlying biology, we utilized two bioinformatics tools: the Disease Association Protein-Protein Link Evaluator (DAPPLE)18 and Genomatix Pathway System (GePS)19. DAPPLE defines genomic regions in LD with variants achieving GWS and suggestive thresholds to determine any potential protein-protein interaction (PPI) that could elucidate the underlying biological perturbations in Sjögren’s syndrome. In assessing the GWS variants outside the HLA region only, we observed no direct PPI, and while indirect PPI was observed, it was not more likely than expected by chance (Supplementary Figure 26). However, assessing both GWS and suggestive variants outside the HLA region, we observed a direct PPI network consisting of 9 proteins with 6 direct interactions (P=0.0007) and mean associated protein direct connectivity of 1.33 (expected=0.63, P=0.048) in addition to an indirect network that exhibited a mean associated protein indirect connectivity trending toward significance (P=0.082) (Supplementary Figures 27 and 28). Pathway analysis in GePS for the 6 GWS loci outside HLA found enrichment in immune signal transduction pathways (3/269 genes; P=1.54×10−3) and IL-12 signaling pathways (2/7 genes; P=3.80×10−5), primarily due to the influence of IL12A and STAT4. Adding the remaining 23 loci demonstrating suggestive association yielded similar results, with dominance of IL-12- and STAT4-dependent signaling (P=1.63×10−6). Assessing gene involvement in disease processes using Medical Subject Headings (MeSH) showed significant enrichment in multiple immunologically-mediated diseases, including non-Hodgkin lymphoma (16/2910 genes; P=1.7×10−6), systemic lupus erythematosus (SLE) (12/1691 genes; P=4.74×10−6), Epstein-Barr virus infections (11/1421 genes; P=6.18×10−6), dry eye syndrome (8/868 genes; P=4.99×10−5), and Sjögren’s syndrome (6/756 genes; P=1.15×10−3).
Discussion
In this study, associations previously identified in the HLA region with Sjögren’s syndrome were replicated and represent the strongest genetic risk factors with OR ≈ 3.5. Our results are consistent with a trans-racial meta-analysis of 23 case-control studies evaluating Class II loci in Sjögren’s syndrome, which also identified HLA-DQA1*0501, HLA-DQB1*0201, and HLA-DRB*0301 as disease risk factors11. Associated variants in this study with a Pmeta<5×10−5 were found to be enriched within 100 bp of RFX5 ChIP-seq peaks identified by ENCODE17. Mutations in RFX5 can lead to the inability to transcribe Class II HLA molecules and bare lymphocyte syndrome20. In Sjögren’s syndrome, several risk alleles in or near the RFX5 ChIP-Seq peaks in the Class II region were eQTLs (Figure 3a-e), indicating that this TF may play a critical pathogenic role. Moreover, eQTLs were identified in the Class I region with additional HLA molecules, including HLA-C, HLA-A, HLA-H, and HLA-G, and previous studies have found that RFX5 regulates the expression of these loci21. Extensive LD between variants in the HLA region may influence the enrichment analysis performed in GenomeRunner when using all variants as background; however, we continued to observe enrichment when focusing only on the HLA region. Although we cannot completely rule out the influence of LD on these results, the observation that several of these variants are eQTLs further indicates biological relevance.
Although HLA Class II molecules have classically been implicated in Sjögren’s syndrome risk, HLA Class I loci have also been reported22. This suggests that risk attributed to the HLA region may involve altered expression of multiple genes residing in this region and important to the immune system through altered binding of RFX5 to DNA motifs and/or accessory molecules. However, it is likely that other mechanisms, such as amino-acid changes similar to those identified by Raychaudhuri et al. in rheumatoid arthritis (RA)23, are involved in the genetic pathophysiology of this region. Comparing the effect sizes for the two primary HLA associations with other autoimmune diseases, we find that effect sizes in Sjögren’s syndrome appear stronger than those in SLE and RA, similar to those in MS, and weaker than those in celiac disease, type I diabetes, and psoriasis (Supplementary Figure 29).
This study also established IRF5-TNPO3, STAT4, IL12A, FAM167A-BLK, DDX6-CXCR5, and TNIP1 as risk loci. IRF5 is a transcription factor mediating type I interferon responses in monocytes, dendritic cells, and B cells24 that induces the transcription of interferon-α genes and the production of pro-inflammatory cytokines (including IL-12, p40, IL-6, and TNF-α)25 upon viral infection. IRF5 is an established risk locus in SLE, RA, ulcerative colitis, primary biliary cirrhosis (PBC), and systemic sclerosis (SSc)26-33. In SLE, association with three independent functional alleles was observed34, similar to the effects described in this study. Previous studies in Sjögren’s syndrome have also implicated IRF5 as a risk locus, but only ~15 variants have been tested, with a CGGGG indel polymorphism yielding the most statistically significant result and appearing to be responsible for the promoter region association (Supplementary Table 1)13,15,35,36. While we were unable to impute the CGGGG indel, the variant rs2004640, which is correlated (r2=0.7) with the CGGGG indel36, demonstrated association (Pmeta=8.86×10−15). Moreover, a three variant haplotype, consisting of rs3757385, rs2004640, and rs10954213, perfectly tags the CGGGG indel37,38. Adjusting logistic regression models for these three variants showed significant residual association (P=1.05×10−7) at rs3757387. After adding rs3757387, residual association (P=1.64×10−4) was observed at rs17339836, suggesting that adjusting for the 3 SNP haplotype tagging the CGGGG indel was equivalent to adjusting for only rs10954213. Additional studies are necessary to determine the precise causal variant in the IRF5 promoter region.
STAT4 is critical for cellular responses initiated by type I interferons and is subsequently induced by IL-12 in lymphocytes, leading to the transcription of interferon-γ39. The association of variants in the third intron of STAT4 has been well established in other inflammatory diseases32,40-42 and has been implicated in Sjögren’s syndrome (Supplementary Table 1)12,14,15,43. The variants reported in these studies are highly correlated (r2>0.95) with the associated variant rs10553577 identified in this study.
IL12A encodes the p35 subunit that forms the IL-12 heterodimer with the p40 subunit encoded by IL12B44. IL-12, an immunomodulatory cytokine primarily secreted by monocytes and dendritic cells, plays a critical role in T-helper 1 cell differentiation and the production of interferon-γ by T cells and NK cells45. Although no studies in Sjögren’s syndrome report association to the IL12A region, several studies in related conditions have identified variants near IL12A that increase disease risk. Association of variants in the 3′ end of IL12A have been reported for PBC, while 5′ effects have been described for celiac disease46-48. Interestingly, the variants reported in PBC showed weak correlations (r2<0.35) with the top SNP in Sjögren’s syndrome, and analyses adjusting for variants identified in related diseases continued to exhibit residual association (Presidual<0.05) at rs485497. Moreover, rs574808, rs495499, and rs6441286, the most statistically significant variants in PBC42,46, are highly correlated with the eQTL polymorphism reported in this study. Previous studies observed only suggestive association with the block encompassing the coding region of this gene, which includes the top overall associated variant, rs485497, in this study. Thus, the effect in the region of IL12A in Sjögren’s syndrome appears to be at least partially distinct from PBC.
IRF5, IL12A, and STAT4, all participate in type I interferon pathway signaling. A role for type I interferon pathway dysregulation in Sjögren’s syndrome has been described through mRNA gene expression microarray studies of labial salivary glands and peripheral blood8,9,49,50. Moreover, Emamian et al. has shown that overexpression of type I interferon inducible genes correlates with titers of the classic Sjögren’s syndrome autoantibodies anti-Ro and anti-La9. Genetic effects in Sjögren’s syndrome potentially influence both subunits of IL-12. IRF5 can initiate transcription of IL12B while variants in the region of IL12A have also been identified as risk factors in this study. Once secreted from dendritic or NK cells, IL-12 binds T cell receptors, thereby initiating a signaling cascade through STAT4 phosphorylation44.
BLK is a non-receptor src family tyrosine kinase involved in B cell receptor signaling and B cell development. B cell receptor signaling is essential for proper immune function, deletion of autoreactive B cells, and subsequent receptor editing51,52. Variants in the BLK locus have been reported to influence both FAM167A and BLK mRNA and protein expression and are associated with related diseases, such as SLE53,54. Previous studies have implicated this region in Sjögren’s syndrome risk with peak association observed within FAM167A at rs1254979614, while the current study identifies the shared promoter region and the first intron of BLK as the most statistically significant associations (Figure 2e).
CXCR5, a membrane-bound protein present on some memory B cells and on follicular helper T cells, acts as a receptor for B-lymphocyte chemoattractant (BLC). Variants in the region of CXCR5 have been associated with multiple sclerosis and PBC42,55. Although no genetic associations between Sjögren’s syndrome and CXCR5 have been reported, several studies have found CXCR5 to be dysregulated in B cells in salivary gland tissues and the periphery49,56. Interestingly, IL-12 alone can induce CXCR5 expression, leading to the early commitment of naïve CD4+ T cells to the T follicular helper cell lineage57.
Although the exact function of TNIP1 has not been defined, TNIP1 does bind TNFAIP3, which suppresses TLR-induced apoptosis by negatively regulating NF-κB58. Variants in the region of TNIP1 have been found to increase risk for RA, SLE, psoriasis, and SSc59-63. In SLE, two independent risk haplotypes have been identified, with the H2 haplotype defined by Adrianto et al.62 corresponding to the association identified in this study. Similar to the findings reported above, the H2 haplotype in SLE also shows allele-specific differential expression62.
Considered jointly in the logistic regression model, the genome-wide and suggestively associated variants could explain a significant proportion of the heritable risk (C statistic = 0.811, Supplementary Table 14). This value is higher than that of the SLE model in women of European ancestry (C statistic = 0.67)33. Consideration was also given to the effect size of Sjögren’s syndrome-associated variants in LD with previously established loci achieving GWS in other autoimmune diseases. Of note, the OR and 95% CI for Sjögren’s syndrome variants outside the HLA region (Supplementary Table 15 and Supplementary Figure 30) are comparable to those in related autoimmune diseases, particularly with respect to SLE, with much greater variability in effect size and CI when considering the variants in the HLA region (Supplementary Table 16 and Supplementary Figure 29).
Using bioinformatics tools, the combination of GWS and suggestively associated variants provides some evidence of significant direct and indirect PPI and enrichment of genes involved in immune signaling processes, particularly with respect to IL-12 and STAT4 and many autoimmune-related diseases. The importance of type I interferons has been previously recognized in the pathogenesis of Sjögren’s syndrome. However, the type II interferon pathway, acting through interferon-γ downstream of IL-12 and STAT4, is now clearly implicated by this study. In addition, multiple genes in the NF-κB pathway, including TNIP1 and TNFAIP3, support a role for this pathway in Sjögren’s syndrome pathogenesis. Furthermore, establishing association of several genes important in the adaptive immune response, including CXCR5 and BLK, as well as suggestive associations in loci such as FCGR2A, provides insight into the complex genetic architecture for Sjögren’s syndrome and will facilitate further studies to understand the precise functional consequences of these risk variants. Taken together, this work provides significant progress towards explaining the underlying genetic pathophysiology of Sjögren’s syndrome, which will ultimately provide important new therapeutic targets and potential diagnostic markers, both of which are lacking.
In summary, we performed a large-scale genetic study in Sjögren’s syndrome and established IRF5-TNPO3, STAT4, IL12A, FAM167A-BLK, DDX6-CXCR5, and TNIP1 as risk loci. We have determined that there are likely multiple independent effects within the HLA and IRF5 regions. Our results also strongly implicate 29 suggestive regions in Sjögren’s syndrome etiology warranting further study. Together, these results highlight the importance of the innate and adaptive immune systems in Sjögren’s syndrome etiology. Future work is needed to replicate these additional candidate associations and to characterize the causal variant(s) for the regions established in this study.
Online Methods
Subjects
The samples used in this study were collected through the Sjögren’s Genetics Network (SGENE) and organized at the Oklahoma Medical Research Foundation (OMRF; Supplementary Table 2). In Dataset 1, 438 Sjögren’s syndrome cases and 3917 population controls of European descent were subjected to quality control measures outlined below. Each Sjögren’s syndrome case was genetically matched to five population controls in DS1 using the identity-by-state (IBS) to assess allele sharing as implemented in PLINK64, and the remaining controls were used in DS2 and DS3. In the next phase, 1521 Sjögren’s syndrome cases and 6626 population controls were evaluated in DS2 only for variants that overlapped with DS1; 1169 Sjögren’s syndrome cases and 3078 population controls were evaluated in DS3; and 1897 cases and 3039 population controls were evaluated in DS4 (313 cases overlapped with DS1). Cases fulfilled the American-European Consensus Group (AECG) criteria for primary Sjögren’s syndrome5, and all population controls were evaluated within their respective study. All cases were evaluated by expert clinicians for possible overlap of clinical features or concurrent diagnosis with other autoimmune diseases. Written informed consent was obtained by each participant following protocols approved by the Institutional Review Boards of each institution where the samples were collected.
Genotyping and Quality Control
Genotypes for DS1 were obtained using the Illumina Omni1-Quad array using Infinium chemistry at OMRF following the manufacturer’s protocol (Illumina, Inc., San Diego, CA). Similarly, DS2/DS4 genotyping was performed with the ImmunoChip (an Illumina iSelect custom array designed by the ImmunoChip Consortium)65. DS3 was genotyped on iSelect arrays containing 1536 variants overlapped with DS1 that were not available on the ImmunoChip. Strict quality control procedures were implemented before SNPs were used in this analysis, including requirements for: well-defined cluster scatter plots; minor allele frequency >1%; SNP call rate >95%; sample call rate >95%; Hardy-Weinberg proportion (HWP) test with a P > 0.001 in controls; and P > 0.001 for differential missingness between cases and controls.
Samples with <95% call rate and excessive increased heterozygosity (>5 standard deviations from the mean) were excluded from the analysis. Relatedness within the remaining samples was determined using identity-by-descent (IBD) estimates as determined by PLINK v1.0764. One individual from each pair was removed if the proportion of the alleles shared identity by descent was >0.4, with preference towards cases and/or the individual with the highest call rate. Gender was estimated genetically, and discrepancies with self-report were scrutinized. Males were required to be heterozygous at rs2557523 (since the G allele for this SNP is only observed on the Y chromosome and the A allele appears only on the X chromosome) and to have chromosome X heterozygosity ≤10%, while females were required to be homozygous for the A allele at rs2557523 and have chromosome X heterozygosity >10%. After quality control, 648,937 SNPs were available to test for association across all datasets for DS1. Only SNPs present on both DS1 and DS2 arrays (20,055 after quality control) were used for meta-analysis and fine-mapping (after imputation, see below for more details). All base pair positions are given according to the hg19 version of the human genome reference sequence.
Ascertainment of Population Stratification
Genetic outliers were removed from further analysis as determined by principal components analysis (Supplementary Figure 31)66,67. Population substructure was identified within the sample set using EIGENSTRAT66 with 40,073 (DS1) and 2,827 (DS2/DS4) independent markers (r2<0.20 between variants). The resulting Eigenvectors were able to distinguish the four continental ancestral populations in the following HapMap samples: Africans (ASW, LWK, MKK, and YRI), Europeans (CEU and TSI), Hispanic and East Indians (MEX and GIH), and Asians (CHB, CHD, and JPT); Supplementary Figure 30)67,68. We utilized principal components to identify samples output by EIGENSTRAT that were not of European ancestry. Samples >6 standard deviations from the mean were eliminated from further analysis. Samples were then plotted by PC1 and PC2 for only the cases and controls used in DS1, DS2, or DS4 (Supplementary Figure 31). We further scrutinized and removed outliers that resided outside the main cluster of cases and controls. In addition, each Sjögren’s syndrome case was genetically matched to 5 out-of-study population controls for DS1 using the cluster feature in PLINK v1.0764. After quality control, 395 Sjögren’s syndrome cases and 1975 population controls in DS1, 1243 cases and 4779 controls in DS2, 1158 cases and 3071 controls on DS3, and 1541 case and 2634 controls in DS4 were available to test for association (Supplementary Figure 1 and Supplementary Table 2).
Statistical Analysis
To test for SNP-Sjögren’s syndrome association, logistic regression models were computed as implemented in PLINK v1.0764. The additive genetic model was calculated for chromosomes 1-22 while adjusting for the first three principal components (as these accounted for >80% of the variation in the dataset after quality control) and gender. For chromosome X, only females were tested for association with Sjögren’s syndrome. Meta-analysis of the SNPs observed in both DS1 and DS2 or DS3 were calculated using a weighted Z-score in METAL69. Each dataset group was weighted by the square root of its sample size to control for differences between the two phases. For a locus to be considered it must have been significant in both datasets and the Pmeta must have exceeded the suggestive threshold of Pmeta<5×10−5.
To test for meta-analysis heterogeneity, we utilized both the Cochran’s Q test statistic70 and I2 index71 (reported in the Supplementary Tables 3, 7-12). A P < 0.05 was considered significant evidence of heterogeneity for Cochran’s Q test. The I2 index ranges between 0% and 100%, where I2 equal 0% to 25%, 26% to 50%, 51% to 75%, and 76% to 100%, indicating low, moderate, high, and very high heterogeneity, respectively. These data are provided in the Supplementary Tables for each region (Supplementary Tables 3, 7-12).
Linkage disequilibrium and probable haplotypes were determined using HAPLOVIEW ver. 4.272. Haplotype blocks were calculated using the solid-spine of LD algorithms with minimum r2 values of 0.872. To determine the number of independent effects in each region, we used step-wise logistic regression adjusting for the most significant variant with P<0.0001 in the both the adjusted and unadjusted analysis to be considered. Adjusted logistic regression analysis results for each Sjögren’s syndrome-associated region were plotted using LocusZoom73.
We computed the C statistic of the multiple logistic regression model using “rms” package in R. The C statistic is the area under the receiver operator characteristic (ROC) curve that provides a measure on how well the model can discriminate between cases and population controls.
Imputation
To increase informativeness, imputation was conducted in subjects from both the DS1 and DS2 over a 100 kb interval spanning loci that met criteria for genome-wide significance or select loci demonstrating suggestive significance. Imputation of both datasets was performed across 12 regions: FCGR2A (chr1: 161,425,205-161,539,360), STAT4 (chr2: 191,844,302-192,066,322), IL12A (chr3: 159,356,623-159,763,806), DGKQ (chr4: 923,459-1,001,272), TNIP1 (chr5: 150,359,504-150,517,221), PTTG1 (chr5: 159,528,253-159,902,455), HLA (chr6: 28,400,000-33,400,000), TNFAIP3 (chr6: 137,938,325-138,254,451), IRF5 (chr7: 128.527,994-128,740,088), BLK (chr8: 11,251,521-11,472,108), PRDM1 (chr6: 106,484,195-106607,814), and DDX6-CXCR5 (chr11: 118,548,473-118,816,980). Imputation was performed using IMPUTE2 and the European Impute2 1000 Genomes Phase 1 April 2012 reference panel74-76. Imputed genotypes had to meet or exceed a probability threshold of 0.9, an information measure of >0.5, and the quality control criteria described above to be included in the analyses.
The HLA classical allele imputation was performed using HLA Genotype Imputation with Attribute Bagging (HIBAG)77. HIBAG is a highly accurate, computationally tractable package for imputing HLA types using SNP data without a training dataset in various populations, including European population. By using HLA SNPs after QC, we imputed the HLA classical alleles in seven HLA genes in both DS1 and DS2 (Supplementary Figure 6). Individuals with imputation score < 0.8 and alleles with frequency < 0.01 were removed for analysis.
Expression Quantitative Trait Loci (eQTL) Analysis
Gene expression profile analysis: We performed global gene expression profiling (GEP) analysis using peripheral blood on 48,803 probes representing 37,805 loci from the Illumina HumanWG-6 v3.0 BeadChip in 258 subjects: 182 primary Sjögren’s syndrome cases meeting AECG 2002 criteria5 and 76 population controls. Analyses were performed in the R Bioconductor suite. The GEP data were obtained from two separate microarray experiments. Quality control (QC) was applied in each dataset to filter out transcripts that were expressed in less than 10% of the subjects (detection call threshold: P<0.05) and probes with differential missing rates (P<0.001 by Fisher’s exact test) between the two datasets. Re-annotation and Mapping of Oligonucleotide Array Technologies (ReMOAT) was used to determine the quality of Illumina BeadArray probes78. Each dataset was then normalized independently using Robust Multiarray Average (RMA)79 followed by log2 transformation and quantile normalization. ComBat was subsequently applied to the combined dataset to adjust for non-biological experimental variation (batch effect)80. After QC and normalization, 15,063 probes (in 12,248 genes) were kept for further statistical analysis.
Cis-expression quantitative trait loci analysis: Expression levels of the Sjögren’s syndrome-associated genes identified in the meta-analysis of DS1 and DS2 exceeding GWS were selected for use as phenotypic traits in cis-eQTL analyses in 222 European subjects (190 cases and 32 controls). Variants associated with Sjögren’s syndrome (P<5×10−5) in the meta-analysis flanking the target genes were evaluated for genetic association with gene expression levels using both a linear model in Matrix-eQTL81 (using gender and disease status as covariates) and Analysis of Variance (ANOVA) models in Prism 6. An eQTL was considered significant if a false discovery rate (FDR) adjusted P-value, accounting for the number of tests performed in each region, was P<0.05.
Enrichment analysis
GenomeRunner82 was used to search for statistically significant enrichment of the set of 9,394 SNPs showing suggestive (P<5×10−5) or lower association with Sjögren’s syndrome with annotated genomic features. Genome annotations include genes and gene prediction tracks, mRNA and EST tracks, regulation related tracks from ENCODE project, histone modification marks, comparative genomics tracks, structural variation and repeats tracks. GenomeRunner evaluates the null hypothesis that co-localization of a set of Sjögren’s syndrome-associated SNPs with genome annotation features is not statistically significantly different from what would be observed for a random set of SNPs of the same size. Briefly, the Sjögren’s syndrome-associated set of SNPs was tested for co-localization with each of the genome annotation tracks obtained from the UCSC genome database83 for the human GRCh37/hg19 genome assembly. The whole set of 219,958 SNPs after imputation of select regions present on the ImmunoChip array was used to randomly select an equally sized group.
Separate analyses included identification of overrepresented experimentally validated TFBSs transcription factor binding sites, a total of 148 of them extracted from the “Transcription Factor ChIP-seq from ENCODE” track (wgEncodeRegTfbsClustered). Regions of Chromatin State Segmentation by HMM from ENCODE/Broad (wgEncodeBroadHmmGm12878HMM) were also used to assess overrepresentation of defined chromatin states containing Sjögren’s syndrome-associated SNPs. Tracks of Histone Modifications obtained by ChIP-seq from ENCODE/Broad Institute, from ENCODE/Stanford/Yale/USC/Harvard, and from ENCODE/University of Washington were used for assessment of epigenetic marks. Unless otherwise noted, data from the GM12878 lymphoblastoid cell line were used, as the majority of the best quality genome annotation data was obtained from this cell line. The number of Sjögren’s syndrome-associated SNPs co-localized with a genome annotation track was compared with that of obtained from 1,000 random simulations using Chi-square test. Additional analyses of random sets of SNPs showed no statistically significant enrichments (data not shown). For an enrichment to be considered statistically significant, we set a Bonferroni correction threshold for 6000 independent tests at P<8.33×10−6.
RFX5-HLA region enrichment analysis
The HLA region was defined by genomic coordinates chr6:28400295-33398620 (GRCh37/hg19 human genome assembly). The number of significant SNPs in this region overlapping with RFX5 binding sites was calculated using BedTools84. The number of all genome-wide associated SNPs in the HLA region overlapping RFX5 binding sites was calculated and compared with the SNPs significantly associated with Sjögren’s syndrome using Fisher’s exact test (scipy.stats.fisher_exact function).
Protein-protein interactions
The Disease Association Protein-Protein Link Evaluator (DAPPLE)18 was used to assess the potential effects of associated variants on the underlying biology. For DAPPLE, seed variants were converted to genes based on variant overlap and LD structure within the region of interest. Analyses were performed using the following settings: 10,000 permutations; a common interactor binding degree cutoff of 2; gene regulatory region 50kb upstream and downstream of transcription start and end; simplification of the indirect network; and iteration to prioritize genes with a Bonferroni-corrected score of P<0.05. A total of 14 GWS variants and 114 suggestive variants were present in the DAPPLE databases; where variants were not found to be present, attempts to find proxies in high LD were made.
Pathway Analysis in Genomatix
Pathway analysis was carried out using the Genomatix Pathway System (GePS)19. This system utilizes information obtained from public and proprietary annotation databases to characterize input gene lists, determine the representation of these genes in canonical pathways, and create gene networks based on co-citation of genes from the literature. The public annotation data sets incorporated in this software include gene ontology (GO) and medical subject headings (MeSH) in addition to others. GePS also provides a probability statistic using Fisher’s exact test to determine the likelihood of finding the number of co-represented genes with a particular annotation when considering the length of the input gene list. Input gene sets were generated using the gene locus nearest the association signals, which were required to demonstrate suggestive or genome-wide significance, and the pathway analysis was carried out using the gene symbols of the input set.
Supplementary Material
Acknowledgements
We are grateful to all the individuals with Sjögren’s syndrome and those serving as healthy controls who participated in this study. We would like to thank the following individuals for their help in the collection and ascertainment of the samples used in this study: Erin Rothrock, Judy Harris, Sharon Johnson, Sara Cioli, Nicole Weber, Dominique Williams, Wes Daniels, Cherilyn Pritchett-Frazee, Kylia Crouch, Laura Battiest, Justin Rodgers, James Robertson, Thuan Nguyen, Amanda Crosbie, Ellen James, Carolyn Meyer, Amber McElroy, Eshrat Emamian, Julie Ermer, Kristine Rohlf, Joanlise Leon, Anita Petersen, Danielle Hartle, Jill Novizke, Ward Ortman, Carl Espy, Beth Cobb, Gudlaug Kristjansdottir, Marianne Eidsheim and Joelle Benessiano Centre de Ressources Biologiques, Hôpital Bichat, Paris, France. We would also like to thank Stuart Glenn and Jared Ning for their ongoing assistance in developing and maintaining the computational infrastructure used to perform this study. We would like to thank the following funding agencies for their support: This publication was made possible by grants P50 AR0608040 (K.L.S., C.J.L., R.H.S., and A.D.F.), 5R01 DE015223 (K.L.S. and J.B.H.), 5RC2 AR058959 (P.M.G.), 5P01 AR049084-10 (J.B.H), 5P30 AR053483 (J.A.J. and J.M.G.), 5U19 AI082714 (K.L.S., J.A.J., and C.J.L.), 1R01 DE018209-02 (K.L.S and J.B.H.), 5R01 DE018209 (K.L.S.), 8P20 GM103456 (P.M.G., C.J.L., J.D.W, and I.A.), P20 GM103636 (M.G.D. and J.D.W.), 5R37 AI024717-25 (J.B.H), 5P01 AI083194-03 (K.L.S. and J.B.H.), 7S10 RR027190-02 (J.B.H.), 1U01 AI101934 (J.A.J. and J.M.G.), 1RC1 AR058554 (J.A.J. and J.M.G.), and 5P30 GM103510 (J.A.J. AND J.M.G.) from the NIH. The contents are the sole responsibility of the authors and do not necessarily represent the official views of the NIH. Additional funding was obtained from Intramural Research Program of the National Institute of Dental and Craniofacial Research (G.G.I.), US Department of Veterans Affairs IMMA 9 (J.B.H.), USA Department of Defense PR094002 (J.B.H), American College of Rheumatology Research and Education Foundation/Abbott Health Professional Graduate Student Preceptorship Award 2009 (C.J.L. and K.L.S.), Oklahoma Medical Research Foundation (C.J.L. and K.L.S.), Sjögren’s Syndrome Foundation (K.L.S.), Phileona Foundation (K.L.S.), French ministry of health: PHRC N°2006-AOM06133 and French ministry of research: ANR-2010-BLAN-1133 (X.M. and C.M.R.), The Strategic Research Program at Helse Bergen, Western Norway Regional Health Authority (L.G.G., J.G.B., and R.J.), The Broegelmann Foundation (J.G.B. and R.J.), Norwegian Foundation for Health and Rehabilitation (E.H.), KFO 250 TP03, WI 1031/6-1 (T.W.), KFO 250, Z1 (T.W.), Medical Research Council, UK G0800629 (W.F.N. and S.B.), Northumberland, Tyne & Wear CLRN (W.F.N.), The Swedish Research Council (M.W.H. and L.R.), The King Gustaf the V-th 80-year Foundation (M.W.H.), Knut and Alice Wallenberg Foundation (L.R.), and The Swedish Rheumatism Association (M.W.H., G.N., L.R., and P.E.). This study made use of genotypes available through dbGAP, with acknowledgment provided in the supplement.
Footnotes
Data access.
URLs.
HLA Genotype Imputation with Attribute Bagging http://cran.r-project.org/web/packages/HIBAG/
R Bioconductor suite http://www.R-project.org
Prism 6 http://www.graphpad.com/scientific-software/prism/
GenomeRunner http://sourceforge.net/projects/genomerunner
Genomatix Pathway System http://www.genomatix.de
Accession Numbers: Genes and Blood Clotting Study dataset(s) obtained through dbGaP: phs000304.v1.p1
GWAS of Ischemic Stroke dataset(s) obtained through dbGaP: phs000292.v1.p1
CIDR:NGRC Parkinson’s disease study dataset(s) obtained through dbGaP: phs000196.v2.p1
High Density SNP Association Analysis of Melanoma: Case-Control and Outcomes Investigation dataset(s) obtained through dbGaP: phs000187.v1.p1
Competing Financial Interests: The authors declare no competing financial interests.
References
- 1.Helmick CG, et al. Estimates of the prevalence of arthritis and other rheumatic conditions in the United States. Part I. Arthritis Rheum. 2008;58:15–25. doi: 10.1002/art.23177. [DOI] [PubMed] [Google Scholar]
- 2.Pillemer SR, et al. Incidence of physician-diagnosed primary Sjogren syndrome in residents of Olmsted County, Minnesota. Mayo Clin Proc. 2001;76:593–9. doi: 10.4065/76.6.593. [DOI] [PubMed] [Google Scholar]
- 3.Plesivcnik Novljan M, et al. Incidence of primary Sjogren’s syndrome in Slovenia. Ann Rheum Dis. 2004;63:874–6. doi: 10.1136/ard.2003.014027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jonsson R, et al. The complexity of Sjogren’s syndrome: novel aspects on pathogenesis. Immunol Lett. 2011;141:1–9. doi: 10.1016/j.imlet.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 5.Vitali C, et al. Classification criteria for Sjogren’s syndrome: a revised version of the European criteria proposed by the American-European Consensus Group. Ann Rheum Dis. 2002;61:554–8. doi: 10.1136/ard.61.6.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thanou-Stavraki A, James JA. Primary Sjogren’s syndrome: current and prospective therapies. Semin Arthritis Rheum. 2008;37:273–92. doi: 10.1016/j.semarthrit.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 7.Cobb BL, Lessard CJ, Harley JB, Moser KL. Genes and Sjogren’s syndrome. Rheum Dis Clin North Am. 2008;34:847–68. vii. doi: 10.1016/j.rdc.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hjelmervik TO, Petersen K, Jonassen I, Jonsson R, Bolstad AI. Gene expression profiling of minor salivary glands clearly distinguishes primary Sjogren’s syndrome patients from healthy control subjects. Arthritis Rheum. 2005;52:1534–44. doi: 10.1002/art.21006. [DOI] [PubMed] [Google Scholar]
- 9.Emamian ES, et al. Peripheral blood gene expression profiling in Sjogren’s syndrome. Genes Immun. 2009;10:285–96. doi: 10.1038/gene.2009.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ice JA, et al. Genetics of Sjogren’s syndrome in the genome-wide association era. J Autoimmun. 2012;39:57–63. doi: 10.1016/j.jaut.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cruz-Tapias P, Rojas-Villarraga A, Maier-Moore S, Anaya JM. HLA and Sjogren’s syndrome susceptibility. A meta-analysis of worldwide studies. Autoimmun Rev. 2012;11:281–7. doi: 10.1016/j.autrev.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 12.Korman BD, et al. Variant form of STAT4 is associated with primary Sjogren’s syndrome. Genes Immun. 2008;9:267–70. doi: 10.1038/gene.2008.1. [DOI] [PubMed] [Google Scholar]
- 13.Miceli-Richard C, et al. Association of an IRF5 gene functional polymorphism with Sjogren’s syndrome. Arthritis Rheum. 2007;56:3989–94. doi: 10.1002/art.23142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nordmark G, et al. Association of EBF1, FAM167A(C8orf13)-BLK and TNFSF4 gene variants with primary Sjogren’s syndrome. Genes Immun. 2011;12:100–9. doi: 10.1038/gene.2010.44. [DOI] [PubMed] [Google Scholar]
- 15.Nordmark G, et al. Additive effects of the major risk alleles of IRF5 and STAT4 in primary Sjogren’s syndrome. Genes Immun. 2009;10:68–76. doi: 10.1038/gene.2008.94. [DOI] [PubMed] [Google Scholar]
- 16.Kang HI, et al. Comparison of HLA class II genes in Caucasoid, Chinese, and Japanese patients with primary Sjogren’s syndrome. J Immunol. 1993;150:3615–23. [PubMed] [Google Scholar]
- 17.A user’s guide to the encyclopedia of DNA elements (ENCODE) PLoS Biol. 2011;9:e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rossin EJ, et al. Proteins encoded in genomic regions associated with immune-mediated disease physically interact and suggest underlying biology. PLoS Genet. 2011;7:e1001273. doi: 10.1371/journal.pgen.1001273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frisch M, Klocke B, Haltmeier M, Frech K. LitInspector: literature and signal transduction pathway mining in PubMed abstracts. Nucleic Acids Res. 2009;37:W135–40. doi: 10.1093/nar/gkp303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nekrep N, et al. Mutation in a winged-helix DNA-binding motif causes atypical bare lymphocyte syndrome. Nat Immunol. 2002;3:1075–81. doi: 10.1038/ni840. [DOI] [PubMed] [Google Scholar]
- 21.Meissner TB, et al. NLRC5 cooperates with the RFX transcription factor complex to induce MHC class I gene expression. J Immunol. 2012;188:4951–8. doi: 10.4049/jimmunol.1103160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loiseau P, et al. HLA class I and class II are both associated with the genetic predisposition to primary Sjogren syndrome. Hum Immunol. 2001;62:725–31. doi: 10.1016/s0198-8859(01)00253-1. [DOI] [PubMed] [Google Scholar]
- 23.Raychaudhuri S, et al. Five amino acids in three HLA proteins explain most of the association between MHC and seropositive rheumatoid arthritis. Nat Genet. 2012;44:291–6. doi: 10.1038/ng.1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Savitsky D, Tamura T, Yanai H, Taniguchi T. Regulation of immunity and oncogenesis by the IRF transcription factor family. Cancer Immunol Immunother. 2010;59:489–510. doi: 10.1007/s00262-009-0804-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takaoka A, et al. Integral role of IRF-5 in the gene induction programme activated by Toll-like receptors. Nature. 2005;434:243–9. doi: 10.1038/nature03308. [DOI] [PubMed] [Google Scholar]
- 26.Sigurdsson S, et al. Polymorphisms in the tyrosine kinase 2 and interferon regulatory factor 5 genes are associated with systemic lupus erythematosus. Am J Hum Genet. 2005;76:528–37. doi: 10.1086/428480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sigurdsson S, et al. Association of a haplotype in the promoter region of the interferon regulatory factor 5 gene with rheumatoid arthritis. Arthritis Rheum. 2007;56:2202–10. doi: 10.1002/art.22704. [DOI] [PubMed] [Google Scholar]
- 28.Stahl EA, et al. Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat Genet. 2010;42:508–14. doi: 10.1038/ng.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dideberg V, et al. An insertion-deletion polymorphism in the interferon regulatory Factor 5 (IRF5) gene confers risk of inflammatory bowel diseases. Hum Mol Genet. 2007;16:3008–16. doi: 10.1093/hmg/ddm259. [DOI] [PubMed] [Google Scholar]
- 30.Liu X, et al. Genome-wide meta-analyses identify three loci associated with primary biliary cirrhosis. Nat Genet. 2010;42:658–60. doi: 10.1038/ng.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dieude P, et al. Association between the IRF5 rs2004640 functional polymorphism and systemic sclerosis: a new perspective for pulmonary fibrosis. Arthritis Rheum. 2009;60:225–33. doi: 10.1002/art.24183. [DOI] [PubMed] [Google Scholar]
- 32.Radstake TR, et al. Genome-wide association study of systemic sclerosis identifies CD247 as a new susceptibility locus. Nat Genet. 2010;42:426–9. doi: 10.1038/ng.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harley JB, et al. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat Genet. 2008;40:204–10. doi: 10.1038/ng.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Graham RR, et al. Three functional variants of IFN regulatory factor 5 (IRF5) define risk and protective haplotypes for human lupus. Proc Natl Acad Sci U S A. 2007;104:6758–63. doi: 10.1073/pnas.0701266104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miceli-Richard C, et al. The CGGGG insertion/deletion polymorphism of the IRF5 promoter is a strong risk factor for primary Sjogren’s syndrome. Arthritis Rheum. 2009;60:1991–7. doi: 10.1002/art.24662. [DOI] [PubMed] [Google Scholar]
- 36.Sigurdsson S, et al. Comprehensive evaluation of the genetic variants of interferon regulatory factor 5 (IRF5) reveals a novel 5 bp length polymorphism as strong risk factor for systemic lupus erythematosus. Hum Mol Genet. 2008;17:872–81. doi: 10.1093/hmg/ddm359. [DOI] [PubMed] [Google Scholar]
- 37.Dawidowicz K, et al. The interferon regulatory factor 5 gene confers susceptibility to rheumatoid arthritis and influences its erosive phenotype. Ann Rheum Dis. 2011;70:117–21. doi: 10.1136/ard.2010.129171. [DOI] [PubMed] [Google Scholar]
- 38.Kristjansdottir G, et al. Interferon regulatory factor 5 (IRF5) gene variants are associated with multiple sclerosis in three distinct populations. J Med Genet. 2008;45:362–9. doi: 10.1136/jmg.2007.055012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kaplan MH. STAT4: a critical regulator of inflammation in vivo. Immunol Res. 2005;31:231–42. doi: 10.1385/IR:31:3:231. [DOI] [PubMed] [Google Scholar]
- 40.Remmers EF, et al. STAT4 and the risk of rheumatoid arthritis and systemic lupus erythematosus. N Engl J Med. 2007;357:977–86. doi: 10.1056/NEJMoa073003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rueda B, et al. The STAT4 gene influences the genetic predisposition to systemic sclerosis phenotype. Hum Mol Genet. 2009;18:2071–7. doi: 10.1093/hmg/ddp119. [DOI] [PubMed] [Google Scholar]
- 42.Mells GF, et al. Genome-wide association study identifies 12 new susceptibility loci for primary biliary cirrhosis. Nat Genet. 2011;43:329–32. doi: 10.1038/ng.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gestermann N, et al. STAT4 is a confirmed genetic risk factor for Sjogren’s syndrome and could be involved in type 1 interferon pathway signaling. Genes Immun. 2010;11:432–8. doi: 10.1038/gene.2010.29. [DOI] [PubMed] [Google Scholar]
- 44.Watford WT, et al. Signaling by IL-12 and IL-23 and the immunoregulatory roles of STAT4. Immunological reviews. 2004;202:139–56. doi: 10.1111/j.0105-2896.2004.00211.x. [DOI] [PubMed] [Google Scholar]
- 45.Xu M, et al. Regulation of antitumor immune responses by the IL-12 family cytokines, IL-12, IL-23, and IL-27. Clin Dev Immunol. 2010;2010 doi: 10.1155/2010/832454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hirschfield GM, et al. Primary biliary cirrhosis associated with HLA, IL12A, and IL12RB2 variants. N Engl J Med. 2009;360:2544–55. doi: 10.1056/NEJMoa0810440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hunt KA, et al. Newly identified genetic risk variants for celiac disease related to the immune response. Nat Genet. 2008;40:395–402. doi: 10.1038/ng.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lessard CJ, et al. Identification of IRF8, TMEM39A, and IKZF3-ZPBP2 as susceptibility loci for systemic lupus erythematosus in a large-scale multiracial replication study. Am J Hum Genet. 2012;90:648–60. doi: 10.1016/j.ajhg.2012.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gottenberg JE, et al. Activation of IFN pathways and plasmacytoid dendritic cell recruitment in target organs of primary Sjogren’s syndrome. Proc Natl Acad Sci U S A. 2006;103:2770–5. doi: 10.1073/pnas.0510837103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Perez P, et al. Gene expression and chromosomal location for susceptibility to Sjogren’s syndrome. J Autoimmun. 2009;33:99–108. doi: 10.1016/j.jaut.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 51.Cornall RJ, Goodnow CC. B cell antigen receptor signalling in the balance of tolerance and immunity. Novartis Found Symp. 1998;215:21–30. doi: 10.1002/9780470515525.ch3. discussion 30-40. [DOI] [PubMed] [Google Scholar]
- 52.Nemazee D, Weigert M. Revising B cell receptors. J Exp Med. 2000;191:1813–7. doi: 10.1084/jem.191.11.1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hom G, et al. Association of systemic lupus erythematosus with C8orf13-BLK and ITGAM-ITGAX. N Engl J Med. 2008;358:900–9. doi: 10.1056/NEJMoa0707865. [DOI] [PubMed] [Google Scholar]
- 54.Simpfendorfer KR, et al. The autoimmunity-associated BLK haplotype exhibits cis-regulatory effects on mRNA and protein expression that are prominently observed in B cells early in development. Hum Mol Genet. 2012;21:3918–25. doi: 10.1093/hmg/dds220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sawcer S, et al. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature. 2011;476:214–9. doi: 10.1038/nature10251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hansen A, Lipsky PE, Dorner T. B cells in Sjogren’s syndrome: indications for disturbed selection and differentiation in ectopic lymphoid tissue. Arthritis Res Ther. 2007;9:218. doi: 10.1186/ar2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ma CS, et al. Early commitment of naive human CD4(+) T cells to the T follicular helper (T(FH)) cell lineage is induced by IL-12. Immunol Cell Biol. 2009;87:590–600. doi: 10.1038/icb.2009.64. [DOI] [PubMed] [Google Scholar]
- 58.Adrianto I, et al. Association of a functional variant downstream of TNFAIP3 with systemic lupus erythematosus. Nat Genet. 2011;43:253–8. doi: 10.1038/ng.766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Uddin M, Sturge M, Rahman P, Woods MO. Autosome-wide copy number variation association analysis for rheumatoid arthritis using the WTCCC high-density SNP genotype data. J Rheumatol. 2011;38:797–801. doi: 10.3899/jrheum.100758. [DOI] [PubMed] [Google Scholar]
- 60.Allanore Y, et al. Genome-wide scan identifies TNIP1, PSORS1C1, and RHOB as novel risk loci for systemic sclerosis. PLoS Genet. 2011;7:e1002091. doi: 10.1371/journal.pgen.1002091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nair RP, et al. Genome-wide scan reveals association of psoriasis with IL-23 and NF-kappaB pathways. Nat Genet. 2009;41:199–204. doi: 10.1038/ng.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Adrianto I, et al. Two independent functional risk haplotypes in TNIP1 are associated with systemic lupus erythematosus. Arthritis Rheum. 2012 doi: 10.1002/art.34642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gateva V, et al. A large-scale replication study identifies TNIP1, PRDM1, JAZF1, UHRF1BP1 and IL10 as risk loci for systemic lupus erythematosus. Nat Genet. 2009;41:1228–33. doi: 10.1038/ng.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cortes A, Brown MA. Promise and pitfalls of the Immunochip. Arthritis Res Ther. 2011;13:101. doi: 10.1186/ar3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 67.McKeigue PM, Carpenter JR, Parra EJ, Shriver MD. Estimation of admixture and detection of linkage in admixed populations by a Bayesian approach: application to African-American populations. Ann Hum Genet. 2000;64:171–86. doi: 10.1017/S0003480000008022. [DOI] [PubMed] [Google Scholar]
- 68.Halder I, Shriver M, Thomas M, Fernandez JR, Frudakis T. A panel of ancestry informative markers for estimating individual biogeographical ancestry and admixture from four continents: utility and applications. Hum Mutat. 2008;29:648–58. doi: 10.1002/humu.20695. [DOI] [PubMed] [Google Scholar]
- 69.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–1. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cochran WG. The Combination of Estimates from Different Experiments. Biometrics. 1954;10:101–29. [Google Scholar]
- 71.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. Bmj. 2003;327:557–60. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 73.Pruim RJ, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–7. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Frazer KA, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–61. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Via M, Gignoux C, Burchard EG. The 1000 Genomes Project: new opportunities for research and social challenges. Genome Med. 2010;2:3. doi: 10.1186/gm124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zheng X, et al. HIBAG-HLA genotype imputation with attribute bagging. Pharmacogenomics J. 2013 doi: 10.1038/tpj.2013.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Barbosa-Morais NL, et al. A re-annotation pipeline for Illumina BeadArrays: improving the interpretation of gene expression data. Nucleic Acids Res. 2010;38:e17. doi: 10.1093/nar/gkp942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Irizarry RA, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–64. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 80.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–27. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 81.Shabalin AA. Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinformatics. 2012;28:1353–8. doi: 10.1093/bioinformatics/bts163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dozmorov MG, Cara LR, Giles CB, Wren JD. GenomeRunner: Automating genome exploration. Bioinformatics. 2011 doi: 10.1093/bioinformatics/btr666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fujita PA, et al. The UCSC Genome Browser database: update 2011. Nucleic Acids Res. 2011;39:D876–82. doi: 10.1093/nar/gkq963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–2. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.