Abstract
Neutrophils are always surrounded by/interacting with other components of the immune system; however, the current mechanistic understanding of neutrophil function is largely based on how neutrophils respond to a single chemical signal in a simplified environment. Such approaches are unable to recapitulate the in vivo microenvironment; thus, cell behavior may not fully represent the physiological behavior. Herein, we exploit a microfluidic model of the complex in vivo milieu to investigate how cell-cell interactions influence human neutrophil migration and surface marker expression. Neutrophil migration against a bacterially-derived chemoattractant (formyl-met-leu-phe, fMLP), with and without pre-activation by interleukins (interleukin-2 or interleukin-6), was evaluated in the presence and absence of endothelial support cells. Pre-activation by interleukins or interaction with endothelial cells resulted in altered migration rates compared to naïve neutrophils, and migration trajectories deviated from the expected movement toward the fMLP signal. Interestingly, interaction with both interleukins and endothelial cells simultaneously resulted in a slight compensation in the deviation – on endothelial cells, 34.4% of untreated neutrophils moved away from the fMLP signal while only 15.2% or 22.2% (IL-2-activated or IL-6-activated) of pre-activated cells moved away from fMLP. Neutrophils interacting with interleukins and/or endothelial cells were still capable of prioritizing the fMLP signal over a competing chemoattractant, leukotriene B4 (LTB4). Fluorescence imaging of individual human neutrophils revealed that neutrophils treated with endothelial cell-conditioned media showed up-regulation of the surface adhesion molecules CD11b and CD66b upon stimulation. On the other hand, CD11b and CD66b down-regulation was observed in untreated neutrophils. These results leverage single cell analysis to reveal that the interaction between neutrophils and endothelial cells is involved in surface marker regulation, and thus, chemotaxis of neutrophils. This study brings new knowledge about neutrophil chemotaxis in the context of cell-to-cell communications, yielding both fundamental and therapeutically relevant insight.
INTRODUCTION
Because they are the most abundant white blood cell type in the human circulatory system, abnormal behavior of neutrophils has significant impact on human immune response. Neutrophils originate from bone marrow and circulate in search of foreign invaders or dead/dying host cells, playing active roles in both innate and adaptive immunity in humans. When abnormal events such as infection occur, neutrophils are the first cells that migrate to the event site through a process called chemotaxis.1–3 Chemotaxis is regulated by concentration gradients of chemotaxis-inducing chemical mediators; however, in the body, neutrophils co-exist with a variety of non-chemotaxis inducing chemical mediators and other cell types.4–9 As such, it is highly likely that such interactions between neutrophils and other immune system components will have an influence on neutrophil chemotaxis. Unfortunately, however, conventional experimental methods are frequently incapable of adapting critical extracellular environmental aspects while monitoring neutrophil chemotaxis; thus, our understanding of neutrophil chemotaxis is limited to measurements in oversimplified environments. Animal experiments are another genre of studies frequently used to investigate chemotaxis- or neutrophil-related pathophysiology; however, these studies are expensive, slow, labor-intensive, hard to control, and frequently not representative of human physiological response.
Microfluidics is a powerful approach to overcome such limitations.10–12 Microfluidic platforms offer advantages for human cell biology studies by enabling creation of stable but dynamic environments with precise control and small volume sample requirements.10,12–15 Thus, increased experimental complexity, such as multiple chemical signals and/or cell types, can be easily incorporated using a microfluidic platform. Interestingly, the microfluidics-supported microenvironment is simply an altered culture condition; as such, analysis on individual target cells can be done with minimal complication from the added biological complexity. This is a big analytical strength as many studies have pointed out heterogeneity in cellular behaviors and the importance of understanding such heterogeneity in addition to the collective behavior of cells. Our previous study showed disrupted chemotaxis in neutrophils with decreased p38 mitogen-activated protein kinase activity, which was apparent based on the strengths of single cell analysis techniques, and as will become clear below, this microfluidic platform keeps the single cell analysis capability despite the addition of an endothelial cell component.44–47
Thus, in this study, neutrophil chemotaxis is studied in the context of “interaction” (Figure 1). Neutrophils always interact with endothelial cells, the cells lining blood vessel walls, and molecules for which they express receptors. Endothelial cells were chosen because they are ubiquitous in the body and actively interact with neutrophils during neutrophil migration (Interaction C in Figure 1).16–18, 49–56
Figure 1.
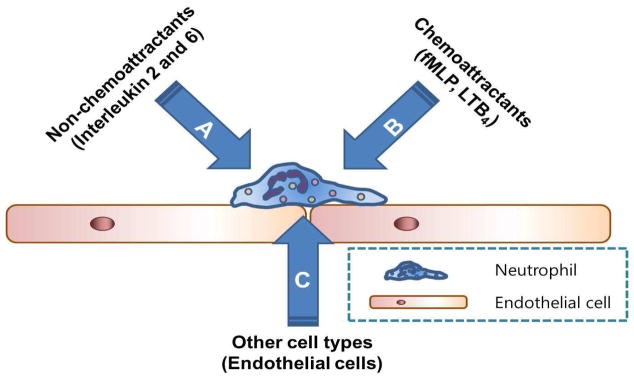
Neutrophil-involved interactions in vivo - neutrophils interacting with chemical mediators that do not directly induce chemotaxis (A), neutrophils interacting with chemoattractants (B), neutrophils interacting with other cell types (C).
In addition to the impact of neutrophil-endothelial cell interaction in neutrophil chemotaxis, the effect of chemical mediators in neutrophil chemotaxis is also considered herein, since chemical mediators are a major cell-cell signaling method. Previous works, including ours, have focused on comprehensive analyses of various chemotaxis-inducing mediators, known as chemoattractants, in neutrophil chemotaxis (Interaction B in Figure 1).19,20 Those studies revealed that neutrophils respond to all present chemoattractants while prioritizing a particular chemoattractant. In this study, the impact of non-chemotaxis inducing mediators is considered (Interaction A in Figure 1). Literature precedent indicates active roles for interleukin-2 and interleukin-6 (IL-2 and IL-6) in neutrophil biology.5,6,8,9,21 Known impacts of IL-2-induced neutrophil activation include enhanced chemical mediator production,5 reduced apoptosis,5 enhanced surface Fcγ expression,4 and altered gene expression.6 On the other hand, IL-6 challenge of neutrophils is known to alter neutrophil production of mediators such as platelet-activating factor (PAF)22 and reactive oxygen species.23 Also, challenging IL-6-activated neutrophils with fMLP is known to enhance the elastase release from neutrophils.6,24 Such IL-2- and IL-6-induced alterations of neutrophil biology likely have an impact on the entire immune system, and as such, abnormal IL-2 and IL-6 levels are frequently reported in the context of infections.25–33 Based on the known IL-2- and IL-6- induced impacts on neutrophil biology, this work explores interleukin-exposed neutrophil behavior in a concentration gradient of the bacterial chemoattractant fMLP. While the impact of IL-2, IL-6, or endothelial cells on neutrophil function has been described by a few studies, there are no comprehensive studies on their contributors to chemotaxis and surface CD11b and CD66b expression, especially in the context of cell-cell interactions. Despite the additional complexities (e.g. endothelial cells and interleukins) in the experimental platform, neutrophils in each condition were individually monitored and the impact of the additional complexities was comprehensively analyzed in this study.
EXPERIMENTAL SECTION
1. Neutrophil Preparation
Whole human blood, anti-coagulated with ethylenediaminetetraacetic acid (EDTA), was obtained from Memorial Blood Center (St. Paul, MN) according to approved IRB protocol E&I ID#07809. All blood samples were collected on the day of experiments from healthy donors and were used immediately after the samples were obtained. Each condition has 6 biological replicates (6 different donors). Polymorphprep (Accurate Chemical & Scientific Corp., Westbury, NY) was used to isolate neutrophils by density gradient centrifugation as recommended by the manufacturer. Hank’s buffered salt solution (HBSS, Sigma-Aldrich, St. Louis, MO) was used while washing and resuspending neutrophils during isolation, and finally, ~5 × 106 isolated neutrophils were resuspended in HBSS containing 2% human serum albumin (HSA, Sigma-Aldrich, St. Louis, MO). The final cell suspension was kept at 37 °C until use.
2. Device Fabrication
Device fabrication and endothelial cell culture in the device were done using standard soft lithography techniques and is described in detail in our previous paper.1 A brief description of the procedure can also be found in the supplemental data.
3. Endothelial Cell Culture in a Microfluidic Channel
A detailed description of how endothelial cells (hy926 human endothelial cell line) were cultured is given in our previous paper.2 A brief description can also be found in the supplemental information. To note, the endothelial cell culture media, denoted as DMEM w/ FBS and PS throughout this article, was Dulbecco’s modified eagle medium (DMEM) with high glucose (formula: 4mM L-glutamine, 4.5g/L L-glucose, and 1.5g/L sodium pyruvate (Gibco®, Carlsbad, CA)), supplemented with 10% fetal bovine serum and 1% penicillin and streptomycin (Sigma Aldrich, Milwaukee, WI).
4. Chemicals
Before introducing isolated human neutrophils into a microfluidic device, they were pretreated with either IL-2 or IL-6 (Sigma Aldrich, St. Louis, MO) in HBSS at the final concentration of 10 ng/mL. Gradient of chemoattractants, fMLP and LTB4, was 0 – 10 ng/mL. 10 ng/mL fMLP, IL2, IL-6, and LTB4 are 22.9 nM, 29.7 nM, 0.6 nM, and 0.4 nM respectively.
5. Sample preparation for fluorescence imaging
Detailed procedure for surface CD11b and CD66b fluorescence imaging can be found in the supplemental information. For single stimulation conditions, cells were challenged with IL-2, IL-6, or fMLP for an hour. For co-stimulation conditions with both interleukins and fMLP, cells were subject to IL-2 or IL-6 for an hour first and then challenged with fMLP for another hour. For co-stimulation conditions with endothelial cell-conditioned medium and fMLP, cells were subject to endothelial cell-conditioned medium for an hour first and then challenged with fMLP for another hour. For co-stimulation conditions with interleukins, endothelial cell-conditioned medium, and fMLP, cells were subject to interleukins with endothelial cell-conditioned medium for an hour and then challenged with fMLP for another hour. After stimulation, cells were washed with HBSS three times and then subject to anti-CD11b conjugated with AlexaFluor 700 (Life Technologies, Grand Island, NY) and anti-CD66b conjugated with AlexaFluor 647 (BD Pharmingen, San Diego, CA) following instructions from manufacturers, incubated for 1 hour, washed with HBSS three times, and then surface expression of those adhesion molecules on individual cells were fluorescently monitored.
6. Analysis of migration data
From each experiment, individual neutrophils were randomly chosen and individually tracked using Metamorph Ver. 7.7.5 software. Once the imaging software extracted the trajectory information for individual cells from the collected image stack (SI Fig. 1), these raw data were imported into Microsoft Excel and further processed to calculate two unitless numeric parameters: motility index (MI) and chemotactic index (CI) as previously described.1,10 Briefly, MI is the ratio of migration displacement (d) and theoretical migration displacement (dmax) of a neutrophil:
where dmax is defined as the average migration speed times the total migration time. As such, this MI is an indicator of how active/mobile individual cells are.
CI quantifies the cells’ orientations:
where x is the final displacement along the direction of the gradient and dtotal is the total migration distance. Thus, this CI is an indicator of how directional individual cells’ migration is.
In this report, movement toward the right side of the observation channel, toward a higher concentration of fMLP, results in positive CI. In addition to these parameters, % population was calculated simply by taking the number of cells with negative CI values divided by the total number of cells. Thus, this % population represents cells with overall migration opposite to the direction of the fMLP gradient. Also, the angular displacement of individual cells were digitized by giving a score of “+1” to cells in each frame that moved with − 90° < angle < 90°, and giving a score of “−1” to cells in each frame moved with angle > 90° or with angle < −90°. The angle value was obtained with respect to the x-axis (parallel to the gradient direction); thus, this parameter is descriptive of how much deviation from the parallel to the fMLP gradient occurred during migration. An angle distribution value of “+1” would indicate that the cell moved straight toward the fMLP signal in every collected image. To assess individual neutrophils’ response in a time-resolved manner, this angle parameter was calculated for the first 1, 4, and 7 minutes, and for the entire duration of the experiment. All angle distribution plots provided in this manuscript contain data for the first 4 minutes and for the entire duration. T-tests were used for statistical comparison, and errors are reported as +/− standard error of mean, SEM, throughout the manuscript.
RESULTS
Neutrophil migration against the fMLP gradient – the impact of the presence of endothelial cells
To investigate the impact of cell-cell interactions between neutrophils and endothelial cells, chemotaxis of neutrophils was assessed when the human cells were introduced onto a layer of endothelial cells (B&C in Figure 1). Figure 2 shows representative trajectories of 20 neutrophils on fibronectin- and endothelial cell-coated devices. The representative trajectories indicate that neutrophils on endothelial cells are not as directed toward the fMLP signal as neutrophils on a simplified fibronectin-coated surface, and their migration patterns are analyzed more in detail using numerical parameters defined in experimental section in the supporting information. As shown in Figure 3, mobility and directionality in neutrophil migration toward the fMLP signal (MI and CI) were significantly lower in neutrophils on endothelial cells (0.44 ± 0.04 and 0.16 ± 0.06 compared to 0.64 ± 0.03 (p < 0.0001) and 0.40 ± 0.03 (p < 0.0001), respectively). Interestingly, rather than diminishing migration speed, the reduced MI and CI seemed to be caused by deviation in migration direction. The average velocity of individual cells on endothelial cells was similar to that on fibronectin (0.26 ± 0.03 compared to 0.25 ± 0.01, p = 0.67, Figure 3-c) while the angular distribution data showed more deviated migration on endothelial cells (0.23 ± 0.07 compared to 0.61 ± 0.06 for the first 4 minutes (p < 0.0001), and 0.25 ± 0.08 compared to 0.47 ± 0.06 for the entire 30 minute duration of the experiment (p = 0.03), Figure 3-b). Again, this angle distribution represents the level of deviation from the gradient direction for individual cells during migration, and a value close to “+1” indicates that the neutrophil moved straight toward the fMLP signal in every collected image. The deviation in migration angle for neutrophils on endothelial cells is also linked to the higher % population data; 34.4% neutrophils moved away from the fMLP signal on endothelial cells while only 10.9% cells moved away from the fMLP signal on a fibronectin-coated surface (Figure S-2).
Figure 2.
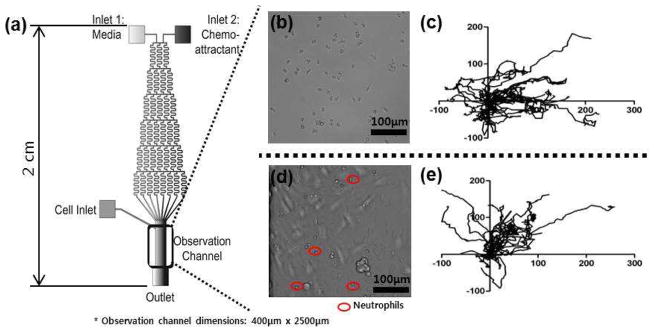
(a) Microfluidic device schematic. Solutions introduced into the two inlets are mixed while traversing the serpentine channels to establish a stable concentration gradient across the observation channel. While a gradient of fMLP is established by introducing media into the inlet 1 and fMLP solution into the inlet 2, a competing gradient of LTB4-fMLP is established by introducing LTB4 solution into the inlet 1 and fMLP solution into the inlet 2. (b) Neutrophils on a fibronectin-coated surface in the observation channel. (c) Trajectories of 20 representative control neutrophils (unactivated) within the fMLP gradient. (d) Neutrophils on an endothelial cell-coated surface in the observation channel. (e) Trajectories of 20 representative control neutrophils (unactivated) within the fMLP gradient.
Figure 3.
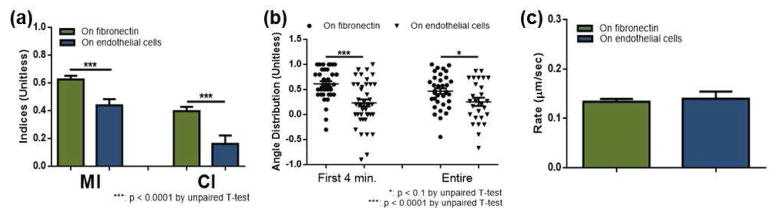
Neutrophil chemotaxis within the fMLP gradient on fibronectin and on endothelial cells. (a) Motility index (MI) and chemotactic index (CI) of neutrophils on fibronectin (green) and on endothelial cells (blue). (b) Angular distribution of neutrophil movement on fibronectin (circles) and on endothelial cells (triangles) with a value of 1 indicating migration of a cell straight toward fMLP. Each point represents the angular displacement of an individual neutrophil, and the solid bar indicates the average. (c) Average rate of neutrophil migration on fibronectin (green) and on endothelial cells (blue).
Neutrophil migration against fMLP gradient – the impact of cytokines
To explore the effect of non-chemotaxis-inducing cytokines on neutrophil chemotaxis, neutrophils were pre-incubated with either 0.6 nM interleukin-2 (IL-2) or 0.4 nM interleukin-6 (IL-6), and then exposed to an fMLP gradient (A&B in figure 1). Challenging neutrophils with 0.6 nM IL-2 or 0.4 nM IL-6 has been shown to be effective in activating neutrophils, but also, few nM level elevation of these interleukins are frequently found in individuals with abnormal immune response.23,33–38 In this section, comparison is performed between neutrophils that were not activated (denoted as “control”) and that were pre-activated by IL-2 or IL-6 (denoted as “IL-2-activated” or “IL-6-activated”). The results indicate that neutrophils move faster when they are pre-incubated with interleukins (0.29 ± 0.02 for IL-2 (p = 0.06) and 0.34 ± 0.02 for IL-6 (p < 0.0001) compared to 0.25 ± 0.01 for control, Figure 4-c) even though MI and CI were the largest in control neutrophils. Remembering that MI and CI have a speed term (multiplied by the time) in the denominator by definition, this indicates that interleukin-exposed cells do not reflect their increased migration speed into the actual motility and directionality.
Figure 4.
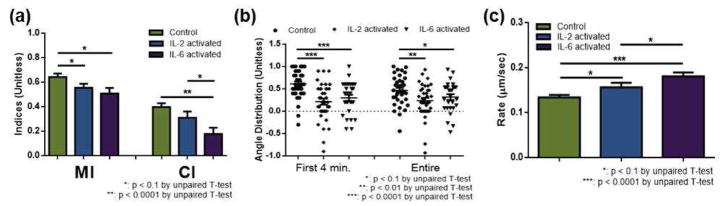
Neutrophil chemotaxis within the fMLP gradient: unactivated (control), IL-2-activated, and IL-6-activated neutrophils on fibronectin. (a) Motility index (MI) and chemotactic index (CI) of neutrophils. (b) Angle distribution of neutrophils, with a value of 1 indicating migration of a cell straight toward fMLP. Each point represents the angular displacement of a cell, and the solid bar indicates the average. (c) Average migration rate of neutrophils.
Specifically, IL-2-activated neutrophils had decreased MI (0.55 ± 0.03 for IL-2-activated compared to 0.64 ± 0.03 for control, p = 0.04, Figure 4-a), and IL-6 pre-activation led to the down-regulation of both motility and directionality (0.51 ± 0.05 and 0.18 ± 0.05 for IL-6-activated compared to 0.64 ± 0.03 (p = 0.012) and 0.40 ± 0.03 (p = 0.01) for control, Figure 4-a). The angle distribution shows that, in both pre-activated conditions, more cells deviated from the fMLP signal during migration (0.23 ± 0.06 (p = 0.01) for IL-2-activated and 0.32 ± 0.07 (p = 0.09) for IL-6-activated compared to 0.47 ± 0.06 for control). % population data also supported this deviated migration pattern: 18.2% IL-2-activated cells and 21.1% IL-6-activated cells moved away from the fMLP signal while only 11% of control cells moved away from the fMLP signal (Figure S-2). The first 4 minutes of movement was especially influenced, with significantly more pre-activated neutrophils moving away from the fMLP signal (0.21 ± 0.07 (p < 0.0001) for IL-2-activated and 0.30 ± 0.07 (p < 0.0001) for IL-6-activated compared to 0.61 ± 0.06 for control, Figure 4-b). Consistent with the neutrophils in contact with endothelial cells, these data indicate that increased neutrophil migration speed was compensated by deviated migration from the direction of fMLP signal.
Neutrophil migration – the synergistic impact of cytokines and endothelial cells
As the microfluidic platform allowed separate study of cytokine impact (A&B in Figure 1) and endothelial cell impact (B&C in Figure 1) on neutrophil chemotaxis (B in Figure 1), we also considered the simultaneous impact of both cytokines and endothelial cells on neutrophil chemotaxis (A&B&C in Figure 1). Comparison in this section was between neutrophils on endothelial cells, and the neutrophils were either unactivated (control), pre-activated with IL-2 (IL-2-activated), or pre-activated with IL-6 (IL-6-activated). Only the MI in IL-2-activated neutrophils on endothelial cells was altered when compared to control neutrophils on endothelial cells (0.59 ± 0.03 compared to 0.44 ± 0.04, p = 0.01, Figure 5-a). The average migration rates for IL-2- and IL-6-activated conditions were not significantly different from the control condition on endothelial cells, although IL-6 pre-incubation resulted in a slightly faster migration rate than IL-2 pre-incubation (0.30 ± 0.04 for IL-6-activated compared to 0.24 ± 0.02 for IL-2-activated, p = 0.1, Figure 5-c). While angle distribution data showed no significant differences among conditions by mean ± SEM comparison, the distribution patterns were noticeably different. The distribution was most focused toward the fMLP signal in IL-2-activated neutrophils on endothelial cells (Figure 5-b). The % population data also supported this deviated migration: on endothelial cells, 15% IL-2-activated cells and 22% IL-6-activated cells moved away from the fMLP signal while 34% control cells moved away from the fMLP signal (Figure S-2).
Figure 5.
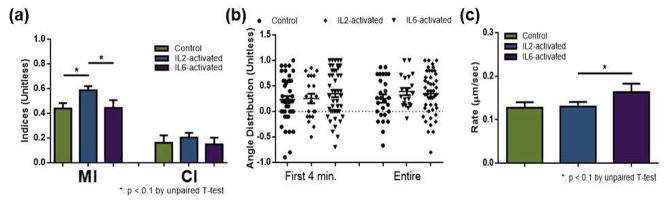
Neutrophil chemotaxis within the fMLP gradient: unactivated (control), IL-2-activated, and IL-6-activated neutrophils on endothelial cells, (a) Motility index (MI) and chemotactic index (CI) of neutrophils. (b) Angle distribution of neutrophils, with a value of 1 indicating migration of a cell straight toward fMLP. Each point represents the angular displacement of a cell, and the solid bar indicates the average. (c) Average migration rate of neutrophils.
Unlike pre-activated neutrophils on a fibronectin-coated surface (Figure 4), directionality of pre-activated neutrophils migrating on endothelial cells seems to be independent of pre-activation (no significant differences in CIs or angle distributions, Figure 5). As such, we further studied chemotaxis of pre-activated neutrophils on endothelial cells by incorporating a competing gradient of chemoattractants. In our previous study,19 we showed that the microfluidic platform allows incorporation of multiple chemoattractant gradients and that neutrophils prioritize a fMLP signal over a LTB4 signal. Herein, we adapted the same competing gradient approach from the previous study into the microfluidic platform and exposed neutrophils on endothelial cells to a competing gradient of LTB4 and fMLP (LTB4-fMLP). A LTB4-fMLP gradient is established by introducing 0.4 nM LTB4 and 22.9 nM fMLP into the left and right inlet, respectively (Figure 2). This achieves a 0.4 – 0 nM LTB4 gradient superimposed on a 0 – 22.9 nM fMLP gradient within the observation channel. The gradient of each chemoattractant was chosen based on previous study; in this concentration gradient, neutrophils exposed to the LTB4 gradient performed a similar chemotaxis level when compared to neutrophils exposed to the fMLP gradient.19 Thus, chemotaxis investigation within such a competing gradient reveals the hierarchy of neutrophil response among the molecular signals. In previous work, it was demonstrated that neutrophils prioritize fMLP over leukotriene B4 (LTB4) on a fibronectin-coated surface; however, we were concerned that other cells or the activation state of neutrophils would influence the neutrophil behavior. Herein, neutrophils were exposed to the LTB4-fMLP competing gradient on endothelial cells. Neutrophils were either unactivated (control), pre-activated by IL-2 (IL-2-activated), or pre-activated by IL-6 (IL-6-activated). Most importantly, the results demonstrate that the major population of neutrophils moved toward fMLP in all conditions (Figure 6). This indicates that neutrophils still prioritized fMLP signal over LTB4 signal, and that the neutrophil interaction with non-chemotaxis inducing contributors (IL-2, IL-6, and endothelial cells) did not change the hierarchy of neutrophil response to these molecular signals. Also, while no noticeable variation was observed from the angle distribution data, the results indicate that IL-6 is the most potent contributor in these conditions: MI, average migration rate, and % population were noticeably different from all other conditions in IL-6 pre-incubated neutrophils on endothelial cells (Figure 6).
Figure 6.
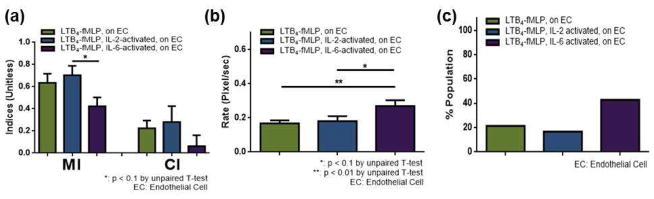
Neutrophil chemotaxis within the competing gradient of LTB4-fMLP: unactivated (control), IL-2-activated, and IL-6-activated neutrophils on endothelial cells. (a) Motility index (MI) and chemotactic index (CI) of neutrophils. (b) Average migration rate of neutrophils. (c) % population of neutrophils moved away from the fMLP signal.
Surface expression of adhesion molecules – the individual and synergistic impact of cytokines and endothelial cells
As shown above, the microfluidic platform employed herein allowed detailed investigation of neutrophil chemotaxis and its relation to surrounding cytokines and different cell types that do not directly induce neutrophil chemotaxis, and revealed the impact of each of these contributors on the outcome chemotaxis. To broaden the mechanistic understanding of the IL-2, IL-6, and endothelial cell impacts on neutrophil chemotaxis, surface expression of the adhesion molecules CD11b and CD66b were fluorescently monitored. Adhesion of neutrophils is the initial step for chemotaxis and CD66b and CD11b are well-known key surface markers for neutrophil adhesion to endothelial cells. To mimic neutrophils migrating on endothelial cells, neutrophils were treated with endothelial cell-conditioned medium (denoted as “EC-treated”). As such, comparison in this section is between two populations of cells, untreated vs. EC-treated, either with no stimulation (control), activation with IL-2 (IL-2-activated), activation with IL-6 (IL-6-activated), stimulation with fMLP (denoted as “fMLP-stimulated”), or activation with IL-2 or IL-6 followed by fMLP stimulation (denoted as “IL-2-fMLP” and “IL-6-fMLP”); all statistical comparisons are shown in Table 1. The CD11b levels in untreated neutrophils had a general trend of down-regulation upon stimulation (Figure 7-a and Figure 8-a). Interestingly, co-stimulation resulted in higher expression of CD11b than individual stimulation. Compared to activation with IL-2 or IL-6 and fMLP stimulation, the CD11b expression was higher in IL-2-fMLP and IL-6-fMLP conditions (Figure 7 and Figure 8, Table 1). CD66b levels in untreated cells had a similar trend; cells expressed less CD66b when stimulated (Figure 7 and Figure 8, Table 1). Unlike CD11b expression, the synergistic impact of interleukins and the chemoattractant fMLP on the CD66b expression was only clear in the IL-6-fMLP condition (Figure 7 and Figure 8, Table 1). On the other hand, both surface CD11b and CD66b expression was significantly different in EC-treated cells (Figure 7 and Figure 8). While stimulation of untreated cells resulted in down-regulation of CD11b, stimulation of EC-treated cells resulted in up-regulation of CD11b (Figure 7 and Figure 8, Table 1). Also, while stimulation of untreated cells resulted in a decreasing trend in surface CD66b expression, stimulation of EC-treated cells resulted in an increasing trend in surface CD66b expression (Figure 7 and Figure 8, Table 1). Only IL-2-activated EC-treated cells resulted in down-regulation of CD11b (Figure 7-b, Table 1) and CD66b (Figure 8-b, Table 1).
Table 1.
Fluorescence results summary for all activation/stimulation conditions. All statistical comparisons are between the experimental condition and the indicated control.
| CD11b | CD66b | ||||
|---|---|---|---|---|---|
|
| |||||
| Fluorescence (A.U.max) | p value | Fluorescence (A.U.max) | p value | ||
| On fibronectin | Control | 1498 ± 29 | 2786 ± 60 | ||
| IL-2-activated | 1183 ± 9 | (p < 0.0001) | 2180 ± 44 | (p < 0.0001) | |
| IL-6-activated | 1169 ± 8 | (p < 0.0001) | 1990 ± 33 | (p < 0.0001) | |
| fMLP-stimulated | 1078 ± 5 | (p < 0.0001) | 2007 ± 45 | (p < 0.0001) | |
| IL-2-fMLP | 1284 ± 15 | (p < 0.0001) | 2010 ± 67 | (p < 0.0001) | |
| IL-6-fMLP | 1387 ± 19 | (p < 0.01) | 2781 ± 64 | (p > 0.1) | |
|
| |||||
| On endothelial cells | Control | 1517 ± 23 | 3926 ± 113 | ||
| IL-2-activated | 1356 ± 20 | (p < 0.0001) | 2231 ± 119 | (p < 0.0001) | |
| IL-6-activated | 1743 ± 24 | (p < 0.0001) | 4494 ± 107 | (p < 0.0001) | |
| fMLP-stimulated | 1866 ± 55 | (p < 0.0001) | 5308 ± 174 | (p < 0.0001) | |
| IL-2-fMLP | 1466 ± 14 | (p < 0.1) | 4061 ± 100 | (p > 0.1) | |
| IL-6-fMLP | 1528 ± 15 | (p > 0.1) | 3972 ± 100 | (p < 0.1) | |
Figure 7.
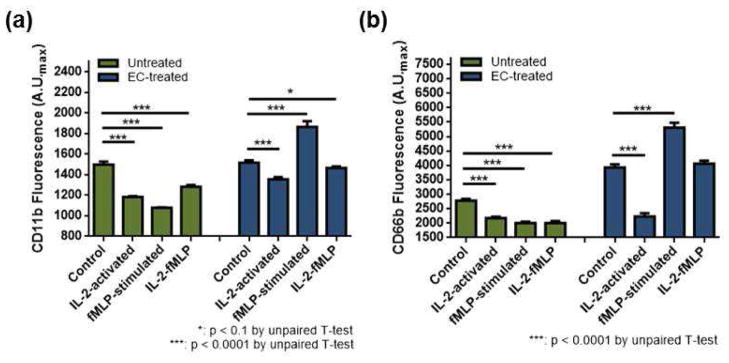
Individual and cooperative impact of IL-2 with endothelial cells on surface CD11b and CD66b expression of neutrophils: un-activated (control), IL-2-activated, fMLP-stimulated, and IL-2-fMLP co-stimulated neutrophils. (a) CD11b expression of untreated (green) and EC-treated (blue) neutrophils. (b) CD66b expression of untreated (green) and EC-treated (blue) neutrophils.
Figure 8.
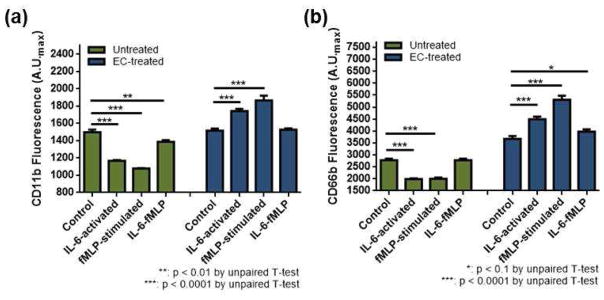
Individual and cooperative impact of IL-6 with endothelial cells on surface CD11b and CD66b expression of neutrophils: unactivated (control), IL-6-activated, fMLP-stimulated, and IL-6-fMLP co-stimulated neutrophils. (a) CD11b expression of untreated (green) and EC-treated (blue) neutrophils. (b) CD66b expression of untreated (green) and EC-treated (blue) neutrophils.
DISCUSSION
Neutrophil Chemotaxis: Cytokines and Endothelial Cells
Neutrophils circulate in the human body seeking abnormalities on the endothelial cells lining the inner wall of blood vessels. They make a contact with endothelium repeatedly, and once they confront an abnormality, they stay in contact with endothelium and seek the exact site of the abnormality. As such, neutrophils inevitably interact with endothelial cells while performing their critical roles in the immune system. Also, neutrophils express surface receptors for IL-2 and IL-6 which are frequently up- regulated in the human body in various abnormal circumstances. Thus, it is highly likely that neutrophils utilize IL-2 and IL-6 before/during migration as a way to communicate with other components of the immune system; however, there have been no comprehensive studies of such interactions due to technical difficulties. Herein, we exploit a simple microfluidic platform to introduce such biological complexity into experimental investigations. The interactions among non-chemotaxis inducing mediators (A in Figure 1), chemoattractants (B in Figure 1), and other cell types (C in Figure 1), that are the critical in generating immune response, are accounted for in our microfluidic platform. While maintaining high fidelity analysis of individual neutrophil behavior, our platform provides mechanistic insight in a resolved manner for each of contributing complexities (endothelial cells and chemical mediators). To assist understanding, discussion below will utilize Figure 1 when necessary, and results described in bar charts above are summarized in Table 2 and Table 3. Again, pre-incubation with IL-2 or IL-6 represents interaction A, fMLP, or LTB4-fMLP gradient exposure represents interaction B (chemotaxis), and added endothelial cell represents interaction C in Figure 1. The neutrophil chemotaxis was significantly disturbed on endothelial cells compared to that on a simple fibronectin coating, which proves that neutrophils behave differently when they interact with endothelial cells (B&C in Figure 1). The interaction of neutrophils with endothelial cells does not seem to significantly impact migration speed. However, neutrophil migration on endothelial cells was not as directed as that on fibronectin. It may be, in part, because in vivo neutrophils migrate through the junctions between endothelial cells rather than over the endothelial cells, which will cause a slight disturbance in the directional movements.39,40 Directional deviation from the chemoattractant gradient direction was also observed from IL-2-activated and IL-6-activated neutrophils (A&B in Figure 1, Table 2). All of these demonstrate that, while IL-2, IL-6, and endothelial cells do not directly induce chemotaxis, they play a role in neutrophil chemotaxis by adjusting both direction and movement of neutrophils.
Table 2.
Summary of neutrophil chemotaxis under the fMLP gradient - C: Control (unactivated), IL-2: IL-2-activated, and IL-6: IL-6-activated.
| On fibronectin-coated surface | On endothelial cell-coated surface | |
|---|---|---|
|
| ||
| MI | Control > IL-2 > IL-6 | IL-2 > Control = IL-6 |
| CI | Control = IL-2 > IL-6 | Control = IL-2 = IL-6 |
| Angle distribution | Control > IL-2 = IL-6 | Control = IL-2 = IL-6 |
| Migration rate | IL-6 > IL-2 > Control | Control = IL-2 = IL-6 |
Table 3.
Summary of surface CD11b and CD66b expressions – Control: unactivated, IL-2: IL-2-activated, IL-6: IL-6- activated, fMLP: fMLP-stimulated, IL-2-fMLP: IL-2-activated and fMLP-stimulated, IL-6-fMLP: IL-6-activated and fMLP- stimulated, EC-treated: endothelial cell culture medium-treated.
| CD11b | CD66b | |
|---|---|---|
|
| ||
| Untreated | Control > IL-2-fMLP > IL-2 > fMLP | fMLP > Control > IL-2-fMLP > IL-2 |
| Control > IL-6-fMLP > IL-6 > fMLP | fMLP > IL-6 > Control = IL-6-fMLP | |
|
| ||
| EC-treated | fMLP > Control > IL-2-fMLP > IL-2 | fMLP > Control = IL-2-fMLP > IL-2 |
| fMLP > IL-6 > Control = IL-6-fMLP | fMLP > IL-6 > IL-6-fMLP > Control | |
To be noted, neutrophils on endothelial cells respond to the interleukins differently than those on fibronectin (All interactions in Figure 1 together, Table 2). As mentioned, IL-2- and IL-6-activated neutrophils on a fibronectin-coated surface had decreased motility and chemotactic indices (MI and CI) and altered migration patterns (angle distribution and % population) despite increased migration rate when compared to control, un-activated, neutrophils. On the other hand, when compared to control neutrophils on endothelial cells, IL-2-activated neutrophils on endothelial cells had an increased MI and more directed migration toward the fMLP signal (focused angle distribution and lower % population). The migration rate of IL-2-activated neutrophils was not significantly different from control neutrophils when they were on endothelial cells. IL-6-activated neutrophils on endothelial cells had similar MI and CI and showed more directed migration toward the fMLP signal (similar angle distribution but lower % population). The migration rate of IL-6-activated neutrophils on endothelial cells was higher than that of control neutrophils on endothelial cells. From these results, it is obvious that IL-2 or IL-6 cooperatively contribute with endothelial cells to neutrophil chemotaxis. It was previously found that neutrophils simultaneously respond to all present chemoattractants,19 and this study demonstrates that non-chemotaxis inducing mediators also make their own individual/synergistic contributions. In this study, neutrophils, whether pre-activated by interleukins or not, exposed to the competing chemoattractant gradients (LTB4-fMLP) prioritize fMLP over LTB4 on endothelial cells, which is consistent with our previous finding from neutrophils on a fibronectin surface. Our results demonstrate that indirect contributors to chemotoxis alter the capability of neutrophils to follow a signal rather than their prioritization of a particular signal.
Neutrophil Surface Marker Expression: Cytokines and Endothelial Cells
Once neutrophils sense a chemical signal, they adhere to the endothelium and start migrating; thus, adhesion is an important component of chemotaxis. To correlate this phenomenon with the measured chemotaxis, individual human neutrophils were analyzed for expression of the surface adhesion molecules CD11b and CD66b via fluorescence imaging. Again, two populations of cells, untreated or EC-treated, were activated/stimulated by IL-2 (IL-2-activated), IL-6 (IL-6-activated), fMLP (fMLP-stimulated), IL-2 followed by fMLP (IL-2-fMLP), or IL-6 followed by fMLP (IL-6-fMLP), and surface expression of CD11b and CD66b of those cells were compared to those from un-stimulated cells (control). All measured results demonstrate that IL-2, IL-6, and endothelial cells influence the expression of neutrophil CD11b and CD66b (Figure 7, Figure 8, Table 3).
The most noteworthy trend in CD11b and CD66b expression was a down-regulation in untreated cells but an up-regulation in EC-treated cells. In untreated neutrophils, exposure to IL-2, IL-6, and fMLP resulted in down-regulation of CD11b and CD66b and this down-regulation may have contributed to the faster migration rate in neutrophils upon stimulation. On the other hand, in EC-treated cells, IL-6 activation and fMLP stimulation resulted in an up-regulation of CD11b and CD66b. To be noted, both untreated and EC-treated neutrophils show a decreased CD11b and CD66b expression when activated by IL-2. This indicates that IL-2 down-regulates surface CD11b and CD66b expression in neutrophils, and its impact is more potent than the neutrophil-endothelial cell interaction. In addition, in general, the impact of individual IL-2, IL-6, or fMLP on CD11b and CD66b expression was different than the cooperative impact of IL-2-fMLP or IL-6-fMLP, indicating a synergistic effect for the interleukins and fMLP.
An interesting aspect in this research is the CD11b and CD66b down-regulation upon neutrophil stimulation. The same trend, down-regulation of these adhesion molecules, was found in our previous study where neutrophils were stimulated by CXC-motif chemokine 8 (CXCL8);20 however, as multiple literature precedents also reported up-regulation of those molecules from neutrophils after stimulation,41–43 this unexpected down-regulation was unexplained in the previous study. Upon close examination of precedent work done in co-culture,41–43 we see that CD11b and CD66b expressions were up-regulated in EC-treated cells. As such, it was hypothesized that the neutrophil-endothelial cell interaction initiates a signaling cascade that converts a down-regulation of CD11b and CD66b upon stimulation to an up-regulation. As the EC-treated condition contained a step that exposes neutrophils to a different medium, the impact of the medium in CD11b and CD66b expression was analyzed to assess this hypothesis. The endothelial cell culture medium was, as described in experimental section, DMEM w/ FBS and PS (DMEM with high glucose supplemented with 10% fetal bovine serum and 1 % penicillin and streptomycin), while the base medium for neutrophil handling was Hank’s balanced salt solution (HBSS). Thus, while keeping all other conditions the same, neutrophils were treated with DMEM with high glucose, and DMEM w/ FBS and PS and then respective stimuli, and the CD11b and CD66b expression was assessed. (Figure S-3) Again, the control condition in each cell population was the un-activated cells. As shown in Figure S-3, both IL-2 and IL-6 stimulation of neutrophils with DMEM with high glucose or DMEM w/ FBS and PS resulted in down-regulated CD11b and CD66b expression when compared to controls. Only the fMLP showed a possible interaction with DMEM with high glucose or DMEM w/ FBS and PS. This demonstrates that our results of CD11b and CD66b up-regulation in EC-treated cells are not attributed to the medium, and thus, neutrophil-endothelial cell interaction plays a role in regulating surface CD11b and CD66b in neutrophils. It will be of particular interest in the near future to profile neutrophil- and endothelial cell-expressed mediators and find out what signaling cascade is responsible for this process.
CONCLUSIONS
This work provides detailed information about neutrophil chemotaxis upon interaction with non-chemotaxis inducing components of the immune system. The microfluidic platform successfully incorporates neutrophils and endothelial cells, enabling the investigation of neutrophil chemotaxis on a layer of endothelial cells. All of various parameters above that describe neutrophil migration and surface marker expressions, each critical in defining neutrophil biology, are obtained at significantly reduced amount of cost, labor, time, and consumption of samples and chemicals, when compared to conventional assays. Moreover, the insight gleaned from the single cell analysis approach is not easily available with conventional assays, proving that this microfluidic approach will provide opportunities to broaden our understanding beyond the traditional level. Our findings also demonstrate the strength of the microfluidic approach by revealing the contribution of chemical and cellular mediators in neutrophil chemotaxis. Specifically, our results show that indirect chemotaxis-inducing mediators (interleukins and endothelial cells) have an active role in the regulation of neutrophil chemotaxis, and the role is, in part, mediated through adjustment of surface adhesion molecule expression. IL-2 and IL-6 have their own individual and synergistic impact on neutrophil chemotaxis, but this impact does not alter the neutrophils’ ability to respond to an fMLP chemoattractant signal. The most noteworthy finding is that, in all conditions, the presence of endothelial cells or endothelial cell-expressed mediators altered neutrophil response to both cytokines and chemoattractants. Every single immune response is managed by cell-to-cell communications; as such, our finding may form the basis for chemotaxis studies in the context of cell-cell communications. It will be of particular interest in the future to profile neutrophil/endothelial cell-expressed mediators to delineate their interaction in more detail, leading to a more complete understanding of pathophysiology of neutrophil-mediated diseases.
Supplementary Material
Acknowledgments
This work was funded by grants from the National Institutes of Health New Innovator Award (DP2 OD004258-01). Device fabrication was done in Minnesota Nano Center (NFC) at the University of Minnesota that is supported by National Science Foundation (NSF). We thank to Ozlem Ersin, an associate professor at Manchester College School of Pharmacy (Fort Wayne, IN, U.S.A.) for valuable discussions regarding neutrophil chemotaxis, and Leah Laux, a NNIN REU awardee from Washington University in St. Louis, MO, for her efforts in fabricating preliminary microfluidic devices.
Footnotes
Additional information as noted in the text. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Cotter TG, Keeling PJ, Henson PM. J Immunol. 1981;127:2241–2245. [PubMed] [Google Scholar]
- 2.Li J, Gyorffy S, Lee S, Kwok CS. Inflammation. 1996;20:361–372. doi: 10.1007/BF01486739. [DOI] [PubMed] [Google Scholar]
- 3.Jacobs AA, Huber JL, Ward RA, Klein JB, McLeish KR. J Leuko Biol. 1995;57:679–686. doi: 10.1002/jlb.57.4.679. [DOI] [PubMed] [Google Scholar]
- 4.Jablons D, Bolton E, Mertins S, Rubin M, Pizzo P, Rosenberg SA, Lotze MT. J Immunol. 1990;144:3630–3636. [PubMed] [Google Scholar]
- 5.Djeu JY, Liu JH, Wei S, Rui H, Pearson CA, Leonard WJ, Blanchard DK. J Immunol. 1993;150:960–970. [PubMed] [Google Scholar]
- 6.Girard D, Gosselin J, Heitz D, Paquin R, Beaulieu AD. Blood. 1995;86:1170–1176. [PubMed] [Google Scholar]
- 7.Cara DC, Kaur J, Forster M, McCafferty DM, Kubes P. J Immunol. 2001;167:6552–6558. doi: 10.4049/jimmunol.167.11.6552. [DOI] [PubMed] [Google Scholar]
- 8.Videm V, Strand E. Scand J Immunol. 2004;59:25–33. doi: 10.1111/j.0300-9475.2004.01351.x. [DOI] [PubMed] [Google Scholar]
- 9.Zarbock A, Ley K. Microcirculation. 2009;16:31–42. doi: 10.1080/10739680802350104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Jeon N, Baskaran H, Dertinger SKW, Whitesides GM, Van De Water L, Toner M. Nat Biotech. 2002;20:826–830. doi: 10.1038/nbt712. [DOI] [PubMed] [Google Scholar]
- 11.Li Z, Dong X, Wang Z, Liu W, Deng N, Ding Y, Tang L, Hla T, Zeng R, Li L, Wu D. Nat Cell Biol. 2005;7:399–404. doi: 10.1038/ncb1236. [DOI] [PubMed] [Google Scholar]
- 12.Toetsch S, Olwell P, Prina-Mello A, Volkov Y. Integ Biol. 2009;1:170–181. doi: 10.1039/b814567a. [DOI] [PubMed] [Google Scholar]
- 13.Atencia J, Morrow J, Locascio LE. Lab Chip. 2009;9:2707–2714. doi: 10.1039/b902113b. [DOI] [PubMed] [Google Scholar]
- 14.Mitchell P. Nat Biotech. 2001;19:717–721. doi: 10.1038/90754. [DOI] [PubMed] [Google Scholar]
- 15.Blow N. Nat Meth. 2009;6:683–686. [Google Scholar]
- 16.Stroka KM, Aranda-Espinoza H. Blood. 2011;118:1632–1640. doi: 10.1182/blood-2010-11-321125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reddy RC, Standiford TJ. Curr Opi Hematol. 2010;17:18–24. doi: 10.1097/MOH.0b013e32833338f3. [DOI] [PubMed] [Google Scholar]
- 18.Schaff UY, Xing MMQ, Lin KK, Pan N, Jeon NL, Simon SI. Lab Chip. 2007;7:448–456. doi: 10.1039/b617915k. [DOI] [PubMed] [Google Scholar]
- 19.Kim D, Haynes CL. Anal Chem. 2012;84:6070–6078. doi: 10.1021/ac3009548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee HL, Yi T, Baek K, Kwon A, Hwang HR, Qadir AS, Park HJ, Woo KM, Ryoo HM, Kim GS, Baek JH. J Cell Physiol. 2013;228:1076–1086. doi: 10.1002/jcp.24256. [DOI] [PubMed] [Google Scholar]
- 21.Alon R, Hammer DA, Springer TA. Nature. 1995;374:539–542. doi: 10.1038/374539a0. [DOI] [PubMed] [Google Scholar]
- 22.Vuoristo MS, KellokumpuLehtinen P, Laine S, Soppi E. Immunopharm Immunotox. 1996;18:337–54. doi: 10.3109/08923979609052740. [DOI] [PubMed] [Google Scholar]
- 23.Schultz MJ, Speelman P, Zaat S, van Deventer SJH, van der Poll T. Antimicrob Agents Chemother. 1998;42:1605–1609. doi: 10.1128/aac.42.7.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zu YL, Qi J, Gilchrist A, Fernandez GA, Vazquez-Abad D, Kreutzer DL, Huang CK, Sha’afi RI. J Immunol. 1998;160:1982–1989. [PubMed] [Google Scholar]
- 25.Neveu WA, Allard JL, Raymond DM, Bourassa LM, Burns SM, Bunn JY, Irvin CG, Kaminsky DA, Rincon M. Respir Res. 2010;11:28–37. doi: 10.1186/1465-9921-11-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kordonowy LL, Burg E, Lenox CC, Gauthier LM, Petty JM, Antkowiak M, Palvinskaya T, Ubags N, Rincon M, Dixon AE, Vernooy JHJ, Fessler MB, Poynter ME, Suratt BT. Am J Respir Cell Mol Biol. 2012;47:120–127. doi: 10.1165/rcmb.2011-0334OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Elgazar-Carmon V, Rudich A, Hadad N, Levy R. Diabetologia. 2008;51:S329–S329. [Google Scholar]
- 28.Singh D, Edwards L, Tal-Singer R, Rennard S. Respir Res. 2010;11:77–88. doi: 10.1186/1465-9921-11-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carpagnano GE, Palladino GP, Lacedonia D, Koutelou A, Orlando S, Foschino-Barbaro MP. BMC cancer. 2011;11:226–233. doi: 10.1186/1471-2407-11-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Welbourn R, Goldman G, Kobzik L, Paterson I, Shepro D, Hechtman HB. Ann Surg. 1991;214:181–186. doi: 10.1097/00000658-199108000-00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Asensi V, Valle E, Meana A, Fierer J, Celada A, Alvarez V, Paz J, Coto E, Carton JA, Maradona JA, Dieguez A, Sarasua J, Ocana MG, Arribas JM. Infection and Immunity. 2004;72:3823–3828. doi: 10.1128/IAI.72.7.3823-3828.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fenton RR, Molesworth-Kenyon S, Oakes JE, Lausch RN. Invest Ophthalmol Vis Sci. 2002;43:737–743. [PubMed] [Google Scholar]
- 33.Munshi UK, Niu JO, Siddiq MM, Parton LA. Pediat Pulmonol. 1997;24:331–336. doi: 10.1002/(sici)1099-0496(199711)24:5<331::aid-ppul5>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 34.Rysz J, Banach M, Cialkowska-Rysz A, Stolarek R, Barylski M, Drozdz J, Okonski P. Cell Mol Immunol. 2006;3:151–154. [PubMed] [Google Scholar]
- 35.Lahat N, Zlotnick AY, Shtiller R, Bar I, Merin G. Clin Exp Immunol. 1992;89:255–260. doi: 10.1111/j.1365-2249.1992.tb06941.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Simpson MA, Madras PN, Cornaby AJ, Etienne T, Dempsey RA, Clowes GHA, Monaco AP. Transplantation. 1989;47:218–223. doi: 10.1097/00007890-198902000-00004. [DOI] [PubMed] [Google Scholar]
- 37.Rysz J, Kocur E, Blaszczak R, Bartnicki P, Stolarek RA, Piechota M. Cent Eur J Med. 2008;3:199–202. [Google Scholar]
- 38.Dong WM, Azconaolivera JI, Brooks KH, Linz JE, Pestka JJ. Toxicol Appl Pharm. 1994;127:282–290. doi: 10.1006/taap.1994.1163. [DOI] [PubMed] [Google Scholar]
- 39.Zahler S, Hoffmann A, Gloe T, Pohl U. J Leuko Biol. 2003;73:118–126. doi: 10.1189/jlb.0402184. [DOI] [PubMed] [Google Scholar]
- 40.Johnson-Leger C, Aurrand-Lions M, Imhof BA. J Cell Sci. 2000;113:921–933. doi: 10.1242/jcs.113.6.921. [DOI] [PubMed] [Google Scholar]
- 41.Fortunati E, Kazemier KM, Grutters JC, Koenderman L, Van den Bosch VMM. Clin Exp Immunol. 2009;155:559–566. doi: 10.1111/j.1365-2249.2008.03791.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Deree J, Lau R, Melbostad H, Loomis W, Hoyt DB, Coimbra R. Surgery. 2006;140:186–191. doi: 10.1016/j.surg.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 43.Auffray C, Fogg D, Garfa M, Elain G, Join-Lambert O, Kayal S, Sarnacki S, Cumano A, Lauvau G, Geissmann F. Science. 2007;317:666–670. doi: 10.1126/science.1142883. [DOI] [PubMed] [Google Scholar]
- 44.Zheng Y, Nguyen J, Wei Y, Sun Y. Lab Chip. 2013;13:2464–2483. doi: 10.1039/c3lc50355k. [DOI] [PubMed] [Google Scholar]
- 45.Chen J, Li J, Sun Y. Lab Chip. 2012;12:1753–1767. doi: 10.1039/c2lc21273k. [DOI] [PubMed] [Google Scholar]
- 46.Kim SI, Jung HI. J Breast Cancer. 2010;13:125–131. [Google Scholar]
- 47.Kim D, Koseoglu S, Manning BM, Meyer AF, Haynes CL. Anal Chem. 2011;83:7242–7249. doi: 10.1021/ac200666c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zare RN, Kim S. Annu Rev Biomed Eng. 2010;12:187–201. doi: 10.1146/annurev-bioeng-070909-105238. [DOI] [PubMed] [Google Scholar]
- 49.Korin N, Kanapathipillai M, Mathews BD, Cresente M, Brill A, Mammoto T, Ghosh K, Jurek S, Bencherif SA, Bhatta D, Coskun AU, Feldman CL, Wagner DD, Ingber DE. Science. 2012;337:738–741. doi: 10.1126/science.1217815. [DOI] [PubMed] [Google Scholar]
- 50.Hwang SY, Kwon KW, Jang KJ, Park MC, Lee JS, Suh KY. Anal Chem. 2010;82:3016–3022. doi: 10.1021/ac100107z. [DOI] [PubMed] [Google Scholar]
- 51.Khan OH, Seffon MV. Biomed Microdevices. 2011;13:69–87. doi: 10.1007/s10544-010-9472-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim D, Lin YS, Haynes CL. Anal Chem. 2011;83:8377–8382. doi: 10.1021/ac202115a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Young EWK, Watson MWL, Srigunapalan S, Wheeler AR, Simmons GA. Anal Chem. 2010;82:808–816. doi: 10.1021/ac901560w. [DOI] [PubMed] [Google Scholar]
- 54.Tolan NV, Genes LI, Subasinghe W, Raththagala M, Spence DM. Anal Chem. 2009;81:3102–3108. doi: 10.1021/ac900084g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martin RS, Root PD, Spence DM. Analyst. 2006;131:1197–1206. doi: 10.1039/b611041j. [DOI] [PubMed] [Google Scholar]
- 56.Genes LI, Tolan NV, Hulvev MK, Martin RS, Spence DM. Lab Chip. 2007;7:1256–1259. doi: 10.1039/b712619k. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


