I. INTRODUCTION
A small scratch on the skin can be a painful experience. Fortunately, this pain is transitory and dissipates quickly once the wound heals. This healing process is facilitated by neutrophils (white blood cells) which remove bacteria and other foreign materials from the wound. These cells normally reside in your circulatory system but are able to leave the bloodstream and efficiently navigate through connective tissue to the injured area when needed. So, how do these cells figure out where to go? The answer is chemotaxis, the process of cells following chemical gradients.
In addition to wound healing, chemotaxis plays a role in many other biological processes. Chemical information can help sperm find the egg cell during fertilization. In embryonic development, cells are often directed to their proper location through gradients. Chemotaxis can also aid the spreading of cancer during metastasis, the process by which cells leave the primary tumor and seed new tumors in other parts of the body. Experiments have shown that an initial step in the metastatic process involving the movement of malignant cells away from the tumor and towards blood vessels, is guided by gradients of growth factors.
All these examples involve eukaryotic cells, cells with a nucleus, that form the basis of multi-cellular life. Chemotaxis, however, is not limited to eukaryotic cells but is also employed by bacterial (prokaryotic) cells. There, chemical gradients are typically used to determine the location of food sources. The motility mechanisms employed by these bacteria, however, are fundamentally different than the ones used by the much larger eukaryotes (see the article by Howard Berg in PHYSICS TODAY, January 2000, page 24). Bacteria utilize a temporal sampling mechanism to determine the direction of the gradient: if a cell senses that the concentration is increasing as it moves, it continues moving in the same direction. Eukaryotic cells, on the other hand, use their size to measure spatial differences across their cell body and their ability to sense a chemotactic gradient does not require cell motion.
In this article, we will review the physics behind eukaryotic chemotaxis using a model system, the crawling motion of the amoeboid Dictyostelium discoideum. Dictyostelium cells are inherently motile cells that use chemotaxis to form large scale aggregates when they are deprived of food. This survival mechanism eventually leads to a fruiting body, containing the majority of cells as spores, on top of a ≈ 1 mm tall stalk. Dictyostelium has become a favorite model system for chemotaxis among biologists and physicists alike for a number of reasons. First, these cells are relatively easy to grow and do not require special temperature or atmospheric conditions. Second, Dictyostelium cells move much faster than most chemotaxing cells, at a rate of approximately one cell diameter per minute, or 10 μm/min. Third, its genome has been fully sequenced and annotated, facilitating the construction of fluorescent markers and mutants that can be used to probe the dynamics of the molecular components that are involved in chemotaxis. As a result, it is possible to generate high-quality quantitative data that can be used to construct accurate mathematical models. Conversely, predictions generated by modeling studies can often be tested directly in experiments. Of course, these advantages would be limited if the chemotactic mechanisms employed by Dictyostelium were unique to this species. Fortunately, the general mechanisms that underlie chemotaxis and motility are similar for many cell types, and many of the molecular components are conserved across different species.
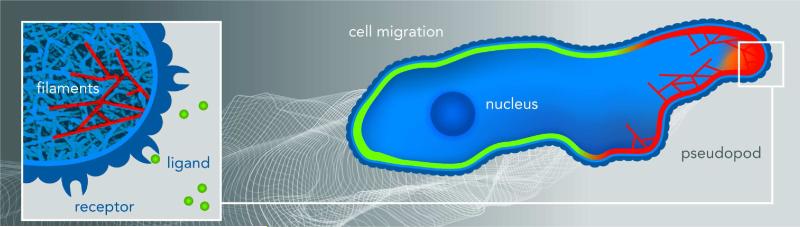
The similarities between different cell types allow us to discuss the relevant processes during eukaryotic chemotaxis in a cell-independent way. As shown schematically in Fig. 1, a cell is able to detect external chemical signals through receptors that are embedded in the cell membrane and reversibly bind the externally diffusing ligands. These receptors are distributed uniformly along the cell body such that there is no a priori cell front or back. If the cell is placed in a gradient, the resulting distribution of bound receptors is asymmetric, with more bound receptors in the membrane at the locations corresponding to the highest local concentration. Subsequent conformational changes of the receptors lead to the excitation of internal signaling pathways. Some of the components of these pathways become localized (or have their activation become localized) at the front or the back of the cells, leading to a breaking of the internal symmetry and polarizing the cell [1]. This symmetry breaking can be clearly seen in Fig. 2 which shows the response of cells to a chemoattractant gradient created by a leaking pipette. These cells have been engineered to contain green fluorescent protein that binds to one of the signaling components of the internal pathway. All cells display a fluorescent crescent at the membrane closest to the pipette, indicating an asymmetric distribution of that signaling component. Importantly, this asymmetry is not permanent and can be reversed following repositioning of the pipette.
FIG. 1.
Cartoon of a cell moving to the right directed by a chemotactic gradient schematically shown by the gray scaled background. The cell is polarized with some signaling components localized at the front, depicted by red membrane, while others are localized at the back (green membrane). The outline of the membrane at previous times is shown by the white lines. Pseudopods (membrane protrusions) are generated mostly at the front of the cell and are associated with certain signaling components, shown by the orange color field. The inset shows the branched actin network that push the membrane forward, with several individual filaments highlighted in red. The external chemical gradient is detected by the cell through the binding of ligands, shown as green balls in the inset, to membrane bound receptors.
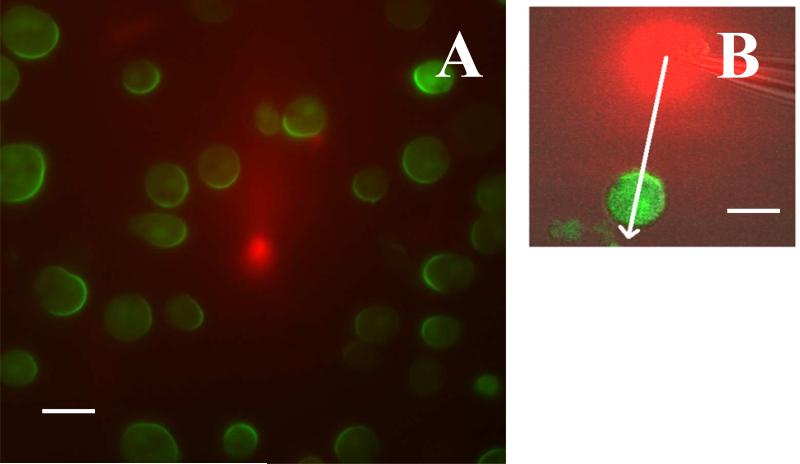
FIG. 2.
Response of cells to steady gradients. (a) The response of a signaling component (PHCrac-GFP) in Dictyostelium cells to a chemoattractant gradient produced by a pipette, placed at the center of the image. The pipette contains a high concentration of chemoattractant (1 μm of cAMP) mixed with red dye to visualize the gradient. The cells were treated with latrunculin, a drug that inhibits actin polymerization, rendering the cells immobile and round (image courtesy of Christopher Janetopoulos, Vanderbilt University). (b) The PHCrac-GFP distribution in a cell exposed to a steady cAMP gradient visualized with a red dye. The asymmetry in the front and back distribution of PHCrac-GFP is larger than the asymmetry in the cAMP concentration, as measured along the white line, indicating a several-fold amplification of the input signal. The scale bar in (a) and (b) represents 10 μm. (Adapted from ref. [13])
The cascade of internal signaling events eventually results in the polymerization of actin filaments at the front of the cell [2]. These filaments push against the membrane, resulting in membrane deformations. In Dictyostelium, this results in localized regions of protrusion called pseudopods (see Fig 1). Obviously, for net translation, protrusive movement at the front needs to be complemented by retraction at the back. This retraction is partially mediated through the motor protein myosin, the same motor responsible for our own muscular contractions. These forces act on the cell and its membrane, a lipid bilayer whose shape is affected by its physical properties, including surface tension and bending rigidity. Forces generated both by actin polymerization and myosin contraction combine with friction with the substratum to determine all mechanical aspects of cell motion.
Physics plays a role at almost every level of the chemotactic process. The binding and unbinding of molecules to the receptors is an inherently stochastic process, leading to a number of interesting sensitivity questions that can be addressed using techniques developed in statistical physics. Understanding the subsequent recognition of the gradient by the internal pathways and the concomitant polarization of the cell requires the development and analysis of spatially extended nonlinear reaction-diffusion models. Obviously, the physics of elasticity is critical for determining how the internally generated forces can create motion. Finally, physics can also play an important role in the development of novel experimental techniques. Microfluidic devices are able to precisely control the experimental conditions experienced by cells and have become an integral component of chemotactic experiments. In the following, we will look more carefully at some of these crucial physical processes.
II. SIGNAL DETECTION
The first requirement for directed motility is the detection of the external gradient. Experiments show that Dictyostelium cells can detect chemical gradients that vary by only a few percent across their body and that this exquisite sensitivity is present for a large range of background concentrations [3]. As already mentioned, cells do this sensing via specialized proteins embedded in their membranes. These surface receptors reversibly bind the particular chemical to be detected and report the results to the cell interior via changes in the shape of their cytoplasmic segments. For our Dictyostelium amoeba model, the protein responsible for chemotaxis is cAR1 (cyclic AMP receptor, type 1) and there are roughly 70,000 receptors per cell. cAR1 is a prototypical G-protein coupled receptor (GPCR) of the type used throughout the biological world to detect external environmental conditions. The precise meaning of this classification will be explained shortly.
Each of these receptor proteins acts as an independent two-state system, either bound (denoted by S = 1) or unbound (S = 0) to the cAMP ligand. The rate of binding of receptor i is linearly proportional to the chemical concentration at its location ci (with coefficient k+), whereas the rate of unbinding (k–) is constant. This means that in equilibrium, the probability of occupation of a single receptor i is
where is the dissociation constant, the concentration at which the occupation probability equals 0.5. If we consider the entire array of N receptors (and neglect for the moment any motion of these proteins), the overall distribution is just the product of these independent binomials.
Sampling the receptor distribution clearly conveys information about the gradient's direction. To see this, we can imagine a two dimensional circular cell of diameter d with receptors located at equidistant angles θi, placed in a linearly varying field. The statistic Φ defined through
| (1) |
acts as an estimator of the gradient direction. This is because the sum on the right-hand-side has the property that the “spins” on the up-gradient side of the cell are more likely to be 1 than those on the down-gradient side; hence those phases are weighted more heavily in the sum. The distribution of our estimator can be explicitly constructed and this distribution determines the accuracy of the estimate.
To compute the signal-to-noise ratio (SNR) we will focus for simplicity on small gradients. Then, the strength of the directional signal is directly proportional to the gradient of occupation probability, . The “noise” is due to the inherent fluctuations of the sum in the definition of Φ. The variance of our estimator can be treated to lowest order by neglecting the gradient, giving (upon neglect of a geometrical factor)
with c̄ is the mean concentration. Combining these two expressions gives the front/back SNR
| (2) |
One can derive more general expressions valid for finite gradients, more complex cell shapes and nonuniform receptor spacings, using tools borrowed from information theory.
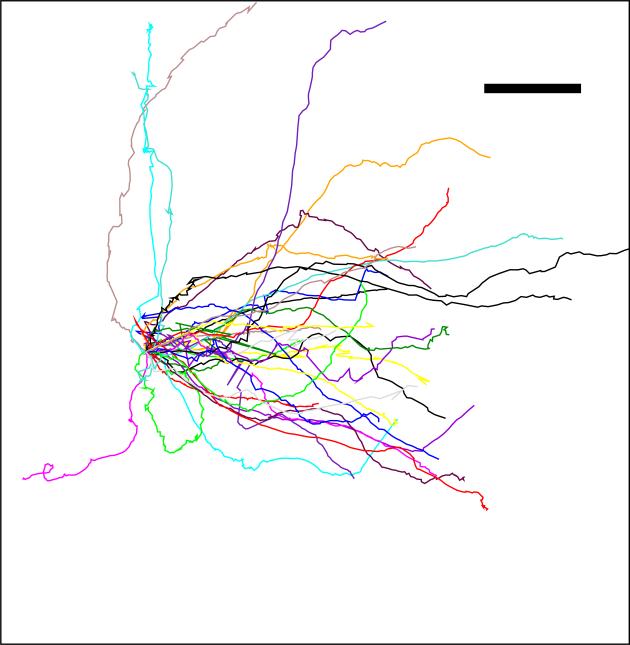
Recently, it has become possible to directly compare this theory with experiments performed via the use of microfluidic technology. These devices can easily be designed and manufactured which allow for precise control of chemical concentrations to be presented to cells. One can obtain reproducible distributions of cell behavior as a function of the applied conditions. Examples of cell tracks in such a device are shown in Fig. 3. At small SNR, a reasonable prediction is that cell behavior is limited by the cell's ability to accurately detect gradient directionality. Data shows that this hypothesis is indeed consistent with careful quantitative measurements [4]. To carry out this comparison, we had to assume that the cell's action is based on single snapshots of the array of detectors on the cell surface. Better performance could be obtained if the cell integrates its measurement over times longer than the natural refresh time of the array. Since Dictyostelium cells are able to decide their motility direction within a few seconds after being exposed to a gradient and this time is the same order as the refresh rate (e.g. the receptor off-rate is 1s–1), we believe this is a reasonable assumption.
FIG. 3.
Cell tracks, with their origins brought to a common point, shown for a gradient with a steepness of 10 %, expressed as the fractional difference in the concentration across a typical cell diameter (10 μm). Note that these tracks do not represent a straight line as the cell shows considerable motile variability. Even for very steep gradients a typical maximal chemotactic index, defined as the ratio of the distance traveled in the direction gradient and the total distance traveled, is at most around 0.7. (Scalebar: 20 μm, arrow indicates gradient direction ; Adapted from ref. [4]).
Before ending this part, it is worth mentioning that there are sources of noise not directly due to the stochastic binding-unbinding process. The cell is moving (and changing shape as it does) and the receptors themselves can diffuse in the membrane, at least over short distances. However, given the time-scale of the detection (1 s), these effects are rather small. For example, the change over one second of an occupation probability due to cell motion (at a speed of .1μm/s) in a gradient of 1nM/μm with c̄ = Kd = 50nM is roughly 10–3 as compared to the standard deviation of the binomial binding/unbinding process . Finally, there are the fluctuations of the ligand concentration itself, as studied in the seminal work of Berg and Purcell on chemosensing in bacteria [5], and extended by others [6]. These eventually limit the usefulness of long-time averaging [7] and similarly increases the amount of time it takes to equilibrate the array to a changed chemical field.
III. SIGNAL PROCESSING
After detection comes processing. As we will see below, interesting questions already arise for the case of large SNR where the input signal can be treated deterministically. The difference in receptor occupation probability couples to chemical networks in the cell interior that “decide” on the gradient direction and polarize the cell accordingly. Such chemical networks and their role in cellular information processing have become popular of late, and have been studied extensively using techniques borrowed from the physics of nonlinear dynamical systems. For example, stem cells (cells that retain maximal flexibility in terms of their functional capabilities) can decide to become more specialized (“differentiate”) in response to external signals. But, the chemotaxis network is necessarily more complex. A spatially local circuit can only respond to a local value of the bound receptor concentration, but a gradient response must arise via comparison of values from different parts of the cell. This requires communication of information between different parts of the cell, which ensures that only that part of the cell that experiences the highest local concentration will be identified as the front. This communication could be chemical, mechanical or both,
The simplest assumption, originally due to Parent and Devreotes, is that the communication takes the form of a diffusing messenger which inhibits the formation of the “front” state that includes the actin polymerization responsible for protrusions [8]. This assumption has been used in numerous subsequent modeling studies and, although the details differ from model to model, the basic idea is that the cell's response is governed by a competition between this global signal and a locally produced membrane bound activator. The two-dimensional LEGI (local excitation - global inhibition) equations introduced by Levchenko and Iglesias show how this works [9]. Here, there is an input variable S(s) (reflecting the bound receptor number which depends on position s along the membrane), which directs the production of a membrane-resident activator A and cytosolic inhibitor I, which affect a downstream (normalized) reactant R in opposite directions:
| (3) |
together with a boundary condition for the outward pointing normal derivative of the cytosolic component:
| (4) |
where D is the cytosolic diffusion constant of the inhibitor. The key mechanism embodied in this model can be seen by first considering a uniform signal for S. It is easy to see that in the solution both Aeq and Ieq at the membrane are linearly proportional to the assumed value of S and therefore the equilibrium value of R is independent of the signal. If the input signal is changed, the output R will respond, as the excitation kinetics is typically faster than that of the inhibitor, but eventually return to its baseline level (see the Box). In other words, the system perfectly adapts.
In order to test this basic property of the LEGI proposal, one needs to identify specific molecules related to the abstract activator, inhibitor and response variables in the model. Above, we referred to cAR1 as a G-protein coupled receptor. This refers to the first stages of the intracellular processing cascade, which here happens via the dissociation of a specific protein (called a G-protein) into separate parts that can catalyze downstream effects. One of the earliest such effects in Dictyostelium is the activation of the Ras protein [10]. This regulatory protein is activated by exchanging a low-energy cyclic nucleotide (guanosine diphosphate - GDP) with a high-energy one (guanosine triphosphate - GTP). The exchange is catalyzed by a guanosine exchange factor (a RasGEF) and the reverse process by a GTP-ase activating protein (a RasGAP). The simplest possibility is to associate the activator with the RasGEF, the inhibitor with the RasGAP, and the response readout with the percentage of activated Ras.
Armed with this dictionary, we and our colleagues set out to quantify the activated Ras response of Dictyostelium cells to abrupt changes in the chemoattractant concentration [11]. These changes were achieved using a microfluidic device which was able to quickly (< 1 s) and reproducibly alter the global concentration. The results show that the response did indeed perfectly adapt after roughly 30 seconds [11]. Furthermore, we were able to argue that of all possible means of achieving perfect adaptation, only a pathway of the LEGI type is consistent with the observed transient behavior (see the Box for a discussion).
There is one more step necessary to use LEGI for gradient inputs, and that has to do with amplification. The simple model given above will zero out the mean and makes the high-end of the cell show a larger than baseline (and the low-end of the cell a smaller than baseline) response value. But, the overall amplitude of this difference is no larger than that of the external gradient itself. The fact that Dictyostelium cells can detect very shallow gradients suggests that one key early steps of the processing machinery is the amplification of the external gradient. Indeed, experiments using fluorescent markers have revealed a amplification that can be at least 5-fold [12, 13]. Differing models of chemotaxis incorporate this amplification in a variety of ways. One of the challenges in this modeling is that the amplification cannot be so strong as to preclude the possibility of the cell changing its motion if the external directionality of the signal changes; Dictyostelium cells actually do quite a good job of continuously tracking changing gradients and avoid getting completely locked in to a fixed polarization.
One speculative idea that shows promise is that of having the response reaction operate in a parameter regime which exhibits zero-order ultrasensitivity [14]. This involves replacing the R equation in the LEGI model by the more general Michaelis-Menten enzyme kinetics form
| (5) |
The equilibrium value of R can be strongly amplified in the case where the constants KI and KA are small, and β = krAeq/k–rIeq ≃ 1 since then the small location-dependent change in the ratio of A and I will dramatically change its value. Once this is done, the LEGI model can be used to fit the type of data shown in Fig 2b, the amplified asymmetric response of immobilized cells to an imposed chemical gradient.
IV. PSEUDOPOD FORMATION
Movies of chemotaxing cells (see SI) reveal that pseudopods come and go stochastically; this occurs even at large gradients and hence is not caused by the sensing noise discussed above. One important fact is that the pseudopodal protrusions mirror the stochastic appearance and disappearance of patches of Ras activation (see Fig. 4a) [15]. Thus, the LEGI mechanism explains how the gradient limits activation to the front but the model outlined so far needs to be augmented by additional dynamical processes for it to be quantitatively applied to moving cells. The most compelling hypothesis that has arisen to date is that the formation of pseudopods is an excitable process due to the positive feedback present in the dynamics of actin polymerization [15, 16]. An excitable process is one for which the system will strongly amplify an above-threshold perturbation of a linearly stable state, but which will eventually return back to that state. In biology, the most well-known example of such a process is the propagation of electrical waves down the axons of neuronal cells, due to positive feedback between the voltage and the conductance. Actin at the leading edge of a motile cell forms a branched network which is constantly growing via the addition of monomers and via the catalyzed formation of new branches. As the network grows, it creates more free ends which further accelerate the growth, hence the notion of positive feedback. This concept accounts in a general way for the observations of traveling waves of filamentous actin (F-actin) in a variety of cell types.
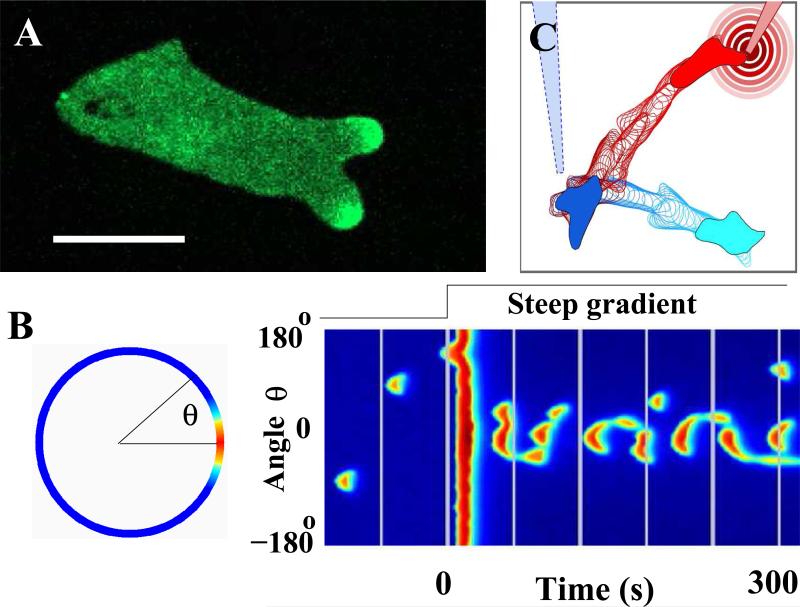
FIG. 4.
Analyzing and modeling the dynamics of pseudopods in a migrating cell. (a) Snapshot of a chemotaxing cell, moving to the right, that contains a fluorescent marker for activated Ras. The marker displays dynamic patches (regions of elevated fluorescence intensity that come and go) at the leading edge that are correlated with pseudopods. (Adapted from ref. [15]). (b) The response of an excitable model to a spatial gradient, computed in a circular geometry and visualized along the perimeter in a color coded fashion, where red represents large values and blue represents small values of one of the model components. The excitable dynamics of the model are driven by the output of a LEGI module together with a noise source. The left panel shows a patch at angle θ = 00. A spatiotemporal representation of the cell's response is shown in the right panel in the form of a space-time plot. Here, the model component along the perimeter is plotted vertically while time runs from left to right. Before the application of the gradient at t = 0, patches appear stochastically along the entire membrane of the computational cell. Immediately thereafter, the membrane responds in a spatially uniform fashion, followed by patches that are mostly aligned with the external gradient. (Adapted from ref. [16]) (c) Response of a simple motility model, described by Eqns. 6, to spatially changing gradients. The gradient is created by a diffusing chemoattractant from a point source, mimicking the experimental use of a pipette. The simulation was started with a gradient produced by the blue “pipette” and the initial position of the cell is shown in solid light blue. The outline of the cell after regular intervals is shown in blue. After the cell has migrated several diameters up the gradient, the pipette is repositioned to a new location, creating a new gradient schematically shown as concentric circles. The location of the cell at that instance is shown in solid blue. Subsequent outlines of the cell are shown at regular intervals in red.
Several groups have attempted to use these concepts to build models of pseudopods by combining the LEGI model with excitable dynamics (Fig. 4b). The excitable nature guarantees that a pseudopod has a finite lifetime, as is typically observed. The coming and going of individual protrusions implies that a cell will be able to quickly re-direct its response following the a change in the gradient direction.
How does one start to test some of these ideas and couple them to actual cell motion? A fully comprehensive cell motility model would include the physical properties of the membrane (surface tension and bending rigidity), a description of the adhesive forces between the cell and the substrate, and a description of the spatially extended internal signaling pathways. Furthermore, experiments have demonstrated that the cytoskeleton is not stationary but flows within the cell, necessitating a fluid dynamics description of the actin network. This is a major challenge for the future. In the interim, one can get insight by studying “toy” motility models which relate protrusion forces (as determined by the excited patches) to actual cell shape and motion via simple geometrical assumptions regarding membrane dynamics. The basic equation that governs such models has the general form
| (6) |
Here, f(R) represents the driving force that is associated with patches of the “biochemical” component R, vn is the (normal) membrane velocity, κ is the curvature, γ represents the membrane rigidity, λ is a friction coefficient, and C is a Lagrange multiplier that is used to implement the constraint that the total enclosed area remains equal to A0. This type of model can produce quite realistic motion [15], an example of which is shown in Fig. 4c. What is more important is that it can be used to develop insight into how the different dynamical processes involved in cell motility can interact and thereby determine the time course of the system. Such insights can then be tested on direct experimental data and on more quantitatively realistic models of cell mechanics.
Before concluding this section, we should emphasize that in our approach the gradient ”instructs” the cell as to where to place pseudopods. As we have discussed, this notion is supported by the observation of gradient sensing in immobilized cells (see Fig 2) and by measurements of adaptation (see Box). An alternative conceptual approach, explored by Meinhardt and others [17] starts with pseudopods as the result of spontaneous self-organization and the gradient ”selects” ones that orient up-gradient by, for example, modulating their lifetime. Once we incorporate non-linear actin dynamics into the LEGI model or conversely add adaptation to the front end of the self-organization approaches, these ideas begin to converge to each other and, we truly hope, to the actual behavior of this complex biological process.
V. REPRISE
As physicists struggle to understand the fundamental principles underlying biological function, it is important to have examples that can be studied in quantitative detail and for which identifiable progress can be made. We have argued here that eukaryotic chemotaxis is one such critical example. Combined efforts by physicists and biologists and by theorists and experimentalists have revealed a fascinating set of dynamical processes that allow the cell to go from the molecular input to the utilitarian output. Lessons learned here may have practical uses (how to prevent tumor cells from navigating to the vasculature, how to enhance wound healing, etc.), although this is never certain. What is certain is that we have learned and will continue to learn valuable lessons about the myriad of ways in which physics sets up the framework inside which biological systems manage to accomplish the art of living.
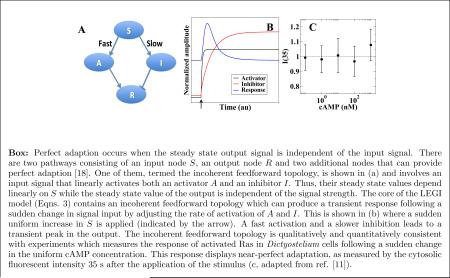
Box: Perfect adaption occurs when the steady state output signal is independent of the input signal. There are two pathways consisting of an input node S, an output node R and two additional nodes that can provide perfect adaption [18]. One of them, termed the incoherent feedforward topology, is shown in (a) and involves an input signal that linearly activates both an activator A and an inhibitor I. Thus, their steady state values depend linearly on S while the steady state value of the output is independent of the signal strength. The core of the LEGI model (Eqns. 3) contains an incoherent feedforward topology which can produce a transient response following a sudden change in signal input by adjusting the rate of activation of A and I. This is shown in (b) where a sudden uniform increase in S is applied (indicated by the arrow). A fast activation and a slower inhibition leads to a transient peak in the output. The incoherent feedforward topology is qualitatively and quantitatively consistent with experiments which measures the response of activated Ras in Dictyostelium cells following a sudden change in the uniform cAMP concentration. This response displays near-perfect adaptation, as measured by the cytosolic fluorescent intensity 35 s after the application of the stimulus (c, adapted from ref. [11]).
ACKNOWLEDGMENTS
We would like to thank our many experimental and theoretical collaborators for providing data and useful comments. This work was supported by the National Institutes of Health (P01 GM078586) and by the NSF Physics Frontier Center program.
References
- 1.Swaney KF, Huang CH, Devreotes PN. Eukaryotic chemotaxis: a network of signaling pathways controls motility, directional sensing, and polarity. Annu Rev Biophys. 2010;39:265–289. doi: 10.1146/annurev.biophys.093008.131228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carlsson AE, Sept D. Mathematical modeling of cell migration. Methods Cell Biol. 2008;84:911–937. doi: 10.1016/S0091-679X(07)84029-5. [DOI] [PubMed] [Google Scholar]
- 3.Song L, Nadkarni SM, Bödeker HU, Beta C, Bae A, Franck C, Rappel W-J, Loomis WF, Bodenschatz E. Dictyostelium discoideum chemotaxis: threshold for directed motion. Eur J Cell Biol. 2006;85:981–989. doi: 10.1016/j.ejcb.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 4.Fuller D, Chen W, Adler M, Groisman A, Levine H, Rappel WJ, Loomis WF. External and internal constraints on eukaryotic chemotaxis. PNAS. 2010;107(21):9656–9659. doi: 10.1073/pnas.0911178107. See also Ueda M, Shibata T. Stochastic signal processing and transduction in chemotactic response of eukaryotic cells. Biophysical Journal. 2007;93:11–20. doi: 10.1529/biophysj.106.100263.
- 5.Berg HC, Purcell EM. Physics of chemoreception. Biophysical Journal. 1977;20:193–219. doi: 10.1016/S0006-3495(77)85544-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bialek W, Setayeshgar S. Physical limits to biochemical signaling. PNAS. 2005;102(29):10040–10045. doi: 10.1073/pnas.0504321102. [DOI] [PMC free article] [PubMed] [Google Scholar]; Endres RG, Wingreen NS. Accuracy of direct gradient sensing by single cells. PNAS. 2008;105:15749–15754. doi: 10.1073/pnas.0804688105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang K, Rappel WJ, Kerr R, Levine H. Quantifying noise levels of intercellular signals. Phys Rev E. 2007;75:061905. doi: 10.1103/PhysRevE.75.061905. [DOI] [PubMed] [Google Scholar]
- 8.Parent CA, Devreotes PN. A cell's sense of direction. Science. 1999;284:765–770. doi: 10.1126/science.284.5415.765. [DOI] [PubMed] [Google Scholar]
- 9.Levchenko A, Iglesias PA. Models of eukaryotic gradient sensing: application to chemotaxis of amoebae and neutrophils. Biophys J. 2002;82:50–63. doi: 10.1016/S0006-3495(02)75373-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang S, Charest PG, Firtel RA. Spatiotemporal regulation of Ras activity provides directional sensing. Curr. Biol. 2008 Oct;18(20):1587–1593. doi: 10.1016/j.cub.2008.08.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takeda K, Shao D, Adler M, Charest PG, Loomis WF, Levine H, Groisman A, Rappel WJ, Firtel RA. Incoherent feedforward control governs adaptation of activated ras in a eukaryotic chemotaxis pathway. Sci Signal. 2012 Jan;5(205):ra2. doi: 10.1126/scisignal.2002413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Janetopoulos C, Ma L, Devreotes PN, Iglesias PA. Chemoattractant-induced phosphatidylinositol 3,4,5-trisphosphate accumulation is spatially amplified and adapts, independent of the actin cytoskeleton. PNAS. 2004;101(24):8951–8956. doi: 10.1073/pnas.0402152101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu X, Meier-Schellersheim M, Jiao X, Nelson LE, Jin T. Quantitative imaging of single live cells reveals spatiotemporal dynamics of multistep signaling events of chemoattractant gradient sensing in Dictyostelium. Mol. Biol. Cell. 2005 Feb;16(2):676–688. doi: 10.1091/mbc.E04-07-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goldbeter A, Koshland DE. An amplified sensitivity arising from covalent modification in biological systems. Proc. Natl. Acad. Sci. U.S.A. 1981 Nov;78(11):6840–6844. doi: 10.1073/pnas.78.11.6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hecht I, Skoge ML, Charest PG, Ben-Jacob E, Firtel RA, Loomis WF, Levine H, Rappel WJ. Activated membrane patches guide chemotactic cell motility. PLoS Comput. Biol. 2011 Jun;7(6):e1002044. doi: 10.1371/journal.pcbi.1002044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiong Y, Huang CH, Iglesias PA, Devreotes PN. Cells navigate with a local-excitation, global-inhibition-biased excitable network. Proc. Natl. Acad. Sci. U.S.A. 2010 Oct;107(40):17079–17086. doi: 10.1073/pnas.1011271107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meinhardt H. Orientation of chemotactic cells and growth cones: models and mechanisms. J.Cell Science. 1999;112:2867. doi: 10.1242/jcs.112.17.2867. [DOI] [PubMed] [Google Scholar]; Bosgraaf L, van Haastert PJ. Navigation of chemotactic cells by parallel signaling to pseudopod persistence and orientation. PLoS One. 2009;4:e6842. doi: 10.1371/journal.pone.0006842. [DOI] [PMC free article] [PubMed] [Google Scholar]; King JS, Insall RH. Chemotaxis: finding the way forward with Dictyostelium. Trends in Cell Biology. 2009;19:523–530. doi: 10.1016/j.tcb.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 18.Ma W, Trusina A, El-Samad H, Lim WA, Tang C. Defining network topologies that can achieve biochemical adaptation. Cell. 2009 Aug;138(4):760–773. doi: 10.1016/j.cell.2009.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]