Abstract
BACKGROUND
The homeobox gene HOXB7 is overexpressed across a range of cancers and promotes tumorigenesis through varying effects on proliferation, survival, invasion, and angiogenesis. Although published microarray data suggest HOXB7 is overexpressed in pancreatic ductal adenocarcinoma (PDAC), its function in pancreatic cancer has not been studied.
METHODS
HOXB7 message and protein levels were examined in PDAC cell lines and patient samples, as well as in normal pancreas. HOXB7 protein expression in patient tumors was determined by immunohistochemistry and correlated with clinicopathologic factors and survival. The impact of HOXB7 on cell proliferation, growth, and invasion was assessed by knockdown and overexpression in PDAC cell lines. Candidate genes whose expression levels were altered following HOXB7 knockdown were determined by microarray analysis.
RESULTS
HOXB7 message and protein levels were significantly elevated in PDAC cell lines and patient tumor samples relative to normal pancreas. Evaluation of a tissue microarray of 145 resected PDACs found high HOXB7 protein expression was correlated with lymph node metastasis (P = .034) and an independent predictor of worse overall survival in multivariate analysis (hazard ratio = 1.56, 95% confidence interval = 1.02–2.39). HOXB7 knockdown or overexpression in PDAC cell lines resulted in decreased or increased invasion, respectively, without influencing proliferation or cell viability.
CONCLUSIONS
HOXB7 is frequently overexpressed in PDAC, specifically promotes invasive phenotype, and is associated with lymph node metastasis and worse survival outcome. HOXB7 and its downstream targets may represent novel clinical biomarkers or targets of therapy for inhibiting the invasive and metastatic capacity of PDAC.
Keywords: HOXB7, invasion, metastasis, pancreatic adenocarcinoma, CCBP2, homeobox genes
INTRODUCTION
Pancreatic ductal adenocarcinoma (PDAC) is the fourth leading cause of cancer mortality in the United States.1 Most often presenting as an advanced stage, nonresectable disease, PDAC is notorious for its rapid clinical progression and resistance to chemotherapy and radiation.2 Even after surgical resection, most patients eventually succumb to local recurrence or distant metastasis.3 It is known that invasion and metastasis are heavily dictated by the ability of cancer to subvert normal developmental programs that mediate cell adhesion, motility and tissue morphogenesis during embryogenesis.4,5 Epithelial-mesenchymal transition (EMT) is widely viewed as an initiating event for tumor cell spread and is a subject of intense investigation. In addition, a larger and more diverse spectrum of developmentally regulated genes and signaling pathways also contribute to tumor invasion and metastasis, including various members of the homeobox supergene family.6
The HOX subgroup of the homeobox supergene family encompasses 39 genes located in 4 contiguous clusters (HOXA through HOXD). HOX genes encode transcription factors with the capacity to activate or repress downstream target gene expression. HOX genes regulate growth and differentiation during embryonic development and maintain adult tissue homeostasis.6,7 They are also frequently deregulated in cancer where they variably influence tumor cell proliferation, apoptosis, stem cell renewal, differentiation, motility, and angiogenesis.6–8 Although published microarray data indicate multiple HOX genes are deregulated in PDAC,9 it is not well understood whether these changes in expression are causally linked to pancreatic tumor progression.10
HOXB7 is widely overexpressed in various cancers,11–16 including PDAC, based on published microarray data.17,18 Although its function in PDAC has not been specifically addressed, HOXB7 has been shown to have various protumorigenic activities in other cancers, including promotion of cell proliferation,12–14,19,20 survival,11 anchorage-independent growth,13,20,21 angiogenesis,22 invasion,15 metastasis,14,15,19 transformation and radiation resistance.21 We chose to directly examine HOXB7 and its role in pancreatic tumorigenesis. Here, we show that HOXB7 message and protein are overexpressed in a majority of PDAC, including both established cell lines and patient tumor samples. Higher levels of HOXB7 protein expression were also found to correlate with regional lymph node metastasis and worse overall survival in a large patient cohort of resected PDACs. Finally, we present evidence that HOXB7 specifically augments pancreatic cancer cell invasiveness, an activity which may explain its link to aggressive clinical behavior in PDAC.
MATERIALS AND METHODS
Cell Lines
Human pancreatic cell lines AsPC-1, BxPC-3, MiaPaCa-2, and PANC-1 were obtained from the ATCC (Manassas, Va) and cultured as described.23 HPDE, an immortalized, nontransformed human pancreatic ductal epithelial cell line, was kindly provided by Dr. Ming-Sound Tsao (Princess Margaret Hospital, Toronto, Canada) and cultured as described.24
Human Pancreatic Tissue Samples and Tissue Microarray
All patient tissues and data were obtained in accordance with institutional guidelines and prior institutional review board approval. Remnant fresh human tumor and normal pancreas tissue samples were snap-frozen immediately after surgical resection. The previously described PDAC tissue microarray (TMA) includes 3 separate 1.0-mm tumor cores from each of 145 treatment-naive, American Joint Committee on Cancer (AJCC) stage I or II PDACs resected at the University of California Los Angeles (UCLA) Medical Center from 1990 to 2005.25
RNA Extraction and Quantitative PCR
All RNA extractions were performed using the Qiagen RNeasy Mini Kit (Qiagen, Valencia, Calif). For first-strand complementary DNA (cDNA) synthesis, total RNA was reverse-transcribed with oligo-dT primer using the Superscript First Strand Synthesis System (Invitrogen, Carlsbad, Calif). Quantitative PCR (qPCR) reactions were carried out in 20 μL final volumes including cDNA, 300 nM forward/reverse primers, and 10 μL 2× SYBR green master mix (Diagenode, Denville, NJ) using an ABI-Prism 7700 sequence detector (Applied Biosystems, Foster City, Calif). Reaction parameters were 50°C for 5 seconds, 95°C for 15 minutes, followed by 40 cycles of 95°C for 15 seconds, 60°C for 30 seconds, and 72°C for 30 seconds. ACTB was used as normalization control. Primer sequences are available upon request.
Western Blots
Triton X-100 whole-cell lysates (50 μg per sample) were resolved by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and electroblotted to nitrocellu-lose membranes. Membranes were blocked with 5% nonfat milk in 1% Tween-TBS and then probed overnight with 1:500 anti-HOXB7 rabbit polyclonal antibody (Invitrogen), 1:500 anti-D6/CCBP2 (chemokine binding protein 2) rat monoclonal (R&D Systems, Minneapolis, Minn), or 1:2000 anti-ACTB antibody (Sigma, St. Louis, Mo). ECL Plus detection (GE Healthcare Biosciences, Piscataway, NJ) was performed following 1 hour incubation with 1:1000 to 1:5000 horseradish peroxidase (HRP)-linked secondary antibodies (Cell Signaling Technology, Beverly, Mass).
Immunohistochemistry
Following heat-induced epitope retrieval in a vegetable steamer (10 mM sodium citrate buffer, pH 6.0), sections were incubated with 0.5 μg/mL anti-HOXB7 monoclonal antibody (clone 4C6; Novus Biologicals, Littleton, Colo) for 1 hour at room temperature. The LSAB+ HRP System (Dako, Carpinteria, Calif) was used for visualization. Semiquantitative evaluation of nuclear HOXB7 levels for each tumor core was determined by histoscore (score from 0–300), the product of staining intensity (ranging from 0–3; 0 = absent, 1 = weak, 2 = moderate, 3 = strong), and percent tumor cell staining (range from 0%–100%). Histoscores were generated by 2 surgical pathologists (D.D. and S.F.) who were blinded to clinicopathologic and outcome variables. Average histoscores from both pathologists were used for analysis.
HOXB7 siRNA Gene Knockdown and Stable Gene Overexpression
For HOXB7 gene knockdown, cell lines were transfected with 20 nM of duplexed small interfering RNA (siRNA; IDT, Coralville, Iowa) using Invitrogen Lipofectamine 2000. Duplex siRNAs included: HOXB7 siRNA #1 (sense 5′-CUAUUCGAUUUGAGUUUCCdTdT-3′, antisense 5′-GGAAACUCAAAUCGAAUAGdTdT-3′, as described11); HOXB7 siRNA #2 (sense 5′-GGAACUGACCGCAAA CGAGGCCGdCdC-3′, antisense 5′-GGCGGCCUCG UUUGCGGUCAGUUCCUG-3′); HOXB7 siRNA #3 (sense 5′-CCUAUUUAAAUGAAAGGAGUUUAdAdA-3′, antisense 5′-UUUAAACUCCUUUCAUUUAAAUAG GGU-3′); CCBP2 siRNA (sense 5′-CCAGGUAACAGA GAGCAUCGCCUdTdC-3′, antisense 5′-GAAGGCGA UGCUCUCUGUUACCUGGAG-3′); and IDT siRNA negative control sequence (NC1, sense 5′-CGUUAAUC GCGUAUAAUACGCGUdAdT-3′; antisense 5′-AUAC GCGUAUUAUACGCGAUUAACGAC-3′). For HOXB7 overexpression, full-length human HOXB7 cDNA was PCR-cloned from a previously described pCDNA3 construct26 (gift from Dr. Judith Gasson, UCLA) using primers incorporating AgeI and NotI restriction sites at 5′ and 3′ ends, respectively. This PCR product was directly cloned into MSCV-GFP-IRES-PURO retroviral vector with the same restriction sites, thus replacing the green fluorescent protein (GFP) cassette. HOXB7- or control GFP-MSCV retroviral supernatants collected from 293T Phoenix packaging cell line were used to transduce MiaPaCa-2 cells with puromycin selection.
Migration and Invasion Assays
Tumor cell invasion was assessed using BD BioCoat Matrigel Invasion Chambers with 8 μM porous membranes (BD Biosciences, San Jose, Calif). Cells (2.5 × 104) were placed in the top well (media plus 1% fetal bovine serum) and allowed to migrate for 18 to 24 hours toward a chemoattractant gradient in the bottom well (media plus 10% fetal bovine serum). Cells were removed from the top side of the membrane, after which migrating cells on the bottom side were fixed, stained, and counted by light microscopy. Tumor cell migration assays were performed in identical fashion using uncoated membranes in place of Matrigel membranes.
Cell Growth and Proliferation Assays
Cells were plated at equal density in 96-well plates for 6 hours and then transiently transfected with indicated siR-NAs (4 biological replicates per condition). MTT assays (ATCC, Manassas, Va) were performed per manufacturer’s protocol at 48 and 96 hours. For cell proliferation, bromodeoxyuridine was added 24 hours after initial siRNA transfection and further incubated with cells for 16 hours prior to assay completion per manufacturer’s instructions (BrdU Cell Proliferation Assay; EMD Biosciences, Rockland, Mass).
Gene Expression Microarray and Data Analysis
Gene expression microarray data for comparing HOXB7 expression in whole-patient sections of PDAC and normal pancreas samples have been described27 and are deposited in the NCBI Gene Expression Omnibus database (GSE32688). Oncomine (Compendia Bioscience, Ann Arbor, Mich) was used to obtain and analyze Affymetrix probe-set data for HOXB7 (204779_s_at) and CCBP2 (206887_at) in the published Grutzmann et al gene expression data set of laser-capture–microdissected normal pancreatic ductal samples and PDAC.28 For comparison of control versus HOXB7 siRNA-transfected MiaPaCa-2 cells, total RNA was extracted 48 hours after transfection with microarray analysis performed in the UCLA Clinical Microarray Core Facility, using Affymetrix U133 plus 2.0 oligonucleotide arrays. Data analysis was performed in the dChip Analysis software package using invariant set normalization. The signal intensity was summarized using the MBEI (model-based expression index) algorithm with mismatch probe option for background subtraction. HOXB7-regulated genes were selected by comparative analysis between control samples and each of 3 distinct siRNA transfectants (selection criteria > 1.25-fold change and P < .05). Gene functional analysis, pathway analysis and network analysis were performed using Ingenuity Pathway Analysis (IPA) software (Ingenuity Systems, Redwood, Calif).
Statistical Analyses
All clinicopathologic and survival information was extracted from a prospectively maintained UCLA surgical and pathology institutional database of pancreatic cancer. Overall survival time was measured from the date of surgery to the date of death due to any cause or last clinical follow-up as determined by review of electronic medical records and US social security death index. Patient dichotomization using nuclear HOXB7 histoscore was performed by k-means clustering whereby each patient tumor was segregated into low or high groups based on its relationship to the nearest of 2 means (a histoscore of 110 was the cut-point for dichotomization). Student t tests were used to compare continuous variables. Chi-square tests were used to compare dichotomized HOXB7 groups and baseline clinicopathologic factors. Kaplan-Meier survival curves were evaluated by log-rank test. Univariate Cox regression analyses were performed to determine the prognostic significance of individual clinicopathologic variables. Multivariate analysis to test statistical independence and significance of multiple predictors was performed by stepwise Cox analysis with backward selection using the Akaike Information Criterion. The level of significance for all tests was defined as α = 0.05. All statistics were performed using SPSS, version 20.0, for Windows.
RESULTS
HOXB7 Is Overexpressed in Pancreatic Cancer
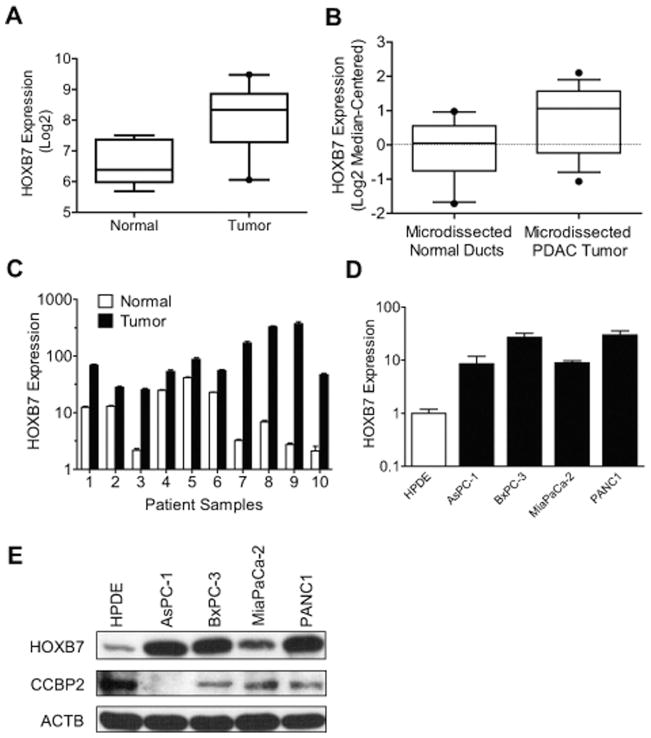
Published cDNA microarrays from cell lines, whole tissues or microdissected tumors suggest HOXB7 is frequently overexpressed in pancreatic adenocarcinoma (PDAC).17,18 Analyzing our own recently published oligonucleotide microarray data set of 43 PDAC tumors and 7 normal pancreas samples,27 we confirmed a statistically significant 4-fold median increase in HOXB7 expression in tumor samples (Fig. 1A; P = .0002; unpaired t test). Analysis of a separately published microarray data set of laser-capture–microdissected normal pancreatic ducts and PDAC patient samples28 further confirmed up-regulation of HOXB7 specifically within malignant versus benign ductal epithelium (Fig. 1B; P = .03; unpaired t test). Quantitative PCR (qPCR) on a separate set of patient samples independently verified a 2- to 50-fold increase in HOXB7 expression across all PDAC tumors relative to patient-matched normal pancreas (Fig. 1C; P = .026; paired t test). Increased HOXB7 message (Fig. 1D) and protein (Fig. 1E) levels were also observed in PDAC cell lines relative to a control, nontransformed pancreatic ductal epithelial cell line (HPDE).
Figure 1.
(A) HOXB7 levels in whole sections of human pancreatic ductal adenocarcinoma (PDAC) (N = 43) and normal pancreas (N = 7) patient samples are shown, based on oligonucleotide microarray analysis. (B) HOXB7 levels are shown in a published gene expression data set of laser-capture–microdissected normal pancreatic duct (N = 11) and PDAC (N = 14) patient samples. Levels of HOXB7 message relative to housekeeping gene (ACTB) are shown as determined by quantitative polymerase chain reaction for (C) patient-matched normal pancreas and PDAC tumors and (D) PDAC cell lines versus HPDE cells, an immortalized, nontransformed human pancreatic ductal cell line. (E) Western blots for HOXB7, CCBP2, and ACTB in the same lines.
HOXB7 Expression Correlates With Poor Prognosis
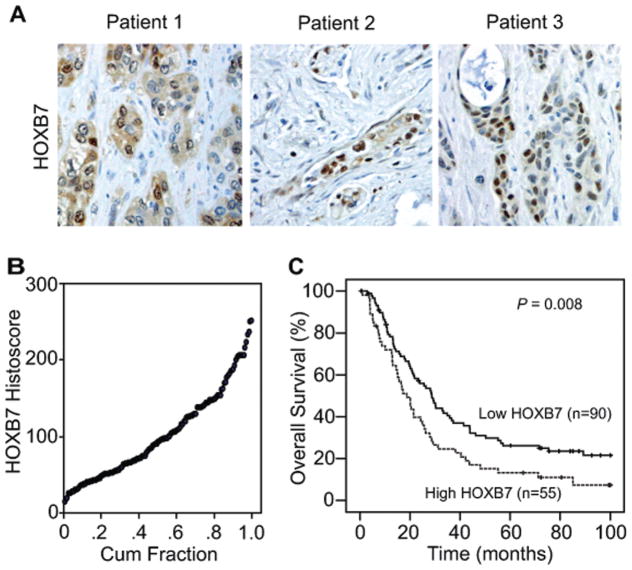
The potential significance of HOXB7 protein expression was examined by immunohistochemistry on a TMA of 145 AJCC stage I or II resected PDAC tumors. Immunohistochemical staining for HOXB7 varied across the TMA with tumor cells showing predominantly nuclear expression and occasional weaker cytoplasmic expression (Fig. 2A). Semiquantitative histoscores of nuclear HOXB7 expression (Fig. 2B) were determined and used to dichotomize patients into groups with either low or high HOXB7 expression. Dichotomized groups were evaluated in relation to clinicopathologic factors and overall survival. No significant associations were noted between HOXB7 expression and baseline clinicopathologic parameters with the exception of a significant correlation between high HOXB7 expression and positive lymph node status (P = .034; Table 1). In relation to clinical outcome, high HOXB7 expression was significantly associated with worse overall survival by univariate Cox regression analysis (hazard ratio = 1.63; 95% CI = 1.12–2.36, P = .009; Table 2) and a median survival of 18.7 months ± 2.5 months versus 28.8 months ± 2.9 months as visualized on Kaplan-Meier curves (log-rank P = .01; Fig. 2C). Multivariate Cox regression analysis also indicated high HOXB7 expression was an independent predictor of worse overall survival in a model including other significant clinicopathologic variables (Table 3).
Figure 2.
(A) Representative micrographs show variable nuclear staining for HOXB7 in pancreatic ductal adenocarcinoma (PDAC) tumors. (B) Distribution of semiquantitative histoscores of nuclear HOXB7 expression is shown for all PDAC tumors in the tissue microarray (N = 145). (C) Kaplan-Meier curves for overall survival are shown based on low (histoscore ≤ 110) versus high (histoscore > 110) HOXB7 protein expression.
Table 1.
Clinicopathologic Characteristics of Pancreatic Ductal Adenocarcinoma Tissue Microarray and Correlation With HOXB7 Expression
| Characteristic | Full Cohort (n = 145) | Low HOXB7 (n = 90) | High HOXB7 (n = 55 ) | P | |||
|---|---|---|---|---|---|---|---|
| No. of Patients | % | No. of Patients | % | No. of Patients | % | ||
| Age, y | |||||||
| ≤60 | 49 | 33.7 | 35 | 38.9 | 14 | 25.5 | .10 |
| >60 | 96 | 66.3 | 55 | 61.1 | 41 | 74.5 | |
| Sex | |||||||
| Female | 70 | 48.3 | 45 | 50.0 | 25 | 45.5 | .59 |
| Male | 75 | 51.7 | 45 | 50.0 | 30 | 54.5 | |
| Greatest tumor dimension | |||||||
| <3 cm | 86 | 59.3 | 51 | 56.7 | 35 | 63.6 | .33 |
| ≥3 cm | 59 | 40.7 | 39 | 43.3 | 20 | 36.4 | |
| Histologic grade | |||||||
| Well + moderate | 83 | 57.2 | 54 | 60.0 | 29 | 52.7 | .39 |
| Poor | 62 | 42.8 | 36 | 40.0 | 26 | 47.3 | |
| pT-stage | |||||||
| pT1 | 26 | 17.9 | 18 | 20.0 | 8 | 14.5 | .41 |
| pT2 + pT3 | 119 | 82.1 | 72 | 80.0 | 47 | 85.5 | |
| N-stage (categorized) | |||||||
| N0 | 69 | 47.6 | 49 | 54.4 | 20 | 36.4 | .034 |
| N1 | 76 | 52.4 | 41 | 45.6 | 35 | 63.6 | |
| AJCC stage | |||||||
| Stage I | 40 | 27.6 | 30 | 33.3 | 10 | 22.2 | .048 |
| Stage II | 105 | 72.4 | 60 | 66.6 | 45 | 81.8 | |
| Margin status | |||||||
| Negative (R0) | 126 | 86.9 | 78 | 87.7 | 48 | 87.3 | .70 |
| Positive (R1) | 19 | 13.1 | 12 | 13.3 | 7 | 12.7 | |
Abbreviation: AJCC, American Joint Commission on Cancer.
P value from 2-sided chi-square test evaluating low versus high HOXB7 for each characteristic.
Table 2.
Univariate Analysis for Overall Survival in Pancreatic Ductal Adenocarcinoma Tissue Microarray
| Variable | No. of Patients Who Died | Total No. of Patients | Hazard Ratio (95% CI) | P |
|---|---|---|---|---|
| HOXB7 expression | ||||
| Low | 67 | 90 | 1.00 | .01 |
| High | 48 | 55 | 1.63 (1.12–2.36) | |
| Age, y | ||||
| ≤60 | 40 | 49 | 1.00 | .89 |
| >60 | 75 | 96 | 1.03 (0.70–1.51) | |
| Sex | ||||
| Female | 60 | 70 | 1.00 | .11 |
| Male | 55 | 75 | 0.74 (0.52–1.07) | |
| Greatest tumor dimension | ||||
| <3 cm | 65 | 87 | 1.00 | .14 |
| ≥3 cm | 50 | 58 | 1.33 (0.92–1.92) | |
| Histologic grade | ||||
| Well + moderate | 60 | 83 | 1.00 | .001 |
| Poor | 55 | 62 | 1.88 (1.30–2.74) | |
| pT-stage | ||||
| pT1 | 23 | 26 | 1.00 | .52 |
| pT2 + pT3 | 92 | 119 | 0.86 (0.55–1.36) | |
| N-stage | ||||
| N0 | 50 | 69 | 1.00 | .002 |
| N1 | 65 | 76 | 1.80 (1.24–2.60) | |
| AJCC stage | ||||
| Stage I | 28 | 40 | 1.00 | .02 |
| Stage II | 87 | 105 | 1.67 (1.09–2.55) | |
| Margin status | ||||
| Negative (R0) | 102 | 126 | 1.00 | .53 |
| Positive (R1) | 13 | 18 | 1.21 (0.68–2.15) | |
Abbreviations: AJCC, American Joint Commission on Cancer; CI, confidence interval.
Table 3.
Multivariate Analysis for Overall Survival in Pancreatic Ductal Adenocarcinoma Tissue Microarray
| Variable | Comparison | Hazard Ratioa (95% CI) | P |
|---|---|---|---|
| HOXB7 expression | High vs. low | 1.50 (1.01–2.22) | .04 |
| Sex | Male vs female | 0.72 (0.50–1.04) | .08 |
| Histologic grade | Poor vs well + moderate | 1.82 (1.24–2.66) | .002 |
| pT status | pT2 + pT3 vs pT1 | 0.60 (0.37–0.98) | .04 |
| AJCC stage | II vs I | 1.61 (1.08–2.40) | .02 |
Abbreviations: AJCC, American Joint Commission on Cancer; CI, confidence interval.
HR > 1 indicates a greater risk of death for the first level of the variable listed. All covariates in Table 2 were addressed, with only those variables retained in the model after backward selection shown here.
HOXB7 Promotes Pancreatic Cancer Cell Invasion
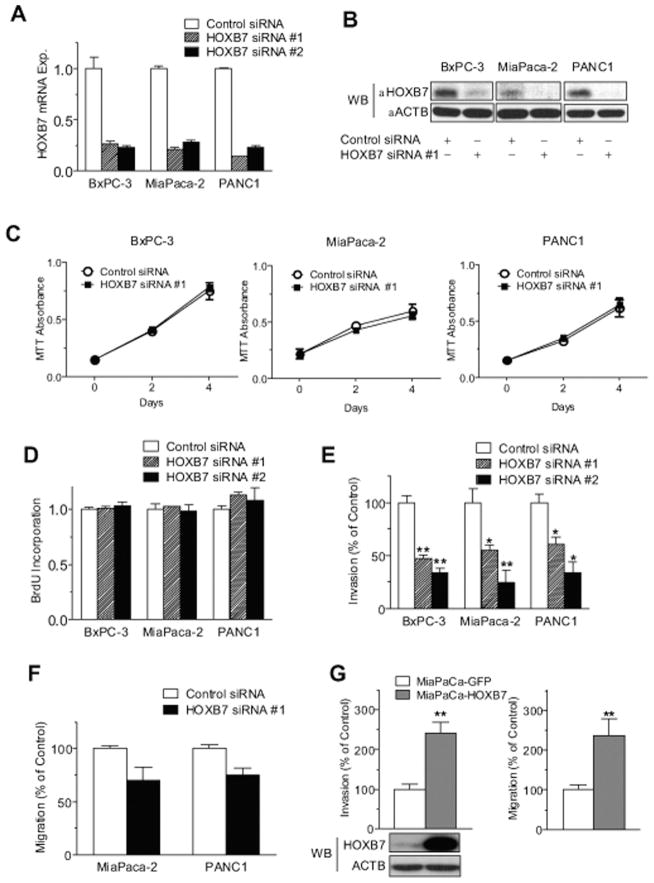
Given its described functions in other cancer types, we investigated the effects of HOXB7 gene knockdown in PDAC cell line on the in vitro phenotypes of growth, survival and invasion. Each of 3 distinct HOXB7 siRNAs effectively reduced HOXB7 mRNA and protein expression by 65% to 80% across multiple PDAC cell lines, but had no significant effect on cell proliferation or growth (Fig. 3A–D and data not shown). In contrast, HOXB7 knockdown significantly inhibited the invasiveness of all 3 cell lines tested (Fig. 3E) and showed a nonsignificant trend toward an inhibition of cell migration toward a chemotactic gradient (Fig. 3F). The effects of exogenous HOXB7 overexpression were also examined. HOXB7 overexpression augmented both the invasiveness and migration of MiaPaCa-2 cells (Fig. 3G) without altering their growth (data not shown). HOXB7 has been previously shown to regulate the expression of various genes with the potential to influence invasion in other tumor types, including FGF2, VEGF, IL-8 and MMP9.12,20,22 Quantitative PCR revealed no change in the expression of any of these genes in BxPC-3, MiaPaCa-2 or PANC1 cells following HOXB7 siRNA transfection (data not shown). HOXB7 has also been shown to confer features of EMT in breast cancer.15 Quantitative PCR again revealed no change in the expression of multiple EMT transcription factors (SNAI1, SNAI2, TWIST1, ZEB1, and ZEB2) in BxPC-3, MiaPaCa-2 and PANC1 following HOXB7 siRNA transfection (data not shown).
Figure 3.
(A) HOXB7 expression was determined by quantitative PCR in PDAC cell lines following transfection with control small interfering RNA (siRNA), HOXB7 siRNA #1, or HOXB7 siRNA #2, as well as (B) parallel western blots. Corresponding in vitro assays are shown for (C) cell growth by MTT, (D) cell proliferation by bromodeoxyuridine (BrdU) incorporation, (E) invasiveness through Matrigel-coated transwell membrane inserts, or (F) migration through uncoated transwell membrane inserts toward chemoattractant. (G) Migration and matrigel invasion assays and western blots are shown with MiaPaCa-2 cells stably overexpressing GFP or HOXB7. Each sample condition was evaluated in triplicate or quadruplicate, with all assays repeated at least twice. Similar results were obtained using HOXB7 siRNA #3 (data not shown). *P < .05, **P < .005. Abbreviations: mRNA exp., messenger RNA expression; WB, western blot.
Gene Expression Profile Regulated by HOXB7 in Pancreatic Cancer
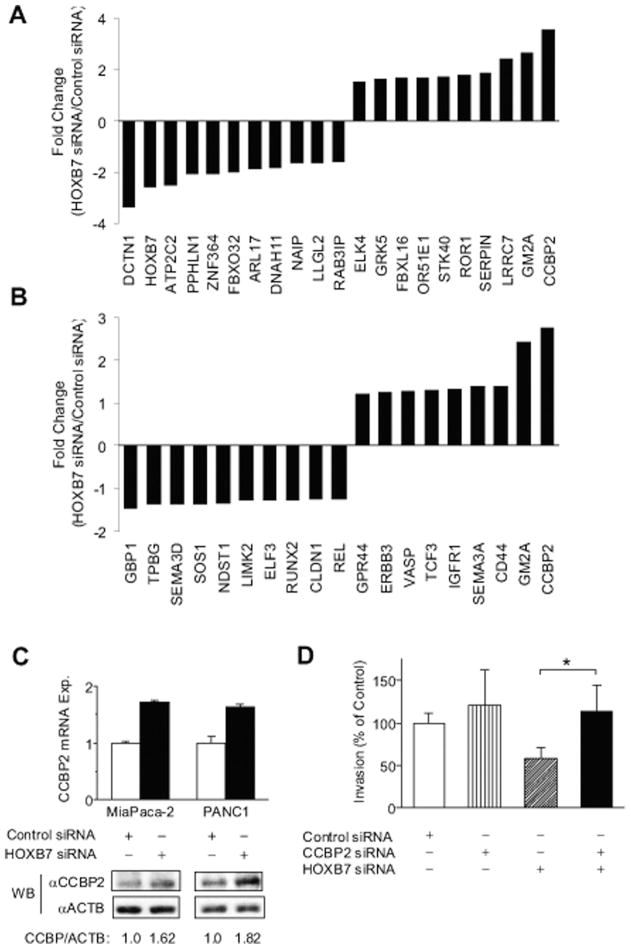
We sought to identify a HOXB7 target gene profile in PDAC with the potential to uncover links between HOXB7 expression and its effects on invasion and aggressive clinical behavior. Gene expression microarrays were performed on MiaPaCa-2 cells 48 hours after transfection with either control or each of 3 separate HOXB7 siRNAs. Initial qPCR confirmed a 70%–75% reduction in HOXB7 expression in each of the HOXB7 siRNA transfectants, whereas subsequent gene expression microarray analysis confirmed a corresponding 2.58-fold average decrease in HOXB7 expression. Pairwise comparisons of control versus HOXB7 siRNA-transfected samples revealed 311 altered transcripts (112 up-regulated and 199 down-regulated) following HOXB7 knockdown (absolute fold change ≥ 1.25; P < .05; median false discovery rate, 5.8%). The top 10 up- and down-regulated genes following HOXB7 siRNA transfection are listed in Fig. 4A.
Figure 4.
(A) Top 10 up-regulated and down-regulated genes on Affymetrix U133 2.0 plus microarray analysis of MiaPaCa-2 cells are shown 48 hours after HOXB7 small interfering RNA (siRNA) transfection. (B) Expression changes are seen with HOXB7 siRNA transfection for genes annotated in the category of cellular movement by Ingenuity Pathway Analysis. (C) CCBP2 levels are shown as measured by quantitative polymerase chain reaction and western blot in separate HOXB7 siRNA transfections of Mia-PaCa-2 and PANC1. (D) Matrigel invasion assay with PANC1 is shown 48 hours after transfection with the indicated combinations of siRNA. *P < .05. Abbreviations: mRNA exp., messenger RNA expression; WB, western blot.
IPA was performed on the full set of altered transcripts to gain functional insight into genes or pathways whose altered expression and biological effects might be linked to HOXB7 in PDAC. The top 5 molecular and cellular functions significantly linked to HOXB7 knockdown included lipid metabolism, small molecule biochemistry, cell death, “cell-to-cell signaling and interaction,” and cellular movement (P values = .002–.047). Genes listed in the IPA category of cellular movement (Fig. 4B) were further considered given their potential to explain observed proinvasive phenotype linked to HOXB7. Of these, CCBP2 (also known as D6) represented the most highly up-regulated gene with HOXB7 knockdown. CCBP2 is a chemokine decoy receptor previously shown to negatively regulate the invasion and metastasis of breast cancer cells.29 Given that HOXB7 repression of a negative regulator of invasive and metastatic phenotype could explain its proinvasive activity, we sought to validate the inverse relationship between HOXB7 and CCBP2 expression. First, a statistically significant strong negative correlation was noted between HOXB7 and CCBP2 expression (Pearson’s r = −0.51, 95% confidence interval = −0.75 to −0.15, P = .009) in the aforementioned published28 microarray data set of laser-capture microdissected normal pancreatic duct and PDAC tumor samples. Likewise, a negative correlation was also observed between HOXB7 and CCBP2 protein expression on western blots of non-transformed HPDE cells and multiple PDAC cell lines (Fig. 1D). Both CCBP2 message and protein levels were up-regulated in response to HOXB7 knockdown in separate gene knockdown experiments performed in Mia-PaCa-2 and PANC1 cell lines (Fig. 4C). Finally, we addressed the potential relationship between invasive phenotype and regulation of CCBP2 by HOXB7 by examining the effects of individual and combined gene knockdown in a Matrigel invasion assay. CCBP2 siRNA transfection resulted in an 85% ± 14% reduction in CCBP expression (P = .0002, unpaired t test) but did not significantly alter invasive phenotype in PANC1 cells (Fig. 4D), possibly due to its already low baseline levels of expression. However, concurrent CCBP2 knockdown rescued the inhibition of invasion resulting from HOXB7 knockdown (Fig. 4D).
DISCUSSION
HOX genes play integral roles in normal embryonic development and adult tissue homeostasis.8 They are also frequently deregulated in cancer, primarily through altered levels of expression as opposed to genetic mutation.8 Here, we have demonstrated that HOXB7 is frequently overexpressed in PDAC, adding it to a growing number of solid tumors shown to have elevated HOXB7 expression, including melanoma12 and carcinomas of the colorectum,13 oral squamous cavity,14 breast,15 and ovary.16 Abnormal patterns of HOX gene expression in cancer generally appear to disrupt normal cellular homeostasis in favor of tumor progression, whether by loss of tumor suppressor function or gain in oncogenic activity.8 Although the vast majority of the published data indicate HOXB7 functions to promote tumor progression in melanoma and epithelial malignancies,12–15,19–22 it is not uniformly pro-oncogenic under all circumstances. For instance, targeted overexpression of HOXB7 has dual roles in a HER-2/neu–induced mouse model of breast carcinogenesis, inhibiting initial tumor onset but promoting subsequent tumor growth and metastasis.19 Therefore, the function of HOXB7 in cancer appears to be dictated by its levels of expression and temporospatial context, a concept in keeping with data presented here. Despite its multiple effects in promoting growth, survival, and invasion in other solid tumors, the in vitro effects of HOXB7 in PDAC appears largely restricted to invasive phenotype. Compatible with this observation, we noted a positive correlation between higher HOXB7 protein expression and regional lymph node metastasis whereby increased invasiveness may lead to greater access to and spread via lymphatics.
HOXB7 presumably mediates its effects by functioning as a master transcriptional regulator. In this capacity, HOXB7 could function as a “rheostat” that incrementally modulates the expression of one or more genes that facilitate invasiveness. Microarray data presented here provide several candidate genes whose direct or indirect regulation by HOXB7 may explain its proinvasive effects in PDAC cell lines (Fig. 4B). Among genes positively correlated with HOXB7 expression was GBP1, which has been previously shown to be a positive effector of tumor cell invasion in glioblastoma30 and oral squamous cell carcinoma.31 Likewise, among those genes inversely correlated with HOXB7 expression were several candidates with known or proposed roles in antagonizing tumor invasion and metastasis, including CCBP2,29 GM2,32 and others. CCBP2 message and protein levels were separately confirmed to be inversely correlated with HOXB7 expression and increase upon HOXB7 knockdown in PDAC lines, although it is not yet known whether this effect is mediated directly or indirectly by HOXB7. Concurrent knockdown of CCBP2 also countered the inhibition of invasion seen in the context of HOXB7 knockdown. Notably, CCBP2 blocks breast cancer cell proliferation and invasion both in vitro and in vivo and correlates negatively with lymph node metastasis and positively with disease-free survival.29 CCBP2 is a nonsignaling chemokine decoy receptor with broad specificity for inflammatory CC chemokines.33 HOXB7, by inhibiting CCBP2 expression, may therefore alter the availability and function of multiple chemokines in the PDAC milieu. Such an effect could influence the recruitment and activity of inflammatory cells and other chemokine-responsive cell types interacting with cancer cells in the tumor microenvironment. Work is underway to better establish the functional relevance of CCBP2 and other candidate genes in mediating these and other phenotypes associated with HOXB7 in PDAC.
We have shown HOXB7 protein expression correlates with positive lymph node status and predicts poor overall survival in a large patient cohort of early-stage, resected PDAC. Data here parallel those of other studies that show HOXB7 protein expression correlates with tumor progression and/or worse clinical outcomes for patients with breast, oral, or colorectal cancers.13–15,19,34 Our data and these other studies are based on retrospective analyses of patient tumors without adjuvant treatment information. Thus, further prospective validation is needed in larger and more diverse patient cohorts with consideration of adjuvant therapy. Nevertheless, they collectively provide compelling circumstantial evidence that HOXB7 functions dominantly to facilitate tumor progression in many solid tumor types. This data could be leveraged clinically where HOXB7 serves as a generalized biomarker of malignant progression and/or survival for multiple tumor types. Furthermore, these data provide strong rationale for future mechanistic studies investigating HOXB7 as a causal factor in promoting cancer progression, as well as its utility as a therapeutic target in PDAC and other tumors where it is deregulated.
Acknowledgments
FUNDING SOURCES
This work was supported by funding from the Hirshberg Foundation for Pancreatic Cancer Research and an American Association for Cancer Research/Pancreatic Cancer Action Network Seena Magowitz Career Development Award for Pancreatic Cancer Research.
Footnotes
CONFLICT OF INTEREST DISCLOSURE
The authors made no disclosure.
References
- 1.American Cancer Society. Cancer Facts & Figures 2010. Atlanta, GA: American Cancer Society; 2010. [Google Scholar]
- 2.Stathis A, Moore MJ. Advanced pancreatic carcinoma: current treatment and future challenges. Nat Rev Clin Oncol. 2010;7:163–172. doi: 10.1038/nrclinonc.2009.236. [DOI] [PubMed] [Google Scholar]
- 3.Neoptolemos JP, Stocken DD, Bassi C, et al. Adjuvant chemotherapy with fluorouracil plus folinic acid vs gemcitabine following pancreatic cancer resection: a randomized controlled trial. JAMA. 2010;304:1073–1081. doi: 10.1001/jama.2010.1275. [DOI] [PubMed] [Google Scholar]
- 4.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 5.Mihaljevic AL, Michalski CW, Friess H, Kleeff J. Molecular mechanism of pancreatic cancer--understanding proliferation, invasion, and metastasis. Langenbecks Arch Surg. 2010;395:295–308. doi: 10.1007/s00423-010-0622-5. [DOI] [PubMed] [Google Scholar]
- 6.Samuel S, Naora H. Homeobox gene expression in cancer: insights from developmental regulation and deregulation. Eur J Cancer. 2005;41:2428–2437. doi: 10.1016/j.ejca.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 7.Abate-Shen C. Deregulated homeobox gene expression in cancer: cause or consequence? Nat Rev Cancer. 2002;2:777–785. doi: 10.1038/nrc907. [DOI] [PubMed] [Google Scholar]
- 8.Shah N, Sukumar S. The Hox genes and their roles in oncogenesis. Nat Rev Cancer. 2010;10:361–371. doi: 10.1038/nrc2826. [DOI] [PubMed] [Google Scholar]
- 9.Harsha HC, Kandasamy K, Ranganathan P, et al. A compendium of potential biomarkers of pancreatic cancer. PLoS Med. 2009;6:e1000046. doi: 10.1371/journal.pmed.1000046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gray S, Pandha HS, Michael A, Middleton G, Morgan R. HOX genes in pancreatic development and cancer. JOP. 2011;12:216–219. [PubMed] [Google Scholar]
- 11.Raval A, Tanner SM, Byrd JC, et al. Downregulation of death-associated protein kinase 1 (DAPK1) in chronic lymphocytic leukemia. Cell. 2007;129:879–890. doi: 10.1016/j.cell.2007.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caré A, Silvani A, Meccia E, et al. HOXB7 constitutively activates basic fibroblast growth factor in melanomas. Mol Cell Biol. 1996;16:4842–4851. doi: 10.1128/mcb.16.9.4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liao WT, Jiang D, Yuan J, et al. HOXB7 as a prognostic factor and mediator of colorectal cancer progression. Clin Cancer Res. 2011;17:3569–3578. doi: 10.1158/1078-0432.CCR-10-2533. [DOI] [PubMed] [Google Scholar]
- 14.De Souza Setubal Destro MF, Bitu CC, Zecchin KG, et al. Overexpression of HOXB7 homeobox gene in oral cancer induces cellular proliferation and is associated with poor prognosis. Int J Oncol. 2010;36:141–149. [PubMed] [Google Scholar]
- 15.Wu X, Chen H, Parker B, et al. HOXB7, a homeodomain protein, is overexpressed in breast cancer and confers epithelial-mesenchymal transition. Cancer Res. 2006;66:9527–9534. doi: 10.1158/0008-5472.CAN-05-4470. [DOI] [PubMed] [Google Scholar]
- 16.Naora H, Yang YQ, Montz FJ, Seidman JD, Kurman RJ, Roden RB. A serologically identified tumor antigen encoded by a homeobox gene promotes growth of ovarian epithelial cells. Proc Natl Acad Sci U S A. 2001;98:4060–4065. doi: 10.1073/pnas.071594398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lowe AW, Olsen M, Hao Y, et al. Gene expression patterns in pancreatic tumors, cells and tissues. PLoS One. 2007;2:e323. doi: 10.1371/journal.pone.0000323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakamura T, Furukawa Y, Nakagawa H, et al. Genome-wide cDNA microarray analysis of gene expression profiles in pancreatic cancers using populations of tumor cells and normal ductal epithelial cells selected for purity by laser microdissection. Oncogene. 2004;23:2385–2400. doi: 10.1038/sj.onc.1207392. [DOI] [PubMed] [Google Scholar]
- 19.Chen H, Lee JS, Liang X, et al. Hoxb7 inhibits transgenic HER-2/neu-induced mouse mammary tumor onset but promotes progression and lung metastasis. Cancer Res. 2008;68:3637–3644. doi: 10.1158/0008-5472.CAN-07-2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carè A, Silvani A, Meccia E, Mattia G, Peschle C, Colombo MP. Transduction of the SkBr3 breast carcinoma cell line with the HOXB7 gene induces bFGF expression, increases cell proliferation and reduces growth factor dependence. Oncogene. 1998;16:3285–3289. doi: 10.1038/sj.onc.1201875. [DOI] [PubMed] [Google Scholar]
- 21.Rubin E, Wu X, Zhu T, et al. A role for the HOXB7 homeodomain protein in DNA repair. Cancer Res. 2007;67:1527–1535. doi: 10.1158/0008-5472.CAN-06-4283. [DOI] [PubMed] [Google Scholar]
- 22.Carè A, Felicetti F, Meccia E, et al. HOXB7: a key factor for tumor-associated angiogenic switch. Cancer Res. 2001;61:6532–6539. [PubMed] [Google Scholar]
- 23.Angst E, Dawson DW, Stroka D, et al. N-myc downstream regulated gene-1 expression correlates with reduced pancreatic cancer growth and increased apoptosis in vitro and in vivo. Surgery. 2011;149:614–624. doi: 10.1016/j.surg.2010.11.002. [DOI] [PubMed] [Google Scholar]
- 24.Liu N, Furukawa T, Kobari M, Tsao MS. Comparative phenotypic studies of duct epithelial cell lines derived from normal human pancreas and pancreatic carcinoma. Am J Pathol. 1998;153:263–269. doi: 10.1016/S0002-9440(10)65567-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manuyakorn A, Paulus R, Farrell J, et al. Cellular histone modification patterns predict prognosis and treatment response in resectable pancreatic adenocarcinoma: results from RTOG 9704. J Clin Oncol. 2010;28:1358–1365. doi: 10.1200/JCO.2009.24.5639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yaron Y, McAdara JK, Lynch M, Hughes E, Gasson JC. Identification of novel functional regions important for the activity of HOXB7 in mammalian cells. J Immunol. 2001;166:5058–5067. doi: 10.4049/jimmunol.166.8.5058. [DOI] [PubMed] [Google Scholar]
- 27.Donahue T, Tran L, Hill R, et al. Intergrative survival-based molecular profiling of human pancreatic cancer. Clin Cancer Res. 2012;18:1352–1363. doi: 10.1158/1078-0432.CCR-11-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grutzmann R, Pilarsky C, Ammerpohl O, et al. Gene expression profiling of microdissected pancreatic ductal carcinomas using high-density DNA microarrays. Neoplasia. 2004;6:611–622. doi: 10.1593/neo.04295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu FY, Ou ZL, Feng LY, et al. Chemokine decoy receptor d6 plays a negative role in human breast cancer. Mol Cancer Res. 2008;6:1276–1288. doi: 10.1158/1541-7786.MCR-07-2108. [DOI] [PubMed] [Google Scholar]
- 30.Li M, Mukasa A, Inda Md, et al. Guanylate binding protein 1 is a novel effector of EGFR-driven invasion in glioblastoma. J Exp Med. 2011;208:2657–2673. doi: 10.1084/jem.20111102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu CJ, Chang KP, Chang YJ, et al. Identification of guanylate--binding protein 1 as a potential oral cancer marker involved in cell invasion using omics-based analysis. J Proteome Res. 2011;10:3778–3788. doi: 10.1021/pr2004133. [DOI] [PubMed] [Google Scholar]
- 32.Todeschini AR, Dos Santos JN, Handa K, Hakomori SI. Ganglioside GM2-tetraspanin CD82 complex inhibits met and its cross-talk with integrins, providing a basis for control of cell motility through glycosynapse. J Biol Chem. 2007;282:8123–8133. doi: 10.1074/jbc.M611407200. [DOI] [PubMed] [Google Scholar]
- 33.Hansell CA, Hurson CE, Nibbs RJ. DARC and D6: silent partners in chemokine regulation? Immunol Cell Biol. 2011;89:197–206. doi: 10.1038/icb.2010.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bitu CC, Carrera M, Lopes MA, Kowalski LP, Soares FA, Coletta RD. HOXB7 expression is a prognostic factor for oral squamous cell carcinoma. Histopathology. 2012;60:662–665. doi: 10.1111/j.1365-2559.2011.04102.x. [DOI] [PubMed] [Google Scholar]