SUMMARY
A primary function of males for many species involves mating with females for reproduction. Drosophila melanogaster males respond to the presence of other males by prolonging mating duration to increase the chance of passing on their genes. To understand the basis of such complex behaviors, we examine the genetic network and neural circuits that regulate rival-induced longer mating duration (LMD). Here we identify a small subset of clock neurons in the male brain that regulate LMD via neuropeptide signaling. LMD requires the function of pigment-dispersing factor (PDF) in four s-LNv neurons and its receptor PDFR in two LNd neurons per hemisphere, as well as the function of neuropeptide F (NPF) in two neurons within the sexually dimorphic LNd region and its receptor NPFR1 in four s-LNv neurons per hemisphere. Moreover, rival exposure modifies the neuronal activities of a subset of clock neurons involved in neuropeptide signaling for LMD.
INTRODUCTION
A primary function of the male sex is to mate with the female to propagate their genes. Male sexual behavior includes copulation and preceding behaviors to locate and detect a female, assess her suitability, and induce her to respond receptively. Notably, D. melanogaster males react to the presence of other males by prolonging the mating duration to increase their paternity share in the progeny (Bretman et al., 2009; Bretman et al., 2010). Our previous study has revealed that rival-induced prolonged mating, namely Longer-Mating-Duration (LMD), is a plastic behavior dependent on a male’s social context. LMD requires visual input and depends on the activity of a subset of circadian clock neurons that are known to express neuropeptides (Kim et al., 2012), raising the question about the role of neuropeptide signaling in LMD.
Neuropeptides regulate development, growth, feeding, metabolism, reproduction, homeostasis and longevity, and modulate olfaction, locomotor control, learning and memory (Nassel, 2000; Nassel and Winther, 2010). Studies of neuropeptides and their receptors as mediators of social behaviors provide one entry point for exploring the “dark matter” – the underlying cellular basis – of social neuroscience (Insel, 2010). Moreover, neuropeptide modulation of behavioral plasticity is likely an ancient form of neuronal signaling conserved among vertebrates and invertebrates (Beets et al., 2012; Choi et al., 2013; Garrison et al., 2012).
Clock neurons in the fly brain secrete several kinds of neuropeptides (see Figures 1A and S1A for neurons relevant to our study). The first output messenger identified in insect clock neurons is the neuropeptide PDF, utilized by five large lateral ventral clock neurons (l-LNvs) and four of the five small lateral ventral neurons (s-LNvs) in the adult brain hemisphere of Drosophila (Renn et al., 1999). The fifth s-LNv is in close proximity to the l-LNvs and lacks PDF expression. The lateral dorsal neurons (LNds) are located more dorsally, and represent a heterogeneous six-cell cluster per hemisphere expressing different neurotransmitters, such as acetylcholine (indicated by the presence of the choline acetyltransferase), ion transporter peptide (ITP), and the long or short form of neuropeptide F (NPF and sNPF).
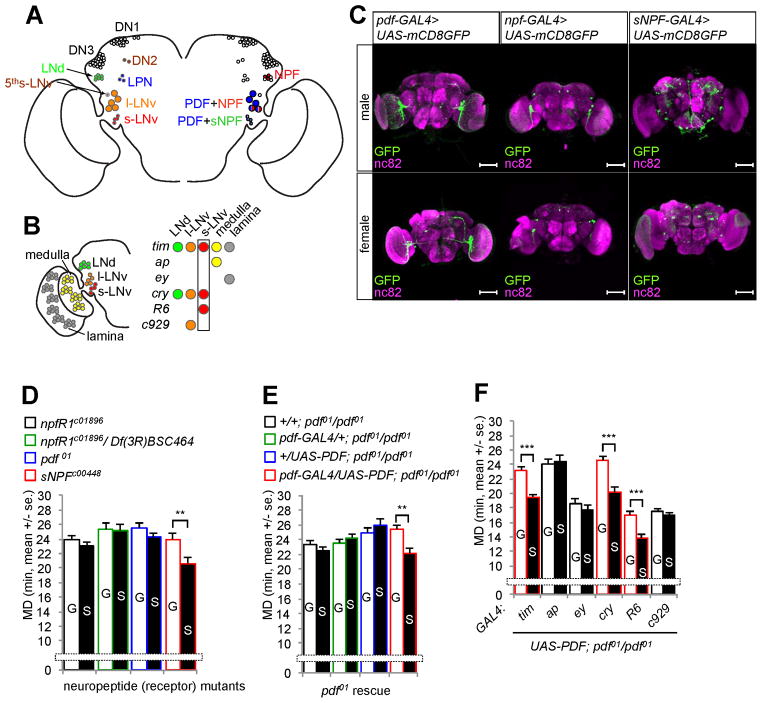
Figure 1.
PDF and NPF are required to generate LMD behavior. (A) Clock neurons (Choi and Nitabach, 2010) and their neuropeptide expression. (B) PDF expression in s-LNv neurons is necessary and sufficient for LMD. Subsets of neurons labeled by GAL4 drivers named in italic are color coded. See Figures S1A and S1B for summary of GAL4 drivers. (C) Male (top) and female (bottom) flies expressing pdf-GAL4, npf-GAL4 or sNPF-GAL4 together with UAS-mCD8GFP were immunostained with anti-GFP (green) and nc82 (magenta) antibodies. Scale bars represent 100 μm. (D) Mating duration assays of neuropeptide or receptor mutants. G for group-reared and S for singly reared males. See Figures S1D and S1E for controls revealing normal courtship latency and indices of these mutants. (E, F) Mating duration assays of pdf rescue experiments. The genotypes are color coded in D and E as specified above the bar graphs, and indicated below the bar graphs in E and F (red marks significant rescue in F). Bars represent mean of the mating duration (MD) with error bars representing the s.e.m. Asterisks represent significant differences revealed by Student’s t test (* p<0.05, ** p<0.01, *** p<0.001). The same symbols for statistical significance are used in all other figures.
Neuropeptide F (NPF or dNPF) shares sequence similarities with the vertebrate neuropeptide Y (NPY) family of peptides that end with tyrosine (Y) amide; NPFs end with phenylalanine (F). In Drosophila, the most prominent NPF-expressing neurons are two large neurons in the protocerebrum per hemisphere that project extensive processes all over the dorsal brain (Krashes et al., 2009). There are 10 NPF neurons in the female fly brain hemisphere and 13 NPF neurons in the male fly brain hemisphere including 3 LNds with male-specific NPF expression; there appear to be no NPF-expressing neurons in the ventral nerve cord (Hermann et al., 2012; Lee et al., 2006).
Neuropeptides bind to receptors of high affinity, hence it is possible for neurons to release neuropeptides and signal other neurons that express the appropriate receptors even if there are no synaptic contacts (Jan and Jan, 1982; Nassel and Winther, 2010). Therefore, for the elucidation of neuropeptide signaling for a specific behavior, it is necessary to identify not only the relevant neuropeptide and receptor but also the neurons that mediate the neuropeptide signaling for the behavior regardless whether they are in synaptic contacts.
The PDF receptor PDFR belongs to the class II (class B) G-protein coupled receptors (GPCRs) (Hyun et al., 2005; Lear et al., 2005; Mertens et al., 2005). In the Drosophila brain, subsets of clock neurons express PDFR (Im et al., 2011; Im and Taghert, 2010). One receptor for NPF (CG10342; npfR1) in Drosophila (Garczynski et al., 2002; Wen et al., 2005) regulates foraging, feeding, alcohol sensitivity, aggression and sexual dimorphism in circadian locomotor activity (Chen et al., 2008; Dierick and Greenspan, 2007; Krashes et al., 2009; Lee et al., 2006; Lingo et al., 2007; Shen and Cai, 2001; Shohat-Ophir et al., 2012; Wen et al., 2005; Wu et al., 2003; Wu et al., 2005a; Wu et al., 2005b). An important aspect of neuropeptide modulation that has not been extensively studied concerns how neuropeptides such as PDF contribute to socially modifiable behaviors (Taghert and Nitabach, 2012).
In this study, we show that both PDF and NPF neuropeptides regulate Longer-Mating-Duration of male flies. In addition to delineating the requisite circuitry by identifying the neurons that express neuropeptide receptors PDFR and NPFR1 to mediate rival-induced prolongation of mating duration, our study has uncovered a number of sexually dimorphic arrangements in the neuronal circuitries, as well as a strong influence of rival exposure over the activities of central neurons, including those implicated in neuromodulation of rival-induced prolonged mating duration.
RESULTS
LMD requires the neuropeptides PDF and NPF, but not sNPF
Longer-Mating-Duration (LMD) is a plastic response of Drosophila males to rivals. When males are housed together with rivals for 5 d before mating, they prolong their mating duration as compared to socially isolated males. Our previous study has found that LMD requires a subset of PDF-expressing neurons (Kim et al., 2012), including s-LNvs and l-LNvs that express not only PDF but also NPF or sNPF (Figure 1A). The relevant GAL4 driver for each of these three neuropeptides labels a subset of cells in the fly brain (Figures 1B and S1B). Unlike pdf-GAL4 and npf-GAL4 drivers, sNPF-GAL4 expression displayed sexual dimorphism (Figure 1C). To test for possible involvement of these neuropeptides, we performed mating duration assays with mutants lacking the neuropeptide or its receptor. Since no npf mutant alleles are available (Nitabach and Taghert, 2008), we tested npfR1 mutants (Burke et al., 2012; Krashes et al., 2009). sNPF mutant males (Lee et al., 2008) but not npfR1 mutant males or pdf mutant males displayed LMD (Figure 1D). In light of the normal courtship index and latency of pdf and npfR1 mutant males (Figures S1D and S1E), PDF and NPF signaling specifically affects LMD rather than the general courtship behavior. These results reveal that the LMD behavior requires the neuropeptides PDF and NPF, but not sNPF.
LMD requires PDF expression in four s-LNv neurons
To identify the neurons that express PDF to mediate LMD, we used different GAL4 drivers to express PDF in various subsets of the ~150 clock neurons in pdf mutants (Figure S1B). LMD was restored in pdf01 mutants expressing the UAS-PDF transgene via pdf-GAL4 driver (Figure 1E), as expected. Expressing PDF broadly in clock neurons via tim-GAL4 or cry-GAL4 also reinstated LMD in pdf01 mutants (Figure 1F), whereas expressing the UAS-PDF transgene within the compound eye via ap-GAL4 or ey-GAL4 was insufficient to restore LMD in pdf01 mutants (Figure 1F). Moreover, expression of UAS-PDF transgene in s-LNvs via R6-GAL4, but not in l-LNvs via c929-GAL4, restored LMD in pdf01 mutants (Figure 1F). These studies reveal that PDF expression in s-LNvs is sufficient to generate LMD (Figure 1B).
LMD requires PDFR expression in two LNd neurons that express CRY
To identify those pdfR-expressing neurons required to generate LMD, we used two independent pdfR-siRNA lines (UAS-pdfR-dsRNA or UAS-pdfR-IR) in combination with various GAL4 drivers to knock down pdfR expression in the GAL4 expressing cells (Figure S1B). LMD was abolished by expression of pdfR-siRNA in all neurons (via the pan-neuronal elav-GAL4 driver) but not in the ellipsoid body (EB) neurons (Figures 2A and S2C), notwithstanding the expression of pdfR-GAL4 drivers in the EB (Im and Taghert, 2010; Parisky et al., 2008). Whereas LMD remained intact with expression of pdfR-siRNA in the PDF-expressing neurons (via pdf-GAL4) or in a subset of dorsal neurons (via Clk4.1M-GAL4 or Clk4.5F-GAL4 drivers), LMD was abolished by expression of pdfR-siRNA in CRY-positive cells, which include most of the lateral neurons and a small subset of dorsal neurons (Figures 2A and S2C). Furthermore, knockdown of pdfR expression in CRY-positive and PDF-negative cells using cry-GAL4; pdf-GAL80 driver was sufficient to disrupt LMD (cry; pdf-GAL80 in Figures 2A and S2C). Thus, PDFR expression in neurons that express CRY but not PDF is required for LMD.
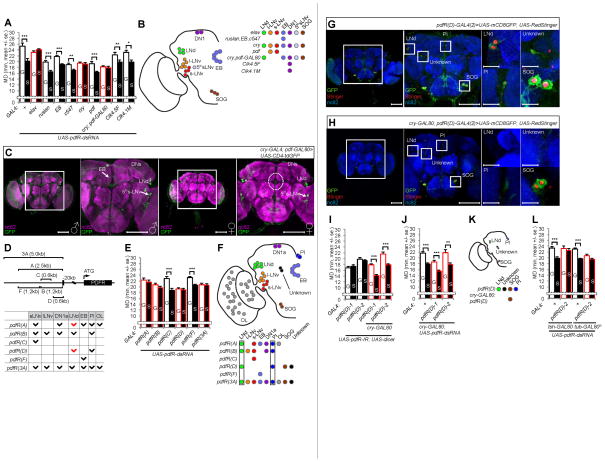
Figure 2.
pdfR expression in a subset of clock neurons is required to generate LMD. (A) Mating duration assays for GAL4 driven knockdown of PDFR via UAS-pdfR-dsRNA. The GAL4 drivers are specified below the bars, and those that affect LMD are marked with red outline, for panels A and E. See Figure S2C for similar results with UAS-pdfR-IR; UAS-dicer. (B) Schematic diagram of GAL4 drivers employed in panel A to identify cells with PDFR expression important for LMD. Subsets of neurons labeled by GAL4 drivers (in italic) are color coded. (C) Flies bearing cry-GAL4; pdf-GAL80 and UAS-CD4-tdGFP flies were immunostained with anti-GFP (green) and nc82 (magenta) antibodies to reveal neuronal projections. Enlarged images of the boxed regions are shown in the panels to the right. Scale bars represent 100 μm. See Figures S2A, S2B and S2D for further characterization of expression patterns. (D) A schematic of the captured promoter region in each pdfR-GAL4 construct (Im and Taghert, 2010; Parisky et al., 2008) (top) and a summary of expression patterns (bottom, known expression marked in black and expression determined in this study marked in red). (E) Mating duration assays for pdfR-GAL4 mediated knockdown of PDFR via UAS-pdfR-dsRNA. See Figure S2Q for similar results with UAS-pdfR-IR; UAS-dicer. GAL4 control experiments are shown in Figure S2P. (F) Implication of PDFR expression in LNd and PI neurons for LMD. Subsets of neurons labeled by GAL4 drivers (in italic) used in panel E are color coded. (G–H) Flies expressing pdfR(D)-GAL4(2) or cry-GAL80; pdfR(D)-GAL4(2) together with UAS-mCD8GFP, UAS-RedStinger were immunostained with anti-GFP (green), anti-DsRed (red) and nc82 (blue) antibodies. Scale bars represent 100 μm in the left 2 panels and 10 μm in the magnified right panels. See Figures S2F–G for additional characterization of expression patterns. (I) Mating duration assays for pdfR(D)-GAL4 mediated knockdown of PDFR via UAS-pdfR-IR; UAS-dicer. GAL4 and Names of the GAL4 drivers and GAL80 are indicated below the bars and GAL80 combinations that did not affect LMD are marked with red outline for panels I and J. GAL4 control experiments are shown in Figure S3I. (J) Mating duration assays for pdfR(D)-GAL4 mediated knockdown of PDFR via UAS-pdfR-dsRNA. (K) Schematic of cells tested in panels I and J. (L) Mating duration assays for pdfR(D)-GAL4(2) mediated knockdown of PDFR via UAS-pdfR-dsRNA together with tsh-GAL80 or tub-GAL80ts. Names of the GAL4 drivers and GAL80 are indicated below the bars and those combinations that affect LMD are marked with red outline. Mean ± standard error (s.e.m) are shown.
To identify those neurons that must express PDFR to mediate the LMD, we characterized the expression pattern driven by the cry-GAL4; pdf-GAL80 driver using the reporter UAS-CD4-tdGFP (a cell membrane marker) (Figure 2C) (Han et al., 2011), or UAS-Denmark (a dendritic marker) together with UAS-sytGFP (a presynaptic marker) (Figure S2A) (Nicolai et al., 2010), or UAS-RedStinger (a nuclear marker) (Figure S2B). We identified ~20–25 cells that are labeled by the cry-GAL4; pdf-GAL80 driver, including 4 LNds, the 5th s-LNv, a subset of dorsal cluster neurons, neurons in the SOG (subesophageal ganglion) and the EB in the male fly brain (Figures 2C and S2A), indicating the pdfR activity in these ~20–25 clock neurons per hemisphere is important for LMD (Figure 2B).
These CRY-positive but PDF negative neurons appear to be in brain regions interconnected via their processes. The processes of neurons expressing reporters driven by cry-GAL4; pdf-GAL80 bridged the LNds and the 5th s-LNv (Figures 2C and S2A). The UAS-CD4-tdGFP expression in the EB of male fly brains (Figure 2C) corresponds to pre-synaptic terminals of neurons labeled by the cry-GAL4; pdf-GAL80 driver, because the UAS-sytGFP expression via this driver gave a similar pattern in the EB (Figure S2A). Sexual dimorphism is evident from the absence of such neuronal projection in the EB of the female brain (Figure 2C), and from the presence of 4 CRY-positive LNd neurons in the male brain but only 2 in the female brain (Figure S2B). The male-specific projection of CRY-positive neurons to the EB is of particular interest, in light of the requirement of the EB for visual memory to generate LMD (Kim et al., 2012).
To further narrow down the pdfR expressing neurons involved in LMD, we used different pdfR-GAL4 lines to knock down the pdfR expression in small subsets of neurons (Figure 2D). RNAi-mediated pdfR knock-down via pdfR(C)-GAL4 or pdfR(F)-GAL4, which drives expression in the s-LNv or EB (Figures S2M and S2N), had no effect on LMD (Figures 2E and S2Q). Of the GAL4 drivers that were effective in suppressing pdfR expression to eliminate LMD, the pdfR(D)-GAL4 driver contains a minimal region of the pdfR-promoter (Figure 2D). Among two independent insertion lines, pdfR(D)-GAL4(1) and pdfR(D)-GAL4(2), we chose pdfR(D)-GAL4(2) since this driver labels a smaller number of cells (Figure S2O). The 15 cells per brain that are labeled by pdfR(D)-GAL4(2) can be divided into 4 different subsets of cells in both the male (Figure 2G) and female brain (Figure S2E): 2 LNds, 3 SOG neurons and 2 uncharacterized neurons near the antennal lobe (AL) per hemisphere, and one neuron in the central PI (pars intercerebralis) (Figure 2G). When this GAL4 driver was combined with cry-GAL80, UAS-mCD8GFP and UAS-RedStinger, expression was restricted to the SOG region (Figure 2H). Expression of pdfR-siRNA by pdfR(D)-GAL4(1) or pdfR(D)-GAL4(2) combined with cry-GAL80 did not affect LMD (Figures 2I and 2J), indicating the PDFR expression in the SOG is not required for LMD. Using tsh-GAL80 to block GAL4 expression in the ventral nerve cord (VNC), we confirmed that expressing pdfR-siRNA with the pdfR(D)-GAL4(2) driver within the brain is sufficient to impair LMD (Figure 2L). Adult-specific expression of pdfR-siRNA with pdfR(D)-GAL4(2) via tub-GAL80ts was also sufficient to impair LMD (Figure 2L). These data suggest that PDFR activity in LNd or PI cells labeled by pdfR(D)-GAL4(2) might modulate LMD (Figure 2F).
To validate that PDFR activity in a subset of PDFR expressing cells is required for LMD, we used various GAL4 drivers to express PDFR in different subsets of clock neurons in pdfR mutants to see which ones can rescue the LMD phenotype. With the pdfR gene on the X-chromosome and the w− mutation in pdfRhans5304 mutants, we introduced the white gene from a short X-chromosome duplication inserted in the third chromosome to rescue the visual deficiency (Popodi et al., 2010) and the LMD defects of w− mutants (Figure S3B). LMD was restored in pdfRhans5304 mutants expressing the UAS-pdfR transgene in broad clock neuron populations via tim-GAL4 or cry-GAL4 drivers (Figure 3A) or in a subset of PDFR expressing neurons via the pdfR(D)-GAL4(2) driver (Figure 3B). However, there was no rescue of LMD if the UAS-pdfR transgene was expressed in PDF-expressing neurons or the visual system, by using a pdf-GAL4 or ap-GAL4 driver respectively (Figures 3A and S1B). Adult specific expression of UAS-pdfR transgene in CRY-positive cells using tub-GAL80ts together with cry-GAL4 was sufficient to rescue LMD in pdfRhans5304 mutants (Figure 3C). Thus, PDFR function in cells that express both PDFR and CRY in the adult male brain, including LNd and PI neurons, is sufficient to generate LMD (Figure 2K).
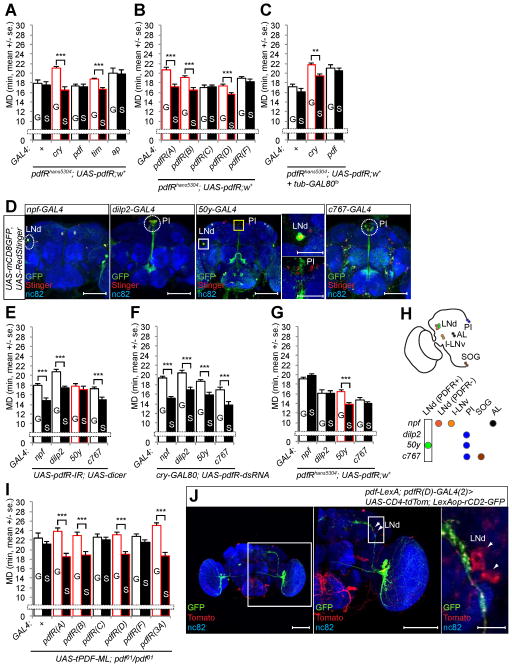
Figure 3.
pdfR expression in LNd is required to generate LMD. (A–B) Mating duration assays of pdfR rescue experiments. Names of the GAL4 drivers are indicated below the bars and those that rescue LMD in pdfRhans5304 mutant are marked with red outline for panels A–C. (C) Mating duration assays of pdfR rescue experiments in adult specific expression of UAS-pdfR transgene. (D) Flies expressing each GAL4 drivers together with UAS-mCD8GFP, UAS-RedStinger were immunostained with anti-GFP (green), anti-DsRed (red) and nc82 (blue) antibodies. White dashed circles indicate LNd and PI neurons. Scale bars represent 100 μm. See Figures S2K–L for additional characterization of expression patterns. (E) Mating duration assays for LNd and PI expressing GAL4 drivers mediated knockdown of PDFR via UAS-pdfR-IR; UAS-dicer. Names of the GAL4 drivers are indicated below the bars and those that affect LMD are marked with red outline. GAL4 control experiments are shown in Figure S3J. (F) Mating duration assays for LNd and PI expressing GAL4 drivers mediated knockdown of PDFR in CRY-negative neurons via cry-GAL80; UAS-pdfR-dsRNA. (G) Mating duration assays of pdfR rescue experiments with LNd and PI expressing GAL4 drivers. Names of the GAL4 drivers are indicated below the bars and the GAL4 driver that rescues LMD in pdfRhans5304 mutant is marked with red outline. (H) Identification of LNd neurons with PDFR expression that is necessary and sufficient for LMD. Subsets of neurons labeled by GAL4 drivers (in italic) are color coded. (I) Mating duration assays of pdf rescue experiments with membrane-tethered UAS-tPDF transgene. GAL4 drivers that rescue LMD in pdf01 mutant are marked with red outline. (J) Flies expressing pdf-LexA; pdfR(D)-GAL4(2) drivers together with UAS-CD4-tdTomato; LexAop-rCD2-GFP were immunostained with anti-GFP (green), anti-DsRed (red) and nc82 (blue) antibodies. White arrowheads indicate LNd neurons labeled by pdfR(D)-GAL4(2) driver. Scale bars represent 100 μm in the left and middle panels and 10 μm in the magnified right panel. See Figure S3F for the absence of synapses detectable via GRASP. Mean ± standard error (s.e.m) are shown.
To determine which subset of cells labeled by pdfR(D)-GAL4(2) are required to generate LMD, we used several GAL4 drivers which label LNd neurons or PI cells (Figure S1B). Expression of UAS-mCD8GFP or UAS-RedStinger via these GAL4 drivers confirmed that npf-GAL4 labels a subset of LNd neurons, but not PI cells, dilp2-GAL4 specifically labels PI neurons but not LNd neurons, 50y-GAL4 labels a subset of LNd neurons and PI cells, and c767-GAL4 labels a subset of PI cells but not LNd neurons (Figures 3D and S1B). Since LMD was not affected by expressing pdfR-siRNA via GAL4 drivers that label only PI cells (dilp2-GAL4, c767-GAL4), and UAS-pdfR expression via these GAL4 drivers did not rescue the impaired LMD of pdfRhans5304 mutants (Figures 3E and 3G), PDFR activity in PI cells is dispensable for LMD. Only 50y-GAL4 combined with pdfR-siRNA was effective in abolishing LMD (Figure 3E), and expression of UAS-pdfR transgene with 50y-GAL4 was sufficient to rescue the impaired LMD in pdfRhans5304 mutants (Figure 3G). Adding cry-GAL80 to 50y-GAL4 and pdfR-siRNA had no effect on LMD (Figure 3F), indicating pdfR expression in CRY-positive neurons is crucial for LMD. We further confirmed that two of the LNd neurons labeled by the 50y-GAL4 driver are also CRY-positive by using cry-GAL80 (Figures S2K and S2L). Thus, PDFR function in two CRY-positive LNd neurons labeled by the 50y-GAL4 driver but not PI neurons labeled by dilp2-GAL4, 50y-GAL4 and c767-GAL4 drivers is necessary and sufficient to generate LMD (Figure 3H).
PDF likely diffuses to reach PDFR expressing neurons not in synaptic contact with PDF expressing neurons required for LMD
PDF could be released in a paracrine fashion to activate PDFR (Hyun et al., 2005; Lear et al., 2005; Mertens et al., 2005). Since PDFR activity in two LNd neurons is necessary and sufficient to generate LMD via PDF signals from four s-LNv neurons per hemisphere, we designed additional rescue experiments with a membrane-tethered form of PDF (UAS-tPDF-ML) (Choi et al., 2009). Interestingly, expression of UAS-tPDF-ML in pdfR(D)-GAL4(2) expressing cells in pdf01 mutants rescued the impaired LMD (Figure 3I), indicating that PDFR activation by membrane-tethered PDF expressed in a subset of PDFR expressing cells is sufficient to generate LMD. Whereas these cells normally do not express PDF (Figure S1B), a subset of LNd neurons labeled with red fluorescence by pdfR(D)-GAL4(2) are in the vicinity of the PDF-expressing s-LNv neurons labeled with green fluorescence (Figure 3J). To look for evidence of synaptic contacts between LNd neurons and PDF-expressing neurons, we adopted the GFP reconstitution across synaptic partners (GRASP) approach involving the expression of two halves of a split-GFP on the cell membrane of distinct neuronal populations (Feinberg et al., 2008). Expression of one half of the split-GFP (UAS-CD4::spGFP1-10) under the control of pdfR(D)-GAL4(2) and the other half (LexAop-CD4::spGFP11) under the control of pdf-LexA did not yield GFP-positive signals (Figure S3F) in spite of the physical proximity of the dorsal horn of PDF-expressing s-LNv neurons and the cell bodies of pdfR(D)-GAL4(2)-labeled LNd (Figures 3J and S2G). In accordance with the hypothesis that PDF is released in a paracrine manner from varicosities (Helfrich-Forster et al., 2007), our findings suggest that PDFR in a small subset of LNd neurons may be activated by PDF released by nearby s-LNv axons that do not make direct synaptic contacts with them to generate LMD.
LMD requires NPFR1 expression in the four PDF-positive s-LNv neurons
Next, we asked which subsets of neurons with npfR1 expression are required to generate LMD, by expressing npfR1-siRNA (UAS-npfR1-dsRNA) via various GAL4 drivers (Figure S1B). Expression of npfR1-siRNA in all neurons (via the pan-neuronal elav-GAL4 driver) but not in the EB (via the c547-GAL4 driver) or a subset of dorsal neurons (via the Clk4.1M-GAL4 driver) eliminated LMD (Figure 4A). LMD was abolished by expression of npfR1-siRNA in the CRY-positive cells, which include most of the lateral neurons and a small subset of dorsal neurons (cry in Figure 4A), and by expression of npfR1-siRNA in the PDF-expressing neurons (pdf in Figure 4A). However, knockdown of npfR1 expression in the CRY-positive and PDF-negative cells had no effect on LMD (cry-GAL4; pdf-GAL80 in Figure 4A). Thus, NPFR1 activity in PDF-expressing neurons is important for LMD.
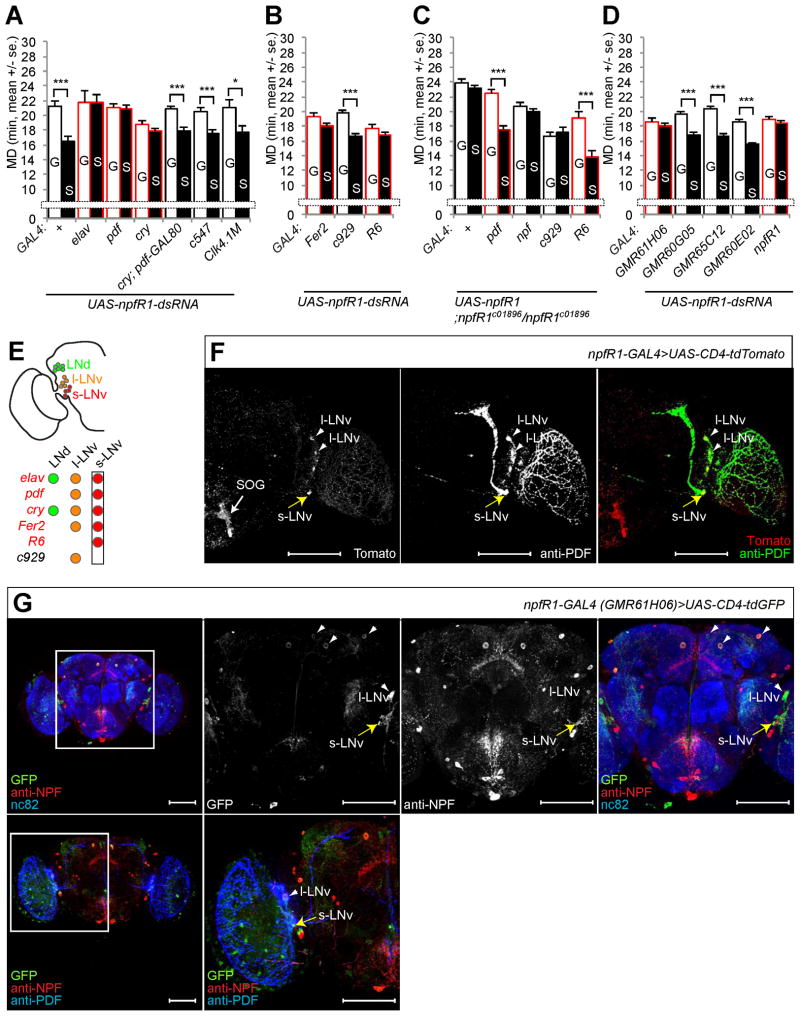
Figure 4.
npfR1 expression in s-LNv is required to generate LMD. (A) Mating duration assays for GAL4 mediated knockdown of NPFR1 via UAS-npfR1-dsRNA. GAL4 drivers that affect LMD are marked in red. Names of the GAL4 drivers are indicated below the bars and those that affect LMD are marked with red outline for panels A, B and D. (B) Mating duration assays for GAL4 drivers for NPFR1 knockdown in LNv via UAS-npfR1-dsRNA. GAL4 control experiments are shown in Figure S4F. (C) Mating duration assays of npfR1 rescue experiments. Names of the GAL4 drivers are indicated below the bars and those that rescue LMD in npfR1c01896 mutant are marked with red outline. (D) Mating duration assays for npfR1-GAL4s mediated knockdown of NPFR1 via UAS-npfR1-dsRNA. GAL4 control experiments are shown in Figure S4F. (E) NPFR1 expression in s-LNv neurons is necessary and sufficient for LMD. Subsets of neurons labeled by GAL4 drivers (in italic) are color coded. (F) Flies expressing npfR1-GAL4 driver (Wen et al., 2005) together with UAS-CD4-tdTomato were immunostained with anti-PDF (green) and anti-DsRed (red) antibodies. White arrowheads indicate l-LNv region labeled by both npfR1-GAL4 driver and anti-PDF antibodies. Yellow arrow indicates s-LNv region labeled by both npfRF1-GAL4 and anti-PDF antibodies. White arrow indicates SOG region labeled by npfRF1-GAL4. (G) Flies expressing GMR61H06-GAL4 driver together with UAS-CD4-tdTomato were immunostained with anti-GFP (green), anti-NPF (red) and anti-nc82 (blue) antibodies (top panels) or with anti-GFP (green), anti-NPF (red) and anti-PDF (blue) antibodies (bottom panels). White arrowheads indicate l-LNv region labeled by both GMR61H06-GAL4 driver and anti-PDF antibodies. Yellow arrow indicates s-LNv region labeled by both GMR61H06-GAL4 and PDF antibodies. Scale bars represent 100 μm. See Figures S4A–D for additional characterizations of expression patterns. Mean ± standard error (s.e.m) are shown.
To identify the PDF-expressing neurons that are important to generate LMD, we used three different GAL4 drivers which specifically label both s-LNv and l-LNv (Fer2-GAL4, Fer2 expression is enriched in lateral ventral neurons) (Nagoshi et al., 2010), only l-LNv (c929-GAL4) (O’Brien and Taghert, 1998), or only s-LNv (R6-GAL4) (Helfrich-Forster et al., 2007) (Figure S1B). We found that npfR1-siRNA expression in s-LNv (Fer2 and R6 in Figure 4B) but not in l-LNv (c929 in Figure 4B) compromised LMD, indicating that the NPFR1 activity in s-LNv is necessary for LMD (Figure 4E).
To determine whether the NPFR1 activity in s-LNv is sufficient to generate LMD, we used different GAL4 drivers to express NPFR1 in various subsets of circadian clock neurons in npfR1 mutants. LMD was restored in npfR1c01896 mutants expressing the UAS-npfR1 transgene in s-LNv neurons via pdf-GAL4 or R6-GAL4 (Figure 4C). In contrast, expression of the UAS-npfR1 transgene in NPF-expressing neurons or only in l-LNv via npf-GAL4 or c929-GAL4 could not rescue the LMD (Figure 4C). Thus, the NPFR1 function in s-LNv neurons is sufficient to generate LMD (Figure 4E).
To validate the expression of various GAL4 drivers in s-LNv neurons, we expressed UAS-CD4-tdGFP via the npfR1-GAL4 driver (Wen et al., 2005). Although this GAL4 driver produces weak expression of the fluorescence protein reporter, we confirmed the expression in s-LNv and l-LNv by using an anti-PDF antibody (Wen et al., 2005) (Figure 4F). We also collected the Janelia Farm GAL4 lines with GAL4 driven by the npfR1 promoter and coding regions (Pfeiffer et al., 2008). Among these GAL4 lines, GMR61H06 labeled s-LNv in both male brains (Figure 4G) and female brains (Figure S4A). The other GAL4 drivers did not label s-LNv (Figures S4B, S4C, and S4D). LMD was eliminated by expressing npfR1-siRNA via npfR1-GAL4 or GMR61H06, but not the other GAL4 drivers without s-LNv expression (Figure 4D). In summary, expression of NPFR1 in 4 PDF-positive s-LNv neurons is necessary and sufficient to generate LMD (Figure 4E).
Two male-specific NPF expressing LNd neurons are required for LMD
Sexual dimorphism of brain structure and function generates neural circuitries important for gender-specific behaviors. In Drosophila, fruitless (fru) is an essential neural sex determinant responsible for male-specific behaviors (Ryner et al., 1996). It is also well known that a subset of clock neurons is sexually dimorphic (Kadener et al., 2006; Lee et al., 2006). We therefore tested whether LMD depends on the function of a subset of sexually dimorphic LNd neurons.
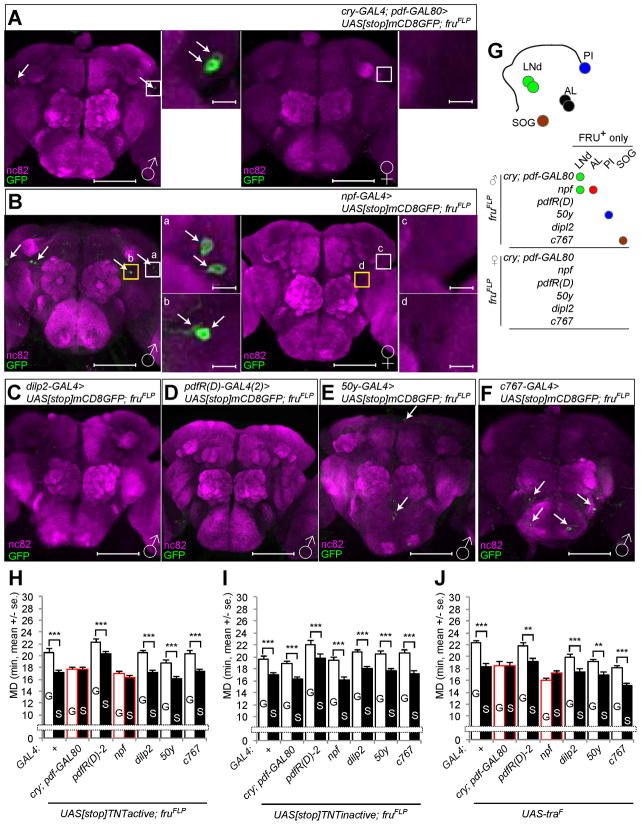
To determine whether sexually dimorphic neurons are involved in LMD, we used intersectional methods to genetically dissect ~1500 fru neurons into smaller subsets. We used a combination of the fruFLP allele that drives FLP-mediated recombination specifically in fru neurons with UAS[stop]X (X could be various reporters or effector transgenes) to express a UAS transgene in only those cells that are labeled by the GAL4 driver and also fru-positive, due to FLP-mediated excision of the stop cassette ([stop]). Using cry-GAL4; pdf-GAL80 to identify sexually dimorphic cells, we found 2 male-specific cells in the LNd region per hemisphere (Figure 5A). We also found that npf-GAL4 labeled 2 fru-positive LNd neurons and 2 fru-positive cells in the antenna lobe of the male but not female brain (Figure 5B). In contrast, we could not find any fru-positive central neurons that are labeled by dilp2-GAL4 or pdfR(D)-GAL4(2) drivers (Figures 5C and 5D). A subset of cells labeled by 50y-GAL4 and c767-GAL4 drivers were fru-positive, however, these cells seemed unlikely to be involved given their location far from those PDF, PDFR, NPF and NFR1 expressing neurons implicated in LMD (Figures 5E and 5F). Since LNd neurons labeled by 50y-GAL4 (Figure 3D) did not emerge in the fruFLP experiment, we concluded that the 50y-GAL4 labeled LNd neurons are different from the NPF- and FRU-positive LNd neurons (Figure 5G).
Figure 5.
Male specific FRU-positive subsets of LNd neurons are required to generate LMD. (A–F) Flies expressing each of the GAL4 drivers together with UAS[stop]mCD8GFP; fruFLP were immunostained with anti-GFP (green) and nc82 (magenta) antibodies. White arrows indicate fruitless-positive neurons. Scale bars represent 100 μm in the left panels and 10 μm in the magnified right panels. See Figures S6F–G for dendrites and synaptic terminals labeled both by npf-GAL4 and fruFLP. (G) Schematic diagram of sexually dimorphic cells. Subsets of neurons labeled by GAL4 drivers (in italic) in combination with fruFLP are color coded. (H–I) Mating duration assays for GAL4 drivers for inactivation of synaptic transmission via UAS[stop]TNTactive; fruFLP (H) or control experiments with UAS[stop]TNTinactive; fruFLP (I). Names of the GAL4 drivers are indicated below the bars and those that affect LMD are marked with red outline for panels H–J. (J) Mating duration assays for GAL4 drivers for feminization of neurons via UAS-traF. See Figures S5A–G for expression pattern characterizations. Mean ± standard error (s.e.m) are shown.
To test whether the small subset of fru-positive LNd cells are involved in LMD, we expressed tetanus toxin light chain (UAS[stop]TNTactive) with various GAL4 drivers along with fruFLP to inhibit synaptic transmission in sexually dimorphic subsets of fru-positive cells. As a control, we found that LMD was unaffected when we used each of these GAL4 drivers in combination with UAS[stop]TNTinactive to express an inactive form of tetanus toxin light chain (Figure 5I). LMD was abolished by inhibition of transmitter release from cry-GAL4; pdf-GAL80 or npf-GAL4 labeled fru-positive cells, whereas blocking synaptic transmission of the other GAL4 labeled fru-positive cells did not affect LMD (Figure 5H). Similar results (Figure S3C) were obtained using the cell autonomous toxin Ricin A (UAS[stop]RicinA) which encodes only the catalytic subunit of the toxin that can enter the cell to mediate cell ablation (Hidalgo et al., 1995). These findings indicate that LMD requires a subset of sexually dimorphic neurons labeled by cry-GAL4; pdf-GAL80 or npf-GAL4 drivers (Figure 5G).
Systemic expression of a female form of tra cDNA (UAS-traF) in a male during development elicits female characteristics (Belote and Baker, 1987). Moreover, male-specific NPF expression is eliminated when npf-GAL4 labeled cells are feminized by UAS-traF (Lee et al., 2006). In an attempt to express UAS-traF under the control of various GAL4 drivers to feminize different subsets of neurons (Figure S5A–G), we found that LMD was eliminated by such feminization of cry-GAL4; pdf-GAL80 or npf-GAL4 labeled cells (Figure 5J), but not by expression of UAS-traF in other neuronal subsets (Figure 5J) or in PDF-expressing neurons or EB neurons via the pdf-GAL4 or c547-GAL4 driver (Figure S3D). These data suggest that LMD requires the masculine functions of a subset of neurons labeled by cry-GAL4; pdf-GAL80 or npf-GAL4 drivers.
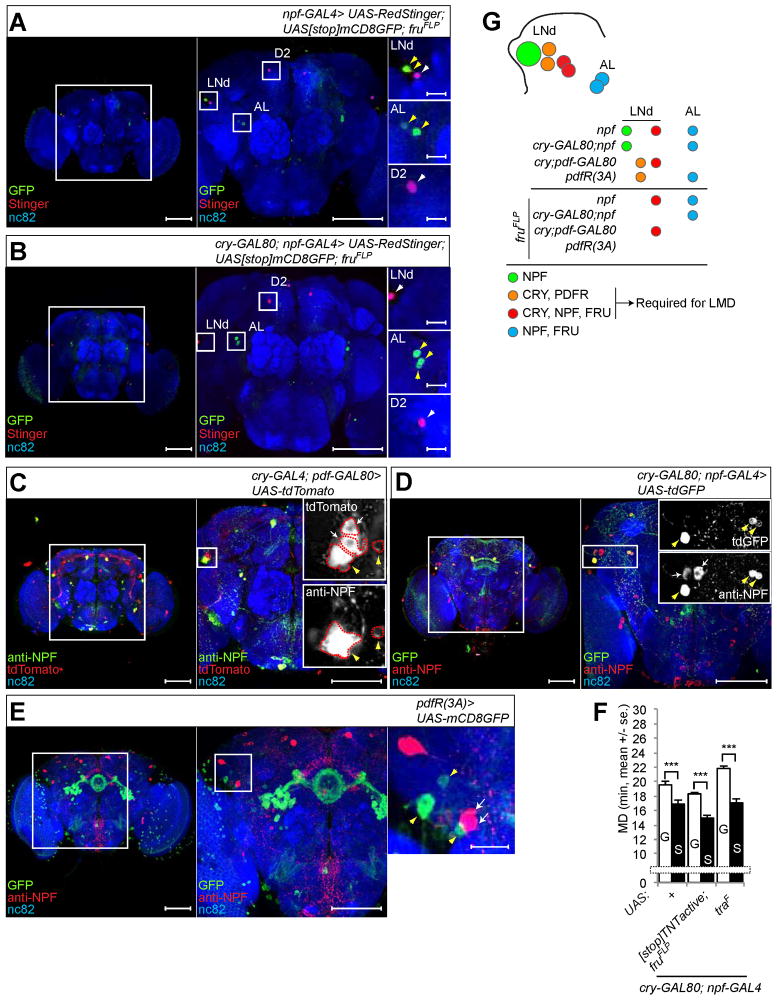
Next, we asked whether npf-GAL4 labeled fru-positive LNd neurons are cry-positive. First, we expressed UAS-RedStinger, UAS[stop]mCD8GFP in combination with fruFLP, so that the nuclear RedStinger labels all cells expressing a particular GAL4 driver, but the mCD8GFP labels only those GAL4 expressing cells that are fru-positive. Among the ~12 cells labeled by npf-GAL4 in each hemisphere, only two LNd neurons and two cells near the antenna lobe were fru-positive (Figure 6A). Combining npf-GAL4 with cry-GAL80 reduced the number of cells expressing RedStinger and eliminated fru-positive cells in the LNd region (Figure 6B). To further validate that some of the LNd neurons are cry-positive and npf-positive, we expressed UAS-CD4-tdTomato via the cry-GAL4; pdf-GAL80 driver and then used anti-NPF antibody to identify two NPF-expressing cells among the four cells that were labeled by cry-GAL4; pdf-GAL80 per hemisphere (Figure 6C). Only one npf-positive cell in the LNd region was cry-negative (Figure 6D). Since this cell was not labeled in the fruFLP experiments, we conclude that only two cells are both npf-positive and cry-positive, and they are the fru-positive sexually dimorphic neurons in the LNd cluster (Figure 6G). We could not detect any cells that are both npf-positive and pdfR-positive in the LNd region (Figure 6E). Since pdfR(D)-GAL4(2) labeled cells in the LNd region were not fru-positive (Figure 5D), we conclude that the two fru-positive, cry-positive and npf-positive cells are distinct from the two cry-positive and pdfR-positive cells in the LNd region (Figure 6G). In summary, we observe at least 3 types of cells in the LNd region: one cell positive for npf but negative for cry, fru and pdfR (Figures 6B and 6D), two cells positive for pdfR and cry but negative for npf and fru (Figures 5D and 6E), two cells positive for npf, cry and fru but negative for pdfR (Figures 5A and 6B–E, and Figure 6G for schematic diagram).
Figure 6.
NPF- and FRU-positive neurons that are required for LMD are CRY-positive but PDFR-negative. (A–B) FRU-positive neurons labeled by npf-GAL4 (A) or cry-GAL80; npf-GAL4 driver (B). Flies expressing each GAL4 drivers together with UAS-RedStinger; UAS[stop]mCD8GFP; fruFLP were immunostained with anti-GFP (green) anti-DsRed (red) and nc82 (blue) antibodies. White arrowheads indicate GAL4 labeled LNd neurons and yellow arrowheads indicate additional FRU-positive neurons labeled by GAL4 driver. Scale bars represent 100 μm in the left 2 panels and 10 μm in the magnified right panels. (C) Flies expressing cry-GAL4; pdf-GAL80 driver and UAS-CD4-tdTomato were immunostained with anti-NPF (green) anti-DsRed (red) and nc82 (blue) antibodies. White arrows indicate CRY-positive, NPF-negative neurons labeled by cry-GAL4; pdf-GAL80 driver among the LNd neurons and yellow arrowheads indicate CRY- and NPF-positive neurons labeled by the same driver. LNd in the boxed region is magnified to clearly show two CRY- and NPF-positive neurons (yellow arrowheads) and CRY-negative and NPF-positive neurons (white arrows) with red dotted outlines. Scale bars represent 100 μm. (D) Flies expressing cry-GAL80; npf-GAL4 driver and UAS-CD4-tdGFP were immunostained with anti-GFP (green) anti-NPF (red) and nc82 (blue) antibodies. White arrows indicate CRY- and NPF-positive neurons labeled by cry-GAL80; npf-GAL4 driver among the LNd neurons and yellow arrowhead indicates CRY-negative and NPF-positive neurons labeled by the same driver, in the enlarged images corresponding to the boxed region in the right panel. Scale bars represent 100 μm. (E) Flies expressing pdfR(3A)-GAL4 and UAS-mCD8GFP were immunostained with anti-GFP (green) anti-NPF (red) and nc82 (blue) antibodies. White arrows indicate NPF-positive and PDFR-negative neurons labeled by pdfR(3A)-GAL4 driver among LNd neurons and yellow arrowheads indicate NPF-negative and PDFR-positive neurons labeled by the same driver. Scale bars represent 100 μm in the left 2 panels and 10 μm in the magnified right panel. See Figures S2H–J, S3E, S4E, S5J–L and S6F, S6G and S6I for additional characterization of expression patterns. (F) Mating duration assays for different UAS transgenes expressed via the cry-GAL80; npf-GAL4 driver. Names of the UAS transgenes are indicated below the bars. (G) Schematic of LNd cells regarding their expression of NPF, CRY, PDFR and FRU, showing LMD requires two neurons positive for CRY and PDFR, and two other neurons positive for CRY, NPF and FRU. Subsets of neurons labeled by GAL4 drivers (in italic) are color coded. Mean ± standard error (s.e.m) are shown.
Next, we tested for the possible involvement of these three types of cells in LMD. LMD was abolished by UAS[stop]TNTactive expression driven by npf-GAL4 (Figure 5H), but not by the more restricted expression imposed by the addition of cry-GAL80 (Figure 6F). Likewise, LMD was not affected by the expression of UAS-traF via npf-GAL4; cry-GAL80 (Figure 6F). We also confirmed that NPF expression in cry- and fru-positive neurons is important for LMD by npf-siRNA (Figure S3A). These studies reveal that LMD does not require the cell in the LNd region that is positive for npf but negative for cry, fru, and pdfR nor does it require the two cells located near the antennal lobe of each hemisphere that are positive for npf and fru but negative for cry and pdfR (see Figures 6G and S7C for schematic diagrams). Thus, LMD requires two NPF expressing neurons in the LNd region that are sexually dimorphic, positive for CRY but negative for PDFR (Figure 6G).
Rival exposure affects the activities of neurons involved in neuropeptide modulation of LMD
To find out whether neuronal activities are altered in neurons involved in association with LMD, we used the CaLexA (calcium-dependent nuclear import of LexA) system (Masuyama et al., 2012), based on the activity dependent nuclear import of the nuclear factor of activated T cells (NFAT), a transcription factor. Since LMD requires chronic exposure to rivals for at least 6–12 h (Kim et al., 2012), the repeated sensory inputs could conceivably result in the accumulation of the engineered transcription factor in the nuclei of active neurons in vivo.
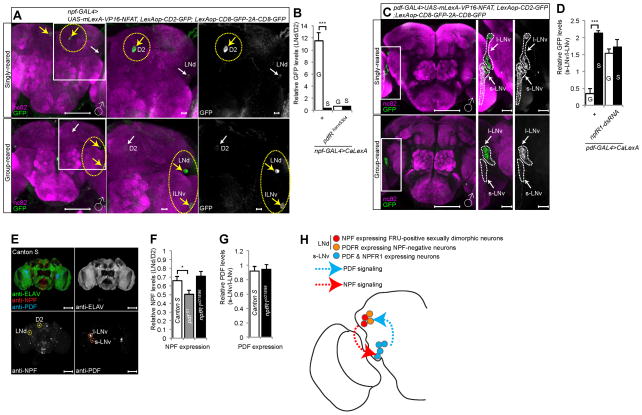
We first examined NPF expressing neurons since they have been implicated in LMD. Indeed, neuronal activities of some npf-GAL4 labeled neurons were altered by the exposure to rivals. Singly reared male flies harboring npf-GAL4 and LexAop-CD2-GFP; UAS-mLexA-VP16-NFAT, LexAop-CD8-GFP-2A-CD8-GFP showed robust fluorescence in the D2 dorsomedial neurons after 5 days of social isolation. In contrast, no such signals could be detected in group-reared males. Instead, we observed fluorescent signals in a subset of LNd and l-LNv neurons in male flies exposed to rivals for 5 days (Figures 7A and 7B). Notably, group rearing increased the fluorescent signals of LNd neurons normalized by those of D2 neurons (LNd/D2 fluorescence) in males (Figures 7A and 7B) but not females (Figures S6A and S6B). Moreover, group rearing did not increase the LNd/D2 fluorescence in pdfRhans5304 mutant males (Figure 7B), indicating that PDF signaling is crucial for this experience-dependent modulation of the activity of NPF expressing neurons. We also failed to detect the experience-dependent increase of LNd/D2 fluorescence when CaLexA was expressed via the cry-GAL80; npf-GAL4 driver (Figures S7D and S7E), indicating that rival exposure enhanced the activity of LNd neurons that are positive for both CRY and NPF.
Figure 7. Rival experience alters the neural activity of neuropeptide expressing neurons.
(A) and (C) show different levels of neural activity of LNd, l-LNv and s-LNv neurons as revealed by the CaLexA system in group-reared versus singly reared male flies. Flies expressing npf-GAL4 (A) or pdf-GAL4 (C) along with LexAop-CD2-GFP; UAS-mLexA-VP16-NFAT, LexAop-CD8-GFP-2A-CD8-GFP were singly reared or group reared for five days. Brains were immunostained with anti-GFP (green) and anti-nc82 (magenta) antibodies. Yellow arrows indicate the subset of neurons showing elevated fluorescence and white arrows indicate the subset of neurons showing reduced fluorescence in that rearing condition (A). Scale bars represent 100 μm in the left panel and 10 μm in the two magnified right panels. (B) and (D) show quantification of GFP fluorescence. GFP fluorescence of LNd region is normalized by that in D2 region (B), and GFP fluorescence in the s-LNv region is normalized by that in the l-LNv region (D). Genotypes of flies were described below the bars. G for group-reared flies and S for singly reared flies. Bars represent mean of the normalized GFP fluorescence level with error bars representing the s.e.m. Asterisks represent significant differences revealed by Student’s t test (* p<0.05, ** p<0.01, *** p<0.001). See Figures S6A–E for expression in female brains and Figures S6H and S7D–E for additional controls. (E) CS male fly brain was immunostained with anti-ELAV (green), anti-NPF (red) and anti-PDF (blue) for quantification of neuropeptide expression levels. Scale bars represent 100 μm. (F–G) Quantification of NPF (F) and PDF (G) expression levels in various fly strains. Genotypes are described inside the bars. (H) Schematic diagram of PDF and NPF signaling amongst clock neurons that are required to generate LMD.
Since PDF-expressing neurons are also critical for generating LMD (Kim et al., 2012), we next used the pdf-GAL4 driver to assess their neuronal activity. The fluorescence intensity of s-LNv normalized by that of l-LNv neurons in male flies harboring pdf-GAL4 and LexAop-CD2-GFP; UAS-mLexA-VP16-NFAT, LexAop-CD8-GFP-2A-CD8-GFP was decreased in group-reared males as compared to singly reared males (Figures 7C and 7D), whereas flies with npfR1 knocked down in these neurons showed no such activity dependence on rearing conditions (Figure 7D), indicating that NPF signaling is essential for the experience-dependent modulation of the activity of PDF expressing neurons. Female flies that were group-reared or singly reared also showed no difference in neuronal activity (Figures S6C and S6D). We could not detect any fluorescence in male (Figure S6H) and female flies (Figure S6E) with the CaLexA system combined with cry-GAL4; pdf-GAL80 or pdfR(D)-GAL4(2) (data not shown), most likely because of the weak GAL4 expression. In summary, we found that exposure to rival males caused an increase of activity of the LNd neurons expressing both NPF and CRY that are required for LMD. Rival exposure also decreased the activity of s-LNv neurons expressing PDF and NPFR, when normalized by the activity of nearby l-LNv neurons.
PDF and NPF signaling for LMD is unlikely to involve modulation of the expression of these neuropeptides
Given that both PDF and NPF neuropeptides are involved in LMD, it is of interest to explore the possibility of potential crosstalk at the level of PDF expression in s-LNv neurons or NPF expression in LNd neurons. The NPF expression levels of LNd neurons normalized by those in D2 neurons were mildly reduced in pdf mutants compared to wild type animals (Figure 7E–F). There was no change of NPF expression levels in npfR1 mutants (Figure 7F). The PDF expression levels of s-LNv neurons normalized by those in l-LNv neurons in npfR1 mutants were comparable to those in wild type controls (Figure 7G). Moreover, we could not observe any differences in the expression levels of NPF or PDF between group-reared and singly reared animals (data not shown). These observations taken together with the CaLexA data support the notion that each neuropeptide modulates the activity of neurons including those expressing the other neuropeptide, without drastically altering the expression of the other neuropeptide.
DISCUSSION
In this study we provide evidence for the crucial involvement of two neuropeptides, PDF and NPF, in the modulation of reproductive behavior by the male’s prior experience with other males. By identifying neurons required for this neuropeptide modulation, we delineate the central neuronal circuitry and find that the crucial neurons expressing a neuropeptide are not in synaptic contact with the crucial neurons expressing its receptor, providing further evidence for the long-range influence of neuropeptides. Remarkably, sharing housing with male rivals alters the activity of a subset of clock neurons including those neurons expressing PDF and NPF that are crucial for this behavioral modulation. We also found that these altered neuronal activities of PDF- and NPF-expressing neurons in group-reared males are dependent on the signaling by NPF and PDF, respectively.
Neuropeptides and their receptors in clock neurons are crucial for rival-induced prolongation of mating duration
LMD requires PDF expression in four s-LNv neurons (Figure 1), and it also requires the expression of the NPF receptor, NPFR1, in those four s-LNv neurons (Figure 4). These four s-LNv neurons thus appear to act in the LMD generation as a relay station to receive NPF neuropeptide signaling and to transmit PDF neuropeptide signaling to neurons expressing the PDF receptor PDFR.
Unlike PDF-expressing neurons with well known functions for circadian rhythm behavior, much less is known about neurons expressing PDFR (Im et al., 2011; Im and Taghert, 2010; Lear et al., 2005; Mertens et al., 2005; Parisky et al., 2008). To search for the PDFR expressing cells involved in LMD, we began by identifying a small number of CRY-positive but PDF-negative neurons required to generate LMD (Figures 2 and S2). We then used various pdfR-GAL4 lines to identify LNd neurons and PI neurons as candidate PDFR expressing neurons (Figure 2). After we ruled out the involvement of PI neurons, we demonstrated that expressing PDFR in LNd neurons of pdfR mutants was sufficient to rescue the LMD deficits (Figure 3). Among this small group of CRY-positive but PDF-negative LNd neurons, two cells that express PDFR but not NPF (Figure 5) and another distinct group of two sexually dimorphic cells that express NPF but not PDFR (Figure 6) in each hemisphere are required for LMD. Moreover, the neuronal activities of these male-specific LNd neurons that express NPF were increased by the exposure to rivals whereas the neuronal activity of PDF expressing s-LNv neurons appeared to be decreased by rival exposure (Figure 7). These four s-LNv neurons also express NPFR1, which is coupled to Gi to mediate inhibition of adenylyl cylcase (Garczynski et al., 2002). Given that the rival exposure-induced alteration of s-LNv neuronal activity requires NPFR1 function (Figure 7D), one plausible scenario is that rival exposure increases the activity of NPF expressing LNd neurons, which release NPF to activate NFPR1 on s-LNv neurons so as to reduce the activity of these PDF expressing s-LNv neurons.
LMD requires PDFR expression in neurons that are not in synaptic contact with crucial neurons with PDF expression
PDF appears to be released in a paracrine fashion to activate the G-protein-coupled receptor PDFR. PDFR is not found in the four s-LNv neurons that express PDF (Hyun et al., 2005; Lear et al., 2005; Mertens et al., 2005). One LNd neuron is known to be PDFR-positive (Hyun et al., 2005), though its PDFR signaling has not been characterized. We found that the two PDFR expressing LNd neurons per hemisphere are crucial for LMD (Figures 2 and 3) and they do not form direct synaptic contact with the s-LNv dorsal projections (Figure 3J and Figure S3F), consistent with the previous report that presynaptic terminals of PDF-expressing neurons have no direct contact with LNd neurons (Helfrich-Forster et al., 2007). Expression of the secreted form of PDF via an s-LNv specific GAL4 driver in pdf01 mutant could rescue the disrupted LMD (Figure 1E), however, expression of a membrane-tethered form of PDF could not (Figure S3H). In contrast, expression of a membrane-tethered form of PDF via pdfR(D)-GAL4(2), with restricted expression in LNd and PI neurons, could rescue the disrupted LMD phenotype of pdf01 mutants (Figure 3I). These results indicate that PDF secreted from s-LNv neurons can activate PDFR in LNd neurons to generate LMD. The dendrites of PDFR-positive LNd neurons labeled by pdfR(D)-GAL4(2) are located near the dorsal projections of PDF-expressing neurons (Figures S2F and S2G). The dendrites of LNd neurons labeled by 50y-GAL4, which could impair LMD when it drives the expression of pdfR-siRNA to reduce PDFR activity in LNd neurons, also are located near these PDF-expressing neuronal projections (Figures 3E and S2K). It has been reported that neuropeptide signaling does not require synaptic contacts. The released peptide may diffuse over tens of micrometers to reach its receptors (Jan and Jan, 1982), and the action of a peptide is limited by dilution as well as degradation/inactivation by membrane-bound peptidases (Nassel and Winther, 2010). Thus, PDF released from s-LNv neuronal projections may signal nearby PDFR-positive LNd neurons via diffusion rather than direct synaptic contact. In summary, we identified 2 PDFR-positive LNd neurons per hemisphere that are responsible for generating LMD via PDF/PDFR signaling. We suggest that PDF released from s-LNv is responsible for PDFR signaling in these LNd neurons (Figures 6G and 7H).
Rival influences the central neuronal activity of subset of neuropeptide expressing neurons dependent on the signaling by the other neuropeptide
Our study reveals that PDF and NPF signaling is crucial for the mating duration that is controlled by the male’s experience with rivals. Moreover, rival exposure greatly reduced the activity of the s-LNv neurons normalized by that of l-LNv neurons both expressing PDF, but increased the activity of LNd neurons normalized by that of D2 neurons both expressing NPF (Figure 7). Interestingly, this increase in neuronal activity of NPF-positive LNd neurons in group-rearing conditions is not observed in pdfR mutant animals. Given that PDFR and NPF are expressed by two distinct populations of LNd neurons, the requirement of PDFR function for the rival-induced modulation of NPF expressing neuronal activity in the LNd region raises the intriguing question whether neuronal signaling perhaps involving another as yet unidentified neuropeptide is involved in LMD.
A recent study has identified four abdominal ganglion (AG) interneurons (INs) that contain the neuropeptide corazonin (Crz) and modulate copulation duration (Tayler et al., 2012). These neurons might play a role as a final set of effectors for the convergent effects of acute (Garbaczewska et al., 2013) and chronic (Bretman et al., 2011; Kim et al., 2012) rival competition on the copulation duration. Elucidating the neural circuitry between these AG neurons and clock neurons would be helpful to further our understanding as to how male flies regulate mating duration in response to rivals.
Recent studies have shown that sexually dimorphic responses to pheromones in the nematode Caenorhabditis elegans may arise from differences in the balance of neural circuits during development (White and Jorgensen, 2012) or in the adult via neuromodulation (Jang et al., 2012). Our study adds to this emerging body of literature illustrating the importance of sexually dimorphic neuromodulation via neuropeptide signaling in social behavior.
EXPERIMENTAL PROCEDURES
Mating Duration Assays
Mating duration assay was described previously (Kim et al., 2012). For group rearing, 4 males from the same strain were placed into a vial with food. Five CS females were collected from bottles and placed into a vial for 5 days. These females provide mating partners for mating duration assays.
Mating duration assay is performed as previously described (Yang et al., 2009). At the fifth day after eclosion, males of the appropriate strain and CS females were mildly anaesthetized by CO2. After placing a single female in to the mating chamber, we inserted a transparent film then placed a single male to the other side of the film in each chamber. After allowing for 1 h of recovery in the mating chamber in a 25°C incubator, we removed the transparent film and recorded the mating activities. Only those males that succeeded to mate within 1 h were included for analyses. Initiation and completion of copulation were recorded with an accuracy of 10 sec, and total mating duration was calculated for each couple. All assays were performed from noon to 4 pm.
Courtship assay
Courtship assay was performed as previously described (Ejima and Griffith, 2007), under normal light conditions in circular courtship arenas 11 mm in diameter, from noon to 4 pm. Courtship latency is the time between female introduction and the first obvious male courtship behavior such as orientation coupled with wing extensions. Once courtship began, courtship index was calculated as the fraction of time a male spent in any courtship-related activity during a 10 min period or until mating occurred.
Immunostaining and antibodies
As described before (Lee and Luo, 1999), brains dissected from adults 5 days after eclosion were fixed in 4% formaldehyde for 30 min at room temperature, washed with 1% PBT three times (30 min each) and blocked in 5% normal donkey serum for 30 min. The brains were then incubated with primary antibodies in 1% PBT at 4°C overnight followed with fluorophore-conjugated secondary antibodies for 1 hour at room temperature. Brains were mounted with anti-fade mounting solution (Invitrogen, catalog #S2828) on slides for imaging. Primary antibodies: chicken anti-GFP (Aves Labs, 1:1000), rabbit anti-DsRed express (Clontech, 1:250), mouse anti-Bruchpilot (nc82) (DSHB, 1:50), mouse anti-PDF (DSHB, 1:100), and rabbit anti-NPF (gift from Dr. Ping Shen, 1:2000). Fluorophore-conjugated secondary antibodies: Alexa Fluor 488-conjugated goat anti-chicken (Invitrogen, 1:100), Alexa Fluor 488-conjugated donkey anti-rabbit (Invitrogen, 1:100), RRX-conjugated donkey anti-rabbit (Jackson Lab, 1:100), RRX-conjugated donkey anti-mouse (Jackson Lab, 1:100), Dylight 649-conjugated donkey anti-mouse (Jackson Lab, 1:100).
Quantitative Analysis of GFP Fluorescence
To quantify PDF and NPF levels in brain regions, we measured PDF or NPF immunofluorescence using the histogram tool of ImageJ (National Institutes of Health, Bethesda, MD; http://rsb.info.nih.gov/ij). Fluorescence was quantified in a manually set region of interest (ROI) of the D2 region or LNd region (for NPF expression) and s-LNv region or l-LNv region (for PDF expression). To compensate for differences in immunofluorescence between different ROI, PDF or NPF fluorescence was normalized to the fluorescence of anti-ELAV staining. GFP fluorescence for CaLexA was normalized to nc82 staining, and then the fluorescence of ROI was quantified using the histogram tool of ImageJ. Both hemispheres of six fly brains were analyzed (total 12) for statistical analysis. All specimens were imaged under identical conditions.
Statistical Analysis
Statistical analysis of mating duration assay was described previously (Kim et al., 2012). More than 36 males (group- or singly reared) were used for mating duration assay. Our experience suggests that the relative mating duration differences between group- and singly reared are always consistent; however, both absolute values and the magnitude of the difference in each strain can vary. So we always include internal controls for each treatment as suggested by previous studies (Bretman et al., 2011). Therefore, statistical comparisons were made between groups that were exposed or not exposed to rivals within each experiment. As mating duration of males showed normal distribution (Kolmogorov-Smirnov tests, p > 0.05), we used two-sided Student’s t tests. Each figure shows the mean ± standard error (s.e.m) (*** = p < 0.001, ** = p < 0.01, * = p < 0.05).
Supplementary Material
Acknowledgments
We thank Drs. Alex Keene, Justin Blau, Charles Choi, Michael N Nitabach, Seol Hee Im, Paul H Taghert, Leslie C. Griffith, Gero Miesenböck, Ravi Allada, Eric Rulifson, Michael Rosbash, Barry Dickson, Jing Wang, J. Douglas Armstrong, Chun Han and Peter Soba for kindly providing valuable flies. We are grateful to Dr. Ping Shen for providing their valuable rabbit anti-NPF antibody and unpublished UAS-npfR1 construct. We thank Dr. Kassandra Ori-McKenney and Dr. Matthew Klassen for helpful comments on this paper. The work was supported by NIH grant 2R37NS040929 to YNJ. LYJ and YNJ are investigators of Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Beets I, Janssen T, Meelkop E, Temmerman L, Suetens N, Rademakers S, Jansen G, Schoofs L. Vasopressin/oxytocin-related signaling regulates gustatory associative learning in C. elegans. Science. 2012;338:543–545. doi: 10.1126/science.1226860. [DOI] [PubMed] [Google Scholar]
- Belote JM, Baker BS. Sexual behavior: its genetic control during development and adulthood in Drosophila melanogaster. Proceedings of the National Academy of Sciences of the United States of America. 1987;84:8026–8030. doi: 10.1073/pnas.84.22.8026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretman A, Fricke C, Chapman T. Plastic responses of male Drosophila melanogaster to the level of sperm competition increase male reproductive fitness. Proc Biol Sci. 2009;276:1705–1711. doi: 10.1098/rspb.2008.1878. Epub 2009 Feb 1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretman A, Fricke C, Hetherington P, Stone R, Chapman T. Exposure to rivals and plastic responses to sperm competition in Drosophila melanogaster. Behavioral Ecology. 2010;21:317–321. [Google Scholar]
- Bretman A, Westmancoat JD, Gage MJ, Chapman T. Males use multiple, redundant cues to detect mating rivals. Curr Biol. 2011;21:617–622. doi: 10.1016/j.cub.2011.03.008. [DOI] [PubMed] [Google Scholar]
- Burke CJ, Huetteroth W, Owald D, Perisse E, Krashes MJ, Das G, Gohl D, Silies M, Certel S, Waddell S. Layered reward signalling through octopamine and dopamine in Drosophila. Nature. 2012;492:433–437. doi: 10.1038/nature11614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Zhang Y, Shen P. A protein kinase C activity localized to neuropeptide Y-like neurons mediates ethanol intoxication in Drosophila melanogaster. Neuroscience. 2008;156:42–47. doi: 10.1016/j.neuroscience.2008.07.008. [DOI] [PubMed] [Google Scholar]
- Choi C, Fortin JP, McCarthy E, Oksman L, Kopin AS, Nitabach MN. Cellular dissection of circadian peptide signals with genetically encoded membrane-tethered ligands. Current biology: CB. 2009;19:1167–1175. doi: 10.1016/j.cub.2009.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi C, Nitabach MN. Circadian biology: environmental regulation of a multi-oscillator network. Curr Biol. 2010;20:R322–324. doi: 10.1016/j.cub.2010.02.036. [DOI] [PubMed] [Google Scholar]
- Choi S, Chatzigeorgiou M, Taylor KP, Schafer WR, Kaplan JM. Analysis of NPR-1 Reveals a Circuit Mechanism for Behavioral Quiescence in C. elegans. Neuron. 2013;78:869–880. doi: 10.1016/j.neuron.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dierick HA, Greenspan RJ. Serotonin and neuropeptide F have opposite modulatory effects on fly aggression. Nat Genet. 2007;39:678–682. doi: 10.1038/ng2029. [DOI] [PubMed] [Google Scholar]
- Ejima A, Griffith LC. Measurement of Courtship Behavior in Drosophila melanogaster. CSH Protoc. 2007;2007 doi: 10.1101/pdb.prot4847. pdb prot4847. [DOI] [PubMed] [Google Scholar]
- Feinberg EH, Vanhoven MK, Bendesky A, Wang G, Fetter RD, Shen K, Bargmann CI. GFP Reconstitution Across Synaptic Partners (GRASP) defines cell contacts and synapses in living nervous systems. Neuron. 2008;57:353–363. doi: 10.1016/j.neuron.2007.11.030. [DOI] [PubMed] [Google Scholar]
- Garbaczewska M, Billeter JC, Levine JD. Drosophila melanogaster males increase the number of sperm in their ejaculate when perceiving rival males. Journal of insect physiology. 2013;59:306–310. doi: 10.1016/j.jinsphys.2012.08.016. [DOI] [PubMed] [Google Scholar]
- Garczynski SF, Brown MR, Shen P, Murray TF, Crim JW. Characterization of a functional neuropeptide F receptor from Drosophila melanogaster. Peptides. 2002;23:773–780. doi: 10.1016/s0196-9781(01)00647-7. [DOI] [PubMed] [Google Scholar]
- Garrison JL, Macosko EZ, Bernstein S, Pokala N, Albrecht DR, Bargmann CI. Oxytocin/vasopressin-related peptides have an ancient role in reproductive behavior. Science. 2012;338:540–543. doi: 10.1126/science.1226201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han C, Jan LY, Jan YN. Enhancer-driven membrane markers for analysis of nonautonomous mechanisms reveal neuron-glia interactions in Drosophila. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:9673–9678. doi: 10.1073/pnas.1106386108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfrich-Forster C, Shafer OT, Wulbeck C, Grieshaber E, Rieger D, Taghert P. Development and morphology of the clock-gene-expressing lateral neurons of Drosophila melanogaster. The Journal of comparative neurology. 2007;500:47–70. doi: 10.1002/cne.21146. [DOI] [PubMed] [Google Scholar]
- Hermann C, Yoshii T, Dusik V, Helfrich-Forster C. Neuropeptide F immunoreactive clock neurons modify evening locomotor activity and free-running period in Drosophila melanogaster. The Journal of comparative neurology. 2012;520:970–987. doi: 10.1002/cne.22742. [DOI] [PubMed] [Google Scholar]
- Hidalgo A, Urban J, Brand AH. Targeted ablation of glia disrupts axon tract formation in the Drosophila CNS. Development. 1995;121:3703–3712. doi: 10.1242/dev.121.11.3703. [DOI] [PubMed] [Google Scholar]
- Hyun S, Lee Y, Hong ST, Bang S, Paik D, Kang J, Shin J, Lee J, Jeon K, Hwang S, et al. Drosophila GPCR Han is a receptor for the circadian clock neuropeptide PDF. Neuron. 2005;48:267–278. doi: 10.1016/j.neuron.2005.08.025. [DOI] [PubMed] [Google Scholar]
- Im SH, Li W, Taghert PH. PDFR and CRY signaling converge in a subset of clock neurons to modulate the amplitude and phase of circadian behavior in Drosophila. PLoS One. 2011;6:e18974. doi: 10.1371/journal.pone.0018974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im SH, Taghert PH. PDF receptor expression reveals direct interactions between circadian oscillators in Drosophila. J Comp Neurol. 2010;518:1925–1945. doi: 10.1002/cne.22311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insel TR. The challenge of translation in social neuroscience: a review of oxytocin, vasopressin, and affiliative behavior. Neuron. 2010;65:768–779. doi: 10.1016/j.neuron.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan LY, Jan YN. Peptidergic transmission in sympathetic ganglia of the frog. J Physiol. 1982;327:219–246. doi: 10.1113/jphysiol.1982.sp014228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang H, Kim K, Neal SJ, Macosko E, Kim D, Butcher RA, Zeiger DM, Bargmann CI, Sengupta P. Neuromodulatory state and sex specify alternative behaviors through antagonistic synaptic pathways in C. elegans. Neuron. 2012;75:585–592. doi: 10.1016/j.neuron.2012.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadener S, Villella A, Kula E, Palm K, Pyza E, Botas J, Hall JC, Rosbash M. Neurotoxic protein expression reveals connections between the circadian clock and mating behavior in Drosophila. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:13537–13542. doi: 10.1073/pnas.0605962103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim WJ, Jan LY, Jan YN. Contribution of visual and circadian neural circuits to memory for prolonged mating induced by rivals. Nat Neurosci. 2012 doi: 10.1038/nn.3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krashes MJ, DasGupta S, Vreede A, White B, Armstrong JD, Waddell S. A neural circuit mechanism integrating motivational state with memory expression in Drosophila. Cell. 2009;139:416–427. doi: 10.1016/j.cell.2009.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lear BC, Merrill CE, Lin JM, Schroeder A, Zhang L, Allada R. A G protein-coupled receptor, groom-of-PDF, is required for PDF neuron action in circadian behavior. Neuron. 2005;48:221–227. doi: 10.1016/j.neuron.2005.09.008. [DOI] [PubMed] [Google Scholar]
- Lee G, Bahn JH, Park JH. Sex- and clock-controlled expression of the neuropeptide F gene in Drosophila. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:12580–12585. doi: 10.1073/pnas.0601171103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KS, Kwon OY, Lee JH, Kwon K, Min KJ, Jung SA, Kim AK, You KH, Tatar M, Yu K. Drosophila short neuropeptide F signalling regulates growth by ERK-mediated insulin signalling. Nat Cell Biol. 2008;10:468–475. doi: 10.1038/ncb1710. [DOI] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- Lefranc A, Bundgaard J. The influence of male and female body size on copulation duration and fecundity in Drosophila melanogaster. Hereditas. 2000;132:243–247. doi: 10.1111/j.1601-5223.2000.00243.x. [DOI] [PubMed] [Google Scholar]
- Lingo PR, Zhao Z, Shen P. Co-regulation of cold-resistant food acquisition by insulin- and neuropeptide Y-like systems in Drosophila melanogaster. Neuroscience. 2007;148:371–374. doi: 10.1016/j.neuroscience.2007.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuyama K, Zhang Y, Rao Y, Wang JW. Mapping neural circuits with activity-dependent nuclear import of a transcription factor. J Neurogenet. 2012;26:89–102. doi: 10.3109/01677063.2011.642910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertens I, Vandingenen A, Johnson EC, Shafer OT, Li W, Trigg JS, De Loof A, Schoofs L, Taghert PH. PDF receptor signaling in Drosophila contributes to both circadian and geotactic behaviors. Neuron. 2005;48:213–219. doi: 10.1016/j.neuron.2005.09.009. [DOI] [PubMed] [Google Scholar]
- Nagoshi E, Sugino K, Kula E, Okazaki E, Tachibana T, Nelson S, Rosbash M. Dissecting differential gene expression within the circadian neuronal circuit of Drosophila. Nat Neurosci. 2010;13:60–68. doi: 10.1038/nn.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassel DR. Functional roles of neuropeptides in the insect central nervous system. Naturwissenschaften. 2000;87:439–449. doi: 10.1007/s001140050756. [DOI] [PubMed] [Google Scholar]
- Nassel DR, Winther AM. Drosophila neuropeptides in regulation of physiology and behavior. Prog Neurobiol. 2010;92:42–104. doi: 10.1016/j.pneurobio.2010.04.010. [DOI] [PubMed] [Google Scholar]
- Nicolai LJ, Ramaekers A, Raemaekers T, Drozdzecki A, Mauss AS, Yan J, Landgraf M, Annaert W, Hassan BA. Genetically encoded dendritic marker sheds light on neuronal connectivity in Drosophila. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:20553–20558. doi: 10.1073/pnas.1010198107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitabach MN, Taghert PH. Organization of the Drosophila circadian control circuit. Current biology: CB. 2008;18:R84–93. doi: 10.1016/j.cub.2007.11.061. [DOI] [PubMed] [Google Scholar]
- O’Brien MA, Taghert PH. A peritracheal neuropeptide system in insects: release of myomodulin-like peptides at ecdysis. J Exp Biol. 1998;201:193–209. doi: 10.1242/jeb.201.2.193. [DOI] [PubMed] [Google Scholar]
- Parisky KM, Agosto J, Pulver SR, Shang Y, Kuklin E, Hodge JJ, Kang K, Liu X, Garrity PA, Rosbash M, et al. PDF cells are a GABA-responsive wake-promoting component of the Drosophila sleep circuit. Neuron. 2008;60:672–682. doi: 10.1016/j.neuron.2008.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer BD, Jenett A, Hammonds AS, Ngo TT, Misra S, Murphy C, Scully A, Carlson JW, Wan KH, Laverty TR, et al. Tools for neuroanatomy and neurogenetics in Drosophila. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:9715–9720. doi: 10.1073/pnas.0803697105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popodi E, Kaufman TC, Holtzman SL, Park S, Carlson JW, Hoskins RA, Schulze KL, Venken KJT, Bellen HJ. Small X duplications for the stock center collection. 2010 doi: 10.1534/genetics.110.121285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renn SC, Park JH, Rosbash M, Hall JC, Taghert PH. A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila. Cell. 1999;99:791–802. doi: 10.1016/s0092-8674(00)81676-1. [DOI] [PubMed] [Google Scholar]
- Ryner LC, Goodwin SF, Castrillon DH, Anand A, Villella A, Baker BS, Hall JC, Taylor BJ, Wasserman SA. Control of male sexual behavior and sexual orientation in Drosophila by the fruitless gene. Cell. 1996;87:1079–1089. doi: 10.1016/s0092-8674(00)81802-4. [DOI] [PubMed] [Google Scholar]
- Shen P, Cai HN. Drosophila neuropeptide F mediates integration of chemosensory stimulation and conditioning of the nervous system by food. J Neurobiol. 2001;47:16–25. doi: 10.1002/neu.1012. [DOI] [PubMed] [Google Scholar]
- Shohat-Ophir G, Kaun KR, Azanchi R, Mohammed H, Heberlein U. Sexual deprivation increases ethanol intake in Drosophila. Science. 2012;335:1351–1355. doi: 10.1126/science.1215932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taghert PH, Nitabach MN. Peptide neuromodulation in invertebrate model systems. Neuron. 2012;76:82–97. doi: 10.1016/j.neuron.2012.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tayler TD, Pacheco DA, Hergarden AC, Murthy M, Anderson DJ. A neuropeptide circuit that coordinates sperm transfer and copulation duration in Drosophila. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:20697–20702. doi: 10.1073/pnas.1218246109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen T, Parrish CA, Xu D, Wu Q, Shen P. Drosophila neuropeptide F and its receptor, NPFR1, define a signaling pathway that acutely modulates alcohol sensitivity. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:2141–2146. doi: 10.1073/pnas.0406814102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JQ, Jorgensen EM. Sensation in a single neuron pair represses male behavior in hermaphrodites. Neuron. 2012;75:593–600. doi: 10.1016/j.neuron.2012.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Wen T, Lee G, Park JH, Cai HN, Shen P. Developmental control of foraging and social behavior by the Drosophila neuropeptide Y-like system. Neuron. 2003;39:147–161. doi: 10.1016/s0896-6273(03)00396-9. [DOI] [PubMed] [Google Scholar]
- Wu Q, Zhang Y, Xu J, Shen P. Regulation of hunger-driven behaviors by neural ribosomal S6 kinase in Drosophila. Proceedings of the National Academy of Sciences of the United States of America. 2005a;102:13289–13294. doi: 10.1073/pnas.0501914102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Zhao Z, Shen P. Regulation of aversion to noxious food by Drosophila neuropeptide Y- and insulin-like systems. Nat Neurosci. 2005b;8:1350–1355. doi: 10.1038/nn1540. [DOI] [PubMed] [Google Scholar]
- Yang CH, Rumpf S, Xiang Y, Gordon MD, Song W, Jan LY, Jan YN. Control of the postmating behavioral switch in Drosophila females by internal sensory neurons. Neuron. 2009;61:519–526. doi: 10.1016/j.neuron.2008.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.