Abstract
Aging is characterized by a widespread loss of homeostasis in biological systems. An important part of this decline is caused by age-related deregulation of regulatory processes that coordinate cellular responses to changing environmental conditions, maintaining cell and tissue function. Studies in genetically accessible model organisms have made significant progress in elucidating the function of such regulatory processes and the consequences of their deregulation for tissue function and longevity. Here, we review such studies, focusing on the characterization of processes that maintain metabolic and proliferative homeostasis in the fruitfly Drosophila melanogaster. The primary regulatory axis addressed in these studies is the interaction between signaling pathways that govern the response to oxidative stress, and signaling pathways that regulate cellular metabolism and growth. The interaction between these pathways has important consequences for animal physiology, and its deregulation in the aging organism is a major cause for increased mortality. Importantly, protocols to tune such interactions genetically to improve homeostasis and extend lifespan have been established by work in flies. This includes modulation of signaling pathway activity in specific tissues, including adipose tissue and insulin-producing tissues, as well as in specific cell types, such as stem cells of the fly intestine.
KEY WORDS: Aging, Drosophila, Homeostasis
Loss of homeostasis during aging
Aging is associated with a widespread loss of function at all levels of biological organization: molecular, cellular, tissue and organismic (Greer and Brunet, 2008; Murphy and Partridge, 2008; Schumacher et al., 2008; Broughton and Partridge, 2009; Campisi and Sedivy, 2009; Hayes, 2010; Richardson, 2009; Schumacher, 2009; Kapahi, 2010; Katewa and Kapahi, 2010; Liu and Rando, 2011; Rando and Chang, 2012). The proximal cause of this loss is the progressive decline of processes that maintain biological order and complexity, and thus ensure homeostasis in the young. Studies using model organisms have generated significant insight into the genetic factors and environmental conditions that influence, cause and prevent this age-related decline (Guarente and Kenyon, 2000; Tissenbaum and Guarente, 2002; Kenyon, 2005; Greer and Brunet, 2008; Katewa and Kapahi, 2010). It is clear that a combination of stress-induced damage to biological macromolecules, side effects of energy metabolism, deregulation of processes ensuring cellular and tissue integrity, and loss of coordination of systemic physiological control mechanisms are involved in the age-related loss of homeostasis. At the same time, a number of conserved genetic mechanisms preventing or counteracting this decline have evolved. Understanding the integration of these diverse processes, and thus gaining comprehensive insight into the causes and mechanisms of the aging process, remains a challenge. In recent years, however, the study of aging has progressed from the identification of single-gene mutations that influence lifespan toward more complex integrative analysis of the aging process. This work has also been facilitated by genetically accessible model systems and includes detailed analysis of the interaction between environmental factors and genetics in the control of longevity, as well as assessment of tissue-specific contributions to lifespan regulation (Guarente and Kenyon, 2000; Tissenbaum and Guarente, 2002; Kenyon, 2005; Greer and Brunet, 2008; Mair and Dillin, 2008; Broughton and Partridge, 2009; Panowski and Dillin, 2009; Toivonen and Partridge, 2009; Taylor and Dillin, 2011; Conboy and Rando, 2012). An important emerging insight is the complex role that various regulatory processes play during aging: signaling interactions required for the maintenance of cellular, tissue or organismal integrity in the young can cause dysfunction in the aging organism (Kahn and Olsen, 2010; Karpac and Jasper, 2009; Toivonen and Partridge, 2009; Kapahi, 2010; Biteau et al., 2011b). This ‘antagonistic pleiotropy’ of biological regulation is an important cause for many age-related metabolic and proliferative diseases, and its causes and immediate consequences are only beginning to be understood.
A particularly successful model in the characterization and elucidation of these regulatory interactions is Drosophila melanogaster, which has been used to characterize signaling mechanisms that control cellular damage responses and metabolic activity; coordinate tissue responses to damage, infection and inflammation; regulate tissue regeneration; and promote longevity in a variety of environmental conditions (Tatar et al., 2003; Lemaitre and Hoffmann, 2007; Leopold and Perrimon, 2007; Broughton and Partridge, 2009; Karpac and Jasper, 2009; Kapahi et al., 2010; Biteau et al., 2011a; Biteau et al., 2011b; Jones and Rando, 2011). Classic studies have established genetic interventions that can promote longevity, including a reduction in insulin signaling activity, increased expression of antioxidant proteins and protein chaperones, and activation of stress response signaling pathways (Parkes et al., 1998; Clancy et al., 2001; Tatar et al., 2001; Wang et al., 2003; Sun et al., 2004; Tower, 2011). These studies have been complemented in recent years with work addressing the complex interactions between tissue damage, metabolic regulation, innate immune processes and regenerative decline (Leopold and Perrimon, 2007; Biteau et al., 2011a). A comprehensive overview of causes and consequences of aging is emerging from this work, and new insight into possible therapeutic interventions against age-related diseases, is being generated.
Here, we review recent findings in the D. melanogaster model that are the basis for emerging concepts explaining the breakdown of homeostasis in the aging organism. Two specific areas will be discussed: signaling mechanisms regulating metabolic responses to tissue damage, and the control of proliferative homeostasis in the aging intestinal epithelium. The unifying concept emerging from these areas of research is the importance of signaling interactions between stress signaling pathways, innate immune signaling and growth signaling through the insulin and Tor signaling pathways in the maintenance of homeostasis in the young, and the deregulation of these interactions in the old organism. Because the signaling interactions discussed here are evolutionarily conserved, these paradigms provide important insight into the development of age-related metabolic and proliferative diseases, and may lead the way towards development of integrative therapies against such diseases.
Insulin signaling: a central regulator of longevity
The insulin signaling pathway is a central regulator of longevity in both invertebrates and vertebrates (Wolkow et al., 2000; Tatar et al., 2003; Kenyon, 2005; Greer and Brunet, 2008; Broughton and Partridge, 2009; Kenyon, 2010; Barzilai et al., 2012). Single-gene loss-of-function mutations in insulin signaling components extend lifespan in Caenorhabditis elegans and D. melanogaster, and loss of insulin signaling activity in specific tissues of mice can also promote longevity. Insight into the role of insulin signaling in regulating lifespan was first obtained from genetic screens in worms, in which mutations in the PI3K homologue AGE-1 were identified that extend lifespan by more than 100% (Friedman and Johnson, 1988). Further genetic studies identified loss-of-function mutations in the insulin receptor daf-2 and in the Akt and SGK kinases as long-lived (Kenyon et al., 1993; Kimura et al., 1997; Hertweck et al., 2004). Similar genetic studies in flies identified long-lived strains mutant in the insulin receptor and the insulin receptor substrate Chico (Clancy et al., 2001; Tatar et al., 2001). In mice, heterozygosity for the insulin-like growth factor (IGF) 1 receptor increases lifespan, while knockdown of the insulin receptor in adipose tissue or knockdown of IRS2 in the brain is sufficient to increase lifespan (Blüher et al., 2003; Holzenberger et al., 2003; Taguchi et al., 2007).
These longevity effects of reducing insulin signaling are caused by activation of the transcription factor Foxo (Forkhead box protein O), which is negatively regulated downstream of Akt by insulin signaling, and is essential for the lifespan extension observed in insulin loss-of-function conditions in both worms and flies (Kenyon et al., 1993; Giannakou et al., 2004; Hwangbo et al., 2004; Wang et al., 2005; Greer and Brunet, 2008). Foxo translocates to the nucleus and activates transcription when insulin signaling activity is low, inducing the expression of a number of stress defense and repair proteins in addition to metabolic enzymes and regulators (Murphy et al., 2003; Greer and Brunet, 2008; Biteau et al., 2011b). This activity of Foxo promotes cellular stress resistance and damage repair, and can regulate apoptosis (Brunet et al., 1999; Kops et al., 2002; Brunet et al., 2004; Wang et al., 2005; Luo et al., 2007; Demontis and Perrimon, 2010).
Through Foxo-induced gene expression, the lifespan extension observed by reduced IIS activity is thus linked to metabolic regulation, but also to Foxo-mediated cellular stress response and repair functions.
IIS/Foxo function and metabolic adaptation
Metabolic regulation by Foxo has been described in several organisms, and is likely tied to its role in longevity. Adaptation to dietary conditions is required to maintain metabolic and energy homeostasis (Roberts and Rosenberg, 2006; Luca et al., 2010), and deregulation of adaptive responses because of modern (high sugar/high fat) diets or age-related dysfunction has been proposed as a cause for metabolic diseases (Odegaard and Chawla, 2013). This notion is supported by the increased incidence of metabolic diseases in the elderly (Roberts and Rosenberg, 2006).
The role of insulin signaling in metabolic processes is well understood, but many questions about the link between metabolic regulation by insulin and lifespan remain. Understanding the complex network of tissue-specific functions of insulin/IGF signaling (IIS) in the control of metabolic homeostasis is likely to be required to fully grasp the causal relationship between the regulation of metabolic adaptation and longevity. IIS governs energy homeostasis systemically by mediating tissue interactions that control lipid and carbohydrate metabolism, stabilizing blood glucose levels and serving anabolic purposes (Saltiel and Kahn, 2001). The health consequences of reducing insulin signaling activity, however, are complex and tissue-specific. While loss of the insulin receptor in adipose tissue of mice can have positive effects on lifespan (Blüher et al., 2003), IIS inhibition and Foxo activation in other tissues (such as liver, brain and muscle) result in metabolic and degenerative diseases (Kido et al., 2000; Michael et al., 2000; Suzuki et al., 2010; Mihaylova et al., 2011). These phenotypes are reminiscent of many age-related metabolic pathologies in humans that are associated with chronic insulin resistance (Barzilai et al., 2012). In these conditions, Foxo activation contributes to the breakdown of lipid and glucose homeostasis, promoting diabetes (Samuel and Shulman, 2012; Eijkelenboom and Burgering, 2013).
Many of the metabolic consequences of altered IIS and Foxo signaling are caused by changes in glucose and lipid metabolism. The function of insulin signaling in regulating glucose uptake and glycogen synthesis in the muscle and liver is well understood (Saltiel and Kahn, 2001; Accili and Arden, 2004; Samuel and Shulman, 2012; Eijkelenboom and Burgering, 2013). At the same time, altered IIS activity is also associated with abnormal lipid metabolism (Kitamura et al., 2003; Teleman, 2010), yet the cell- and tissue-specific mechanisms by which IIS regulates lipid metabolism are only partially understood. In both Drosophila and mammals, Foxo regulates the transcription of lipases in adipose tissue required for the lipolysis of stored lipids (Vihervaara and Puig, 2008; Chakrabarti and Kandror, 2009; Wang et al., 2011), as well as the expression of enzymes and other transcription factors involved in lipid catabolism (Deng et al., 2012; Xu et al., 2012). Accordingly, changes in IIS/Foxo regulated lipases (such as adipose triglyceride lipase) and lipogenic transcription factors (such as SREBP-1c) have been linked to the dyslipidemia associated with type 2 diabetes and other metabolic syndromes (Shimomura et al., 2000; Schoenborn et al., 2006; Badin et al., 2011).
Linking stress signaling and metabolic homeostasis
In addition to genes that promote metabolic homeostasis, Foxo also regulates genes encoding stress-response and repair proteins. The activation of Foxo in conditions of low insulin signaling can thus shift energy resources from anabolic functions to cellular preservation and repair. This central role as a switch between anabolism and repair is reflected in the importance of the C. elegans Foxo homologue Daf16 in both dauer formation (where developing worms enter developmental and metabolic arrest under nutrient deprivation or stress) and longevity (Kenyon et al., 1993; Giannakou et al., 2004; Hwangbo et al., 2004; Wang et al., 2005; Greer and Brunet, 2008).
Highlighting the importance of Foxo in regulating cellular stress responses, its activity is not only controlled by insulin signaling, but can also be stimulated by stress-response signaling pathways. A well-understood interaction between IIS and stress signaling is mediated by the Jun-N-terminal kinase (JNK) pathway, which regulates gene expression in response to a wide range of environmental stressors. JNK can promote nuclear translocation of Foxo independently of insulin signaling, providing an avenue to induce Foxo-mediated gene expression programs independently of the metabolic status of the cell (Hirosumi et al., 2002; Karpac and Jasper, 2009; Biteau et al., 2011b; Samuel and Shulman, 2012).
JNK activity is induced by a range of intrinsic and environmental insults (e.g. UV irradiation, reactive oxygen species, DNA damage, heat and inflammatory cytokines) through a MAP kinase cascade that is initiated by a number of stress-specific upstream kinases (JNK kinase kinases). JNK commonly regulates transcription factors, such as Foxo and AP-1, leading to changes in gene expression. Its activation has highly context-dependent consequences for the cell, ranging from apoptosis over morphogenetic changes to increased survival (Davis, 2000; Weston and Davis, 2002; Manning and Davis, 2003; Karin and Gallagher, 2005). Mirroring the lifespan extension observed in insulin signaling mutants, moderate activation of JNK signaling results in increased stress tolerance and extended lifespan in flies and worms. Flies heterozygous for the JNK phosphatase Puckered (puc), or in which the JNK kinase Hemipterous (JNKK/Hep) is overexpressed in neuronal tissue, are resistant to the oxidative stress-inducing compound Paraquat (Wang et al., 2003). Similarly, JNKK/Hep mutant flies exhibit increased stress sensitivity and are deficient in the induction of a transcriptional stress response (Wang et al., 2003). Supporting the protective role for JNK signaling in flies, puc heterozygotes or Hep-overexpressing animals are long-lived under normal conditions (Wang et al., 2003; Libert et al., 2008). Similar consequences of JNK activation have been described in C. elegans. Worms with increased JNK activity due to inhibition of the JNK phosphatase VHP-1 (VH1 dual-specificity phosphatase) are protected against heavy metal toxicity, and overexpression of JNK increases lifespan under normal conditions (Mizuno et al., 2004; Oh et al., 2005).
Cell-autonomous JNK/IIS interactions
Genetic interaction studies reveal that all these protective effects of JNK signaling are mediated by DAF-16/Foxo (Oh et al., 2005; Wang et al., 2005). In response to JNK signaling, Foxo promotes the expression of genes that inhibit growth and promote repair, but also of genes that induce apoptosis (Wang et al., 2005; Luo et al., 2007). Accordingly, activation of JNK dominantly suppresses IIS-induced cell growth, and Foxo acts downstream of JNK signaling to extend lifespan in flies and worms, but also to mediate apoptotic responses to excessive DNA damage (Matsumoto and Accili, 2005; Oh et al., 2005; Wang et al., 2005; Luo et al., 2007; Wolf et al., 2008). Activation of Foxo proteins by JNK has further been described in mammalian cells, suggesting that the interaction between IIS and JNK signaling is conserved (Essers et al., 2004; Jaeschke and Davis, 2007).
Interestingly, activation of Foxo by JNK signaling is achieved through multiple cell-autonomous and non-autonomous mechanisms and has been implicated not only in lifespan extension, but also in pathological disruptions of metabolic homeostasis. In fact, JNK-mediated repression of insulin signaling activity has been proposed as a major cause for insulin resistance in obese patients (Waeber et al., 2000; Hirosumi et al., 2002; Solinas et al., 2007; Yang et al., 2007; Emanuelli et al., 2008). JNK can suppress insulin signaling activity and activate Foxo by phosphorylating the insulin receptor substrate (thus preventing signal transduction through this molecule), by phosphorylating 14-3-3 proteins and disrupting their inhibition of Foxo, as well as by phosphorylating Foxo proteins directly (Aguirre et al., 2000; Mamay et al., 2003; Essers et al., 2004; Tsuruta et al., 2004; Sunayama et al., 2005; Yoshida et al., 2005). While C. elegans JNK was shown to phosphorylate DAF-16 (Oh et al., 2005), sequence alignment shows that the JNK phosphorylation sites in mammalian IRS or Foxo4 are not conserved in the Drosophila proteins, while the phosphorylation sites on 14-3-3 are conserved in both flies and C. elegans (Essers et al., 2004; Tsuruta et al., 2004; Sunayama et al., 2005; Yoshida et al., 2005; Nielsen et al., 2008; Karpac and Jasper, 2009). The antagonism between JNK and IIS thus appears to be evolutionarily ancient and critical for survival.
Systemic regulation of IIS by JNK
In addition to the cell-autonomous mechanisms of IIS inhibition described above, JNK can promote the expression of cytokines and adipokines that suppress insulin signal transduction at a distance (such as IL-6, which is secreted in response to JNK activity from the adipose tissue and macrophages and prevents insulin signaling in the liver) (Sabio et al., 2008; Sabio and Davis, 2010; Han et al., 2013). JNK thus mediates important cell-autonomous and systemic metabolic adaptations to changing environmental conditions. While chronic engagement of this signaling system can cause metabolic diseases, the acute JNK-mediated repression of insulin signaling activity is an adaptive process that decreases metabolic activity under stress conditions (Karpac and Jasper, 2009).
This adaptive process is exemplified by dauer formation in C. elegans, an extreme form of adaptation in which development and metabolic activity are curtailed under unfavorable conditions. Repression of insulin signaling activity is central to dauer formation and can be achieved by stress-induced activation of Foxo, as well as by stress-induced repression of insulin-like peptides. Accordingly, the regulation of insulin-like peptide expression in specific chemosensory neurons in the head (ASI neurons) is crucial for dauer formation. These neurons express the insulin-like peptide DAF-28, which is repressed in response to starvation and dauer pheromone, activating DAF-16 in the periphery (Bargmann and Horvitz, 1991; Li et al., 2003). This peripheral activation of DAF-16, in turn, is sufficient to increase lifespan (Libina et al., 2003). The intestine, the major fat-storing tissue in the worm, has an important role in this lifespan extension, but Foxo activity in other peripheral cells can also extend lifespan (Libina et al., 2003; Zhang et al., 2013).
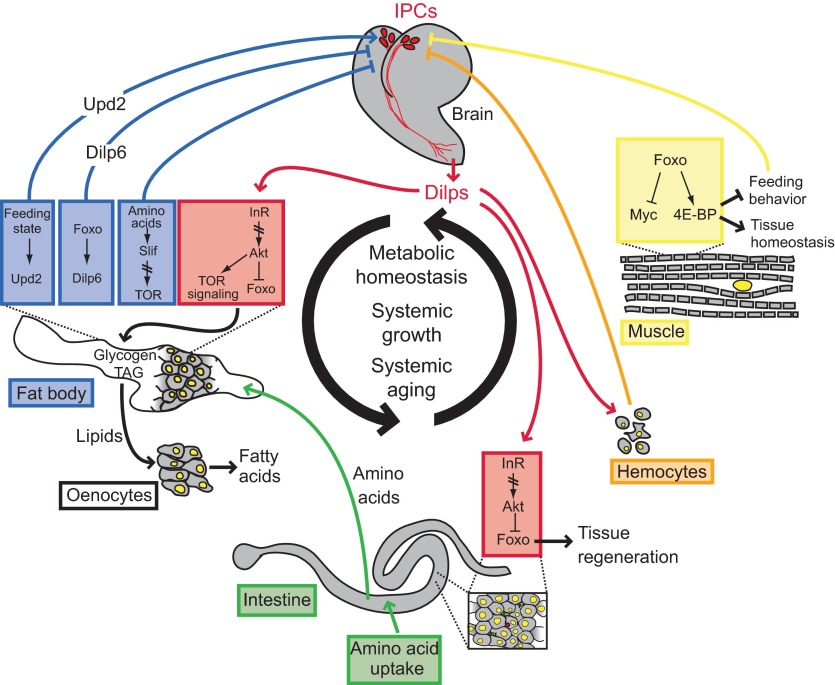
In Drosophila, similar endocrine regulation of IIS activity by stress signaling has been described (Fig. 1). JNK activation in insulin-producing cells (IPCs) is crucial for metabolic and growth adaptation under stress conditions (Wang et al., 2005; Karpac et al., 2009). This is achieved by repression of Drosophila insulin-like peptide 2 (Dilp2) transcription, and involves Foxo activation in IPCs. As in worms, JNK signaling thus systemically represses IIS activity, allowing coordinated metabolic adaptation of insulin target tissues to environmental changes, and extending the lifespan of flies (Wang et al., 2005; Karpac et al., 2009). This mechanism is part of a complex system of endocrine interactions between insulin-producing tissues, fat-storing tissues and peripheral insulin target tissues that regulate lifespan and metabolic homeostasis (Fig. 1) (Leopold and Perrimon, 2007; Russell and Kahn, 2007; Karpac and Jasper, 2009; Rajan and Perrimon, 2012). Thus, ablation of IPCs is sufficient to extend lifespan in flies (Wessells et al., 2004; Broughton et al., 2005), and overexpression of Foxo in the peripheral fat body also leads to lifespan extension. Interestingly, Foxo activation in this tissue is accompanied by feedback repression of insulin-like peptide gene expression in IPCs (Giannakou et al., 2004; Hwangbo et al., 2004). A recent study has identified a fat-body-derived insulin-like peptide, Dilp6, as a candidate for this feedback signal (Okamoto et al., 2009; Slaidina et al., 2009; Bai et al., 2012). Dilp6 is a peptide that is secreted in non-feeding and fasting states from the fat body, and sustains insulin-dependent growth under such conditions during development (Okamoto et al., 2009; Slaidina et al., 2009). The induction of Dilp6 under starvation conditions in the adult, the repression of brain insulin-like peptides, and the lifespan extension observed when Dilp6 is overexpressed from this tissue highlight the complexity of the regulation of insulin signaling activity in peripheral tissues by a diversity of locally and systemically acting insulin-like peptides. Other examples include insulin-like peptides acting locally in the brain (Chell and Brand, 2010; Sousa-Nunes et al., 2011) and in the intestine (O'Brien et al., 2011), and systemically regulating growth in response to tissue overgrowth and tumor formation (Fig. 1) (Colombani et al., 2012; Garelli et al., 2012).
Fig. 1.
Summary of endocrine interactions regulating metabolic and proliferative homeostasis in Drosophila. Tissue systems and endocrine signals that mediate specific responses to dietary changes and stress to maintain homeostasis in the adult animal are summarized. Drosophila insulin-like peptides (Dilps) play a central role in the regulation of metabolic and proliferative homeostasis in flies by coordinating multiple metabolic and regenerative responses to nutritional and stress conditions. This includes regulation of metabolic activity in the fat body, survival in hemocytes, and regeneration in the intestinal epithelium. Dilps secreted from peripheral tissues (Dilp6 in the fat body and Dilp3 in the intestine) control metabolic and proliferative activity locally, and may interact with insulin-producing cells (IPCs) in the brain to regulate the expression and secretion of brain-derived Dilps. Secretion of brain-derived Dilps is also regulated by stress signals (Jun-N-terminal kinase, JNK) and by fat-body-derived hormones such as Unpaired2 (Upd2). 4E-BP, eukaryotic translation initiation factor 4E binding protein; Foxo, Forkhead box protein O; InR, insulin receptor; Slif, slimfast.
These findings support the notion that complex, evolutionarily conserved endocrine interactions between insulin-producing and insulin-target tissues influence metabolic homeostasis and lifespan. Stress signaling through JNK appears to be an integral part of this regulatory network. Recent studies have explored this regulation in more detail, and have established the spatiotemporal coordination of stress, innate immune and inflammatory signals that govern this regulatory process in flies (Karpac et al., 2011): local tissue damage (induced, for example, by UV irradiation) initiates a transient systemic repression of insulin signaling activity by inducing inflammatory cytokines (including Upd3, a functional analogue of IL-6) that activate the JAK/Stat signaling pathway in adipose tissue and hemocytes. This activation is required to repress insulin transcription in IPCs. Subsequent activation of Foxo in adipose tissue leads to the activation of NFkB/Relish signaling in these cells, which in turn is required to initiate a recovery phase of the adaptive response. This recovery restores insulin transcription in IPCs and is crucial to resume growth and anabolic functions after damage has been repaired (Karpac et al., 2011).
The central role of the fat body in coordinating these stress responses is reminiscent of the importance of adipocyte-derived signals in the maintenance of metabolic homeostasis in vertebrates. A recent study has identified Upd2, a cytokine with significant similarity to Upd3, as a factor that is secreted from adipose tissue in the fed state and activates JAK/Stat signaling in a population of GABAergic neurons in the brain that have direct connections to IPCs (Rajan and Perrimon, 2012). This interaction controls Dilp secretion in IPCs, and thus regulates systemic growth and anabolic activity. Strikingly, human leptin can functionally replace Upd2 in this system, highlighting the evolutionary conservation of mechanisms that maintain metabolic and growth homeostasis in metazoans. Furthermore, it has also been shown in flies that amino acids, sensed by the fat body, can relay humoral signals to control insulin release from the brain (Géminard et al., 2009). Slimfast (an amino acid transporter) is required upstream of the amino-acid-sensing and IIS-responsive TOR signaling pathway in the fat body to link dietary amino acid levels with circulating Dilps during metabolic adaptation. It is interesting to speculate that TOR signaling may couple nutrient sensing in the fat body to insulin levels in flies through the regulation of Upd2.
Taken together, these findings show that Drosophila is emerging as model for the study of organ–organ communication (Fig. 1). Studies of physiological cross-talk between fly tissues are revealing mechanistic insight into the regulation of IIS and the control of systemic growth, aging and energy homeostasis, as well as the regulation of tissue homeostasis and innate immunity (reviewed below). The similarity of fly organs to vertebrate organ systems, the evolutionarily conserved signaling pathways encoded in the Drosophila genome, and the highly developed genetic tool kit available for the fly make Drosophila a crucial model organism in which to study the signaling mechanisms and tissue interactions that regulate IIS.
An interesting example of the similarity between the described processes in flies and vertebrates is the repression of insulin/IGF signaling activity in response to DNA damage, which has also been observed in mice. Mutations in genes involved in DNA repair cause repression of IGF signaling, resulting in growth attenuation (Niedernhofer et al., 2006; van der Pluijm et al., 2007; Garinis et al., 2008; Schumacher et al., 2008). The repression of the growth hormone/IGF axis is believed to promote tissue repair in organs afflicted by increased DNA damage, but paradoxically, is also associated with progeroid (or accelerated aging) phenotypes. The complexity of tissue-specific responses to the attenuation of growth hormone/IGF signaling remains to be explored in detail and is likely to provide new insight into the acute as well as chronic mechanisms promoting metabolic adaptation and tissue dysfunction in response to DNA damage and injury.
Chronic inflammatory diseases – too much of a good thing?
The antagonistic interaction between stress signaling and insulin signaling is thus an evolutionarily conserved adaptive mechanism to promote cell survival and tissue repair when the organism experiences stressful and damaging conditions. Why does this mechanism then contribute to age-related metabolic diseases? It seems likely that the duration of JNK-mediated IIS repression is crucial for the ultimate outcome of this signaling event. While acute suppression of IIS activity by JNK is likely beneficial, the system can cause significant damage when the organism is stressed continuously, or when stress signaling is otherwise chronically activated (Fig. 2). Such conditions include inflammation that is associated with obesity and aging. In fact, JNK signaling has been identified as a central mediator of insulin resistance in genetic and dietary-induced models of obesity and insulin resistance (Waeber et al., 2000; Hirosumi et al., 2002; Solinas et al., 2007; Yang et al., 2007; Emanuelli et al., 2008). This effect involves complex interactions between JNK activity in the liver, muscle, adipocytes and macrophages (Waeber et al., 2000; Hirosumi et al., 2002; Solinas et al., 2007; Yang et al., 2007; Emanuelli et al., 2008; Han et al., 2013). In obese models, JNK is activated by endoplasmic reticulum stress in adipocytes, as well as by adipose-derived inflammatory signals such as free fatty acids and TNF-α and the concurrent activation of Toll-like receptors (Tsukumo et al., 2007), and results in phosphorylation and inhibition of IRS-1 (Hotamisligil et al., 1996; Yin et al., 1998; Hotamisligil, 1999; Aguirre et al., 2000; Hirosumi et al., 2002; Ozcan et al., 2004; Hotamisligil, 2005; Hotamisligil, 2006; Bouzakri and Zierath, 2007). Furthermore, JNK activation in adipose tissue has been shown to induce inflammatory cytokines such as IL-6, which in turn promotes insulin resistance in the liver by activating JAK/Stat signaling (Sabio et al., 2008; Sabio and Davis, 2010). New therapeutic approaches that specifically target these signaling interactions are expected to be required to restore metabolic and growth homeostasis in these conditions.
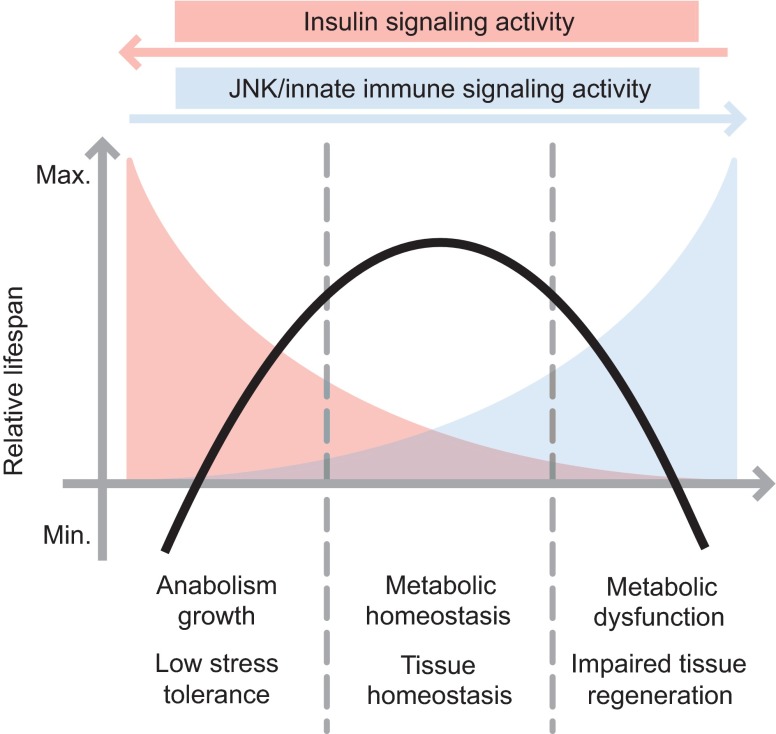
Fig. 2.
Relationship between insulin signaling activity and stress/innate immune signaling in the control of homeostasis and lifespan. Stress signaling through Jun-N-terminal kinase (JNK) and innate immune signaling though NFkB can inhibit insulin signaling activity both systemically and locally. This antagonism between stress and insulin signaling influences both metabolic and proliferative tissue homeostasis in flies, significantly impacting lifespan (Biteau et al., 2010; Karpac et al., 2011). In conditions in which insulin signaling is high and JNK signaling is low (for example, in conditions of abundant nutrient intake in wild-types or in genetic conditions optimizing insulin signaling), longevity (black line) is low. When insulin signaling is moderately reduced or JNK signaling is activated (under calorie restriction or in genetic conditions leading to moderate increases of JNK or reduced insulin signaling capacity), tissue and metabolic homeostasis are maximized, increasing lifespan. When JNK is chronically or excessively activated (in conditions of stress or inflammation), or when insulin signaling is strongly impaired (strong loss-of-function mutations in insulin signaling components), lifespan declines again.
Maintenance of proliferative homeostasis in the aging Drosophila intestine
The interaction between stress and insulin signaling described above not only influences lifespan by promoting metabolic homeostasis, but also has a major impact on proliferative homeostasis in regenerating tissues. Regeneration is crucial for renewal and maintenance of many tissues in vertebrates, and aging, as well as deregulation of insulin/IGF signaling, has important deleterious consequences for regenerative capacity (Jasper and Jones, 2010; Jang et al., 2011; Jones and Rando, 2011). Of particular interest are processes that maintain proliferative homeostasis in high-turnover tissues, as deregulation of such processes is likely to promote cancer or degenerative diseases. In fact, the universal impact of the interaction between stress and metabolic signaling on tissue health is highlighted by recent insights into the development and progression of age-related tissue dysfunction in the intestinal epithelium of Drosophila. This tissue has emerged as a very productive and accessible model system in which to explore the cell-autonomous and systemic regulation of stem cell function, tissue regeneration and the loss of proliferative homeostasis. In the following, we will discuss the implications of this research for our understanding of human diseases and the establishment of therapeutic approaches for age-related proliferative and degenerative diseases.
The Drosophila intestine as a model system for intestinal dysplasia
Drosophila has only recently been recognized as a model in which to study proliferative homeostasis. While flies were long considered post-mitotic animals, two elegant studies in 2006 demonstrated that the intestinal epithelium regenerates in the adult animal (Micchelli and Perrimon, 2006; Ohlstein and Spradling, 2006). Using lineage tracing techniques, Ohlstein, Micchelli and colleagues identified and characterized a population of pluripotent tissue stem cells in the fly midgut that continuously regenerate this tissue in a process that mechanistically and morphologically resembles cell turnover in the intestinal epithelium of mammals (Micchelli and Perrimon, 2006; Ohlstein and Spradling, 2006; Radtke et al., 2006; Ohlstein and Spradling, 2007) (Fig. 3). This work thus introduced the fly intestine as an ideal model system in which to characterize the regulation of intestinal stem cell function as well as age-related changes in regenerative capacity of high-turnover tissues.
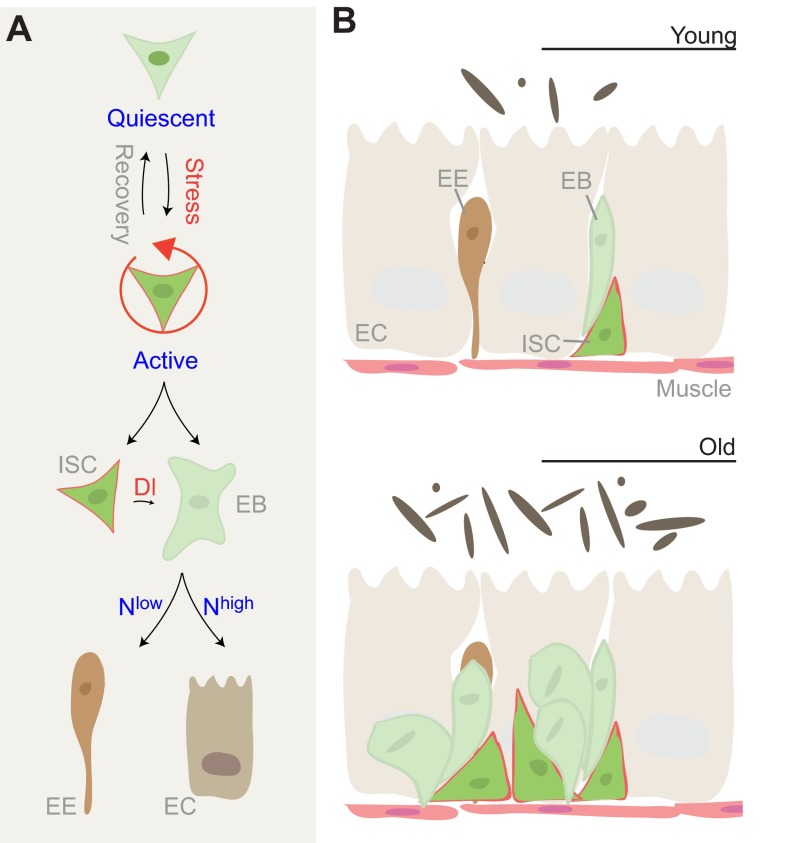
Fig. 3.
Control of tissue regeneration in the Drosophila intestine. (A) The intestinal stem cell lineage in Drosophila. In response to stress, intestinal stem cells (ISCs) divide asymmetrically to give rise to a new ISC and an enteroblast (EB), which differentiates into either an enterocyte (EC) or an enteroendocrine cell (EE). Notch (N) signaling, initiated by a Delta (Dl) signal from the ISC, drives EB differentiation. Depending on the levels of N signaling, EBs differentiate into either EEs or ECs. (B) Age-related changes in homeostasis of the intestinal epithelium. In young animals, the epithelium consists of a monolayer of ECs with interspersed EEs and basally located ISCs. In aging flies, ISCs overproliferate, resulting in the accumulation of misdifferentiated EB-like cells that disrupt structure and function of the intestinal epithelium. This phenotype correlates with an expansion of the commensal bacterial population in the lumen (dark ovals).
Interestingly, this tissue also performs metabolic and innate immune functions, and serves as a crucial barrier epithelium, allowing detailed analysis of complex interactions between tissue regeneration, metabolic regulation, innate immunity and inflammation (Biteau et al., 2008; Choi et al., 2008; Buchon et al., 2009a; Biteau et al., 2010; Biteau et al., 2011a; Rera et al., 2012; Sieber and Thummel, 2012; Rera et al., 2013). We have recently shown that the regenerative capacity of stem cells strongly influences lifespan in Drosophila, and that improving intestinal tissue homeostasis also rescues the age-dependent systemic breakdown of lipid homeostasis (Biteau et al., 2010).
Intestinal stem cells (ISCs) are the only dividing cells in the intestinal epithelium, and can give rise to at least two differentiated intestinal cell types: enteroendocrine cells (EEs) and enterocytes (ECs) (Micchelli and Perrimon, 2006; Ohlstein and Spradling, 2006; Ohlstein and Spradling, 2007) (Fig. 3). When ISC division is triggered, these cells divide asymmetrically, generating a new stem cell that remains basally, while a newly formed progenitor cell, the enteroblast (EB), emerges apically and differentiates into either ECs or EEs (Ohlstein and Spradling, 2007). The asymmetry of the ISC/EB pair is likely to be determined by integrin-mediated adhesion of ISCs to the basement membrane and the underlying visceral muscle (Goulas et al., 2012). The muscle plays an important role not only in maintaining asymmetry in this cell pair, but also in supplying growth factors that promote ISC maintenance and proliferative competence, and has thus been described as the ‘niche’ of the ISC population (Lin et al., 2008; Biteau et al., 2011a; Biteau and Jasper, 2011).
ISC self-renewal and differentiation is controlled by the Notch and Tor signaling pathways. ISCs express the Notch ligand Delta, which in turn activates Notch signaling in the EB, driving their differentiation. The lineage decision between EE and EC fates of the EB is determined by the level of Dl expression in the ISC, and accordingly by the level of Notch signaling activity in the EB, with high Notch signaling promoting EC fates, and low Notch activity promoting EE fates. Differentiation of ECs requires activation of Tor signaling, which is differentially activated in ISCs and EBs. High expression of tuberous sclerosis complex 2 (TSC2), the Rheb GTPase activating protein and inhibitor of the Tor pathway, is required in ISCs to prevent their differentiation, while Notch-mediated suppression of TSC2 expression in EBs is required for differentiation into ECs. Interestingly, TSC2 buffers ISCs from dietary effects, promoting long-term maintenance of ISCs (Amcheslavsky et al., 2011; Kapuria et al., 2012).
Growth and metabolic signaling in the control of regeneration
Dietary conditions and metabolic signals not only influence differentiation of ISCs through the Tor signaling pathway, but also determine proliferative activity of ISCs through the insulin signaling pathway, thus influencing overall tissue growth. Interestingly, the insulin receptor has both cell-autonomous and non-autonomous roles in the regulation of ISC function. ISCs mutant for the insulin receptor do not divide, while increased insulin signaling activity can induce proliferation in widely quiescent ISCs (Amcheslavsky et al., 2009; Biteau et al., 2010; McLeod et al., 2010; Choi et al., 2011; Kapuria et al., 2012). Loss of the insulin receptor in EBs, however, impairs EB differentiation, most likely because of reduced Tor signaling in these cells. Unresolved adhesion of undifferentiated EBs to ISCs, in turn, inhibits ISC proliferation (Choi et al., 2011).
The activity of the insulin signaling pathway in ISCs is determined by both local and systemic Dilps. When insulin-producing cells in the brain are ablated, ISC proliferation is significantly reduced, highlighting the importance of systemic insulin-like signals (Amcheslavsky et al., 2009; Biteau et al., 2010; Kapuria et al., 2012). Dilp3 secreted from the visceral muscle, in contrast, influences ISC proliferation in the young maturing gut and in response to fasting/refeeding cycles, regulating the overall size of the intestinal epithelium (O'Brien et al., 2011). Strikingly, this response can induce ISCs to divide symmetrically, thus managing the overall numbers of available stem cells.
Dietary conditions, and the associated nutrient-responsive signals, thus influence ISC activity and maintenance, and can also significantly influence age-related changes in proliferative homeostasis of the intestinal epithelium. This regulation is integrated into a complex network of regulatory processes that determine compensatory proliferation of ISCs in response to cellular damage, stress and infection. These signaling interactions are complex, yet their impact on tissue homeostasis, health and lifespan suggest that further insight into these processes is crucial to understanding age-related inflammatory and proliferative diseases in vertebrates (Biteau et al., 2011a).
Regenerative response to tissue damage
ISC proliferation rates are regulated by JNK signaling, but also by the Jak/Stat signaling pathway and by p38 MAPK, EGFR and insulin signaling (Biteau et al., 2008; Amcheslavsky et al., 2009; Apidianakis et al., 2009; Buchon et al., 2009a; Buchon et al., 2009b; Chatterjee and Ip, 2009; Jiang et al., 2009; Biteau et al., 2010; Buchon et al., 2010; Biteau and Jasper, 2011; Jiang et al., 2011). These pathways regulate ISC proliferation through a number of local and paracrine interactions involving both ECs and the muscle. Stress, infection, injury or tissue damage leads to JNK activation in ECs, which in turn induces production of the Jak/Stat ligands Upd1–3. These ligands are also induced by activation of Yorkie, which occurs in response to a loss of tissue integrity and repression of the Hippo pathway. Upds activate the Jak/Stat pathway in ISCs, triggering proliferation. At the same time, cell-autonomous activation of JNK signaling in ISCs can promote proliferation of these cells in response to oxidative stress. All these stress-induced pathways determine the activity of the ISC pool by changing the ratio of quiescent to active stem cells. Signaling through receptor tyrosine kinase pathways (including InR and EGFR signaling), however, is crucial to maintaining the proliferative potential of ISCs. The Fos transcription factor integrates JNK signaling with EGFR signaling (which is activated in ISCs by ligands derived from both the muscle and ECs), and thus serves as a crucial node in a signaling network that governs ISC activity (Buchon et al., 2010; Biteau and Jasper, 2011).
Loss of proliferative homeostasis in aging animals: a chronic inflammatory condition?
As regeneration through the ISC lineage is crucial for normal homeostatic tissue turnover as well as for epithelial recovery after damage or infection (Buchon et al., 2009a; Buchon et al., 2009b; Jiang et al., 2009), the activity of ISCs strongly influences stress and infection tolerance, as well as lifespan of flies. Interestingly, ISCs become deregulated in aging flies, overproliferating and producing a large number of cells that undergo incomplete differentiation (Fig. 3). This triggers intestinal dysplasia, which is characterized by loss of apico-basal organization of the epithelium and accumulation of mis-differentiated cells at the basement membrane (Biteau et al., 2008; Choi et al., 2008). The gut then loses both its resorptive and its barrier functions, resulting in increased mortality (Buchon et al., 2009a; Buchon et al., 2009b; Rera et al., 2011; Rera et al., 2012). This phenotype is caused by an age-related increase in global oxidative stress in the intestine that is associated with the presence of commensal bacteria (Buchon et al., 2009a) and results in the widespread activation of stress-responsive signaling pathways (Biteau et al., 2008). This includes p38 MAPK, JNK and the Jak/Stat signaling pathways, which are influenced by the commensal flora and are activated in the gut after exposure to pathogenic bacteria (Apidianakis et al., 2009; Buchon et al., 2009a; Buchon et al., 2009b; Chatterjee and Ip, 2009; Jiang et al., 2009). In the absence of infection, basal activation of JNK and Jak/Stat activities in ISCs by commensal bacteria is sufficient to maintain low levels of epithelial renewal in young flies (Buchon et al., 2009a). In older flies, however, the commensal population seems to cause excessive activation of JNK and JAK/Stat signaling, inducing the intestinal dysplasia described above. Accordingly, midguts from 30-day-old axenically raised flies exhibit a marked decrease in mitotic ISCs, as well as reduced levels of JNK and Jak/Stat signaling pathways compared with control animals (Buchon et al., 2009a).
It is likely that similar interactions between signals that regulate stem cell activity and inflammatory processes cause age-related deregulation of tissue homeostasis in barrier epithelia of vertebrates. Loss of tissue homeostasis and cancer progression is associated with an inflammatory state (Mantovani, 2005; Colotta et al., 2009; Mantovani, 2009), and individuals genetically predisposed to chronic inflammation display a higher incidence of various cancers, while tissue inflammation is present in most tumor microenvironments (Mantovani, 2005; Grivennikov et al., 2010).
This association between inflammation and cancer is particularly significant in barrier epithelia such as the intestinal epithelium, which mount frequent and required immune responses to pathogenic microorganisms (Xavier and Podolsky, 2007; Garrett et al., 2010). Accordingly, patients with inflammatory bowel diseases (including ulcerative colitis or Crohn's disease) have an increased risk of developing colon cancer (Gonda et al., 2009). Interestingly, studies to determine the genetic basis of colon cancer indicate that changes in microflora composition and load might be a prominent etiological factor for chronic inflammation and cancer development (Gonda et al., 2009; Uronis et al., 2009; Kaser et al., 2010; Niwa et al., 2010; Qin et al., 2010). Germ-free mice, accordingly, exhibit reduced intestinal inflammation and are refractory to carcinogen-induced colon tumor formation (Uronis et al., 2009).
Better mechanistic understanding of (1) the interactions between the microflora, (2) the innate immune responses and (3) the regenerative processes in the intestinal epithelium are thus key to the development of models of disease progression and are likely to significantly impact our understanding of the development of age-related tissue dysplasias. Intestinal dysplasia significantly limits Drosophila lifespan, and reducing the rate of stem cell proliferation in the aging intestine is sufficient to extend lifespan of the organism (Biteau et al., 2010; Rera et al., 2011). Similar management of stem cell proliferation in mammalian barrier epithelia is therefore likely to be beneficial for health and lifespan. Fly studies are leading the way to identify strategies that will promote stem cell quiescence, reduce dysplasia and improve health of the tissue. Reducing IIS and Tor activity, limiting JNK activation, promoting activity of the Nrf2 transcription factor, promoting mitochondrial function, as well as overexpressing general antioxidant genes and chaperones are all strategies that will limit ISC proliferation, delay or prevent dysplasia, and increase lifespan in this model (Biteau et al., 2010; Rera et al., 2011).
Conclusions and future directions
The integration of studies characterizing the tissue-specific regulation of metabolic and proliferative homeostasis with studies focusing on systemic control of metabolic homeostasis, fitness and longevity in flies promises to significantly advance our understanding of aging and the development of age-related diseases. It is clear that stress- and nutrient-responsive signaling pathways (including IIS/FOXO, TOR and JNK signaling) play a central role in governing homeostasis and longevity by coordinating metabolic functions, cell growth and proliferation, and stress responses throughout the organism (Figs 1, 2). However, many questions remain: how are these signaling pathways integrated under specific environmental (nutritional and stress) conditions; what are the major determinants that maintain the balance between acute stress responses, tissue repair and metabolic adaptation, and why does this balance degrade with age? It can be expected that further systematic analysis of the role of signaling interactions in the control of stress, regenerative and metabolic responses in the fly will continue to provide new insight and elucidate potential therapeutic avenues against age-associated pathologies.
FOOTNOTES
Competing interests
The authors declare no competing financial interests.
Funding
This work was supported by the National Institute on Aging (NIH RO1 AG028127), the National Institute on General Medical Sciences (NIH RO1 GM100196), the Ellison Medical Foundation (AG-SS-2224-08), and by AFAR/Ellison postdoctoral fellowships to J.K. and L.W. Deposited in PMC for release after 12 months.
References
- Accili D., Arden K. C. (2004). Foxos at the crossroads of cellular metabolism, differentiation, and transformation. Cell 117, 421-426 [DOI] [PubMed] [Google Scholar]
- Aguirre V., Uchida T., Yenush L., Davis R., White M. F. (2000). The c-Jun NH2-terminal kinase promotes insulin resistance during association with insulin receptor substrate-1 and phosphorylation of Ser307. J. Biol. Chem. 275, 9047-9054 [DOI] [PubMed] [Google Scholar]
- Amcheslavsky A., Jiang J., Ip Y. T. (2009). Tissue damage-induced intestinal stem cell division in Drosophila. Cell Stem Cell 4, 49-61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amcheslavsky A., Ito N., Jiang J., Ip Y. T. (2011). Tuberous sclerosis complex and Myc coordinate the growth and division of Drosophila intestinal stem cells. J. Cell Biol. 193, 695-710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apidianakis Y., Pitsouli C., Perrimon N., Rahme L. (2009). Synergy between bacterial infection and genetic predisposition in intestinal dysplasia. Proc. Natl. Acad. Sci. USA 106, 20883-20888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badin P. M., Louche K., Mairal A., Liebisch G., Schmitz G., Rustan A. C., Smith S. R., Langin D., Moro C. (2011). Altered skeletal muscle lipase expression and activity contribute to insulin resistance in humans. Diabetes 60, 1734-1742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai H., Kang P., Tatar M. (2012). Drosophila insulin-like peptide-6 (dilp6) expression from fat body extends lifespan and represses secretion of Drosophila insulin-like peptide-2 from the brain. Aging Cell 11, 978-985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargmann C. I., Horvitz H. R. (1991). Control of larval development by chemosensory neurons in Caenorhabditis elegans. Science 251, 1243-1246 [DOI] [PubMed] [Google Scholar]
- Barzilai N., Huffman D. M., Muzumdar R. H., Bartke A. (2012). The critical role of metabolic pathways in aging. Diabetes 61, 1315-1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biteau B., Jasper H. (2011). EGF signaling regulates the proliferation of intestinal stem cells in Drosophila. Development 138, 1045-1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biteau B., Hochmuth C. E., Jasper H. (2008). JNK activity in somatic stem cells causes loss of tissue homeostasis in the aging Drosophila gut. Cell Stem Cell 3, 442-455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biteau B., Karpac J., Supoyo S., Degennaro M., Lehmann R., Jasper H. (2010). Lifespan extension by preserving proliferative homeostasis in Drosophila. PLoS Genet. 6, e1001159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biteau B., Hochmuth C. E., Jasper H. (2011a). Maintaining tissue homeostasis: dynamic control of somatic stem cell activity. Cell Stem Cell 9, 402-411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biteau B., Karpac J., Hwangbo D., Jasper H. (2011b). Regulation of Drosophila lifespan by JNK signaling. Exp. Gerontol. 46, 349-354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blüher M., Kahn B. B., Kahn C. R. (2003). Extended longevity in mice lacking the insulin receptor in adipose tissue. Science 299, 572-574 [DOI] [PubMed] [Google Scholar]
- Bouzakri K., Zierath J. R. (2007). MAP4K4 gene silencing in human skeletal muscle prevents tumor necrosis factor-alpha-induced insulin resistance. J. Biol. Chem. 282, 7783-7789 [DOI] [PubMed] [Google Scholar]
- Broughton S., Partridge L. (2009). Insulin/IGF-like signalling, the central nervous system and aging. Biochem. J. 418, 1-12 [DOI] [PubMed] [Google Scholar]
- Broughton S. J., Piper M. D., Ikeya T., Bass T. M., Jacobson J., Driege Y., Martinez P., Hafen E., Withers D. J., Leevers S. J., et al. (2005). Longer lifespan, altered metabolism, and stress resistance in Drosophila from ablation of cells making insulin-like ligands. Proc. Natl. Acad. Sci. USA 102, 3105-3110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet A., Bonni A., Zigmond M. J., Lin M. Z., Juo P., Hu L. S., Anderson M. J., Arden K. C., Blenis J., Greenberg M. E. (1999). Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell 96, 857-868 [DOI] [PubMed] [Google Scholar]
- Brunet A., Sweeney L. B., Sturgill J. F., Chua K. F., Greer P. L., Lin Y., Tran H., Ross S. E., Mostoslavsky R., Cohen H. Y., et al. (2004). Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science 303, 2011-2015 [DOI] [PubMed] [Google Scholar]
- Buchon N., Broderick N. A., Chakrabarti S., Lemaitre B. (2009a). Invasive and indigenous microbiota impact intestinal stem cell activity through multiple pathways in Drosophila. Genes Dev. 23, 2333-2344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchon N., Broderick N. A., Poidevin M., Pradervand S., Lemaitre B. (2009b). Drosophila intestinal response to bacterial infection: activation of host defense and stem cell proliferation. Cell Host Microbe 5, 200-211 [DOI] [PubMed] [Google Scholar]
- Buchon N., Broderick N. A., Kuraishi T., Lemaitre B. (2010). Drosophila EGFR pathway coordinates stem cell proliferation and gut remodeling following infection. BMC Biol. 8, 152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campisi J., Sedivy J. (2009). How does proliferative homeostasis change with age? What causes it and how does it contribute to aging? J. Gerontol. A Biol. Sci. Med. Sci. 64A, 164-166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti P., Kandror K. V. (2009). Foxo1 controls insulin-dependent adipose triglyceride lipase (ATGL) expression and lipolysis in adipocytes. J. Biol. Chem. 284, 13296-13300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee M., Ip Y. T. (2009). Pathogenic stimulation of intestinal stem cell response in Drosophila. J. Cell. Physiol. 220, 664-671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chell J. M., Brand A. H. (2010). Nutrition-responsive glia control exit of neural stem cells from quiescence. Cell 143, 1161-1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi N. H., Kim J. G., Yang D. J., Kim Y. S., Yoo M. A. (2008). Age-related changes in Drosophila midgut are associated with PVF2, a PDGF/VEGF-like growth factor. Aging Cell 7, 318-334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi N. H., Lucchetta E., Ohlstein B. (2011). Nonautonomous regulation of Drosophila midgut stem cell proliferation by the insulin-signaling pathway. Proc. Natl. Acad. Sci. USA 108, 18702-18707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy D. J., Gems D., Harshman L. G., Oldham S., Stocker H., Hafen E., Leevers S. J., Partridge L. (2001). Extension of life-span by loss of CHICO, a Drosophila insulin receptor substrate protein. Science 292, 104-106 [DOI] [PubMed] [Google Scholar]
- Colombani J., Andersen D. S., Léopold P. (2012). Secreted peptide Dilp8 coordinates Drosophila tissue growth with developmental timing. Science 336, 582-585 [DOI] [PubMed] [Google Scholar]
- Colotta F., Allavena P., Sica A., Garlanda C., Mantovani A. (2009). Cancer-related inflammation, the seventh hallmark of cancer: links to genetic instability. Carcinogenesis 30, 1073-1081 [DOI] [PubMed] [Google Scholar]
- Conboy I. M., Rando T. A. (2012). Heterochronic parabiosis for the study of the effects of aging on stem cells and their niches. Cell Cycle 11, 2260-2267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis R. J. (2000). Signal transduction by the JNK group of MAP kinases. Cell 103, 239-252 [DOI] [PubMed] [Google Scholar]
- Demontis F., Perrimon N. (2010). FOXO/4E-BP signaling in Drosophila muscles regulates organism-wide proteostasis during aging. Cell 143, 813-825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X., Zhang W., O-Sullivan I., Williams J. B., Dong Q., Park E. A., Raghow R., Unterman T. G., Elam M. B. (2012). Foxo1 inhibits sterol regulatory element-binding protein-1c (SREBP-1c) gene expression via transcription factors Sp1 and SREBP-1c. J. Biol. Chem. 287, 20132-20143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eijkelenboom A., Burgering B. M. (2013). FOXOs: signalling integrators for homeostasis maintenance. Nat. Rev. Mol. Cell Biol. 14, 83-97 [DOI] [PubMed] [Google Scholar]
- Emanuelli B., Eberlé D., Suzuki R., Kahn C. R. (2008). Overexpression of the dual-specificity phosphatase MKP-4/DUSP-9 protects against stress-induced insulin resistance. Proc. Natl. Acad. Sci. USA 105, 3545-3550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essers M. A., Weijzen S., de Vries-Smits A. M., Saarloos I., de Ruiter N. D., Bos J. L., Burgering B. M. (2004). FOXO transcription factor activation by oxidative stress mediated by the small GTPase Ral and JNK. EMBO J. 23, 4802-4812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman D. B., Johnson T. E. (1988). Three mutants that extend both mean and maximum life span of the nematode, Caenorhabditis elegans, define the age-1 gene. J. Gerontol. 43, B102-B109 [DOI] [PubMed] [Google Scholar]
- Garelli A., Gontijo A. M., Miguela V., Caparros E., Dominguez M. (2012). Imaginal discs secrete insulin-like peptide 8 to mediate plasticity of growth and maturation. Science 336, 579-582 [DOI] [PubMed] [Google Scholar]
- Garinis G. A., van der Horst G. T., Vijg J., Hoeijmakers J. H. (2008). DNA damage and ageing: new-age ideas for an age-old problem. Nat. Cell Biol. 10, 1241-1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett W. S., Gordon J. I., Glimcher L. H. (2010). Homeostasis and inflammation in the intestine. Cell 140, 859-870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Géminard C., Rulifson E. J., Léopold P. (2009). Remote control of insulin secretion by fat cells in Drosophila. Cell Metab. 10, 199-207 [DOI] [PubMed] [Google Scholar]
- Giannakou M. E., Goss M., Jünger M. A., Hafen E., Leevers S. J., Partridge L. (2004). Long-lived Drosophila with overexpressed dFOXO in adult fat body. Science 305, 361 [DOI] [PubMed] [Google Scholar]
- Gonda T. A., Tu S., Wang T. C. (2009). Chronic inflammation, the tumor microenvironment and carcinogenesis. Cell Cycle 8, 2005-2013 [DOI] [PubMed] [Google Scholar]
- Goulas S., Conder R., Knoblich J. A. (2012). The Par complex and integrins direct asymmetric cell division in adult intestinal stem cells. Cell Stem Cell 11, 529-540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer E. L., Brunet A. (2008). Signaling networks in aging. J. Cell Sci. 121, 407-412 [DOI] [PubMed] [Google Scholar]
- Grivennikov S. I., Greten F. R., Karin M. (2010). Immunity, inflammation, and cancer. Cell 140, 883-899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L., Kenyon C. (2000). Genetic pathways that regulate ageing in model organisms. Nature 408, 255-262 [DOI] [PubMed] [Google Scholar]
- Han M. S., Jung D. Y., Morel C., Lakhani S. A., Kim J. K., Flavell R. A., Davis R. J. (2013). JNK expression by macrophages promotes obesity-induced insulin resistance and inflammation. Science 339, 218-222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes D. P. (2010). Nutritional hormesis and aging. Dose Response 8, 10-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertweck M., Göbel C., Baumeister R. (2004). C. elegans SGK-1 is the critical component in the Akt/PKB kinase complex to control stress response and life span. Dev. Cell 6, 577-588 [DOI] [PubMed] [Google Scholar]
- Hirosumi J., Tuncman G., Chang L., Görgün C. Z., Uysal K. T., Maeda K., Karin M., Hotamisligil G. S. (2002). A central role for JNK in obesity and insulin resistance. Nature 420, 333-336 [DOI] [PubMed] [Google Scholar]
- Holzenberger M., Dupont J., Ducos B., Leneuve P., Géloën A., Even P. C., Cervera P., Le Bouc Y. (2003). IGF-1 receptor regulates lifespan and resistance to oxidative stress in mice. Nature 421, 182-187 [DOI] [PubMed] [Google Scholar]
- Hotamisligil G. S. (1999). Mechanisms of TNF-alpha-induced insulin resistance. Exp. Clin. Endocrinol. Diabetes 107, 119-125 [DOI] [PubMed] [Google Scholar]
- Hotamisligil G. S. (2005). Role of endoplasmic reticulum stress and c-Jun NH2-terminal kinase pathways in inflammation and origin of obesity and diabetes. Diabetes 54 Suppl., S73-S78 [DOI] [PubMed] [Google Scholar]
- Hotamisligil G. S. (2006). Inflammation and metabolic disorders. Nature 444, 860-867 [DOI] [PubMed] [Google Scholar]
- Hotamisligil G. S., Peraldi P., Budavari A., Ellis R., White M. F., Spiegelman B. M. (1996). IRS-1-mediated inhibition of insulin receptor tyrosine kinase activity in TNF-α- and obesity-induced insulin resistance. Science 271, 665-668 [DOI] [PubMed] [Google Scholar]
- Hwangbo D. S., Gershman B., Tu M. P., Palmer M., Tatar M. (2004). Drosophila dFOXO controls lifespan and regulates insulin signalling in brain and fat body. Nature 429, 562-566 [DOI] [PubMed] [Google Scholar]
- Jaeschke A., Davis R. J. (2007). Metabolic stress signaling mediated by mixed-lineage kinases. Mol. Cell 27, 498-508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang Y. C., Sinha M., Cerletti M., Dall'Osso C., Wagers A. J. (2011). Skeletal muscle stem cells: effects of aging and metabolism on muscle regenerative function. Cold Spring Harb. Symp. Quant. Biol. 76, 101-111 [DOI] [PubMed] [Google Scholar]
- Jasper H., Jones D. L. (2010). Metabolic regulation of stem cell behavior and implications for aging. Cell Metab. 12, 561-565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H., Patel P. H., Kohlmaier A., Grenley M. O., McEwen D. G., Edgar B. A. (2009). Cytokine/Jak/Stat signaling mediates regeneration and homeostasis in the Drosophila midgut. Cell 137, 1343-1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H., Grenley M. O., Bravo M. J., Blumhagen R. Z., Edgar B. A. (2011). EGFR/Ras/MAPK signaling mediates adult midgut epithelial homeostasis and regeneration in Drosophila. Cell Stem Cell 8, 84-95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D. L., Rando T. A. (2011). Emerging models and paradigms for stem cell ageing. Nat. Cell Biol. 13, 506-512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn A., Olsen A. (2010). Stress to the rescue: is hormesis a ‘cure’ for aging? Dose Response 8, 48-52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapahi P. (2010). Protein synthesis and the antagonistic pleiotropy hypothesis of aging. Adv. Exp. Med. Biol. 694, 30-37 [DOI] [PubMed] [Google Scholar]
- Kapahi P., Chen D., Rogers A. N., Katewa S. D., Li P. W., Thomas E. L., Kockel L. (2010). With TOR, less is more: a key role for the conserved nutrient-sensing TOR pathway in aging. Cell Metab. 11, 453-465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapuria S., Karpac J., Biteau B., Hwangbo D., Jasper H. (2012). Notch-mediated suppression of TSC2 expression regulates cell differentiation in the Drosophila intestinal stem cell lineage. PLoS Genet. 8, e1003045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karin M., Gallagher E. (2005). From JNK to pay dirt: Jun kinases, their biochemistry, physiology and clinical importance. IUBMB Life 57, 283-295 [DOI] [PubMed] [Google Scholar]
- Karpac J., Jasper H. (2009). Insulin and JNK: optimizing metabolic homeostasis and lifespan. Trends Endocrinol. Metab. 20, 100-106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpac J., Hull-Thompson J., Falleur M., Jasper H. (2009). JNK signaling in insulin-producing cells is required for adaptive responses to stress in Drosophila. Aging Cell 8, 288-295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpac J., Younger A., Jasper H. (2011). Dynamic coordination of innate immune signaling and insulin signaling regulates systemic responses to localized DNA damage. Dev. Cell 20, 841-854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaser A., Zeissig S., Blumberg R. S. (2010). Inflammatory bowel disease. Annu. Rev. Immunol. 28, 573-621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katewa S. D., Kapahi P. (2010). Dietary restriction and aging, 2009. Aging Cell 9, 105-112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon C. (2005). The plasticity of aging: insights from long-lived mutants. Cell 120, 449-460 [DOI] [PubMed] [Google Scholar]
- Kenyon C. J. (2010). The genetics of ageing. Nature 464, 504-512 [DOI] [PubMed] [Google Scholar]
- Kenyon C., Chang J., Gensch E., Rudner A., Tabtiang R. (1993). A C. elegans mutant that lives twice as long as wild type. Nature 366, 461-464 [DOI] [PubMed] [Google Scholar]
- Kido Y., Burks D. J., Withers D., Bruning J. C., Kahn C. R., White M. F., Accili D. (2000). Tissue-specific insulin resistance in mice with mutations in the insulin receptor, IRS-1, and IRS-2. J. Clin. Invest. 105, 199-205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura K. D., Tissenbaum H. A., Liu Y., Ruvkun G. (1997). daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science 277, 942-946 [DOI] [PubMed] [Google Scholar]
- Kitamura T., Kahn C. R., Accili D. (2003). Insulin receptor knockout mice. Annu. Rev. Physiol. 65, 313-332 [DOI] [PubMed] [Google Scholar]
- Kops G. J., Dansen T. B., Polderman P. E., Saarloos I., Wirtz K. W., Coffer P. J., Huang T. T., Bos J. L., Medema R. H., Burgering B. M. (2002). Forkhead transcription factor FOXO3a protects quiescent cells from oxidative stress. Nature 419, 316-321 [DOI] [PubMed] [Google Scholar]
- Lemaitre B., Hoffmann J. (2007). The host defense of Drosophila melanogaster. Annu. Rev. Immunol. 25, 697-743 [DOI] [PubMed] [Google Scholar]
- Leopold P., Perrimon N. (2007). Drosophila and the genetics of the internal milieu. Nature 450, 186-188 [DOI] [PubMed] [Google Scholar]
- Li W., Kennedy S. G., Ruvkun G. (2003). daf-28 encodes a C. elegans insulin superfamily member that is regulated by environmental cues and acts in the DAF-2 signaling pathway. Genes Dev. 17, 844-858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libert S., Chao Y., Zwiener J., Pletcher S. D. (2008). Realized immune response is enhanced in long-lived puc and chico mutants but is unaffected by dietary restriction. Mol. Immunol. 45, 810-817 [DOI] [PubMed] [Google Scholar]
- Libina N., Berman J. R., Kenyon C. (2003). Tissue-specific activities of C. elegans DAF-16 in the regulation of lifespan. Cell 115, 489-502 [DOI] [PubMed] [Google Scholar]
- Lin G., Xu N., Xi R. (2008). Paracrine Wingless signalling controls self-renewal of Drosophila intestinal stem cells. Nature 455, 1119-1123 [DOI] [PubMed] [Google Scholar]
- Liu L., Rando T. A. (2011). Manifestations and mechanisms of stem cell aging. J. Cell Biol. 193, 257-266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luca F., Perry G. H., Di Rienzo A. (2010). Evolutionary adaptations to dietary changes. Annu. Rev. Nutr. 30, 291-314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X., Puig O., Hyun J., Bohmann D., Jasper H. (2007). Foxo and Fos regulate the decision between cell death and survival in response to UV irradiation. EMBO J. 26, 380-390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mair W., Dillin A. (2008). Aging and survival: the genetics of life span extension by dietary restriction. Annu. Rev. Biochem. 77, 727-754 [DOI] [PubMed] [Google Scholar]
- Mamay C. L., Mingo-Sion A. M., Wolf D. M., Molina M. D., Van Den Berg C. L. (2003). An inhibitory function for JNK in the regulation of IGF-I signaling in breast cancer. Oncogene 22, 602-614 [DOI] [PubMed] [Google Scholar]
- Manning A. M., Davis R. J. (2003). Targeting JNK for therapeutic benefit: from junk to gold? Nat. Rev. Drug Discov. 2, 554-565 [DOI] [PubMed] [Google Scholar]
- Mantovani A. (2005). Cancer: inflammation by remote control. Nature 435, 752-753 [DOI] [PubMed] [Google Scholar]
- Mantovani A. (2009). Cancer: inflaming metastasis. Nature 457, 36-37 [DOI] [PubMed] [Google Scholar]
- Matsumoto M., Accili D. (2005). All roads lead to Foxo. Cell Metab. 1, 215-216 [DOI] [PubMed] [Google Scholar]
- McLeod C. J., Wang L., Wong C., Jones D. L. (2010). Stem cell dynamics in response to nutrient availability. Curr. Biol. 20, 2100-2105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micchelli C. A., Perrimon N. (2006). Evidence that stem cells reside in the adult Drosophila midgut epithelium. Nature 439, 475-479 [DOI] [PubMed] [Google Scholar]
- Michael M. D., Kulkarni R. N., Postic C., Previs S. F., Shulman G. I., Magnuson M. A., Kahn C. R. (2000). Loss of insulin signaling in hepatocytes leads to severe insulin resistance and progressive hepatic dysfunction. Mol. Cell 6, 87-97 [PubMed] [Google Scholar]
- Mihaylova M. M., Vasquez D. S., Ravnskjaer K., Denechaud P. D., Yu R. T., Alvarez J. G., Downes M., Evans R. M., Montminy M., Shaw R. J. (2011). Class IIa histone deacetylases are hormone-activated regulators of FOXO and mammalian glucose homeostasis. Cell 145, 607-621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno T., Hisamoto N., Terada T., Kondo T., Adachi M., Nishida E., Kim D. H., Ausubel F. M., Matsumoto K. (2004). The Caenorhabditis elegans MAPK phosphatase VHP-1 mediates a novel JNK-like signaling pathway in stress response. EMBO J. 23, 2226-2234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy M. P., Partridge L. (2008). Toward a control theory analysis of aging. Annu. Rev. Biochem. 77, 777-798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy C. T., McCarroll S. A., Bargmann C. I., Fraser A., Kamath R. S., Ahringer J., Li H., Kenyon C. (2003). Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature 424, 277-283 [DOI] [PubMed] [Google Scholar]
- Niedernhofer L. J., Garinis G. A., Raams A., Lalai A. S., Robinson A. R., Appeldoorn E., Odijk H., Oostendorp R., Ahmad A., van Leeuwen W., et al. (2006). A new progeroid syndrome reveals that genotoxic stress suppresses the somatotroph axis. Nature 444, 1038-1043 [DOI] [PubMed] [Google Scholar]
- Nielsen M. D., Luo X., Biteau B., Syverson K., Jasper H. (2008). 14-3-3 Epsilon antagonizes Foxo to control growth, apoptosis and longevity in Drosophila. Aging Cell 7, 688-699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa T., Tsukamoto T., Toyoda T., Mori A., Tanaka H., Maekita T., Ichinose M., Tatematsu M., Ushijima T. (2010). Inflammatory processes triggered by Helicobacter pylori infection cause aberrant DNA methylation in gastric epithelial cells. Cancer Res. 70, 1430-1440 [DOI] [PubMed] [Google Scholar]
- O'Brien L. E., Soliman S. S., Li X., Bilder D. (2011). Altered modes of stem cell division drive adaptive intestinal growth. Cell 147, 603-614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odegaard J. I., Chawla A. (2013). Pleiotropic actions of insulin resistance and inflammation in metabolic homeostasis. Science 339, 172-177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S. W., Mukhopadhyay A., Svrzikapa N., Jiang F., Davis R. J., Tissenbaum H. A. (2005). JNK regulates lifespan in Caenorhabditis elegans by modulating nuclear translocation of forkhead transcription factor/DAF-16. Proc. Natl. Acad. Sci. USA 102, 4494-4499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlstein B., Spradling A. (2006). The adult Drosophila posterior midgut is maintained by pluripotent stem cells. Nature 439, 470-474 [DOI] [PubMed] [Google Scholar]
- Ohlstein B., Spradling A. (2007). Multipotent Drosophila intestinal stem cells specify daughter cell fates by differential notch signaling. Science 315, 988-992 [DOI] [PubMed] [Google Scholar]
- Okamoto N., Yamanaka N., Yagi Y., Nishida Y., Kataoka H., O'Connor M. B., Mizoguchi A. (2009). A fat body-derived IGF-like peptide regulates postfeeding growth in Drosophila. Dev. Cell 17, 885-891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozcan U., Cao Q., Yilmaz E., Lee A. H., Iwakoshi N. N., Ozdelen E., Tuncman G., Görgün C., Glimcher L. H., Hotamisligil G. S. (2004). Endoplasmic reticulum stress links obesity, insulin action, and type 2 diabetes. Science 306, 457-461 [DOI] [PubMed] [Google Scholar]
- Panowski S. H., Dillin A. (2009). Signals of youth: endocrine regulation of aging in Caenorhabditis elegans. Trends Endocrinol. Metab. 20, 259-264 [DOI] [PubMed] [Google Scholar]
- Parkes T. L., Elia A. J., Dickinson D., Hilliker A. J., Phillips J. P., Boulianne G. L. (1998). Extension of Drosophila lifespan by overexpression of human SOD1 in motorneurons. Nat. Genet. 19, 171-174 [DOI] [PubMed] [Google Scholar]
- Qin J., Li R., Raes J., Arumugam M., Burgdorf K. S., Manichanh C., Nielsen T., Pons N., Levenez F., Yamada T., et al. (2010). A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59-65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radtke F., Clevers H., Riccio O. (2006). From gut homeostasis to cancer. Curr. Mol. Med. 6, 275-289 [DOI] [PubMed] [Google Scholar]
- Rajan A., Perrimon N. (2012). Drosophila cytokine unpaired 2 regulates physiological homeostasis by remotely controlling insulin secretion. Cell 151, 123-137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rando T. A., Chang H. Y. (2012). Aging, rejuvenation, and epigenetic reprogramming: resetting the aging clock. Cell 148, 46-57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rera M., Bahadorani S., Cho J., Koehler C. L., Ulgherait M., Hur J. H., Ansari W. S., Lo T., Jr, Jones D. L., Walker D. W. (2011). Modulation of longevity and tissue homeostasis by the Drosophila PGC-1 homolog. Cell Metab. 14, 623-634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rera M., Clark R. I., Walker D. W. (2012). Intestinal barrier dysfunction links metabolic and inflammatory markers of aging to death in Drosophila. Proc. Natl. Acad. Sci. USA 109, 21528-21533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rera M., Azizi M. J., Walker D. W. (2013). Organ-specific mediation of lifespan extension: more than a gut feeling? Ageing Res. Rev. 12, 436-444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson R. B. (2009). Ionizing radiation and aging: rejuvenating an old idea. Aging 1, 887-902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts S. B., Rosenberg I. (2006). Nutrition and aging: changes in the regulation of energy metabolism with aging. Physiol. Rev. 86, 651-667 [DOI] [PubMed] [Google Scholar]
- Russell S. J., Kahn C. R. (2007). Endocrine regulation of ageing. Nat. Rev. Mol. Cell Biol. 8, 681-691 [DOI] [PubMed] [Google Scholar]
- Sabio G., Davis R. J. (2010). cJun NH2-terminal kinase 1 (JNK1): roles in metabolic regulation of insulin resistance. Trends Biochem. Sci. 35, 490-496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabio G., Das M., Mora A., Zhang Z., Jun J. Y., Ko H. J., Barrett T., Kim J. K., Davis R. J. (2008). A stress signaling pathway in adipose tissue regulates hepatic insulin resistance. Science 322, 1539-1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saltiel A. R., Kahn C. R. (2001). Insulin signalling and the regulation of glucose and lipid metabolism. Nature 414, 799-806 [DOI] [PubMed] [Google Scholar]
- Samuel V. T., Shulman G. I. (2012). Mechanisms for insulin resistance: common threads and missing links. Cell 148, 852-871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenborn V., Heid I. M., Vollmert C., Lingenhel A., Adams T. D., Hopkins P. N., Illig T., Zimmermann R., Zechner R., Hunt S. C., et al. (2006). The ATGL gene is associated with free fatty acids, triglycerides, and type 2 diabetes. Diabetes 55, 1270-1275 [DOI] [PubMed] [Google Scholar]
- Schumacher B. (2009). Transcription-blocking DNA damage in aging: a mechanism for hormesis. Bioessays 31, 1347-1356 [DOI] [PubMed] [Google Scholar]
- Schumacher B., Garinis G. A., Hoeijmakers J. H. (2008). Age to survive: DNA damage and aging. Trends Genet. 24, 77-85 [DOI] [PubMed] [Google Scholar]
- Shimomura I., Matsuda M., Hammer R. E., Bashmakov Y., Brown M. S., Goldstein J. L. (2000). Decreased IRS-2 and increased SREBP-1c lead to mixed insulin resistance and sensitivity in livers of lipodystrophic and ob/ob mice. Mol. Cell 6, 77-86 [PubMed] [Google Scholar]
- Sieber M. H., Thummel C. S. (2012). Coordination of triacylglycerol and cholesterol homeostasis by DHR96 and the Drosophila LipA homolog magro. Cell Metab. 15, 122-127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaidina M., Delanoue R., Gronke S., Partridge L., Léopold P. (2009). A Drosophila insulin-like peptide promotes growth during nonfeeding states. Dev. Cell 17, 874-884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solinas G., Vilcu C., Neels J. G., Bandyopadhyay G. K., Luo J. L., Naugler W., Grivennikov S., Wynshaw-Boris A., Scadeng M., Olefsky J. M., et al. (2007). JNK1 in hematopoietically derived cells contributes to diet-induced inflammation and insulin resistance without affecting obesity. Cell Metab. 6, 386-397 [DOI] [PubMed] [Google Scholar]
- Sousa-Nunes R., Yee L. L., Gould A. P. (2011). Fat cells reactivate quiescent neuroblasts via TOR and glial insulin relays in Drosophila. Nature 471, 508-512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Molitor J., Tower J. (2004). Effects of simultaneous over-expression of Cu/ZnSOD and MnSOD on Drosophila melanogaster life span. Mech. Ageing Dev. 125, 341-349 [DOI] [PubMed] [Google Scholar]
- Sunayama J., Tsuruta F., Masuyama N., Gotoh Y. (2005). JNK antagonizes Akt-mediated survival signals by phosphorylating 14-3-3. J. Cell Biol. 170, 295-304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki R., Lee K., Jing E., Biddinger S. B., McDonald J. G., Montine T. J., Craft S., Kahn C. R. (2010). Diabetes and insulin in regulation of brain cholesterol metabolism. Cell Metab. 12, 567-579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi A., Wartschow L. M., White M. F. (2007). Brain IRS2 signaling coordinates life span and nutrient homeostasis. Science 317, 369-372 [DOI] [PubMed] [Google Scholar]
- Tatar M., Kopelman A., Epstein D., Tu M. P., Yin C. M., Garofalo R. S. (2001). A mutant Drosophila insulin receptor homolog that extends life-span and impairs neuroendocrine function. Science 292, 107-110 [DOI] [PubMed] [Google Scholar]
- Tatar M., Bartke A., Antebi A. (2003). The endocrine regulation of aging by insulin-like signals. Science 299, 1346-1351 [DOI] [PubMed] [Google Scholar]
- Taylor R. C., Dillin A. (2011). Aging as an event of proteostasis collapse. Cold Spring Harb. Perspect. Biol. 3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teleman A. A. (2010). Molecular mechanisms of metabolic regulation by insulin in Drosophila. Biochem. J. 425, 13-26 [DOI] [PubMed] [Google Scholar]
- Tissenbaum H. A., Guarente L. (2002). Model organisms as a guide to mammalian aging. Dev. Cell 2, 9-19 [DOI] [PubMed] [Google Scholar]
- Toivonen J. M., Partridge L. (2009). Endocrine regulation of aging and reproduction in Drosophila. Mol. Cell. Endocrinol. 299, 39-50 [DOI] [PubMed] [Google Scholar]
- Tower J. (2011). Heat shock proteins and Drosophila aging. Exp. Gerontol. 46, 355-362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukumo D. M., Carvalho-Filho M. A., Carvalheira J. B., Prada P. O., Hirabara S. M., Schenka A. A., Araújo E. P., Vassallo J., Curi R., Velloso L. A., et al. (2007). Loss-of-function mutation in Toll-like receptor 4 prevents diet-induced obesity and insulin resistance. Diabetes 56, 1986-1998 [DOI] [PubMed] [Google Scholar]
- Tsuruta F., Sunayama J., Mori Y., Hattori S., Shimizu S., Tsujimoto Y., Yoshioka K., Masuyama N., Gotoh Y. (2004). JNK promotes Bax translocation to mitochondria through phosphorylation of 14-3-3 proteins. EMBO J. 23, 1889-1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uronis J. M., Mühlbauer M., Herfarth H. H., Rubinas T. C., Jones G. S., Jobin C. (2009). Modulation of the intestinal microbiota alters colitis-associated colorectal cancer susceptibility. PLoS ONE 4, e6026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Pluijm I., Garinis G. A., Brandt R. M., Gorgels T. G., Wijnhoven S. W., Diderich K. E., de Wit J., Mitchell J. R., van Oostrom C., Beems R., et al. (2007). Impaired genome maintenance suppresses the growth hormone–insulin-like growth factor 1 axis in mice with Cockayne syndrome. PLoS Biol. 5, e2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vihervaara T., Puig O. (2008). dFOXO regulates transcription of a Drosophila acid lipase. J. Mol. Biol. 376, 1215-1223 [DOI] [PubMed] [Google Scholar]
- Waeber G., Delplanque J., Bonny C., Mooser V., Steinmann M., Widmann C., Maillard A., Miklossy J., Dina C., Hani E. H., et al. (2000). The gene MAPK8IP1, encoding islet-brain-1, is a candidate for type 2 diabetes. Nat. Genet. 24, 291-295 [DOI] [PubMed] [Google Scholar]
- Wang M. C., Bohmann D., Jasper H. (2003). JNK signaling confers tolerance to oxidative stress and extends lifespan in Drosophila. Dev. Cell 5, 811-816 [DOI] [PubMed] [Google Scholar]
- Wang M. C., Bohmann D., Jasper H. (2005). JNK extends life span and limits growth by antagonizing cellular and organism-wide responses to insulin signaling. Cell 121, 115-125 [DOI] [PubMed] [Google Scholar]
- Wang B., Moya N., Niessen S., Hoover H., Mihaylova M. M., Shaw R. J., Yates J. R., III, Fischer W. H., Thomas J. B., Montminy M. (2011). A hormone-dependent module regulating energy balance. Cell 145, 596-606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessells R. J., Fitzgerald E., Cypser J. R., Tatar M., Bodmer R. (2004). Insulin regulation of heart function in aging fruit flies. Nat. Genet. 36, 1275-1281 [DOI] [PubMed] [Google Scholar]
- Weston C. R., Davis R. J. (2002). The JNK signal transduction pathway. Curr. Opin. Genet. Dev. 12, 14-21 [DOI] [PubMed] [Google Scholar]
- Wolf M., Nunes F., Henkel A., Heinick A., Paul R. J. (2008). The MAP kinase JNK-1 of Caenorhabditis elegans: location, activation, and influences over temperature-dependent insulin-like signaling, stress responses, and fitness. J. Cell. Physiol. 214, 721-729 [DOI] [PubMed] [Google Scholar]
- Wolkow C. A., Kimura K. D., Lee M. S., Ruvkun G. (2000). Regulation of C. elegans life-span by insulinlike signaling in the nervous system. Science 290, 147-150 [DOI] [PubMed] [Google Scholar]
- Xavier R. J., Podolsky D. K. (2007). Unravelling the pathogenesis of inflammatory bowel disease. Nature 448, 427-434 [DOI] [PubMed] [Google Scholar]
- Xu X., Gopalacharyulu P., Seppänen-Laakso T., Ruskeepää A. L., Aye C. C., Carson B. P., Mora S., Orešič M., Teleman A. A. (2012). Insulin signaling regulates fatty acid catabolism at the level of CoA activation. PLoS Genet. 8, e1002478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R., Wilcox D. M., Haasch D. L., Jung P. M., Nguyen P. T., Voorbach M. J., Doktor S., Brodjian S., Bush E. N., Lin E., et al. (2007). Liver-specific knockdown of JNK1 up-regulates proliferator-activated receptor γ coactivator 1β and increases plasma triglyceride despite reduced glucose and insulin levels in diet-induced obese mice. J. Biol. Chem. 282, 22765-22774 [DOI] [PubMed] [Google Scholar]
- Yin M. J., Yamamoto Y., Gaynor R. B. (1998). The anti-inflammatory agents aspirin and salicylate inhibit the activity of IκB kinase-β. Nature 396, 77-80 [DOI] [PubMed] [Google Scholar]
- Yoshida K., Yamaguchi T., Natsume T., Kufe D., Miki Y. (2005). JNK phosphorylation of 14-3-3 proteins regulates nuclear targeting of c-Abl in the apoptotic response to DNA damage. Nat. Cell Biol. 7, 278-285 [DOI] [PubMed] [Google Scholar]
- Zhang P., Judy M., Lee S. J., Kenyon C. (2013). Direct and indirect gene regulation by a life-extending FOXO protein in C. elegans: roles for GATA factors and lipid gene regulators. Cell Metab. 17, 85-100 [DOI] [PMC free article] [PubMed] [Google Scholar]