Abstract
Epigenetic phenomena have sparked much interest resulting in an exponential increase in scientific investigation in the last two decades. While growing, the field of environmental epigenetics remains small when compared to other areas of epigenetic inquiry such as cancer research. In this commentary, our objective is to describe the status of the field of environmental epigenetics and lay out our vision for its future. While environmental epigenetic studies represent fewer than 5% of all epigenetic publications, the National Institute of Environmental Health Sciences ranks second in proportion of dollars spent on epigenetics of all NIH Institutes. Such investment highlights the hypothesis that epigenetic marks are modified by environmental exposures and the hope that interventions targeted at epigenetic mechanisms may ultimately lead to improved health outcomes. The road to achieve this vision will require 1) attention to tissue specificity, 2) focused interventional studies, 3) collaboration among cohorts, 4) inclusion of environmental exposures in new large-scale epigenomic studies, and 5) understanding of multiple mechanisms beyond DNA methylation and histone modifications. The investment in environmental epigenetic inquiry will lead to great rewards if we can understand the biology of how phenotype results from environmental stimuli and genetic code. Understanding the epigenetic implications of our actions and exposures may benefit generations to come.
Keywords: Epigenetics, environmental health, DNA methylation, microRNA, histone modifications
Introduction
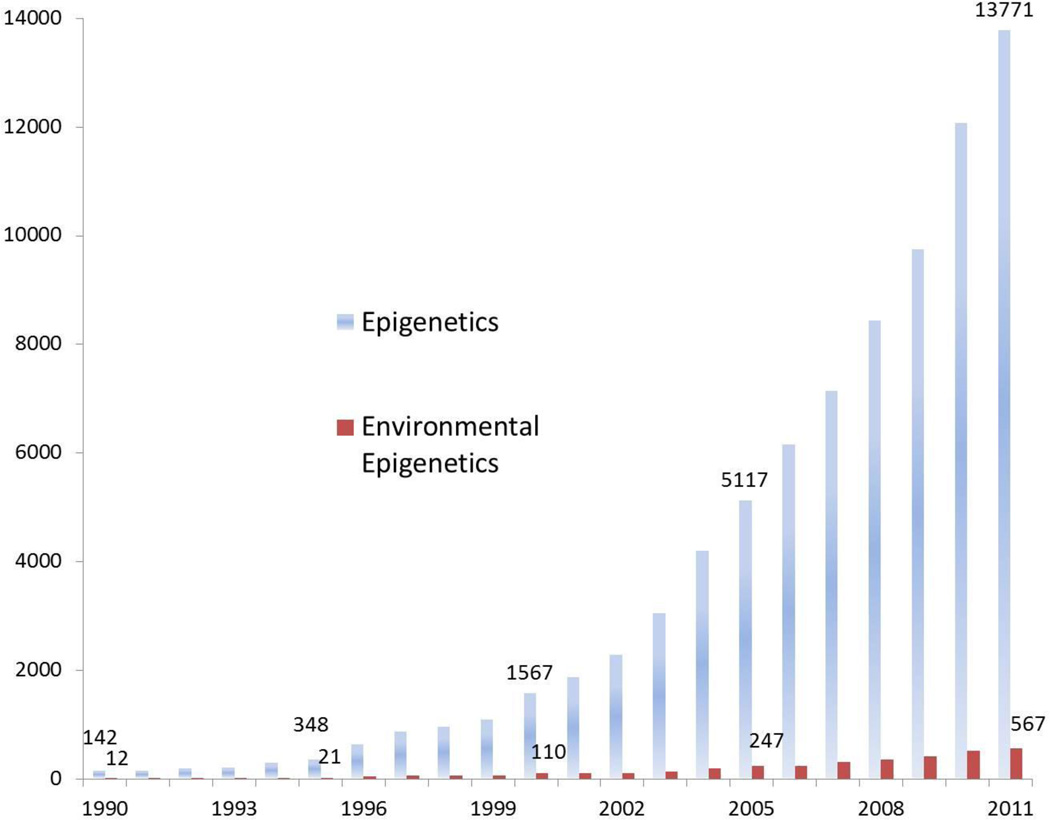
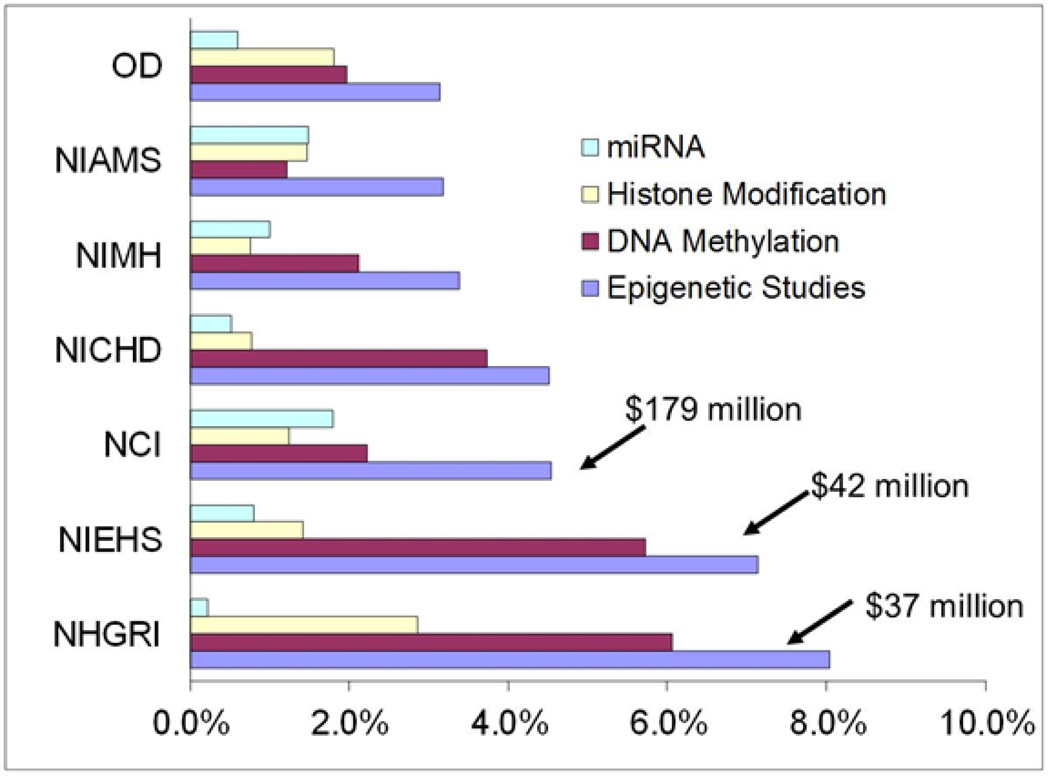
Interest in epigenetic phenomena has dramatically intensified over the last two decades with an exponential increase in PubMed-indexed publications from fewer than 150 in 1990 to over 13,000 in 2011 (figure 1). Despite this surge, proportionally few publications in epigenetics are devoted to assessing the role of the environment, with 12 citations in 1990 and 567 in 2011. Publically available data from the NIH RePORTER reveal that the National Institutes of Health (NIH) spent over $700 million (2.8% of their total costs) on epigenetics in 2012. The National Cancer Institute (NCI) invests the most total dollars (over $179 million or 4.5% of its budget) toward epigenetic studies (Figure 2). However, NIH-wide the National Institute of Environmental Health Sciences (NIEHS) is one of the top two Institutes in proportion of overall spending on epigenetic projects – second only to the National Human Genome Research Institute (NHGRI) (NIEHS’s 7.1% vs. NHGRI’s 8.0%). Such investment highlights the pervasive confidence among NIH and scientific researchers that understanding epigenetic mechanisms, including environmental influences on the epigenome, will result in far-reaching basic science, clinical, and public health implications. In this commentary we aim to discuss and highlight the factors crucial to moving the field of environmental epigenetics forward to deliver high-impact mechanistic results for improved public health..
Figure 1. Number of epigenetica and environmental epigeneticb publications since 1990.
aPubmed Search terms: Epigenesis, Genetic"[Mesh] or "DNA methylation"[mesh] or "microRNAs"[mesh] or "histone modification" or epigenetic;b "Air Pollutants"[Mesh] OR "Water Pollutants, Radioactive"[Mesh] OR "Soil Pollutants, Radioactive"[Mesh] OR "Air Pollutants, Radioactive"[Mesh] OR "Water Pollutants, Chemical"[Mesh] OR "Water Pollutants"[Mesh] OR "Soil Pollutants"[Mesh] OR "Radioactive Pollutants"[Mesh] OR "Environmental Pollutants"[Mesh] OR "Particulate Matter"[Mesh] OR "Water Pollution, Chemical"[Mesh]) OR "Radon"[Mesh] OR "Polychlorinated Biphenyls"[Mesh] OR "Polybrominated Biphenyls"[Mesh] OR "Toxic Actions"[Mesh] OR "Environmental Exposure"[Mesh] OR "Air Pollution"[Mesh] OR "Enviroment"[Mesh] and prior search
Figure 2. Percentage of Institute/Center Total Costs for FY 2012 Used for Epigenetic Studiesa Overall, and by Mechanism (limited to I/Cs spending >3% of their budgets on epigenetics).
aData from NIH RePORTER accessed 10/26/12, search terms: “DNA methylation” or “histone modification” or “microRNA”, Epigenetic Studies” is not the sum of the others because more than one mechanism are included in some projects.
Epigenetic marks are modifiable
Epigenetics refers to heritable changes in phenotype unrelated to differences in underlying DNA sequence. When explaining epigenetics to a broad audience or teaching epigenetics to students, we often describe a process similar to a conductor’s notation of a musical score. The symphony written by the composer represents the DNA sequence. The performance heard in the musical hall is the phenotype. In this musical example, a conductor’s tiny pencil marks, or more permanent ink blots, represent the epigenetic marks that alter any given performance. These epigenetic musical notations do not change the score (i.e., the musical DNA) but they will change the performance (i.e., the musical phenotype) resulting from the score. Akin to conductor’s notations, many epigenetic marks contribute to alterations in gene expression. DNA methylation, histone modifications and microRNAs represent the most-studied mechanisms. These epigenetic phenomena attract epidemiologists, clinical researchers, and basic scientists alike because they can be potentially modified and result in environmental reprogramming of the genome. If environmental toxicants, nutritional factors, and social experiences affect disease risk through epigenetic mechanisms, then a new, wide-open field of diagnostic tests and pharmacotherapeutic/dietary interventions could evolve.
The concept of “developmental programming” of phenotype for exposed individuals and potentially future generations of offspring adds substantial intrigue to gene-environment interactions. Pioneering investigations such as those conducted by Skinner,(Anway et al. 2005; Crews et al. 2012) Jirtle, (Bernal et al. 2012; Waterland and Jirtle 2003) and Dolinoy, (Dolinoy et al. 2006; Dolinoy 2007) have demonstrated through elegant animal models that nutritional (Dolinoy et al. 2006) and environmental (Dolinoy et al. 2007) exposures during pregnancy through lactation can affect disease risk. Dolinoy et al. (2007) further demonstrated in mice that adverse exposures can be mitigated through rescue diets high in methyl donors, such as folic acid or phytoestrogens, such as genistein. Equally fascinating are the findings of Bernal et al. (2012) that low dose radiation elicits a positive adaptive response of increased DNA methylation in similar mouse models that can be interrupted by antioxidant diets. Such enticing findings have prompted human cohort studies to evaluate whether diseases associated with environmental exposures may work through epigenetic phenomena. However, cohort studies are plagued by not only the intrinsic limitations of human observational studies, but also by challenges specific to epigenetic inquiry.
Tissue Specificity
A major challenge in epigenetic research stems from its tissue specificity. While the genome is virtually identical in all diploid cells from the same individual, each tissue and potentially each individual cell or cell type may exhibit a unique epigenomic profile. Cancer scientists have historically led the way into epigenetic studies that took full advantage of the availability of tissue samples after biopsies or tumor resections. Their ability to study large amounts of DNA or other cell products has allowed for massive growth in epigenetic laboratory techniques and has led to the development of clinical applications of epigenetic analyses to help oncologists in fine-tuning prognoses (Lao and Grady 2011) and offering better-tailored therapies for their patients, such as the use of histone deacetylase inhibitors (Harrison et al. 2012). However, even epigenetic studies that focus on the diseased tissue, such as cancer studies, have limitations. While diseased tissue is widely available, appropriate tissue controls are scarce. Investigators often use adjacent non-cancerous tissue as the control which can have correlated DNA methylation patterns due to genetic concordance or systemic inflammation from the disease state. Other studies use tissues obtained from symptomatic patients with benign biopsies. These subjects may be cancer-free but may not be generalizable to healthy controls.
For non-cancer cohort studies examining epigenetics in preclinical populations with specific environmental exposures, epigenetic analyses are even less straightforward. Surrogate tissues that can be easily and non-invasively obtained from individuals who do not need surgical procedures do not always represent the tissue of interest. For example, investigators interested in whether epigenetic factors affect childhood neurodevelopment may not have access to pediatric brain specimens due the invasive techniques necessary to collect such samples. Instead, they use products of conception (placentas), cheek swabs, or circulating leukocytes from blood samples. Surrogate tissues have been shown to have varying correlations with target tissues when evaluated in parallel, but most certainly do not achieve the ultimate goal of direct evaluation of the tissue of interest. Better understanding of which subsets of epigenetic signatures are correlated across tissues may help to delineate which surrogate tissues are appropriate for human studies of difficult to obtain target tissues. However, even within tissues, each human cell has ultimately a unique epigenome that will differ, to a variable extent, from that of even the closest cell. Local paracrine factors, variable proportions of different cell types (e.g., epithelial, inflammatory, and mesenchymal cells in lung tissues; or neutrophils, lymphocytes and their subtypes, monocytes, basophils and eosinophils circulating in blood), cell age and cell location may affect epigenetic marks, introducing many potential sources of variability. Such complexity requires novel bioinformatics techniques to address cell specificity and improve interpretability of epigenetic data (Houseman et al. 2012).
Human environmental studies, a move toward interventions
Despite these inherent challenges, epigenetic studies have generated immense hope for progress and innovation in medicine and public health. Some of the initial findings in environmental health have fueled this hope. For instance, human studies of air pollution have repeatedly demonstrated an association between exposure to air pollution and DNA methylation of circulating leukocytes (Baccarelli et al. 2010; Hou et al. 2012). However, the literature is not yet conclusive (Terry et al. 2011). This has prompted controlled human experiments, in progress at multiple institutions, in which subjects’ DNA methylation is measured before and after exposure to different air pollutants. The use of such controlled studies in environmental health should help to definitively link exposure to epigenetic marks, as well as to related phenotypes. There is a precedent for this type of study design in epigenetic studies from other non-environmental fields. For example, Barrès et al. exposed subjects to exercise and demonstrated DNA methylation changes in skeletal muscle (Barres et al. 2012). Whenever possible and ethical, definitive interventional studies – including controlled human exposure or mitigation studies – should be done to confirm associations between exposures and epigenetic marks.
When controlled studies are not feasible, natural experiments can shed light on mechanisms. One of the best epigenetic examples is that of the Dutch Famine Study which revealed persistent differences in DNA methylation in the insulin-like growth factor-2 (IGF2) imprinted region in offspring exposed early in gestation compared to unexposed siblings (Heijmans et al. 2008). This landmark study gave credence to the hypothesis that epigenetic mechanisms may underpin epidemiologic observation that lead to the Barker Hypothesis of the Developmental Origins of Health and Disease (DOHaD) which proposes that early life exposures can have long lasting health impacts into adulthood (Barker et al. 1989). DNA methylation patterns are largely erased and reestablished early in embryogenesis, and fetal and early life represents a uniquely susceptible window in which environmental exposures can potentially reprogram the epigenome and modify postnatal health trajectories.
A call for consortia
The growing availability of technologies to conduct whole genome-scale scans of large swaths of the human epigenome has lead epigeneticists into a situation familiar to scientists involved in genome-wide association studies (GWAS). These technologies allow for investigating DNA methylation states of hundreds of thousands, if not millions, of individual nucleotides. The risks of false positive findings and technical artifacts, arising from technology- and study- dependent idiosyncrasies, cannot be underestimated. After the current spur of single-cohort studies, consortia of those cohorts may be able to answer epigenetic questions better than any of them in isolation. While this applies to all types of human studies, birth cohorts have been a centerpiece of recent and current epigenetic studies and provide a unique opportunity to build the prototype of epigenetic consortia. We argue that going forward, with each birth cohort contributing 1000–2000 or more samples, consortia should form for the purpose of meta-analyses that would identify signals otherwise undetectable in each individual cohort. Such techniques are not limited to birth cohort studies. Cohorts focused on cardiovascular health, obesity, lung disease, atopic/allergic disorders and autoimmune disease may also benefit from forming consortia to analyze epigenome-wide association study (EWAS), data which should lead to discoveries impossible to make at the individual cohort level.
Expansion of Environmental Epigenetics
An exciting aspect of field of epigenetics is the opportunity to integrate exposures as a type of “exposome.”(Manikkam et al. 2012) Environmental, nutritional and social exposures all affect gene expression and epigenetic mechanisms likely, at least partially, explain how this occurs. However, such exposures are not always known to an individual or a community. In addition to testing for various cancers (e.g. ovarian cancer (Teschendorff et al. 2009)), epigenetic signatures will likely have applications to environmental exposures and disease. To consider epigenetic marks as a complex history taking tool that could uncover exposures early in development and help clinicians to implement preventative clinical interventions or target public health surveillance represents a potential pragmatic applicability to epigenetics. We recognize the challenges associated with obtaining ideal tissues for such inquiries, but even blood born, epigenetic signatures could provide considerable insights into the contributions of environmental exposures to human diseases. While critically important, the Human Reference Epigenome Mapping Project and ENCODE do not necessarily focus on the environment. We call for future studies to incorporate environmental, nutritional, and social factors in addition to addressing sex-specific effects to most completely characterize the epigenetic signatures of an individual’s cumulative exposures.
Expanding Mechanisms
Human studies, thus far, have typically focused on individual epigenetic mechanisms. For example, environmental health investigators have primarily analyzed DNA methylation as shown by funding patterns of different epigenetic studies (Figure 2). NIEHS currently funds analyses of DNA methylation at almost three times that of histone modifications and miRNAs combined. The ease of preserving, storing and transporting DNA compared to the more care-intensive approaches to RNA and chromatin makes DNA methylation the least costly analytic approach. However, more than one mechanism could be studied in concert to better understand cellular function. Additionally, there are exciting developments in other DNA methylation sites beyond traditional methylated CpG sites such as 5-hydroxymethyl-cytosine analyses, mitochondrial DNA methylation, or non-CpG methylation that should be explored. Furthermore, gene-epigene interactions should be examined as demonstrated by studies revealing that DNA methylation differences can be a result of underlying SNPs (Morales et al. 2012). Only by studying how each of these aspects of gene regulation and programming act in conjunction with one another and contribute to gene expression, can we both understand the truths underlying epigenetic phenomena and best position the field to make an impact on human health.
Conclusion
Environmental epigenetics represents a rapidly growing, promising field that is expected to lead to clinical and public health interventions. It relies on intense, and at times challenging, collaboration among basic scientists, epidemiologists, toxicologists and clinicians. We believe toxicologists are uniquely positioned to move this field forward based on the premise that both toxic exposures and our epigenetic responses are potentially modifiable. The rewards will be great if we can understand the biology of how our bodies integrate the world and genetic code we inherit and understand the implications of our actions and exposures on generations to come.
Questions addressed in this mini-review
-
◦
Why do we study epigenetics?
-
◦
What are the limitations of epigenetic studies in human cohorts?
-
◦
How might consortia help us make epigenetic discoveries?
-
◦
Why epigenetics is uniquely suited for trans-disciplinary and translational research?
-
◦
Which epigenetic mechanisms are currently understudied in environmental health?
Acknowledgements
We would like to recognize our colleagues Dana C. Dolinoy, PhD and Allan C. Just, PhD for their thoughtful editing of this piece prior to submission. Dr. Burris is funded by NIH-NIEHS: K23 ES02224 and the Klarman Foundation at Beth Israel Deaconess Medical Center, Dr. Baccarelli is funded through multiple NIEHS grants on epigenetics and environmental health (R01ES020836; R01ES020268; R01ES021357; R21ES020010; R21ES020984) and by the HSPH-NIEHS Center for Environmental Health (P30ES00002).
Footnotes
Competing Financial Interests: We have no competing financial interests to declare.
References
- Anway MD, Cupp AS, Uzumcu M, Skinner MK. Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science. 2005;308:1466–1469. doi: 10.1126/science.1108190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baccarelli A, Tarantini L, Wright RO, Bollati V, Litonjua AA, Zanobetti A, et al. Repetitive element DNA methylation and circulating endothelial and inflammation markers in the va normative aging study. Epigenetics. 2010;5 doi: 10.4161/epi.5.3.11377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker DJ, Osmond C, Golding J, Kuh D, Wadsworth ME. Growth in utero, blood pressure in childhood and adult life, and mortality from cardiovascular disease. BMJ. 1989;298:564–567. doi: 10.1136/bmj.298.6673.564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barres R, Yan J, Egan B, Treebak JT, Rasmussen M, Fritz T, et al. Acute exercise remodels promoter methylation in human skeletal muscle. Cell metabolism. 2012;15:405–411. doi: 10.1016/j.cmet.2012.01.001. [DOI] [PubMed] [Google Scholar]
- Bernal AJ, Dolinoy DC, Huang D, Skaar DA, Weinhouse C, Jirtle RL. Adaptive radiation-induced epigenetic alterations mitigated by antioxidants. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2012 doi: 10.1096/fj.12-220350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crews D, Gillette R, Scarpino SV, Manikkam M, Savenkova MI, Skinner MK. Epigenetic transgenerational inheritance of altered stress responses. Proc Natl Acad Sci U S A. 2012;109:9143–9148. doi: 10.1073/pnas.1118514109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolinoy DC, Weidman JR, Waterland RA, Jirtle RL. Maternal genistein alters coat color and protects avy mouse offspring from obesity by modifying the fetal epigenome. Environ Health Perspect. 2006;114:567–572. doi: 10.1289/ehp.8700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolinoy DC. Epigenetic gene regulation: Early environmental exposures. Pharmacogenomics. 2007;8:5–10. doi: 10.2217/14622416.8.1.5. [DOI] [PubMed] [Google Scholar]
- Dolinoy DC, Huang D, Jirtle RL. Maternal nutrient supplementation counteracts bisphenol a-induced DNA hypomethylation in early development. Proc Natl Acad Sci U S A. 2007;104:13056–13061. doi: 10.1073/pnas.0703739104. 10.1073/pnas.0703739104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison SJ, Bishton M, Bates SE, Grant S, Piekarz RL, Johnstone RW, et al. A focus on the preclinical development and clinical status of the histone deacetylase inhibitor, romidepsin (depsipeptide, istodax((r))) Epigenomics. 2012;4:571–589. doi: 10.2217/epi.12.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heijmans BT, Tobi EW, Stein AD, Putter H, Blauw GJ, Susser ES, et al. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc Natl Acad Sci U S A. 2008;105:17046–17049. doi: 10.1073/pnas.0806560105. 10.1073/pnas.0806560105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou L, Wang S, Dou C, Zhang X, Yu Y, Zheng Y, et al. Air pollution exposure and telomere length in highly exposed subjects in beijing, china: A repeated-measure study. Environment international. 2012;48:71–77. doi: 10.1016/j.envint.2012.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC bioinformatics. 2012;13:86. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lao VV, Grady WM. Epigenetics and colorectal cancer. Nature reviews Gastroenterology & hepatology. 2011;8:686–700. doi: 10.1038/nrgastro.2011.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manikkam M, Guerrero-Bosagna C, Tracey R, Haque MM, Skinner MK. Transgenerational actions of environmental compounds on reproductive disease and identification of epigenetic biomarkers of ancestral exposures. PLoS One. 2012;7:e31901. doi: 10.1371/journal.pone.0031901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales E, Bustamante M, Vilahur N, Escaramis G, Montfort M, de Cid R, et al. DNA hypomethylation at alox12 is associated with persistent wheezing in childhood. Am J Respir Crit Care Med. 2012;185:937–943. doi: 10.1164/rccm.201105-0870OC. [DOI] [PubMed] [Google Scholar]
- Terry MB, Delgado-Cruzata L, Vin-Raviv N, Wu HC, Santella RM. DNA methylation in white blood cells: Association with risk factors in epidemiologic studies. Epigenetics. 2011;6:828–837. doi: 10.4161/epi.6.7.16500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teschendorff AE, Menon U, Gentry-Maharaj A, Ramus SJ, Gayther SA, Apostolidou S, et al. An epigenetic signature in peripheral blood predicts active ovarian cancer. PLoS One. 2009;4:e8274. doi: 10.1371/journal.pone.0008274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterland RA, Jirtle RL. Transposable elements: Targets for early nutritional effects on epigenetic gene regulation. Molecular and cellular biology. 2003;23:5293–5300. doi: 10.1128/MCB.23.15.5293-5300.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]