Abstract
Genes encoding the opioid receptors (OPRM1, OPRD1, and OPRK1) are obvious candidates for involvement in risk for heroin dependence. Prior association studies commonly had samples of modest size, included limited single nucleotide polymorphism (SNP) coverage of these genes, and yielded inconsistent results. Participants for the current investigation included 1459 heroin dependent cases ascertained from maintenance clinics in New South Wales, Australia, 1495 unrelated individuals selected from an Australian sample of twins and siblings as not meeting DSM-IV criteria for lifetime alcohol or illicit drug dependence (non-dependent controls), and 531 controls ascertained from economically-disadvantaged neighborhoods in proximity to the maintenance clinics. A total of 136 OPRM1, OPRD1, and OPRK1 SNPs were genotyped in this sample. After controlling for admixture with principal components analysis, our comparison of cases to non-dependent controls found 4 OPRD1 SNPs in fairly high linkage disequilibrium for which adjusted p values remained significant (e.g., rs2236857; OR 1.25; p=2.95 × 10−4) replicating a previously reported association. A post-hoc analysis revealed that the two-SNP (rs2236857 and rs581111) GA haplotype in OPRD1 is associated with greater risk (OR 1.68; p=1.41 × 10−5). No OPRM1 or OPRK1 SNPs reached more than nominal significance. Comparisons of cases to neighborhood controls reached only nominal significance. Our results replicate a prior report providing strong evidence implicating OPRD1 SNPs and, in particular, the two SNP (rs2236857 and rs581111) GA haplotype in liability for heroin dependence. Support was not found for similar association involving either OPRM1 or OPRK1 SNPs.
Keywords: association study, heroin dependence, OPRD1, OPRK1, OPRM1
INTRODUCTION
Opioid dependence remains a major societal problem worldwide (Degenhardt et al. 2004). Family and twin studies have established that a substantial component of risk for this disorder is attributable to genetic factors [e.g., (Merikangas et al. 1998; Tsuang et al. 1998)]. As with other complex traits, it has proved challenging to determine the specific genes responsible for this contribution.
Single nucleotide polymorphisms (SNPs) in the opioid receptor genes (OPRM1, OPRD1, and OPRK1) are obvious candidates for involvement in liability for heroin dependence. OPRM1, which encodes the mu opioid receptor (MOR) at which heroin and other commonly used opioids exert their primary effects (e.g., analgesia, reward, and dependence), (Le Merrer et al. 2009) has understandably been the most highly investigated of these genes. 118 A >G (rs1799971) is an exonic OPRM1 SNP that results in an amino acid substitution (Bond et al. 1998) which reportedly reduces mRNA expression (Zhang et al. 2005) and alters stress responsivity (Wand et al. 2002). Initial excitement over reported association of this seemingly ideal candidate with heroin dependence [e.g., (Bart et al. 2004)] has been tempered by subsequent findings. Although meta-analyses (Glatt et al. 2007; Coller et al. 2009) have concluded that the preponderance of evidence does not support a significant role in opioid dependence liability for this SNP, its potential involvement in the pathophysiology of other addictive disorders continues to be actively investigated (Ray et al. 2011). Overall, no associations involving OPRM1 SNPs have been consistently replicated; preliminary findings include reports of association with a greater positive response to heroin (Zhang et al. 2007) and risk for heroin dependence (Zhang et al. 2006; Levran et al. 2008).
More recently, investigators have turned their attention to OPRD1 and OPRK1 (Levran et al. 2008; Zhang et al. 2008). Opioid drugs bind delta opioid receptors (DORs), but with a much lower affinity than MORs. DORs play an important role in the development of opioid tolerance (Daniels et al. 2005) and are involved in the rewarding and analgesic effects of opioids (Le Merrer et al. 2009). Overall, kappa opioid receptor (KOR) actions are commonly in opposition to those of MORs. Administration of KOR agonists results in conditioned place aversion rather than conditioned place preference (CPP) (Le Merrer et al. 2009). KORs may play a major role in the dysphoria experienced with opioid withdrawal (Le Merrer et al. 2009).
Several early association studies (e.g, Franke et al. 1999) that focused on coding OPRD1 SNPs in small samples produced largely negative findings. A more recent examination (Zhang et al. 2008) of opioid and other substance dependence observed significant opioid dependence risk associated with rs1042114, a coding SNP in OPRD1, but not with other OPRD1 or OPRK1 SNPs. Another investigation (Levran et al. 2008) that had a larger sample (N=412) of heroin dependent cases and used Goldman’s “addiction chip” (Hodgkinson et al. 2008) reported putative association with three SNPs in OPRD1 and one in OPRK1.
The current investigation examines whether OPRM1, OPRD1, and OPRK1 SNPs are associated with risk for heroin dependence in a very large Australian sample of cases (N=1459) receiving maintenance treatment for heroin dependence in New South Wales (NSW) and non-dependent controls (N=1495 unrelated individuals) ascertained from samples of twins and family members as not meeting DSM-IV criteria for lifetime alcohol or illicit drug dependence. The substantially larger sample size provides an opportunity to investigate more definitively the involvement of these genes. Cases are also compared to a second control group (N=531), termed neighborhood controls, ascertained from economically-disadvantaged neighborhoods in close proximity to maintenance clinics. Because these individuals have higher rates of lifetime drug use and nicotine, alcohol, and non-opioid illicit drug dependence, this comparison can provide insight into the degree to which association findings are specific for heroin dependence or shared with other addiction phenotypes.
MATERIALS AND METHODS
The Comorbidity and Trauma Study (CATS), a collaboration of investigators at Washington University School of Medicine, the Queensland Institute of Medical Research, and the National Drug and Alcohol Research Centre of the University of New South Wales, examined genetic and environmental factors contributing to liability for heroin dependence using a case-control design. A description of the methods used for data collection has been given in previous reports of phenotypic data (Shand et al. 2010). In addition to case and control subjects recruited after funding was obtained, we include here 25 cases and 25 neighborhood controls from the CATS pilot study for whom other protocols were identical and assessment was comparable.
Participants
Cases were recruited from clinics providing opioid replacement therapy (ORT) in the greater Sydney region. Prior to enrollment in ORT, NSW guidelines require an extensive clinical assessment documenting that the individual meets DSM-IV diagnostic criteria for opioid dependence and is suitable for the form of maintenance treatment recommended. For cases, inclusion criteria were age 18 years or older, an adequate understanding of English, and current or past participation in ORT consisting of either methadone or buprenorphine for heroin dependence. Participants reporting recent suicidal intent or known to be currently experiencing psychosis were excluded. Neighborhood controls were recruited from geographic areas in proximity to ORT clinics. The use of opioids recreationally more than ten times lifetime was an exclusion criterion for controls; the inclusion and exclusion criteria except for ORT participation were otherwise identical to cases. Written informed consent was obtained from all participants. Institutional review board (IRB) approval was obtained from the University of New South Wales, Washington University, the Queensland Institute of Medical Research, and the area health service ethics committees governing the participating clinics. Participants were reimbursed AU$50.00 for out-of-pocket expenses.
Interim analyses of phenotypic data revealed high rates of licit and non-opioid illicit drug dependence in neighborhood controls (41.6%, 28.1%, and 36.0% for nicotine, alcohol, and illicit drug dependence respectively in the final sample), which raised concerns that comparisons with these individuals would be ill-suited to identify genetic variants associated with liability that is shared across classes of drug dependence. These concerns prompted a decision to revise the study design to include a non-dependent control group of unrelated individuals selected for the study’s primary genotypic analyses from a large Australian Twin Registry (ATR) pool of twins and family members [see (Hansell et al. 2009) for a description of this sample]. Inclusion criteria for this group included an adequate DNA supply available and existing IRB approval allowing additional genotyping for substance dependence and related phenotypes. Exclusion criteria were lifetime diagnoses of any DSM-IV illicit drug or alcohol dependence at prior interview. Individuals without a lifetime DSM-IV diagnosis of nicotine dependence were also preferentially selected. The resulting non-dependent control sample (N=1500) had a lifetime prevalence of nicotine dependence (12.5%) below that of the Australian general population.
Assessment
All participants completed semi-structured diagnostic interviews. Interviews for case and neighborhood control participants were completed in person; interviews for twin sample controls were completed via telephone. Substance dependence diagnostic sections of the interview were based on the Semi-Structured Assessment for the Genetics of Alcoholism - Australia (SSAGA-OZ) (Bucholz et al. 1994) enabling DSM-IV and DSM-III-R lifetime diagnoses of opioid, cannabis, sedative, stimulant, cocaine, and alcohol abuse and dependence. The nicotine dependence section of the interview was modified from the Nicotine Addiction Genetics Study (Hansell et al. 2009) assessment which was derived from the World Health Organization Composite International Diagnostic Interview (CIDI) (Andrews & Peters 1998). Similar diagnoses were obtained for CATS pilot project participants (25 cases and 25 neighborhood controls) via the CIDI (Andrews & Peters 1998).
Marker selection
The pair-wise option of Tagger (de Bakker et al. 2005), implemented in Haploview (Barrett et al. 2005), was applied to HapMap European ancestry panel data using a threshold of r2≥0.8 for most genes and a higher threshold (≥0.9) for high priority candidates (e.g., opioid receptors) to select a custom set of 1536 SNPs that provided coverage of 72 candidate genes and 47 additional SNPs for which prior studies reported association. A set of 30 ancestry-informative markers (AIMs) distributed physically across the genome was selected from SNPs for which the greatest allele frequency differences were found between populations with European and East Asian ancestry in Hapmap2 data for use in principal components analyses. As summarized above, because SNPs from opioid receptor genes are obvious first-pass candidates, we prioritized examining these SNPs in the first stage of a two stage analytic process (the remaining SNPs will be analyzed in stage two). Data are thus reported here for the 142 SNPs genotyped in the opioid receptor genes: OPRM1, OPRD1, and OPRK1 (for details, see Table 1, Supplementary Tables 1 and 2). We provide data cleaning information below for the entire set of markers since we utilized all retained SNPs to generate the principal components used to control for admixture.
Table 1.
OPRD1 SNP features and association with heroin dependence – comparison of cases (N=1459) to non-dependent controls (N=1495)
| SNP | Location * | Classificatio n |
Minor allele |
Minor allele freq | P value | Odds Ratio
(95% Confidence Interval) |
|
|---|---|---|---|---|---|---|---|
| Cases | Controls | ||||||
| rs569356 | 29136686 | Flank 5’ UTR | G | 0.133 | 0.146 | 0.48 | 0.95 (0.81 – 1.10) |
| rs1042114 | 29138975 | Cod non-syn | C | 0.133 | 0.146 | 0.52 | 0.95 (0.82 – 1.11) |
| rs2236861 | 29139756 | Intron 1 | A | 0.247 | 0.231 | 0.030 | 1.15 (1.01 – 1.30) |
| rs204047 | 29145124 | Intron 1 | A | 0.190 | 0.181 | 0.15 | 1.11 (0.97 – 1.26) |
| rs678849 | 29145188 | Intron 1 | G | 0.473 | 0.458 | 0.053 | 1.11 (1.00 – 1.24) |
| rs419335 | 29151844 | Intron 1 | G | 0.341 | 0.311 | 0.00112 | 1.20 (1.08 – 1.35) |
| rs204055 | 29159373 | Intron 1 | A | 0.473 | 0.458 | 0.00998 | 1.15 (1.03 – 1.28) |
| rs2236857 | 29161609 | Intron 1 | G | 0.273 | 0.243 | 0.000295 | 1.25 (1.11 – 1.41) |
| rs2236855 | 29161999 | Intron 1 | A | 0.273 | 0.243 | 0.000295 | 1.25 (1.11 – 1.41) |
| rs760589 | 29162465 | Intron 1 | A | 0.337 | 0.309 | 0.00109 | 1.20 (1.08 – 1.35) |
| rs2298897 | 29165837 | Intron 1 | C | 0.283 | 0.254 | 0.000469 | 1.24 (1.10 – 1.39) |
| rs3766951 | 29169559 | Intron 1 | G | 0.340 | 0.307 | 0.000411 | 1.22 (1.09 – 1.37) |
| rs529520 | 29174946 | Intron 1 | A | 0.470 | 0.445 | 0.00193 | 1.18 (1.06 – 1.32) |
| rs581111 | 29175373 | Intron 1 | A | 0.276 | 0.263 | 0.045 | 1.13 (1.00 – 1.27) |
| rs680090 | 29175461 | Intron 1 | A | 0.479 | 0.490 | 0.016 | 0.88 (0.79 – 0.98) |
| rs12749204 | 29176213 | Intron 1 | G | 0.194 | 0.186 | 0.09 | 1.12 (0.98 – 1.28) |
| rs2298895 | 29178924 | Intron 1 | T | 0.052 | 0.045 | 0.18 | 1.18 (0.92 – 1.51) |
| rs508448 | 29181525 | Intron 1 | G | 0.457 | 0.460 | 0.18 | 0.93 (0.84 – 1.03) |
| rs4654327 | 29190138 | 3’ UTR | G | 0.477 | 0.471 | 0.24 | 1.07 (0.96 – 1.18) |
| rs204076 | 29190390 | Flank 3’ UTR | T | 0.351 | 0.362 | 0.86 | 1.01 (0.91 – 1.13) |
| rs204069 | 29194818 | Flank 3’ UTR | A | 0.352 | 0.356 | 0.43 | 1.05 (0.94 – 1.17) |
Key: UTR=untranslated region; non-syn=non-synonymous; flank=flanking; cod=coding.
NCBI build 37.2
Genotyping
Genotyping was performed on an Illumina BeadStation using the GoldenGate technology (Peters et al. 2008). DNA samples from CEPH trio 1334 (obtained from the Coriell Cell Repository) served as internal controls for quality of clustering and reproducibility. The primary analysis of the genotyping data with the Illumina BeadStudio software was followed by visual inspection and assessment of data quality and clustering.
Statistical Analyses
Data Cleaning
Data were excluded from one individual (an ATR control) whose DNA did not genotype successfully. Initial quality control found 23 SNPs for which genotyping failed (i.e., GenCall score=0). An additional 9 SNPs that had a call rate below 95% were removed from further analyses. Genotypic data revealed the presence of 32 duplicate samples; further scrutiny found that these individuals had participated in the project more than once. For individuals who had participated both in the pilot and funded phases of the project, interview data from the funded phase were retained. For those who participated multiple times in the funded project, data from the first interview were used for analyses (case-control status was consistent throughout). Data from an additional 3 participants were excluded on the basis of mismatch of genotypic and phenotypic gender. Data from an additional 47 SNPs with minor allele frequency (MAF) less than 2% were not included in analyses. 27 SNPs were removed from further analyses because deviations from Hardy–Weinberg equilibrium in ATR controls exceeded Bonferroni correction for the total number of remaining SNPs (0.05/1457 = 3.43 × 10−5). An examination for cryptic relatedness found 25 instances in which the calculated proportion of shared alleles identical by descent exceeded 0.5, indicative of greater sharing than expected for unrelated individuals; in each case, the participant with the higher project identifier number was excluded. For the final sample, the mean call rate for 136 opioid receptor SNPs that remained after data cleaning exceeded 99.9%.
The current report’s primary analyses compared 1459 heroin dependent cases [888 male; 571 female; mean age 36.5 (SD 8.6)] to 1495 non-dependent controls [972 male; 523 female; mean age 45.0 (SD 9.5)]; cases were compared to 531 neighborhood controls [235 male; 296 female; mean age 34.7 (SD 10.5)] in additional analyses aimed at identifying SNPs with effects specific to heroin dependence liability.
Admixture
Principal components analyses (PCA) were conducted with data from cases and ATR controls using the smartpca program in the Eigensoft 3.0 package (Patterson, Price, & Reich 2006). Due to the dense coverage of high priority candidate genes, the kill r2 option was set at 0.8 for these analyses. Our inclusion of data from AIMs in these analyses prevented using Tracy-Widom statistics to determine the number of principal components (PCs) to be used as covariates. As such, we opted to include all PCs for which case-control differences reached at least a trend level of significance. Post-hoc analyses which included a larger number of PCs (ten) as covariates were performed to demonstrate that our primary findings remained significant despite this additional control for admixture (see limitations).
Association
Single Nucleotide Polymorphism Spectral Decomposition (Nyholt 2004; Li & Ji 2005) was first used to calculate the appropriate correction for multiple testing for analyses of data for the 136 opioid receptor SNPs that remained after data cleaning. As the respective linkage disequilibrium (LD) plots demonstrate (see Figure 1, Supplementary Figures 1 and 2), we genotyped a number of SNPs in high LD for these genes. Based on the calculated effective number of independent loci markers (57.69), the significance threshold necessary to keep the type I error rate at 5% was determined to be 8.89 × 10−4. Association analyses were then performed using PLINK (Purcell et al. 2007). Logistic regression, including the smartpca-derived PCs in the model as independent variables to control for admixture, was used to examine the association between the log-additive effects of minor allele dosage and status (case versus non-dependent control). Although PC covariates were not needed for the secondary comparisons of cases versus neighborhood controls (see PCA results below), logistic regression was again used to obtain results comparable to the above.
Figure 1.
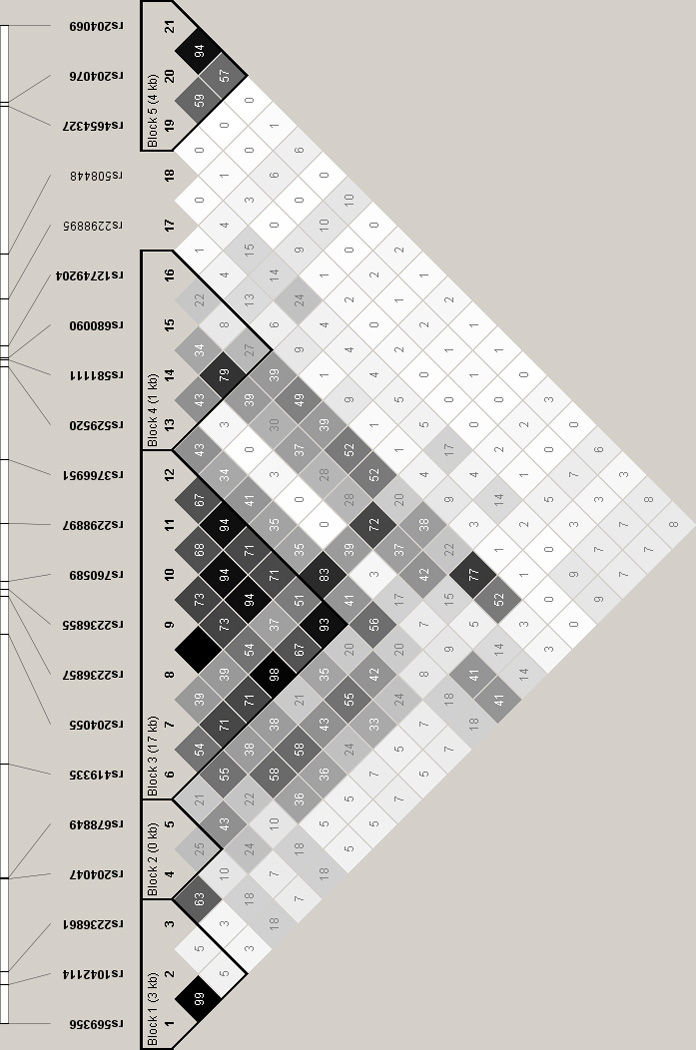
Linkage disequilibrium analysis of OPRD1 SNPs (r2 values are shown)
Several post-hoc analyses were undertaken to provide a better understanding of our primary findings. An additional logistic regression comparing cases and non-dependent controls was performed to control for the allelic dosage of the most significant OPRD1 SNP using the condition option in PLINK to examine whether evidence of more than a single association signal was found. Based on the results of this analysis, the haplotype command in PLINK was used to assess risk associated with a single 2 SNP OPRD1 haplotype. Finally, consistent with the exclusion criteria for the non-dependent controls, we divided neighborhood controls into those who did not meet criteria for dependence on alcohol or a non-opioid illicit (i.e., cannabis, stimulants, sedatives, or cocaine) drug (N=275) and those who did (N=256). We then separately compared cases to each of these groups to estimate the risk for heroin dependence risk associated with the most significant OPRD1 SNP.
RESULTS
The principal components analysis retained data from 1113 of the 1457 SNPs (including AIMs) remaining after data cleaning. As case-non-dependent control differences reached at least a trend level of significance for 4 PCs, a conservative decision was made to include all 4 PCs as covariates in comparisons of these groups. The respective p-values for population differences along these eigenvectors were 1.1 × 10−12, 2.4 × 10−11, 1.1 × 10−4, and 0.064. Although both populations are primarily of European ancestry, the various combinations of PCs 1,2, and 3 (see scatterplots in Supplementary Figures 3A–C) identify individuals of Asian ancestry (i.e., mapping to Han Chinese and Japanese HapMap populations) more prevalent among cases. PC4 appears to be identifying a Northern European ancestry subgroup. A similar examination in cases and neighborhood controls found no significant or trend level population differences and thus no covariates were used for those comparisons.
Unadjusted MAFs and the results of association analyses for OPRD1 SNPs comparing cases to non-dependent controls are displayed in Table 1. Associations of at least nominal significance are seen for 10 of the 21 OPRD1 SNPs. For 4 SNPs (for which pairwise r2 ranged from 0.68–1.0, see Figure 1), the p values are less than the significance threshold calculated to correct for multiple comparisons. Based on the LD relationships of the OPRD1 SNPs, it appears likely that these results could largely be explained by a single strong association signal (minimal p value = 2.95 × 10−4 for rs2236857 and rs2236855). To test this assertion, association for those OPRD1 SNPs of at least nominal significance was reexamined, conditioned on rs2236857 allelic dosage. The only residual signal observed was for rs581111, for which only mild attenuation was found (p=0.059), consistent with the lack of LD (r2=0) for this SNP with either rs2236857 or rs2236855. A post-hoc two locus association analysis of rs2236857 and rs581111 (Table 2) found that the GA haplotype consisting of the coupled minor alleles (prevalence 0.077) is more strongly associated with an odds ratio (OR) of 1.68 (p=1.41 × 10−5).
Table 2.
Association of OPRD1 two SNP (rs2236857 and rs581111) haplotype with heroin dependence – comparison of cases (N=1459) to non-dependent controls (N=1495)
| Haplotype | Frequency | Odds ratio | p value |
|---|---|---|---|
| GA | .077 | 1.68 | 1.41 × 10−5 |
| AA | .193 | 0.98 | 0.73 |
| GG | .181 | 1.13 | 0.10 |
| AG | .550 | 0.84 | 1.85 × 10−3 |
Association analyses comparing cases to non-dependent controls for SNPs in the other opioid receptors are most noteworthy for the dearth of findings reaching even nominal significance. Of the 93 OPRM1 SNPs, only rs10485058 attained this minimal standard [p=0.045; OR=0.86 (95%CI 0.74–1.00)]. Nominal significance was found for only 2 of the 22 OPRK1 SNPs: rs12548098 [p=0.016; OR=0.85 (95%CI 0.74–0.97)] and rs7826614 [p=0.015; OR=0.84 (95%CI 0.74–0.97)].
The results of additional analyses comparing cases and neighborhood controls found no association for an OPRD1 SNP that reached even nominal significance; the ORs for rs2236857 and rs2236855 were 1.09 (p>0.28). For a relatively small number of OPRM1 and OPRK1 SNPs, associations of nominal significance were found (Supplementary Table 3); however, none was within an order of magnitude of the p value threshold required to correct for multiple testing.
Neighborhood controls were divided into those who were dependent on either alcohol or a non-opioid illicit drug (N=256) and those who were not (N=275), consistent with the inclusion criteria for non-dependent control group. The ORs for association of rs2236857 and rs2236855 (identical due to complete LD) with heroin dependence in respective comparisons of cases to neighborhood controls, with and without substance dependence diagnoses, were 1.21 (p=0.08) and 1.00 (p=0.98).
Discussion
Our investigation compared a large sample of heroin dependent cases to individuals with no history of illicit drug or alcohol dependence and found a strong association of four OPRD1 SNPs with heroin dependence. The estimates of risk (odds-ratio 1.25) for the two most highly associated SNPs, rs2236855 and rs2336857, are of similar magnitude to those observed with other complex traits. The degree of LD observed between these SNPs and most of the other nominally-associated OPRD1 polymorphisms is consistent with a single strong signal. These results replicate the findings of a prior report (Levran et al. 2008) which also used Illumina GoldenGate technology to genotype 1536 SNPs (including OPRM1, OPRD1, and OPRK1 polymorphisms) in similarly ascertained cases (412 heroin dependent individuals on methadone maintenance therapy in the U.S. and Israel) and controls (184 individuals screened as having no substantial history of illicit drug or alcohol use). The nine SNPs that were most strongly associated with heroin dependence in their sample included three OPRD1 SNPs, all of which were genotyped in the current report (see Supplementary Table 4). Two (rs2236857 and rs3766951) were among the four SNPs in our sample for which corrected p values were significant; the third (rs2236861) reached only nominal significance. The agreement observed in the OPRD1 findings across these reports is extremely encouraging. Our results contrast with those of another group (Zhang et al. 2008) who observed a significant association with rs1042114 after correction for multiple testing and no significant association involving either of our two most significantly associated SNPs (rs2236857 or rs2236855). Differences in design may have contributed to the divergence in findings. Their sample’s 103 opioid dependent cases included individuals recruited from settings other than ORT clinics including some who were dependent on opioids other than heroin. They genotyped samples using other methods (either polymerase chain reaction restriction fragment length polymorphism or the TaqMan technique). Finally, they report p values for case-control comparisons involving rs2236855 and rs2236857 that, while non-significant, differ substantially in magnitude, surprising given that these two SNPs are in complete LD (i.e., r2=1) in both our sample and in all HapMap populations.
The 4 OPRD1 SNPs that we found to be most strongly associated with heroin dependence are all located in the first intron of this gene. Although these SNPs are not located at a branch point and do not alter either a splice enhancer or silencer, there is some evidence supporting potential epigenetically-mediated functionality. rs2236855 has a high level of evolutionary conservation (PhyloP Score=1.14) and is located within a DNase I hypersensitivity site in many, but not all, cell types (Fujita et al. 2011) suggesting it could be a cell type specific regulatory element. Both rs2236857 and rs2298897 are located within interspersed repeats: the former in a long interspersed element and the latter in a long terminal repeat (Fujita et al. 2011). The post-hoc examination of the two-SNP haplotype (rs2236857 and rs581111), prompted by the limited attenuation of association signal for rs581111 (also located in intron 1) in an analysis conditioned on the most strongly associated SNP, found evidence of an association stronger by more than an order of magnitude (uncorrected p=1.41 × 10−5). These results are suggestive of a non-genotyped SNP with a less prevalent minor allele (the GA haplotype frequency is 0.077), associated with substantial heroin dependence risk (OR 1.68), the identification of which could likely be achieved using currently available approaches such as deep sequencing. Given that this polymorphism is apparently located in an intronic region, its effects may involve altered OPRD1 expression [e.g., see (Lomelin, Jorgenson, & Risch 2010)]. Ongoing research examining the regulation of OPRD1 expression (Tuusa, Leskelä, & Petäjä-Repo 2010) and DOR trafficking (Bie et al. 2010) will also help guide further investigation.
In marked contrast to our OPRD1 findings, our results do not provide support for association with heroin dependence involving OPRM1 and OPRK1 SNPs. The results of our study and multiple meta-analyses do not support an association of rs1799971 with heroin dependence. We also failed to replicate the association of two OPRM1 and one OPRK1 SNP reported by Levran et al (2008) although our comparison with neighborhood controls found a nominal association with rs3778151. Considering the additional power provided by our considerably larger sample and the lack of consistently replicated findings for polymorphisms in these genes, our results suggest that it may be prudent to focus attention preferentially in future association studies on OPRD1 over the other two opioid receptor genes. However, it remains possible that significant heroin dependence risk associated with OPRM1 and OPRK1 SNPs was not detected in the current study due to small effect size, low MAF in our sample, greater variability across populations of differing ethnicity for some of these SNPs (e.g., rs6473797, rs3778151), or our failure to genotype the specific risk-associated SNPs. Given the well-documented involvement of MORs and KORs in the effects of opioids [including important aspects of dependence e.g., see (Christie 2008)] and the close functional interrelationships of these three receptors in which heterodimerization may be integrally involved (Ferré & Franco 2010; von Zastrow 2010) they remain extremely important targets for other avenues of research in order to improve current understanding of the pathophysiology of heroin dependence as well as treatment of this disorder.
Despite decades of active investigation, relatively basic questions about opioid receptors remain unanswered. The distinct pharmacologic profiles of the various opioid receptors are fairly well-characterized; their molecular basis, including whether they are homomeric or heteromeric, remains unclear (van Rijn & Whistler 2009). For example, researchers have variously proposed that the DOR1 is actually a heterodimer composed of DOR and either MOR (van Rijn & Whistler 2009) or KOR (Bhushan et al. 2004) subunits. Similarly, a provocative report (Yekkirala, Kalyuzhny, & Portoghese 2010) found that the affinity of opiate agonists at MOR-DOR heteromers exceeded that at MOR homomers. DORS are primarily intracellular; chronic opioid use results in substantial translocation to cell membranes where they may form heteromers with MORs (von Zastrow 2010). Increased levels of MOR-DOR heteromers have been reported (Gupta A et al. 2010) after chronic morphine administration. The formation of these MOR-DOR heteromers has recently been implicated (He SQ et al. 2011) as playing a major role in opioid tolerance. DOR antagonists have been shown to block the sensitization to the conditioned rewarding effects of morphine that occurs with opioid pretreatment (Shippenberg, Chefer, & Thompson 2009). A recent report (Billa, Xia, & Morón 2010) found that administration of a DOR2 antagonist blocked morphine-induced CPP in rats and resulted in an increase in expression of the DOR dimer in the hippocampal postsynaptic density. These reports support a role for DORs in pathophysiology of opioid dependence which may be at least partially mediated by altered expression. In fact, a recent report proposing the design of an opioid drug that causes reduced tolerance and dependence advocated for drug development focused on DOR/MOR heteromers (Berger & Whistler 2010). Although MORs and DORs are generally considered to have synergistic antinociceptive effects (Zhang & Pan 2010), opposing effects on other behaviors such as impulsivity have been reported in rodents (Olmstead, Ouagazzal, & Kieffer 2009).
A possible alternative interpretation of our findings is suggested by a report (Zeller et al. 2010) that conducted genomewide expression analyses using RNA extracted from peripheral blood monocytes of a community-based German sample in which a genomewide association study (GWAS) had been completed. Among a total of 2745 expression quantitative trait loci (eqtl’s) that had p values less than 5.78 × 10−12, rs2236855 was found to be a cis-eqtl for two genes (p values shown): PHACTR4 (7.57 × 10−27) and ATPIF1 (2.85 × 10−12). Both are expressed in the brain and located in proximity (~300–600 kb) to OPRD1 on chromosome 1. PHACTR4 is thought to be a protein phosphatase-1 (PP-1) inhibitor (Allen PB et al. 2004). cAMP-regulated phosphoprotein (DARPP-32), mediates the action of multiple drugs of abuse (including opioids) by regulating striatal dopaminergic transmission via its actions as a inhibitor of PP-1 and protein kinase A (Svenningsson, Nairn, & Greengard 2005). DARPP-32 has been implicated in maintenance of morphine-induced CPP (Narita et al. 2010) and suggested as a putative therapeutic target for opiate addiction (Mahajan et al. 2009). ATPIF1, a mitochondrial ATPase inhibitor, plays important and diffuse roles in energy regulation. Although the degree to which gene expression data from peripheral blood monocytes correlates with that of brain remains unclear, altered expression of PHACTR4, and perhaps ATPIF1, deserves consideration as an alternative route through which our findings could be explained.
Several limitations must be considered when interpreting our findings. Our cases were ascertained entirely from maintenance clinics in the Greater Sydney Area. Additional study may be needed to determine whether similar results would be seen in samples not currently in treatment or from other areas. Non-dependent controls were interviewed via telephone; cases and neighborhood controls completed an in-person assessment. Given the low general population prevalence and extreme severity of heroin dependence, it seems highly unlikely that telephone administration, used at QIMR for more than 25,000 interviews to date, led to a substantial number of false negative diagnoses. Although both our cases and non-dependent controls included primarily individuals of European ancestry, the groups differed somewhat in ethnic composition with more Asians found among cases. It is possible that population stratification could have contributed to the significant differences that we observed. Since we found an excess of the same alleles among cases as observed in a prior report (Levran et al. 2008) in a predominately Caucasian sample with some Middle Eastern contribution , we consider this possibility unlikely. We also reran analyses increasing the number of principal components to control for admixture to ten (from four), with little effect on results. Despite its widespread use as a method to control for multiple testing, spectral decomposition may be viewed by some as inadequately conservative. It is thus important to note that when a more stringent threshold such as a Bonferroni correction (i.e., 0.05/136= 3.68 × 10−4) is applied, our two most highly associated SNPs remain significant. Despite the considerably larger size of our sample (more than three-fold larger than most prior association studies of heroin dependence), it is possible that we may have failed to detect significant associations because of limited power (i.e. type II error). Similarly, the relatively smaller size of the neighborhood control sample and any sharing of risk with related phenotypes (e.g., other substance dependence or externalizing disorders) more prevalent in this group (than the non-dependent controls) could be contributing to the lack of significant differences found in this comparison. Although not significant, the point estimates (ORs 1.09) for rs2236855 and rs2236857 in the comparison of cases to neighborhood controls were greater than unity suggestive of shared risk. Interestingly, the respective ORs for comparisons of cases to neighborhood controls, with and without substance dependence diagnoses, were 1.21 and 1.00 respectively. Although neither value is significant, the estimate for the comparison to substance dependent neighborhood controls is fairly similar to that found for the case-non-dependent control comparison, a finding more suggestive of specificity of risk for opioid dependence. It is possible that other factors are protective against substance dependence in non-dependent neighborhood controls.
Our investigation provides further evidence that OPRD1 polymorphisms are associated with risk for heroin dependence. Although the strongest observed signal involves intronic SNPs, support is found for potential functionality including either epigenetically-mediated mechanisms (Fujita et al. 2011) or via rs2236855’s status as an eqtl for other genes (Zeller et al. 2010). Our post-hoc finding that greater risk is associated with the less prevalent rs2236857-rs581111 haplotype suggests the importance of additional genotyping to determine the identity of an underlying functional polymorphism. The lack of significant association observed for any SNPs in OPRM1 and OPRK1 provides a striking contrast to the OPRD1 findings. Overall, our results support prioritizing research aimed at increasing current understanding of the important role played by OPRD1 in the pathophysiology of heroin dependence including ongoing efforts focusing on improved opioid drug design (Berger & Whistler 2010).
Supplementary Material
Acknowledgements
This work was supported by the National Institute on Drug Abuse (R01 DA017305 to ECN); support was also received from the National Drug and Alcohol Research Centre and the Australian National Health and Medical Research Council (to LD). The authors would like to thank Anthony Caracella for his work in sample receipt and preparation, Megan Campbell for project coordination, and Lisa Bowdler and Sara Smith for their efforts in sample genotyping.
Footnotes
Author Contributions
ECN, MTL, ACH, LD, NGM, and GWM were responsible for the study concept and design. NW, AA, AJS assisted with data analysis and interpretation of findings. ECN drafted the manuscript. MTL, ACH, NW, AA, FLS, AKH, LW, AAT, AJS, PAFM, LD, NGM, and GWM provided critical revision of the manuscript and contributed to its intellectual content. All authors critically reviewed content and approved the final version of the manuscript.
Conflicts of Interest
None of the authors have a financial or personal conflict of interest.
References
- Allen PB, Greenfield AT, Svenningsson P, Haspeslagh DC, Greengard P. Phactrs 1–4: A family of protein phosphatase 1 and actin regulatory proteins. Proc Natl Acad Sci USA. 2004;101:7187–7192. doi: 10.1073/pnas.0401673101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews G, Peters L. The psychometric properties of the Composite International Diagnostic Interview. Soc Psychiatry Psychiatr Epidemiol. 1998;33:80–88. doi: 10.1007/s001270050026. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Bart G, Heilig M, LaForge KS, Pollak L, Leal SM, Ott J, Kreek MJ. Substantial attributable risk related to a functional mu-opioid receptor gene polymorphism in association with heroin addiction in central Sweden. Mol Psychiatry. 2004;9:547–549. doi: 10.1038/sj.mp.4001504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger AC, Whistler JL. How to design an opioid drug that causes reduced tolerance and dependence. Ann Neurol. 2010;67:559–569. doi: 10.1002/ana.22002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhushan RG, Sharma SK, Xie Z, Daniels DJ, Portoghese PS. A bivalent ligand (KDN-21) reveals spinal delta and kappa opioid receptors are organized as heterodimers that give rise to delta(1) and kappa(2) phenotypes. Selective targeting of delta-kappa heterodimers. J Med Chem. 2004;47:2969–2972. doi: 10.1021/jm0342358. [DOI] [PubMed] [Google Scholar]
- Bie B, Zhang Z, Cai YQ, Zhu W, Zhang Y, Dai J, Lowenstein CJ, Weinman EJ, Pan ZZ. Nerve growth factor-regulated emergence of functional delta-opioid receptors. J Neurosci. 2010;30:5617–5628. doi: 10.1523/JNEUROSCI.5296-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billa SK, Xia Y, Morón JA. Disruption of morphine-conditioned place preference by a delta2-opioid receptor antagonist: study of mu-opioid and delta-opioid receptor expression at the synapse. Eur J Neurosci. 2010;32:625–631. doi: 10.1111/j.1460-9568.2010.07314.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond C, LaForge KS, Tian M, Melia D, Zhang S, Borg L, Gong J, Schluger J, Strong JA, Leal SM, Tischfield JA, Kreek MJ, Yu L. Single-Nucleotide Polymorphism in the Human Mu Opioid Receptor Gene Alters Beta-Endorphin Binding and Activity: Possible Implications for Opiate Addiction. Proc Natl Acad Sci USA. 1998;95:9608–9613. doi: 10.1073/pnas.95.16.9608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucholz KK, Cadoret R, Cloninger CR, Dinwiddie SH, Hesselbrock VM, Nurnberger JI, Jr, Reich T, Schmidt I, Schuckit MA. A new, semi-structured psychiatric interview for use in genetic linkage studies: a report on the reliability of the SSAGA. J Stud Alcohol. 1994;55:149–158. doi: 10.15288/jsa.1994.55.149. [DOI] [PubMed] [Google Scholar]
- Christie MJ. Cellular neuroadaptations to chronic opioids: tolerance, withdrawal and addiction. Br J Pharmacol. 2008;154:384–396. doi: 10.1038/bjp.2008.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller JK, Beardsley J, Bignold J, Li Y, Merg F, Sullivan T, Cox TC, Somogyi AA. Lack of association between the A118G polymorphism of the mu opioid receptor gene (OPRM1) and opioid dependence: a meta-analysis. Pharmgenomics Pers Med. 2009;2:9–19. [PMC free article] [PubMed] [Google Scholar]
- Daniels DJ, Lenard NR, Etienne CL, Law PY, Roerig SC, Portoghese PS. Opioid-induced tolerance and dependence in mice is modulated by the distance between pharmacophores in a bivalent ligand series. Proc Natl Acad Sci USA. 2005;102:19208–19213. doi: 10.1073/pnas.0506627102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bakker PI, Yelensky R, Pe'er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nat Genet. 2005;37:1217–1223. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- Degenhardt L, Hall W, Warner-Smith M, Lynskey M. Illicit drug use. Ch. 13. In: Ezzati M, Lopez AD, Rodgers A, Murray CJL, editors. Comparative Quantification of Health Risks: Global and Regional Burden of Disease Attributable to Selected Major Risk Factors. Volume 1. Geneva, Switzerland: World Health Organization (WHO); 2004. pp. 1109–1175. [Google Scholar]
- Ferré S, Franco R. Oligomerization of G-protein-coupled receptors: a reality. Curr Opin Pharmacol. 2010;10:1–5. doi: 10.1016/j.coph.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke P, Nothen MM, Wang T, Neidt H, Knapp M, Lichtermann D, Weiffenbach O, Mayer P, Holt B, Propping P, Maier W. Human delta-opioid receptor gene and susceptability to heroin and alcohol dependence. Am J Med Genet. 1999;88:462–464. doi: 10.1002/(sici)1096-8628(19991015)88:5<462::aid-ajmg4>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- Fujita PA, Rhead B, Zweig AS, Hinrichs AS, Karolchik D, Cline MS, Goldman M, Barber GP, Clawson H, Coelho A, Diekhans M, Dreszer TR, Giardine BM, Harte RA, Hillman-Jackson J, Hsu F, Kirkup V, Kuhn RM, Learned K, Li CH, Meyer LR, Pohl A, Raney BJ, Rosenbloom KR, Smith KE, Haussler D, Kent WJ. The UCSC Genome Browser database: update 2011. Nucleic Acids Res. 2011;39:D876–D882. doi: 10.1093/nar/gkq963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glatt SJ, Bousman C, Wang RS, Murthy KK, Rana BK, Lasky-Su JA, Zhu SC, Zhang R, Li J, Zhang B, Li J, Lyons MJ, Faraone SV, Tsuang MT. Evaluation of OPRM1 variants in heroin dependence by family-based association testing and meta-analysis. Drug Alcohol Depend. 2007;90:159–165. doi: 10.1016/j.drugalcdep.2007.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A, Mulder J, Gomes I, Rozenfeld R, Bushlin I, Ong E, Lim M, Maillet E, Junek M, Cahill CM, Harkany T, Devi LA. Increased abundance of opioid receptor heteromers after chronic morphine administration. Sci Signal. 2010;3:ra54. doi: 10.1126/scisignal.2000807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansell NK, Agrawal A, Whitfield JB, Morley KI, Gordon SD, Lind PA, Pergadia ML, Montgomery GW, Madden PA, Todd RD, Heath AC, Martin NG. Can we identify genes for alcohol consumption in samples ascertained for heterogeneous purposes? Alcohol Clin Exp Res. 2009;33:729–739. doi: 10.1111/j.1530-0277.2008.00890.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He SQ, Zhang ZN, Guan JS, Liu HR, Zhao B, Wang HB, Li Q, Yang H, Luo J, Li ZY, Wang Q, Lu YJ, Bao L, Zhang X. Facilitation of µ-opioid receptor activity by preventing d-opioid receptor-mediated codegradation. Neuron. 2011;69:120–131. doi: 10.1016/j.neuron.2010.12.001. [DOI] [PubMed] [Google Scholar]
- Hodgkinson CA, Yuan Q, Xu K, Shen PH, Heinz E, Lobos EA, Binder EB, Cubells J, Ehlers CL, Gelernter J, Mann J, Riley B, Roy A, Tabakoff B, Todd RD, Zhou Z, Goldman D. Addictions biology: haplotype-based analysis for 130 candidate genes on a single array. Alcohol Alcohol. 2008;43:505–515. doi: 10.1093/alcalc/agn032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Merrer J, Becker JA, Befort K, Kieffer BL. Reward processing by the opioid system in the brain. Physiol Rev. 2009;89:1379–1412. doi: 10.1152/physrev.00005.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levran O, Londono D, O'Hara K, Nielsen DA, Peles E, Rotrosen J, Casadonte P, Linzy S, Randesi M, Ott J, Adelson M, Kreek MJ. Genetic susceptibility to heroin addiction: a candidate gene association study. Genes Brain Behav. 2008;7:720–729. doi: 10.1111/j.1601-183X.2008.00410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Ji L. Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity. 2005;95:221–227. doi: 10.1038/sj.hdy.6800717. [DOI] [PubMed] [Google Scholar]
- Lomelin D, Jorgenson E, Risch N. Human genetic variation recognizes functional elements in noncoding sequence (Letter) Genome Res. 2010;20:311–319. doi: 10.1101/gr.094151.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan SD, Aalinkeel R, Reynolds JL, Nair BB, Sykes DE, Hu Z, Bonoiu A, Ding H, Prasad PN, Schwartz SA. Therapeutic targeting of "DARPP-32": a key signaling molecule in the dopiminergic pathway for the treatment of opiate addiction. Int Rev Neurobiol. 2009;88:199–222. doi: 10.1016/S0074-7742(09)88008-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merikangas KR, Stolar M, Stevens DE, Goulet J, Preisig MA, Fenton B, Zhang H, O'Malley SS, Rounsaville BJ. Familial transmission of substance use disorders. Arch Gen Psychiatry. 1998;55:973–979. doi: 10.1001/archpsyc.55.11.973. [DOI] [PubMed] [Google Scholar]
- Narita M, Matsushima Y, Niikura K, Narita M, Takagi S, Nakahara K, Kurahashi K, Abe M, Saeki M, Asato M, Imai S, Ikeda K, Kuzumaki N, Suzuki T. Implication of dopaminergic projection from the ventral tegmental area to the anterior cingulate cortex in µ-opioid-induced place preference. Addict Biol. 2010;15:434–447. doi: 10.1111/j.1369-1600.2010.00249.x. [DOI] [PubMed] [Google Scholar]
- Nyholt DR. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am J Hum Genet. 2004;74:765–769. doi: 10.1086/383251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmstead MC, Ouagazzal AM, Kieffer BL. Mu and delta opioid receptors oppositely regulate motor impulsivity in the signaled nose poke task. PLoS One. 2009;4:e4410. doi: 10.1371/journal.pone.0004410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters K, Wiltshire S, Henders AK, Dragovic M, Badcock JC, Chandler D, Howell S, Ellis C, Bouwer S, Montgomery GW, Palmer LJ, Kalaydjieva L, Jablensky A. Comprehensive analysis of tagging sequence variants in DTNBP1 shows no association with schizophrenia or with its composite neurocognitive endophenotypes. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:1159–1166. doi: 10.1002/ajmg.b.30741. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray R, Ruparel K, Newberg A, Wileyto EP, Loughead JW, Divgi C, Blendy J, Logan J, Zubieta J-K, Lerman C. Human Mu Opioid Receptor (OPRM1 A118G) polymorphism is associated with brain mu-opioid receptor binding potential in smokers. Proc Natl Acad Sci USA. 2011;108:9268–9273. doi: 10.1073/pnas.1018699108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shand FL, Degenhardt L, Nelson EC, Mattick RP. Predictors of social anxiety in an opioid dependent sample and a control sample. J Anxiety Disord. 2010;24:49–54. doi: 10.1016/j.janxdis.2009.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shippenberg TS, Chefer VI, Thompson AC. Delta-opioid receptor antagonists prevent sensitization to the conditioned rewarding effects of morphine. Biol Psychiatry. 2009;65:169–174. doi: 10.1016/j.biopsych.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svenningsson P, Nairn AC, Greengard P. DARPP-32 mediates the actions of multiple drugs of abuse. AAPS J. 2005;7:E353–E360. doi: 10.1208/aapsj070235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuang MT, Lyons MJ, Meyer JM, Doyle T, Eisen SA, Goldberg J, True W, Lin N, Toomey R, Eaves L. Co-occurrence of abuse of different drugs in men: the role of drug-specific and shared vulnerabilities. Arch Gen Psychiatry. 1998;55:967–972. doi: 10.1001/archpsyc.55.11.967. [DOI] [PubMed] [Google Scholar]
- Tuusa JT, Leskelä TT, Petäjä-Repo UE. Human delta opioid receptor biogenesis is regulated via interactions with SERCA2b and calnexin. FEBS J. 2010;277:2815–2829. doi: 10.1111/j.1742-4658.2010.07699.x. [DOI] [PubMed] [Google Scholar]
- van Rijn RM, Whistler JL. The delta(1) opioid receptor is a heterodimer that opposes the actions of the delta(2) receptor on alcohol intake. Biol Psychiatry. 2009;66:777–784. doi: 10.1016/j.biopsych.2009.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Zastrow M. Regulation of opioid receptors by endocytic membrane traffic: mechanisms and translational implications. Drug Alcohol Depend. 2010;108:166–171. doi: 10.1016/j.drugalcdep.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wand GS, McCaul M, Yang X, Reynolds J, Gotjen D, Lee S, Ali A. The mu-opioid receptor gene polymorphism (A118G) alters HPA axis activation induced by opioid receptor blockade. Neuropsychopharmacology. 2002;26:106–114. doi: 10.1016/S0893-133X(01)00294-9. [DOI] [PubMed] [Google Scholar]
- Yekkirala AS, Kalyuzhny AE, Portoghese PS. Standard opioid agonists activate heteromeric opioid receptors: evidence for morphine and [d-Ala2-MePhe4-Glyol5]enkephalin as selective µ-d agonists. ACS Chem Neurosci. 2010;1:146–154. doi: 10.1021/cn9000236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller T, Wild P, Szymczak S, Rotival M, Schillert A, Castagne R, Maouche S, Germain M, Lackner K, Rossmann H, Eleftheriadis M, Sinning CR, Schnabel RB, Lubos E, Mennerich D, Rust W, Perret C, Proust C, Nicaud V, Loscalzo J, Hübner N, Tregouet D, Münzel T, Ziegler A, Tiret L, Blankenberg S, Cambien F. Genetics and beyond--the transcriptome of human monocytes and disease susceptibility. PLoS One. 2010;5:e10693. doi: 10.1371/journal.pone.0010693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Shao C, Shao M, Yan P, Wang Y, Liu Y, Liu W, Lin T, Xie Y, Zhao Y, Lu D, Li Y, Jin L. Effect of mu-opioid receptor gene polymorphisms on heroin-induced subjective responses in a Chinese population. Biol Psychiatry. 2007;61:1244–1251. doi: 10.1016/j.biopsych.2006.07.012. [DOI] [PubMed] [Google Scholar]
- Zhang H, Kranzler HR, Yang BZ, Luo X, Gelernter J. The OPRD1 and OPRK1 loci in alcohol or drug dependence: OPRD1 variation modulates substance dependence risk. Mol Psychiatry. 2008;13:531–543. doi: 10.1038/sj.mp.4002035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Luo X, Kranzler HR, Lappalainen J, Yang BZ, Krupitsky E, Zvartau E, Gelernter J. Association between two mu-opioid receptor gene (OPRM1) haplotype blocks and drug or alcohol dependence. Hum Mol Genet. 2006;15:807–819. doi: 10.1093/hmg/ddl024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wang D, Johnson AD, Papp AC, Sadee W. Allelic expression imbalance of human mu opioid receptor (OPRM1) caused by variant A118G. J Biol Chem. 2005;280:32618–32624. doi: 10.1074/jbc.M504942200. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Pan ZZ. Synaptic mechanism for functional synergism between delta- and mu-opioid receptors. J Neurosci. 2010;30:4735–4745. doi: 10.1523/JNEUROSCI.5968-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


